Abstract
Lamin B is a component of the membranous spindle matrix isolated from Xenopus egg extracts, and it is required for proper spindle morphogenesis. Besides lamin B, the spindle matrix contains spindle assembly factors (SAFs) such as Eg5 and dynein which are known to regulate microtubule organization and SAFs known to promote microtubule assembly such as Maskin and XMAP215. Because lamin B does not bind directly to microtubules, it must affect spindle morphogenesis indirectly by influencing the function of spindle matrix-associated SAFs. Using different assays in Xenopus egg extracts, we found that depleting lamin B caused formation of elongated and multipolar spindles, which could be reversed by partially inhibiting the kinesin Eg5, revealing an antagonistic relationship between Eg5 and lamin B. However, lamin B only very weakly antagonizes Eg5 in mediating poleward microtubule-flux based on fluorescence speckle microscopy. Depleting lamin B led to a very small but statistically significant increase in flux. Furthermore, flux reduction caused by partial Eg5 inhibition is only slightly reversed by removing lamin B. Because lamin B does not bind to Eg5, our studies suggest two nonexclusive mechanisms by which lamin B can indirectly antagonize Eg5. It could function in a network that restricts Eg5-driven microtubule sliding only when microtubules come into transient contact with the network. Lamin B could also function to sequester microtubule polymerization activities within the spindle. Without lamin B, increased microtubule assembly caused by the released SAFs would lead to excessive microtubule sliding that results in formation of elongated and multipolar spindles.
Keywords: Cell Division, Kinesin, Lamin, Mitosis, Mitotic Spindle, Eg5, Spindle Matrix
Introduction
The balance of forces between microtubule (MT)3-based motor proteins such as dynein and the tetrameric kinesin Eg5 plays a critical role in mitotic spindle morphogenesis and chromosome segregation. Although such force balancing could theoretically be achieved by antagonizing motor forces exerted on MTs, the precision required makes it unlikely (1, 2). Decades ago it was proposed that a spindle matrix structure could tether spindle assembly factors (SAFs) and/or absorb forces generated on MTs (for review, see Ref. 3). A number of studies using Drosophila embryos (4–8), mammalian tissue culture cells, and Xenopus egg extracts (9, 10) have provided evidence for the existence and tethering function of the spindle matrix. However, the mechanism by which the spindle matrix regulates force balancing and spindle morphogenesis remains unclear.
Among the spindle matrix components identified thus far, lamin B is a conserved structural protein of the type V intermediate filaments (11). In interphase, the lamin B intermediate filaments represent the major structural component of the nuclear lamina found beneath the nuclear envelope (12). Similar to other intermediate filaments, nuclear lamins do not actively produce force, but, together with chromatin, lamins can bear loads exerted on nuclear envelopes by both actin and MTs and enable the nucleus to maintain its shape and integrity (13, 14).
Analogous to its role in the interphase nucleus, insoluble mitotic lamin B in the membranous spindle matrix that surrounds spindle MTs could function to limit motor-driven MT movement and help maintain spindle shape (15). Moreover, both the insoluble and soluble mitotic lamin B might sequester SAFs to inhibit their activity within the spindle. Consistent with the idea that lamin B regulates spindle morphology, we have shown that reduction of lamin B in either Xenopus egg extracts or in HeLa cells results in formation of spindles with defective morphology (10, 16, 17). In addition, spindle defects caused by lamin B depletion in egg extracts can be rescued by purified lamin B (10).
Recent studies of spindle morphogenesis in Drosophila early embryos, which undergo partially open mitosis with incomplete breakdown of nuclear envelope and lamina, have revealed that lamin B exists in an envelope that surrounds prometaphase and metaphase spindles. Either hyperstabilizing or disrupting lamin B in the envelope disrupted spindle length maintenance. Additional studies suggest that the lamin B spindle envelope functions as an elastic spring that helps to increase the robustness of motor balancing in the spindle to stabilize the spindle length (2). However, how the lamin B spindle envelope regulates motor balance and whether the lamin B spindle envelope could affect the motor-driven poleward microtubule flux is unknown.
Unlike the partially open mitosis in Drosophila early embryos, vertebrate mitosis is accompanied by a complete breakdown of nuclear envelope and lamina in prometaphase. Lamin B distributes into both a soluble pool and a membrane-associate fraction in the mitotic cytosol (18, 19). In Xenopus egg extracts, a fraction of lamin B3 (LB3), the major lamin B form in this system (20), is recruited onto MTs and becomes further organized into a membranous network that associates with and surrounds the meiotic spindle (10, 16, 17). LB3 does not directly bind to MTs, yet the isolated LB3 spindle matrix contains a number of SAFs including Eg5, dynein, XMAP215, and Maskin. This suggests that LB3 might affect spindle assembly by influencing the activities of spindle matrix-associated SAFs.
Eg5 is a plus end-directed motor known to slide MTs that drive bipolar spindle assembly and spindle elongation in Xenopus egg extracts. Eg5-mediated MT sliding also drives poleward MT flux in spindles assembled in Xenopus egg extracts (21, 22). If lamin B functions in maintaining spindle morphology, we expect to see functional interactions between lamin B and Eg5 during spindle assembly. We have analyzed the effects of lamin B depletion and Eg5 inhibition on both spindle morphology and MT flux in Xenopus egg extracts. We report here that both the membrane-associated and the soluble LB3 can function indirectly to constrain Eg5-driven spindle pole separation in meiotic spindles in Xenopus egg extracts.
EXPERIMENTAL PROCEDURES
Egg Extract Preparation
Cytostatic factor-arrested egg extracts were prepared as described previously (23). Demembranated sperm chromatin was prepared as described previously and added to extracts to a final concentration of ∼400 sperm nuclei/μl. Aurora A (AurA) beads were prepared as described (24) with modifications. Briefly, 10 μl of protein A-coupled magnetic Dynabead (Invitrogen) slurry was washed using extract buffer (10 mm K-Hepes, pH 7.7, 50 mm sucrose, 100 mm KCl, 1 mm MgCl2, 0.1 mm CaCl2, and 5 mm EGTA) and incubated with 250 μg/ml rabbit polyclonal AurA antibody for 1 h. After washing with extract buffer, the beads were resuspended in 20 μl of extract buffer and incubated at 1:100 with cytostatic factor-arrested egg extracts for 40 min at 4 °C. The extracts used for the AurA bead assay were supplemented with rhodamine-tubulin (∼13 μg/ml final), RanL43E (a constitutively GTP-bound RanGTPase, 1–2 mg/ml final), and energy mix (3.8 mm creatine phosphate, 0.5 mm ATP, 0.5 mm MgCl2, 0.05 mm EGTA). Extracts used for sperm spindle assembly were supplemented as described above, but without RanL43E. Monastrol was dissolved in dimethyl sulfoxide to make a 10 mm stock solution.
Immunodepletion of LB3
100 μl of magnetic protein A-Dynabead slurry was incubated with 100 μl of rabbit polyclonal LB3T antibody (1 mg/ml) (10) or control IgG from unimmunized rabbit (1 mg/ml) for 1 h. The beads were washed in extract buffer and then incubated with 100 μl of egg extracts for 1 h at 4 °C. This immunodepletion procedure routinely removed ∼95% of the LB3 in egg extracts (supplemental Fig. S2A).
Time Lapse and MT Spin-down Assays
For time lapse recording of spindle assembly, 10 μl of extract was placed in a magnetic flow chamber at 4 °C. The magnetic flow chamber was constructed by adhering a glass coverslip to a glass slide using double-stick transparent tape. A 3/8-inch × 1/8-inch axial disc magnet (K&J Magnetics) was affixed to the glass slide by double-stick tape. AurA beads were allowed to settle for 3 min at 4 °C, then the slide was placed on an inverted fluorescence microscope at room temperature. All time lapse recordings were started ∼2 min after beginning the reaction and were taken at 5-s intervals for a total of 15–25 min.
For AurA bead MT spin-down assays, spindle assembly reactions were stopped at the indicated times. Sperm assays were stopped in a similar manner at 90 min. Reactions were stopped and spun down onto coverslips as described (23) and stained with goat anti-rabbit Alexa Fluor 488 to enhance bead visualization. Both time lapse recording and still images were taken using a Nikon Eclipse TE200 fluorescence microscope and a 60 × oil lens or a 40 × dry lens on a Photometrics Coolsnap HQ camera and a LEP Mac5000 automated stage.
Time Lapse Microscopy Analyses
AurA beads in MT structures from the movies were tracked using the Metamorph “Track objects ” application. Tracking data were exported to Excel, and the distance between AurA beads in the process of forming a spindle was calculated in each frame. A Matlab program determined the rate of AurA bead separation in 30-s windows (consisting of six time lapse frames) that stepped forward one frame (5 s) at a time (see supplemental Fig. S1A). Windows were checked to make sure that the beads were undergoing separation across the 30 s. This allowed us to minimize noise due to the loss and reappearance of AurA beads in particular frames. Bead separation rates were averaged across all windows for a given spindle, yielding the spindle elongation rate (code available upon request). Several spindles were measured for each extract. Four independent experiments were performed using different egg extracts made on different dates. AurA beads that interacted but did not separate, started further than 10 μm apart, or left the field of view for more than 1 min were excluded from analysis.
For nearest neighbor assays, 70 images were acquired randomly for each reaction using a Nikon Eclipse TE200 fluorescence microscope with a Photometrics Coolsnap HQ camera and a LEP Mac5000 automated stage driven by Metamorph software. Matlab was used to identify beads and bead clusters by brightness and eccentricity (code available upon request). AurA beads in each image were identified, and the distance between two nearest neighbors (indicated by lines between the beads) was checked for MTs, measured, and exported as a Microsoft Excel file (supplemental Fig. S1B). The measurements were normalized relative to the average of the control dataset to allow the comparison of different egg extracts. At least three independent experiments were carried out and used for the analyses.
Fluorescence Speckle Microscopy
Fluorescence speckle microscopy was performed on a Nikon Eclipse TE200 fluorescence microscope with a Photometrics Coolsnap HQ camera using a 100×/1.4 NA objective. Images were collected using Metamorph software. Egg extract used for fluorescence speckle microscopy was supplemented with rhodamine-tubulin (1:3 dye:protein ratio) to a concentration of 11–16 nm. Small aliquots (5.5 μl) of extracts were placed under a 22 × 22-mm coverslip and sealed with valap (1:1:1 parafin:lanolin:vaseline). Movies were taken at 25 °C at a rate of one frame every 3 s for 4 min. Analysis was performed as described previously (22).
RESULTS
Spindle-like Structures Stimulated by AurA Beads Contain Antiparallel MTs That Drive the Separation of AurA Beads Mediated by Eg5
The mitotic kinase AurA regulates multiple aspects of mitosis, including spindle morphogenesis (for review, see Ref. 25). We have shown previously that AurA beads function as MT-organizing centers to recruit SAFs such as TPX2 (26, 27) and γTuRC (28) to stimulate spindle-like structures in egg extracts (24) in the presence of RanGTP (29–37). We reasoned that because these structures form quickly and robustly, they could be used to approximate interpolar MT interactions and force balancing found during spindle assembly. We therefore characterized the spindle-like structures and found that at the beginning of the reaction AurA beads tended to cluster in groups of two to four, and all beads nucleated microtubule asters. Beads then separate from one another to form bipolar (Fig. 1A top and supplemental Movie S1) or multipolar spindle-like structures (∼53% of total structures; Fig. 1A, bottom, and supplemental Movie S2). MT asters nucleated from single AurA beads remained monopolar (∼47%) until they contacted MTs nucleated from other beads to form highly interconnected MT structures by 15 min into the reaction (supplemental Movie S3). When separate MT structures come in contact with one another, they interact, and the beads in these structures maintain their distance or move further apart. With extended reaction time (20–25 min), some spindle-like structures maintained a steady distance between AurA beads, whereas others continued to elongate or fell apart (data not shown).
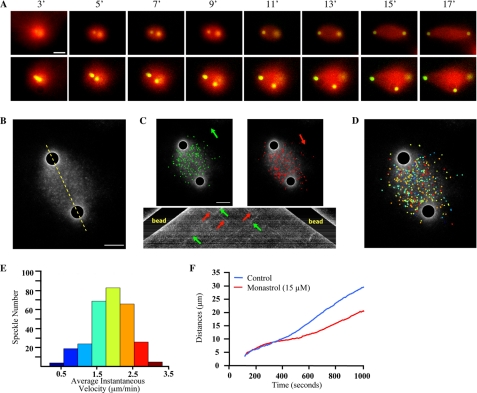
FIGURE 1.
Characterization of spindle-like structures stimulated by AurA beads and RanGTP. A, time lapse frames showing formation of bipolar or multipolar spindle-like structures. Clusters of two to three AurA beads initially nucleate MT asters. The separation of the beads within the asters leads to formation of either bipolar or multipolar spindle-like structures (see supplemental Movies S1 and S2). The time (minutes) above the images corresponds to the time following initiation of the reaction that the images were taken. B, MT speckle images of one bipolar spindle-like structure that is undergoing elongation. AurA beads are masked to visualize the speckles better. The yellow dashed line indicates the region used for kymograph analysis in C. C, speckles, labeled green or red, moving toward each of the AurA beads, are demonstrating speckle movement in each direction. Below, a kymograph shows the speckle movements with respect to the movement of AurA beads. D, speckles color coded according to their average instantaneous speckle velocities shown on the histogram in E (flux rate, 2.229; S.D., 0.532; speckles tracked, 632). The same bipolar spindle-like structure shown in C was used. F, reduction of the rate of AurA bead separation in bipolar spindle-like structures by inhibiting Eg5 with 15 μm Monastrol. The distances between two AurA beads during spindle assembly in the presence or absence of Monastrol were measured and plotted against time. The plot shows average distance for each time point across all four experiments for each condition. Scale bars, 10 μm.
Using GFP-tagged EB1, we tracked plus-end MT behavior in spindle-like structures and found that MTs polymerized from one bead toward the other in an antiparallel fashion (supplemental Movie S4). These MTs underwent flux toward AurA beads (Fig. 1, B–E and supplemental Movie S5) with the average flux rate of 2.229 μm/min (S.D., 0.523; speckles tracked, 632; spindle-like structures tracked, 4), which is similar to the flux rate found in sperm spindles. Kymograph analysis showed that the flux rate of the speckles is similar to the rate of separation of the AurA beads (Fig. 1C).
Given the importance of microtubule sliding to bead separation, we examined the effect of inhibiting Eg5 on bead separation and found that partially inhibiting Eg5 using 15 μm Monastrol allowed separation of AurA beads and formation of spindle-like structures, but the rate of AurA bead separation was reduced (Fig. 1F and supplemental Fig. S1A).
LB3 Limits whereas Eg5 Increases the Distance That Separates AurA Beads in MT Structures
To investigate the effects of LB3 on motor forces, we used the AurA bead system and measured the distance between beads and their nearest neighbors in randomly captured fields (disregarding beads in contact with each other). To check this assay for accuracy, we first made a Monastrol dose-response curve. The average distance between AurA beads decreased as the concentration of Monastrol in the egg extract increased, yielding an estimated EC50 value of 21 μm (Fig. 2, A and B). This EC50 value matches reported values for Monastrol (supplemental Table S1). In contrast to the Monastrol results, depletion of LB3 resulted in a significant increase in the distance between AurA beads in MT structures (Fig. 2, C and D).
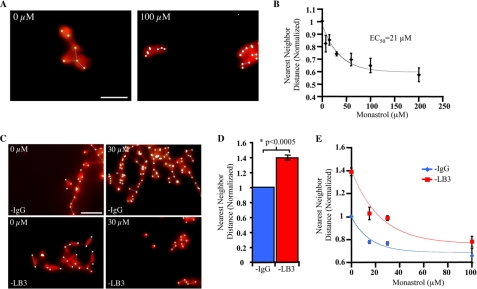
FIGURE 2.
LB3 and Eg5 oppose each other in maintaining the separation of AurA beads in spindle-like structures. A, sample images of MT structures formed in the presence or absence of Monastrol. Images were acquired randomly using an automated stage control. The small white dots and white lines in these and other images in this figure represent AurA beads and the linear distance between nearest neighboring beads connected by MTs, respectively, as characterized by the image recognition software developed in Matlab (see supplemental Fig. S1B). B, dose-dependent decrease of the distances between neighboring AurA beads in MT structures caused by inhibition of Eg5 by Monastrol. The distances are normalized to 0 μm Monastrol (set to 1) to allow comparisons among different egg extracts. C, sample images of MT structures formed in the presence or absence of LB3 and Monastrol. D, increase in the distance between AurA beads caused by LB3 depletion. Quantification of distances between neighboring AurA beads in the presence or absence of LB3 without Monastrol is shown. E, depletion of LB3 relief of the effect of Eg5 inhibition at each concentration of Monastrol tested. Quantification of distances between neighboring AurA beads in the presence or absence of LB3 and an increasing concentration of Monastrol is shown. Error bars, S.E. from at least three experiments performed in different egg extracts on different days with ∼200 measurements of distances in each condition and each experiment. Scale bars, 50 μm.
To determine whether LB3 opposed Eg5 activity, we next measured the average distance of AurA beads in either mock or LB3-depleted egg extracts with increasing concentrations of Monastrol. Depleting LB3 significantly relieved the effect of Monastrol on the average distance between AurA beads in all Monastrol concentrations tested (Fig. , 2C and E). The EC50 values for Monastrol in the presence or absence of LB3 are comparable (supplemental Table S1). This suggests that LB3 does not affect the ability of Eg5 to interact with Monastrol. Moreover, LB3 does not interact with Eg5 in egg extracts based on immunoprecipitation analyses (supplemental Fig. S2B). We confirmed the antagonistic relationship between Eg5 and LB3 using a rabbit antibody raised against the N terminus of Eg5 (amino acids 1–545) to inhibit the activity of Eg5. Results were similar to the Monastrol experiments (supplemental Fig. S3, A and B).
LB3 Opposes Eg5 to Limit the Initial Separation of Clustered AurA Beads
To verify further that LB3 and Eg5 counteract each other, we employed a second assay that measures the initial separation of clustered beads at the beginning of the formation of spindle-like structures. All AurA beads within MT structures were counted and assigned to clustered and unclustered groups (see arrows and arrowheads, respectively, in Fig. 3). Any beads that made contact were defined as clustered, whereas individual beads were defined as unclustered beads. The clustering index, which was calculated by dividing the number of clustered beads by the number of unclustered beads in MT structures, was used to determine the degree of clustering of the beads.
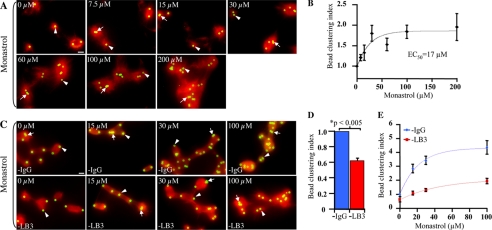
FIGURE 3.
LB3 limits whereas Eg5 promotes the separation of clustered AurA beads during formation of spindle-like structures. A, images of MT structures formed in the presence or absence of various concentrations of Monastrol at 15 min into the reaction. B, increase of AurA bead clustering caused by Eg5 inhibition. Quantification of the bead-clustering index at different concentrations of Monastrol is shown. Values are normalized to 0 μm Monastrol (dimethyl sulfoxide carrier) to allow comparisons among different egg extracts. Monastrol caused a dose-dependent increase in AurA-bead-clustering. C, images of AurA beads in MT structures assembled in the presence or absence of LB3 and Monastrol at 15 min into the reaction. D, decrease in AurA bead clustering caused by LB3 inhibition. Quantification of the bead-clustering index in the presence or absence of LB3 without inhibiting Eg5 is shown. E, depletion of LB3 relief of the clustering effect caused by Monastrol. Quantification of clustering index in the presence or absence of LB3 and an increasing concentration of Monastrol is shown. Arrows and arrowheads in all images indicate clustered and separated AurA beads, respectively. Error bars are S.E. from at least three experiments performed in different egg extracts on different days with 700 beads counted per condition per experiment. Scale bars, 10 μm.
In control reactions without Monastrol, the clustering index decreased as a function of time, indicating that the clustered beads separated continuously from one another during the course of reactions, whereas the clustering index remained relatively constant in the presence of Monastrol during the reaction (supplemental Fig. S2C). This demonstrates that the clustering index is a measure of the degree of separation of initially clustered AurA beads instead of the collapse of the already separated beads during the reaction.
Using the clustering assay at the 15-min time point when most clustered beads had separated, we found that increasing Monastrol concentration caused an increase in the clustering index with an EC50 of 17.3 μm (Fig. 3, A and B), which is similar to published EC50 values for Monastrol measured using different assays (supplemental Table S1). By contrast, we found that depleting LB3 significantly reduced the clustering index compared with mock-depleted egg extracts (Fig. 3, C–E). The difference between the Monastrol curve and the control depletion is due to the immunodepletion procedure, which impaired spindle formation relative to unmanipulated extract (supplemental Fig. S4A). To show that the reduction of the clustering index caused by LB3 depletion was not simply a result of increased “stickiness” of the beads with LB3 present, time course analyses were performed on egg extracts subjected to either control or LB3 IgG depletion. These analyses showed that the AurA bead clustering indices were similar for both conditions at the beginning of the reaction but differed significantly by 6 min with the difference increasing at later time points (supplemental Fig. S4B). Using the Eg5 inhibitory antibody, we found a similar antagonistic relationship between Eg5 and LB3 in this bead clustering assay (supplemental Fig. S3, C and D).
LB3 and Eg5 Antagonize Each Other in Maintaining Sperm Spindle Length and Bipolarity
To establish further that LB3 antagonizes Eg5 to help in maintaining the overall spindle morphology, we used meiotic spindles formed from sperm chromatin. Because LB3 has been shown to facilitate nuclear envelope assembly (38) and DNA replication (39) from Xenopus sperm in egg extracts, we limited our studies to meiotic spindles assembled in cytostatic factor-arrested egg extracts. We examined whether LB3 could antagonize Eg5 in maintaining sperm spindle lengths by quantifying spindles with equivalent amounts of chromatin based on DAPI staining intensity and found that depleting LB3 caused formation of longer spindles, which could be reversed by the addition of 15 μm Monastrol, a concentration that did not severely inhibit spindle assembly induced by sperm (Fig. 4, A and B). LB3 depletion also caused a significant increase of multipolar spindles and a corresponding decrease of bipolar spindles compared with controls (Fig. 4, A and C). This defect could also be rescued by the addition of 15 μm Monastrol.
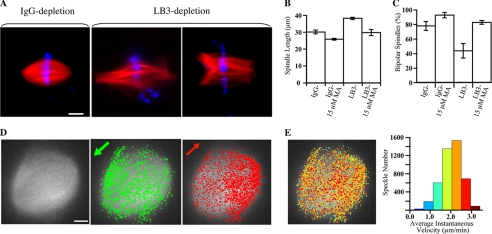
FIGURE 4.
Effects of LB3 and Eg5 on the length, poles, and MT flux of spindles assembled from sperm chromatin. A, images of spindles formed in mock-depleted and LB3-depleted egg extracts. LB3 depletion resulted in larger spindles and an increase in multipolar spindles. B, quantifications of spindle length, excluding half-spindles and monopolar spindles, in both mock-depleted and LB3-depleted extracts in the presence or absence of 15 μm Monastrol. Monastrol rescued the spindle length change caused by LB3 depletion. C, quantification of the percentage of bipolar spindles, excluding half-spindles and monopolar spindles, in mock-depleted extracts and LB3-depleted extracts in the presence or absence of 15 μm Monastrol. LB3 depletion caused a decrease in the percentage of bipolar spindles, which was corrected with the addition of 15 μm Monastrol. Approximately 35 spindles were analyzed per condition per experiment, and three experiments performed on different days were used for quantifications. Error bars, S.E. D, speckle images of sperm spindles. The gray image shows a frame of the speckled spindle. Speckles moving toward opposite spindle poles are labeled as green and red dots. E, speckle trajectories color-coded according to their average instantaneous speckle velocities shown on the histogram to the right (flux rate, 2.031; S.D., 0.488; speckles tracked, 4488). Scale bars, 10 μm.
LB3 Does Not Have a Major Role in MT Flux in Spindles Assembled from Sperm Chromatin
To analyze the effect of LB3 on individual MT behavior, we used fluorescence speckle microscopy to analyze MT flux in sperm spindles assembled in egg extracts depleted with either control or LB3 IgG (Fig. 4, D and E, and supplemental Movie S6). We found that the average MT flux rate in mock-depleted egg extracts to be 2.021 μm/min (S.D., 0.491; 7 spindles, 23,203 speckles). Depleting LB3 resulted in a slight but statistically significant (p < 0.0001 using two-sample two-sided Student's t test) increase of average MT flux rate to 2.159 μm/min (S.D., 0.513; 6 spindles, 20,666 speckles). The addition of 15 μm Monastrol to mock-depleted egg extracts caused an expected large reduction of flux rate to 1.727 μm/min (S.D., 0.445; 8 spindles, 24,192 speckles). This reduction of flux rate was only reversed slightly (p < 0.01 using two-sample two-sided Student's t test) by depleting LB3 (average flux rate, 1.769 μm/min; S.D., 0.438; 8 spindles, 25,593 speckles). Previous studies have shown that the flux rate decreases near spindle poles (22). We also observed this phenomenon, but we found that LB3 depletion had no affect on this regional flux rate difference (data not shown).
DISCUSSION
The length and shape of the mitotic spindle are regulated by a multitude of SAFs. For example, the balance of forces generated by motor proteins plays a critical role in spindle morphogenesis. Plus end-directed motor proteins, such as Eg5, slide antiparallel MTs in the spindle to promote spindle elongation, whereas minus end-directed motor proteins such as dynein function to keep spindle poles together. Besides motor proteins, SAFs that regulate MT polymerization dynamics also affect spindle length and morphology. An increase in MT length in the spindle caused by enhanced MT polymerization has been shown to result in elongated spindles. Deciphering how MT-based motors and MT assembly SAFs coordinate with one another in mitosis is critical to understand spindle morphogenesis. Recent studies have shown that LB3 regulates spindle morphogenesis as a component of the spindle matrix (10, 16, 17). Because the spindle matrix contains both motor proteins and MT assembly regulators, probing how LB3 affects both gross spindle morphology and individual MT flux should provide insights regarding how LB3 might coordinate SAF functions as a component of the spindle matrix.
By analyzing spindle-like structures formed by AurA beads, we found an antagonistic relationship between LB3 and Eg5. We showed that AurA bead separation is driven by MT flux with a lack of MT depolymerization from the minus ends of MTs. The most likely explanation for this is that the high concentration of AurA stably tethered on the beads caused a hyperphosphorylation of mitotic centromere-associated kinesin which is known to reduce its MT depolymerization activity (40, 41). The lack of MT depolymerizing activity near AurA beads could explain why the AurA bead spindles cannot reach a steady state as the sperm spindles do.
Despite the differences between AurA bead and sperm spindles, we showed that LB3 and Eg5 antagonize each other to maintain spindle length and spindle pole morphology in both spindle assembly assays. However, because LB3 does not directly bind to Eg5, the two proteins must antagonize each other indirectly. Because a fraction of LB3 in mitosis is insoluble and remains associated with membranes and because LB3 is found to surround the spindles (2), one possibility is that LB3 functions in this membrane network that ensheaths the spindle to restrict Eg5-driven MT sliding. LB3 depletion could cause defects in the network. The escaping of MTs through the defective network would lead to spindle elongation and multipolarity. Reduction of MT sliding by inhibiting Eg5 should relieve these gross morphological defects. An alternative possibility is that the soluble pool of LB3 could function throughout the spindle to indirectly inhibit Eg5 activity.
The above two possibilities can be differentiated by measuring MT flux because if LB3 functions in a network ensheathing the spindle to restrict Eg5-driven MT movements, only MTs that come into contact with the network would be restricted to move further. Therefore, depletion of LB3 should have a small effect on the overall MT flux rate and there should not be an antagonistic relationship between LB3 and Eg5 as measured by MT flux. On the other hand, if the soluble pool of LB3 indirectly sequesters Eg5 throughout spindles to limit Eg5 activity, the same antagonistic relationship between Eg5 and LB3 would exist for MT flux as those measured for spindle morphology. Our studies of MT flux showed a small but statistically significant increase of MT flux, which is insufficient to antagonize the reduction of MT flux caused by Eg5 inhibition. Therefore, LB3 most likely antagonizes Eg5 in a network to restrict MT sliding driven by Eg5.
Unlike the partially open mitosis in Drosophila early embryos, there is no well defined lamin B-containing spindle envelope that surrounds spindle MTs in vertebrate open mitosis. Yet, vertebrate spindles are surrounded by cytoplasmic membranes that contain lamin B (17). Our findings suggest that this membranous lamin B network in vertebrate mitosis functions similarly to the Drosophila spindle envelope to buffer motor activities (2, 15). LB3 could contribute toward the integrity of the membranous spindle matrix that surrounds the spindle to limit excessive MT sliding and spindle pole separation. Proper organization of this membranous spindle matrix could also allow local positioning of SAFs to limit MT sliding. We have shown that LB3 can bind to Nudel (17). Although depletion of LB3 does not affect dynein recruitment (data not shown), the interaction between LB3 and Nudel could allow LB3 to position dynein properly on the matrix to counteract Eg5.
Because the isolated spindle matrix also contains proteins such as XMAP215 and Maskin that can promote MT growth, it is possible that LB3 could function in the spindle matrix to facilitate the sequestration of MT polymerization activities. In the absence of LB3, the enhanced MT elongation would lead to increased spindle length, which should be relieved by inhibiting Eg5-mediated MT sliding. However, the increase in MT length and spindle elongation would have a small effect on Eg5-driven MT flux. Our analyses of spindle morphology and MT flux suggest that both the membrane-associated and the dynamic soluble pool of LB3 could function together to antagonize Eg5-driven MT sliding indirectly by sequestering MT assembly activities.
In conclusion, by analyzing the effect of Eg5 inhibition and LB3 depletion on the gross spindle morphology and MT flux, we have shown that LB3 indirectly antagonizes Eg5 activity during spindle assembly. Our findings suggest that the membrane-associated LB3 could antagonize Eg5 by functioning in a network to restrain Eg5-driven MT sliding. Alternatively, LB3 could antagonize Eg5 by sequestering activities that promote MT growth. These two functions of LB3 are not mutually exclusive.
Supplementary Material
Acknowledgments
We thank Li Ma, Dan Ducat, Shusheng Wang, and the other members of the Zheng laboratory for comments and suggestions; Ona Martin for technical support; Ming-Ying Tsai and Anna Chan for training; and Mahmud Siddiqi for imaging assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant GM56312 (to Y. Z.). This work was also supported by National Science Foundation Grant 0621740 (to P. I.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4, Table S1, and Movies S1–S6.
- MT
- microtubule
- AurA
- Aurora A
- AurA beads
- magnetic beads coated with the mitotic kinase AurA
- LB3
- Xenopus lamin B3
- LB3T
- COOH-terminal domain (amino acids 383–583) of Xenopus lamin B3
- RanGTP
- RanGTPase bound to GTP
- SAF
- spindle assembly factor.
REFERENCES
- 1.Mogilner A., Wollman R., Civelekoglu-Scholey G., Scholey J. (2006) Trends Cell Biol. 16, 88–96 [DOI] [PubMed] [Google Scholar]
- 2.Civelekoglu-Scholey G., Tao L., Brust-Mascher I., Wollman R., Scholey J. M. (2010) J. Cell Biol. 188, 49–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johansen K. M., Johansen J. (2007) Int. Rev. Cytol. 263, 155–206 [DOI] [PubMed] [Google Scholar]
- 4.Qi H., Rath U., Ding Y., Ji Y., Blacketer M. J., Girton J., Johansen J., Johansen K. M. (2005) J. Cell. Biochem. 95, 1284–1291 [DOI] [PubMed] [Google Scholar]
- 5.Qi H., Rath U., Wang D., Xu Y. Z., Ding Y., Zhang W., Blacketer M. J., Paddy M. R., Girton J., Johansen J., Johansen K. M. (2004) Mol. Biol. Cell 15, 4854–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rath U., Wang D., Ding Y., Xu Y. Z., Qi H., Blacketer M. J., Girton J., Johansen J., Johansen K. M. (2004) J. Cell. Biochem. 93, 1033–1047 [DOI] [PubMed] [Google Scholar]
- 7.Walker D. L., Wang D., Jin Y., Rath U., Wang Y., Johansen J., Johansen K. M. (2000) J. Cell Biol. 151, 1401–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lince-Faria M., Maffini S., Orr B., Ding Y., Cláudia F., Sunkel C. E., Tavares A., Johansen J., Johansen K. M., Maiato H. (2009) J. Cell Biol. 184, 647–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang P., Jacobson M. K., Mitchison T. J. (2004) Nature 432, 645–649 [DOI] [PubMed] [Google Scholar]
- 10.Tsai M. Y., Wang S., Heidinger J. M., Shumaker D. K., Adam S. A., Goldman R. D., Zheng Y. (2006) Science 311, 1887–1893 [DOI] [PubMed] [Google Scholar]
- 11.Herrmann H., Bär H., Kreplak L., Strelkov S. V., Aebi U. (2007) Nat. Rev. Mol. Cell Biol. 8, 562–573 [DOI] [PubMed] [Google Scholar]
- 12.Dechat T., Pfleghaar K., Sengupta K., Shimi T., Shumaker D. K., Solimando L., Goldman R. D. (2008) Genes Dev. 22, 832–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dahl K. N., Kahn S. M., Wilson K. L., Discher D. E. (2004) J. Cell Sci. 117, 4779–4786 [DOI] [PubMed] [Google Scholar]
- 14.Dahl K. N., Scaffidi P., Islam M. F., Yodh A. G., Wilson K. L., Misteli T. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10271–10276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng Y. (2010) Nat. Rev. Mol. Cell Biol. 11, 529–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng Y., Tsai M. Y. (2006) Cell Cycle 5, 2345–2347 [DOI] [PubMed] [Google Scholar]
- 17.Ma L., Tsai M. Y., Wang S., Lu B., Chen R., Iii J. R., Zhu X., Zheng Y. (2009) Nat. Cell Biol. 11, 247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lourim D., Krohne G. (1998) J. Cell Sci. 111, 3675–3686 [DOI] [PubMed] [Google Scholar]
- 19.Adam S. A., Sengupta K., Goldman R. D. (2008) J. Biol. Chem. 283, 8462–8468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lourim D., Kempf A., Krohne G. (1996) J. Cell Sci. 109, 1775–1785 [DOI] [PubMed] [Google Scholar]
- 21.Miyamoto D. T., Perlman Z. E., Burbank K. S., Groen A. C., Mitchison T. J. (2004) J. Cell Biol. 167, 813–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang G., Cameron L. A., Maddox P. S., Salmon E. D., Danuser G. (2008) J. Cell Biol. 182, 631–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murray A. W. (1991) Methods Cell Biol. 36, 581–605 [PubMed] [Google Scholar]
- 24.Tsai M. Y., Zheng Y. (2005) Curr. Biol. 15, 2156–2163 [DOI] [PubMed] [Google Scholar]
- 25.Ducat D., Zheng Y. (2004) Exp. Cell Res. 301, 60–67 [DOI] [PubMed] [Google Scholar]
- 26.Wittmann T., Wilm M., Karsenti E., Vernos I. (2000) J. Cell Biol. 149, 1405–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai M. Y., Wiese C., Cao K., Martin O., Donovan P., Ruderman J., Prigent C., Zheng Y. (2003) Nat. Cell Biol. 5, 242–248 [DOI] [PubMed] [Google Scholar]
- 28.Zheng Y., Wong M. L., Alberts B., Mitchison T. (1995) Nature 378, 578–583 [DOI] [PubMed] [Google Scholar]
- 29.Carazo-Salas R. E., Gruss O. J., Mattaj I. W., Karsenti E. (2001) Nat. Cell Biol. 3, 228–234 [DOI] [PubMed] [Google Scholar]
- 30.Wilde A., Lizarraga S. B., Zhang L., Wiese C., Gliksman N. R., Walczak C. E., Zheng Y. (2001) Nat. Cell Biol. 3, 221–227 [DOI] [PubMed] [Google Scholar]
- 31.Carazo-Salas R. E., Guarguaglini G., Gruss O. J., Segref A., Karsenti E., Mattaj I. W. (1999) Nature 400, 178–181 [DOI] [PubMed] [Google Scholar]
- 32.Kalab P., Pu R. T., Dasso M. (1999) Curr. Biol. 9, 481–484 [DOI] [PubMed] [Google Scholar]
- 33.Wilde A., Zheng Y. (1999) Science 284, 1359–1362 [DOI] [PubMed] [Google Scholar]
- 34.Goodman B., Zheng Y. (2006) Biochem. Soc. Trans. 34, 716–721 [DOI] [PubMed] [Google Scholar]
- 35.Wiese C., Wilde A., Moore M. S., Adam S. A., Merdes A., Zheng Y. (2001) Science 291, 653–656 [DOI] [PubMed] [Google Scholar]
- 36.Gruss O. J., Carazo-Salas R. E., Schatz C. A., Guarguaglini G., Kast J., Wilm M., Le Bot N., Vernos I., Karsenti E., Mattaj I. W. (2001) Cell 104, 83–93 [DOI] [PubMed] [Google Scholar]
- 37.Nachury M. V., Maresca T. J., Salmon W. C., Waterman-Storer C. M., Heald R., Weis K. (2001) Cell 104, 95–106 [DOI] [PubMed] [Google Scholar]
- 38.Lopez-Soler R. I., Moir R. D., Spann T. P., Stick R., Goldman R. D. (2001) J. Cell Biol. 154, 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moir R. D., Spann T. P., Herrmann H., Goldman R. D. (2000) J. Cell Biol. 149, 1179–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lan W., Zhang X., Kline-Smith S. L., Rosasco S. E., Barrett-Wilt G. A., Shabanowitz J., Hunt D. F., Walczak C. E., Stukenberg P. T. (2004) Curr. Biol. 14, 273–286 [DOI] [PubMed] [Google Scholar]
- 41.Zhang X., Ems-McClung S. C., Walczak C. E. (2008) Mol. Biol. Cell 19, 2752–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.