SUMMARY
Formation of microtubule architectures, required for cell shape maintenance in yeast, directional cell expansion in plants and cytokinesis in eukaryotes, depends on antiparallel microtubule crosslinking by the conserved MAP65 protein family. Here, we combine structural and single molecule fluorescence methods to examine how PRC1, the human MAP65, crosslinks antiparallel microtubules. We find that PRC1's microtubule binding is mediated by a structured domain with a spectrin-fold and an unstructured Lys/Arg-rich domain. These two domains, at each end of a homodimer, are connected by a linkage that is flexible on single microtubules, but forms well-defined crossbridges between antiparallel filaments. Further, we show that PRC1 crosslinks do not substantially resist filament sliding by motor proteins in vitro. Together, our data show how MAP65s, by combining structural flexibility and rigidity, tune microtubule associations to establish compliant crosslinks that selectively `mark' antiparallel overlap in dynamic cytoskeletal networks.
INTRODUCTION
The dynamic reorganization of microtubule networks plays a critical role in diverse biological processes, including cell migration, neuronal transport and cell division. It is now clear that different cytoskeletal architectures arise from the interplay between motor proteins, which can crosslink and move microtubules relative to one another, and non-motor microtubule associated proteins (MAPs), which can crosslink microtubules to stabilize specific orientations (Glotzer, 2009; Manning and Compton, 2008). While we have good biophysical and structural models for motor proteins that crosslink microtubules, much less is known about non-motor microtubule crosslinking proteins.
Several non-motor MAPs that crosslink microtubules (e.g. MAP65, NuMA, NuSAP and Mia1p) are now known to play important roles in dividing and non-dividing cells (Ribbeck et al., 2006; Sasabe and Machida, 2006; Schuyler et al., 2003; Thadani et al., 2009; Zeng, 2000). Current models for the functions of these proteins are based on cellular localizations and loss-of-function studies. However, we lack any structural data to explain how microtubule crosslinking is achieved by these MAPs. Recently, there have been several advances in our understanding of the structure of non-motor MAPs. Among the best characterized class of MAPs are the +TIP proteins (e.g. XMAP215, EB1 and CLIP170), which can dynamically track the growing end of a microtubule. Microtubule binding in these proteins is mediated by calponin-homology, CAP/Gly or TOG domains (Slep and Vale, 2007). Similarly, structural work on Ndc80, a conserved mitotic MAP, has revealed how a calpolin-homology domain may be used to establish kinetochore-microtubule associations during cell division (Ciferri et al., 2008; Wei et al., 2007; Wilson-Kubalek et al., 2008). However, due to lack of similarity in primary sequence, it is not very likely that these structural models will shed light on non-motor MAPs that can crosslink two microtubules.
As a step towards developing structural models for how non-motor MAPs may crosslink microtubules, we focused on the conserved MAP65 family, which plays key roles in microtubule organization in eukaryotes. Since their initial discovery in budding yeast, microtubule crosslinking functions of the MAP65 proteins have been shown to be required for cell shape maintenance in yeast cells, directional cell expansion in plants and formation of the central spindle in eukaryotes (Chan et al., 1999; Jiang et al., 1998; Loiodice et al., 2005; Yamashita et al., 2005). Currently, at least three activities have been ascribed to these proteins. First, MAP65s can selectively crosslink microtubules in an anti-parallel orientation (Gaillard et al., 2008; Loiodice et al., 2005). Second, these non-motor crosslinkers can oppose filament movements driven by motor proteins. For example, Ase1, the fungal MAP65, is proposed to antagonize kinesin-14 driven filament sliding required for organizing microtubules during interphase (Janson et al., 2007). Third, these crosslinking proteins can recruit signaling proteins or kinesins to the microtubule structures they stabilize. For example, the recruitment of Polo-like kinase to the central spindle during cytokinesis is mediated via interactions with PRC1, the human MAP65 (Neef et al., 2007) and kinesin-5 driven microtubule sliding during anaphase depends on Ase1 (Khmelinskii, 2009). Currently, we do not have a structural framework to explain how these MAPs specifically crosslink anti-parallel microtubules. Moreover, the activities of MAP65s have not been reconstituted in the presence of motor proteins to test if MAP65s resist filament sliding by motor proteins or if their main function is act as `marks' that recruit other proteins to regions of antiparallel microtubule overlap in dynamic networks.
Here we show, using single molecule fluorescence microscopy assays, X-ray crystallography and electron microscopy that PRC1 uses structured and unstructured domains to bind microtubules. These domains at each end of a PRC1 homodimer are connected by a linker that adopts a rigid conformation only when crosslinking microtubules. We also show, in assays combining TIRF and fluorescent speckle microscopy (FSM), that PRC1 does not substantially resist filament sliding by kinesin-5. Based on these results, we propose a model for how a crosslinking MAP can achieve specific and compliant crosslinking of microtubules by balancing structural rigidity and flexibility.
RESULTS
Structured and unstructured domains mediate microtubule binding in PRC1
PRC1, like other Map65 family proteins, has a modular architecture with an N-terminal coiled-coil domain, a central region that can mediate microtubule binding, and a C-terminal regulatory domain (Figure 1A). While the central domain is thought to be required for microtubule binding, the contributions of the C-terminal domain remain poorly characterized. To address this, we analyzed the microtubule binding activity of PRC1 using two approaches, a TIRF microscopy assay to examine the properties of single molecules and a microtubule cosedimentation assay to analyze equilibrium binding.
FIGURE 1.
Single molecule analysis of microtubule binding by PRC1. (A) Schematic of PRC1's domain organization and a guide for constructs used in the fluorescence microscopy assays (purple: coiled-coil domain; green: microtubule binding domain; black: C-terminal domain). (B) Fluorescence intensity analysis of two PRC1 constructs, GFP-PRC1-FL (aa: 1–620; intensity =2.5×104 ± 0.9×104, N = 469) and GFP-PRC1-NS (aa: 1–466; intensity =2.0×104 ± 0.8×104, N = 156). Dimeric-Eg5-GFP (Intensity = 2.5×104 ± 1.0×104, N = 377) and tetrameric-Eg5-GFP (Intensity = 4.2×104 ± 2.2×104, N = 290) were used as references. (C–H) Single molecule TIRF assay was used to examine the association of PRC1 constructs (green) with microtubules (orange) immobilized on a glass surface. (C) Schematic for assay showing the two constructs, GFP-PRC1-FL and GFP-PRC1-NSΔC (aa.1–486). Single frames showing two-color overlays (top) and associated kymographs (below) of GFP-PRC1-FL (D,E) or GFP-PRC1-NSΔC (F,G). (H) Distribution of microtubule association lifetimes for GFP-PRC1-FL (blue) and GFP-PRC1-NSΔC (red). (I–K) Microtubule association of GFP-PRC1-NSΔC under different ionic strength conditions. Representative kymographs from assays at 0.75× motility buffer (I), motility buffer (J), motility buffer+20 mM KCl. (K) Scale bars: 1.5 μm, 10 s. See also Figures S1 and S3
For visualizing PRC1 molecules by fluorescence microscopy we expressed and purified recombinant GFP-tagged full-length PRC1 in bacteria. We found that C-terminal tags on PRC1 resulted in constructs that were highly unstable and therefore used a construct with GFP fused to the N-terminus of PRC1 (GFP-PRC1-FL). We first examined the oligomerization state of single GFP-PRC1-FL molecules. Analysis of the intensities of single fluorescent spots of GFP-PRC1-FL immobilized on a glass coverslip, when compared to intensities of known dimeric and tetrameric reference constructs, indicated that GFP-PRC1-FL is a dimer (Figure 1B), similar to the yeast homolog, Ase1 (Kapitein et al., 2008a; Schuyler et al., 2003). We also confirmed that untagged PRC1 is a homo-dimer and has an extended conformation (see below, and data not shown). Previous studies have suggested that PRC1 exists as a homo-tetramer, but gel filtration chromatography alone can sometimes be unreliable for such analyses (Zhu et al., 2006). To examine the interaction of single PRC1 molecules with a microtubule, we used streptavidin to attach X-rhodamine-labeled, biotinylated microtubules to a glass surface. After blocking the glass surface, we added low concentrations of GFP-PRC1-FL (20 pM) (Figure 1C). Similar to what is observed for Ase1, we found that single GFP-PRC1-FL molecules diffuse in 1-D along the microtubule surface (Figure 1D, E; (Kapitein et al., 2008a)). Analysis of time-lapse sequences using kymographs showed that individual GFP-PRC1-FL molecules can maintain associations with the microtubule lattice that can last several seconds (t = 7±0.4 s; Figure 1H).
To examine the contribution of the C-terminus of PRC1 to microtubule binding, we first generated a construct comprised of just the N-terminal dimerization and the microtubule binding domain (aa. 1–466, hereafter GFP-PRC1-NS). Intensity distribution of GFP-PRC1-NS indicated that this construct, like the full-length protein, is a dimer (Figure 1B). We were unable to detect microtubule associations of single molecules of GFP-PRC1-NS at the concentrations needed to resolve single molecules using TIRF microscopy (< 10 nM; data not shown), suggesting that residues in the C-terminus domain contribute to microtubule binding. Secondary structure algorithms predict that the C-terminal domain in PRC1 is likely to be unstructured. Consistent with this prediction, we find that PRC1's C-terminus is prone to proteolysis. A highly susceptible cleavage site resides between two Lys/Arg-rich basic clusters in this domain (aa. Asn 500; Figure S1). As basic amino acids are often implicated in mediating interactions with the negatively charged microtubules surface, we generated a GFP-fused PRC1 construct with one of the two basic-residue clusters (aa. 1–486, hereafter GFP-PRC1-NSΔC). Under TIRF microscopy conditions we could readily observe microtubule binding by single GFP-PRC1-NSΔC molecules. Diffusion on the microtubule was apparent for the subset of binding events that lasted several seconds (Figure 1F, G). Analysis of the lifetimes of microtubule association indicated that this construct has a higher microtubule unbinding rate than the full-length protein (Figure 1H, lifetime is less that the lower limit of reliable event detection, <3 s). Increasing salt concentrations reduced the GFP-PRC1-NSΔC binding lifetimes, consistent with a charge-dependent microtubule-binding interaction mediated by PRC1's C-terminus (Figure 1I, J, K).
We next carried out microtubule co-sedimentation assays for an independent analysis of the contribution of the C-terminal domain to PRC1's microtubule interaction. For these experiments we generated PRC1 constructs lacking the dimerization domain so that we could exclude the potentially complex effects of filament crosslinking/bundling on this analysis (Figure 2A). These PRC1 constructs were confirmed to be monomeric by size exclusion chromatography (data not shown). The microtubule binding affinity (Kd) of a construct comprising of the central domain and the C-terminus (aa 341–620, named PRC1-SC) was 0.6 ± 0.3 μM (Figure 2B, D). A construct comprising of the central domain alone (aa. 341–466, hereafter named PRC1-S) had a ~5-fold weaker binding (Kd: 3.3 ± 1.8 μM; Figure 2C, D). Interestingly, we observe cooperative microtubule binding by PRC1-SC (Hill coefficient of 3±1). This suggests that the C-terminus may be responsible for interactions between PRC1 molecules on the microtubule lattice analogous to what has been previously reported for Ase1 (Kapitein et al., 2008a). Together, these data indicate that PRC1's microtubule binding is mediated by two regions, one that is predicted to be structured and another, unstructured region, rich in Lys/Arg residues.
FIGURE 2.
Microtubule co-sedimentation assays to determine equilibrium dissociation constants for PRC1's microtubule binding domains. (A) Schematic for constructs used in this assay: PRC1-S (aa: 341–466) and PRC1-SC (aa: 341–620). SDS-PAGE analysis of co-sedimentation assays for PRC1-SC (B) and PRC1-S (C). Arrows in (B) indicate C-terminus proteolysis products in PRC1-SC. Bands marked with red boxes in (B) indicate the relative tubulin concentration at which 50% of the different PRC1-SC truncation products co-sediment with microtubules. (D) Band intensities from the gels in (C) were used to determine fraction protein bound, and plotted against microtubule concentration (n=3, mean ± SD). The data were fit to a modified Hill equation to determine Kd's (PRC1-S: 3.3 ± 1.8 μM; PRC1-SC: 0.6 ± 0.3 μM).
PRC1's microtubule binding domain has a spectrin fold
To determine the structural basis of microtubule binding by PRC1, we focused on its central microtubule binding domain. As this domain has no obvious homology to any known microtubule binding motif, we used X-ray crystallography to determine its structure. Non-isomorphous crystals of the PRC1-S construct were obtained from both the native protein, which diffracted to 1.75 Å resolution, and the selenomethionyl-substituted protein, which diffracted to 2.0 Å resolution. The structure of the selenomethionyl-substituted protein was solved by single-wavelength anomalous dispersion, refined, and, subsequently used as a model to solve the structure of the native protein using molecular replacement (Table-S1). We found that the microtubule binding domain of PRC1 is an ~70 Å long 3-helix bundle (labeled helix-1, -2, and 3), with connecting loops (labeled loop1–2 and loop2–3) and N- and C- termini at opposite ends (Figure 3A and Figure S2A, B). Hydrophobic residues from all three helices contribute to a hydrophobic core which dominates the interface between the three helices (Figure 3B). This core is flanked on both ends of the helix bundle by two salt bridges mediated by conserved residues (Figure 3C).
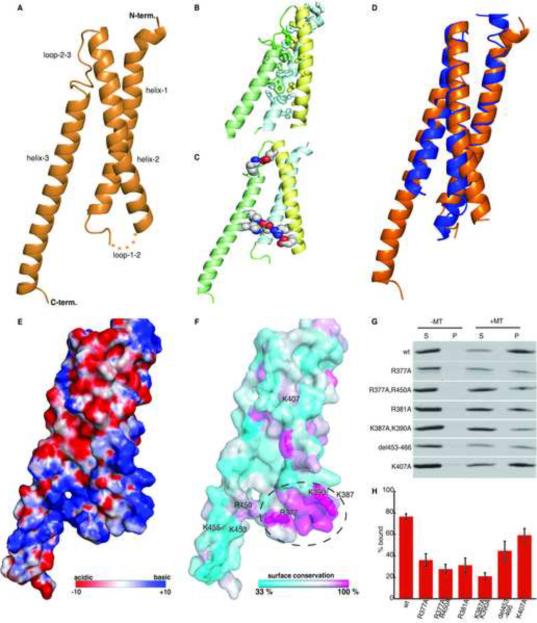
FIGURE 3.
PRC1's conserved microtubule binding domain (PRC1-S; aa: 341–466) adopts a spectrin fold. (A) Ribbon diagram shows the overall structure. Five disordered residues in the loop between helix-1 and 2 are indicated by dots. (B) Hydrophobic residues from helix-1 (cyan), helix-2 (yellow) and helix-3 (green), which form the core of the triple helix bundle, are highlighted (sticks). (C) Salt bridges between charged residues from helix-1 (cyan), helix-2 (yellow) and helix-3 (green) are indicated (spheres). (D) Overlay of PRC1's spectrin domain (orange) with its closest structural homolog, which is a spectrin repeat in Plactin (blue, PDB = 2odu-A, Z-score = 7.7, rmsd = 3.4 Å calculated using DALI). (E) Surface representation showing electrostatic potential of the spectrin domain in PRC1 (Red to blue is −10 kbT to +10 kbT, as calculated using APBS). (F) Residues with low (cyan), intermediate (white), and high (magenta) conservation on the surface of the PRC1's spectrin domain. The labeled residues were selected for mutagenesis studies. (G) SDS-PAGE analysis of microtubule co-sedimentation assays of PRC1-S mutants at 27 μM tubulin. (H) Band intensities from (C) were quantified to determine the fraction of PRC1 bound to microtubules (n=3, error bars indicate SE). See also Figure S2
Using DALI, we found that PRC1-S has high structural similarity to spectrin domains (Figure 3D), motifs commonly found in proteins associated with the actin cytoskeleton (e.g. alpha-actinin, spectrin and dystrophin; (Djinovic-Carugo et al., 2002)). However, a spectrin fold has not thus far been found in any other known MAP. Interestingly, in actin binding proteins, the elongated and rigid triple-helix of a spectrin domain does not directly mediate actin binding, but acts as a spacer between canonical actin binding motifs, such as calponin homology domains. Therefore, it appears that the spectrin fold has been evolutionarily `recycled' and re-engineered to act as a microtubule interacting domain in the Map65 protein family. These findings establish a new role for the spectrin motif as a microtubule binding domain.
A conserved basic region forms the microtubule binding surface in the spectrin domain of PRC1
As it was unclear how the spectrin fold would bind microtubules, we analyzed the electrostatic surface potential and the conservation of surface residues for this domain. A map of the electrostatic surface potential shows a positively charged region comprising of loop1–2 and residues from all three helices that are proximal to this loop in the three dimensional structure (Figure 3E). Interestingly, a cluster of highly conserved residues is also located within this region (Figure 3F), suggesting that the junction of helix-1 and helix-2 could form the microtubule binding region in the spectrin fold. To test this hypothesis we generated constructs of PRC1's spectrin domain in which basic residues in this region were mutated to alanine (Figure S2C). Each of these constructs was characterized to be monomeric by size-exclusion chromatography (Figure S2D). We used microtubule co-sedimentation assays to compare microtubule binding of mutant and wild type PRC1-S constructs (Figure 3G). We found that mutation of each of the basic residues in the most conserved region reduced microtubule binding 2- to 4-fold, though none of the individual mutations abolished microtubule binding (Figure 3H). To test that the observed microtubule binding is not simply a non-specific electrostatic interaction, we tested two other constructs. First, we made a C-terminus truncation in PRC1-SC (labeled del453–466; aa. 341–452) to remove three surface exposed lysines on helix-3. Second, we generated a construct in which a lysine residue on helix-2 (K407), distal to the potential microtubule binding site, was mutated to alanine. Both of these constructs show less than 50% reduction in microtubule binding. From this analysis, we propose that the spectrin domain in PRC1 uses a basic surface comprising of conserved residues at one end of the triple helix bundle to mediate microtubule binding (Figure 3F, circle).
Spectrin domain fits with an optimal orientation into the cryo-EM density map of the PRC1-microtubule complex
To investigate the interaction of PRC1 with the microtubule lattice we examined the structure of dimeric PRC1 bound to single microtubules by cryo-electron microscopy (Cryo-EM) and helical image analysis. As PRC1-FL extensively bundles microtubules at concentrations needed for this analysis, we used our findings from single molecule experiments to engineer a construct PRC1-NSΔC (aa. 1–486; Figure 4A), which was more suitable for this analysis (Figure 1A). This construct was confirmed to be dimeric by HPLC Size Exclusion Chromatography/Laser Light Scattering Analysis (Figure S3), and found to retain sufficient binding to fully decorate single microtubules without extensive bundling.
FIGURE 4.
The spectrin domain fits into the Cryo-EM density map of the PRC1-microtubule complex with an optimal orientation. (A) Schematic comparing full-length PRC1 to the construct PRC1-NSΔC (aa: 1–486) used for Cryo-EM analysis. (B) Surface rendered side-view of 3D EM density map of the microtubule-PRC1-NSΔC complex. (C) Top view of cryo-EM density map of undecorated tubulin (gold) superimposed with the microtubule-PRC1-NSΔC complex (purple mesh). (D) Side- and (E) top-view of the crystal structure (cyan ribbon diagram) docked into the PRC1 density (purple mesh) protruding from an α/β tubulin dimer (gold). Residues R377 and K387, which are involved in microtubule binding, are indicated in red and black respectively. Microtubule polarity indicated in (B) was determined by comparison with a 3D EM structure of a dynein-microtubule complex, which has a well defined polarity (Carter et al., 2008). See also Figure S4
Diffraction of individual cryo-EM images of PRC1-NSΔC bound to microtubules showed an 80 Å layer line, indicating that one PRC1-NSΔC molecule binds a α/β-tubulin heterodimer. The 3-D reconstruction of microtubule bound PRC1-NSΔC showed a single rod-shaped density corresponding to PRC1-NSΔC, protruding approximately perpendicular to the microtubule lattice (microtubule side view, Figure 4B; top view, Figure 4C). Interestingly, the PRC1 density in the reconstruction is of the same size as PRC1's spectrin domain. The optimal fit of the spectrin domain crystal structure to the reconstructions oriented the conserved, basic residues at the junction of helix-1 and helix-2 towards the microtubule lattice (Figure 4D and 4E). This binding model is also consistent with our site-directed mutagenesis analysis (Figure 3H). Comparison of the 3-D reconstruction of this complex with cryo-EM structures of motor proteins (e.g kinesin and dynein's microtubule-binding domain) bound to microtubules suggests that the binding site for PRC1 partially overlaps with, but is not identical to, the microtubule-binding surface used by motor proteins ((Carter et al., 2008); Figure S4B). Surprisingly, we see no extra density distal to the microtubule surface that would correspond to the N-terminus coiled-coil dimerization domain in PRC1-NSΔC. This indicates that the coiled-coil domain is likely to be flexible and can have more than one conformation when bound to a single microtubule.
To further understand the PRC1-microtubule interaction, we obtained a second EM reconstruction with a monomeric construct that lacked most of the oligomerization domain but contained the spectrin domain and the entire C-terminus (aa. 303–620). The observed density for this construct overlaps with the PRC1-NSΔC density close to the microtubule lattice (Figure S4C, D, E). We also observe some additional density distal to the microtubule surface which is not observed in PRC1-NSΔC. It is possible that this density corresponds to PRC1's C-terminus folding back to interact with the spectrin domain or to the amino acids at PRC1's N-terminus that may be ordered when this construct binds microtubules. As we do not observe any additional density past the C-terminus of the spectrin domain that is close to the microtubule lattice, we favor the possibility that the Lys/Arg-rich domain is disordered even when PRC1 is bound to a microtubule. Together, these data show that PRC1's spectrin domain is highly ordered and less flexible than the other domains when associated with a single microtubule.
Cryo-electron tomography reveals ordered PRC1 crosslinks between two anti-parallel microtubules
Helical reconstructions suggest that PRC1 may not be an inherently rigid molecule when bound to one microtubule. It is difficult to explain how a flexible protein with two microtubule interaction surfaces at opposite ends of dimer can achieve the crosslinking specificity reported for MAP65 proteins (Gaillard et al., 2008; Janson et al., 2007). The only other structural analysis of crosslinking by the MAP65 family is a cryo-EM study of the plant homolog of PRC1, which show that the protein can form dense crossbridges between two microtubules (Gaillard et al., 2008). However, the conformational differences between crosslinking and non-crosslinking molecules could not be visualized in this study. It also remains unknown whether establishing ordered crossbridges is a property of single MAP65 molecules or results from protein-protein interactions that may be involved in PRC1's co-operative microtubule binding (Figure 2D). To address these questions, we employed cryo-electron tomography to obtain a higher resolution structure of two microtubules crosslinked by PRC1 (Figure 5A). For this analysis we used the PRC1 construct we had used for helical reconstructions (PRC1-NSΔC; Figure 4A). 3-D reconstructions from cryo-electron tomography of microtubules densely crosslinked by PRC1-NSΔC shows rod-like striations connecting two microtubules (Figure 5B and inset). From analyzing the overall direction of crosslinking protein density, we estimated that 90% (n = 20) of the crosslinked microtubules were anti-parallel. This shows that the C-terminus truncation construct has anti-parallel crosslinking specificity similar to full-length MAP65 and Ase1 (Gaillard et al., 2008; Schuyler et al., 2003). We find that the average distance between two crosslinked filaments was 35 nm (± 2 nm; n = 20) and the crossbridge angle of PRC1 linkages was ~70° (± 5; n = 15) relative to the microtubule surface. As helical reconstruction shows that the spectrin domain in PRC1 protrudes almost perpendicular to the microtubule surface, the crossbridge orientation likely results from ~ 20° hinges between the oligomerization and the spectrin domains.
FIGURE 5.
The dimerization domain in PRC1 is ordered when crosslinking two microtubules. (A) Schematic highlights the section viewed in the cryotomographic reconstructions shown in (B) and (C), which encompasses the two microtubules and the PRC1 crosslinks. (B–C) Slice through cryotomographic 3-D reconstruction of microtubules crosslinked by PRC1-NSΔC. Slices represent the central area of the microtubules and the bound PRC1-NSΔC. The top and bottom of the microtubules are excluded in these views. Examples of microtubule pairs with dense (B) and sparse (C) PRC1 occupancy. Insets show 4-fold enlargements of the crosslinks between microtubules. Red arrows highlight individual cross-bridges. Blue arrow in (B) indicates a PRC1 molecule that does not form crosslinks with another microtubule. Long red arrows in (B) indicate the polarity of microtubules, portions of which are highlighted in blue for clarity. The top two microtubules in (B) share PRC1 crossbridges as do the bottom two microtubules, however, the middle two microtubules are separated by another microtubule (dotted blue line) that is positioned below the plane and only the top of its tubulin lattice can be observed.
Consistent with what is seen from the helical reconstruction analysis, the tomograms show that the PRC1 densities observed on the microtubule surface distal to crosslinked microtubule (i.e., the non-crosslinking PRC1 molecules, Figure 5B, blue arrow) were less ordered than those between two filaments. This indicates that non-crosslinking and crosslinking PRC1 molecules have distinct conformations.
We next examined if the regular crossbridge geometry and the inter-microtubule distances observed are retained in regions of sparse PRC1 decoration between crosslinked microtubules. We find that single PRC1 molecules still form crossbridges with orientations similar to those observed in regions of dense crosslinks (Figure 5C and inset). Interestingly, we found that PRC1 crosslinks were not observed in regions where filament spacing was significantly greater than 35 nm, indicating that PRC1 molecules exhibit specificity for a narrow range of inter-microtubule distances. These findings agree with our observations in fluorescence microscopy assays in which GFP-PRC1-FL accumulation is seen only at a narrow range of microtubule crosslinking angles (unpublished data). Together, our data indicate that although the dimerization domain is not entirely rigid when PRC1 is bound to a single microtubule, this domain in individual PRC1 molecules can adopt a specific conformation upon crosslinking two microtubules. The rigidity in the linker, together with oriented binding by the spectrin domain, can allow PRC1 to be a selective crosslinker of anti-parallel microtubules aligned with a narrow range of inter-filament spacing.
PRC1 is a compliant crosslinker of anti-parallel microtubules
Our analysis of microtubule binding by the structured and unstructured domains in PRC1 shows that these interactions have moderate affinity. Together with the observed 1-D diffusion of single PRC1 molecules on microtubules, these data predict that PRC1 crosslinks may not substantially resist filament sliding by motor proteins. To test this hypothesis, we devised an in vitro assay to visualize near-simultaneously PRC1 localization and motor-protein driven sliding of pairs of crosslinked microtubules. We attached biotinylated microtubules to a glass surface via streptavidin. These microtubules also incorporated low levels of fluorescent tubulin for analysis of relative microtubule motion using fluorescent speckle microscopy (FSM). GFP-PRC1-FL was added to the immobilized microtubules, followed by non-biotinylated microtubules to generate a `sandwich' comprised of two microtubules crosslinked by PRC1 (Figure 6A). As would be expected, based on studies of Ase1, we find that GFP-PRC1-FL shows a 10±4 - fold preference for regions where two microtubule overlap relative to regions of single microtubules (N = 15; Figure S5A, B, C; (Kapitein et al., 2008a)). To drive the relative sliding of microtubules in this `sandwich', we added kinesin-5, a well characterized motor protein needed for cell division.
FIGURE 6.
PRC1 crosslinks do not substantially resist the relative sliding of two microtubules by kinesin-5. (A) Schematic illustrating the assay used to examine the effect of PRC1 (green) on kinesin-5 (cyan)-mediated relative sliding of two microtubules (orange), when extent of overlap between microtubules is unchanged. Near-simultaneous dual-mode microscopy was used to image microtubules (via wide-field fluorescent speckle microscopy) and GFP-PRC1-FL (via Total Internal Reflection Fluorescence (TIRF) microscopy). (B) Frames from a time-lapse sequence (1 min interval) show GFP-PRC1-FL (green) enriched at regions where two crosslinked microtubules (red) overlap. (C) Corresponding kymograph shows the surface-attached static microtubule (vertical streaks) and a moving microtubule (diagonal streaks, 16.5 nm/s). (D) Kymograph shows that the GFP-PRC1-FL decorated region moves at the velocity of the moving microtubule. (E) Schematic illustrating the assay when overlap between microtubules decreases during relative microtubule sliding. (F) Frames from a time-lapse sequence (2 min interval) and corresponding kymographs showing microtubule movement (7 nm/s) (G) and GFP-PRC1-FL localization to overlap region that reduces due to relative sliding of filaments (H). (I) Velocity distributions for kinesin-5 driven microtubule sliding at 1.8 nM kinesin-5 and 0.034 (V = 23.1 ± 6.3 nm/s, N = 43), 0.14 nM (V = 17.7 ± 3.5 nm/s, N = 39), and 0.54 nM GFP-PRC1-FL (V = 9.7 ± 3.1 nm/s, N = 41). Scale bars: 1.5 μm, 100 s. See also Figure S5
Under these conditions we observed two types of events. First, kinesin-5 could move a shorter microtubule relative to a longer filament in the `sandwich' such that the amount of overlap tracked by GFP-PRC1, remained unchanged (Figure 6B, C, D). Second, we observed events in which the microtubule overlap in the `sandwich' reduced at the rate of filament motion (Figure 6E and Figure S5D, E) and GFP-PRC1 dynamically tracked the microtubule overlap zone (Figure 6F). Remarkably, the velocity of relative filament sliding by kinesin-5 did not increase as the extent of microtubule overlap reduced (Figure 6G, H). Fluorescence intensity analysis indicated that the number of PRC1-crosslinks in the overlap region decreased proportionately (data not shown). Analysis of these data show that kinesin-5's sliding velocity reduced only ~2.4 fold over an 18-fold change in PRC1 concentration (Figure 6I). At higher PRC1 concentrations in this assay extensive microtubule bundling was observed and it was very difficult to reliably detect pairs of crosslinked microtubules. The few events that could be analyzed under these conditions indicated that a further reduction in kinesin-5 driven filament sliding was not observed as PRC1 concentration was increased (Figure S5F–I). We estimated the ratio of PRC1 and kinesin-5 at microtubule overlap zones in our assays. Using GFP-kinesin-5 at concentrations similar to the un-tagged kinesin-5 used in the above assays, we found that microtubule sliding was driven by a few motor protein molecules, and unlike PRC1, kinesin-5 did not show a preference for microtubule overlap zones (data not shown and (Hentrich and Surrey, 2010)). The average fluorescence intensity of single GFP-PRC1-FL molecules was then used to determine the number of GFP-PRC1-FL molecules at microtubule overlap regions. These data indicate that the ratio of GFP-PRC1-FL to kinesin-5 molecules at microtubule overlap zones was ~25 (total solution concentration: GFP-PRC1-FL = 0.54 nM, kinesin-5 = 1.8 nM). This shows that crosslinks formed by multiple PRC1 molecules do not substantially resist the relative microtubule movement driven by fewer kinesin-5 molecules.
DISCUSSION
Members of the MAP65 family organize microtubules by preferentially crosslinking filaments with an anti-parallel orientation. In this study, we provide a structural framework for how three structurally distinct domains in human MAP65 (PRC1) are combined to achieve selective and efficient crosslinking of anti-parallel microtubules, as would be needed during the self-organization of dynamic cytoskeletal networks.
The conserved microtubule binding domain in PRC1 has a spectrin fold. First identified in spectrin, a constituent of the membrane skeleton, this fold has since been seen in several actin crosslinking proteins such as alpha-actinin and dystrophin (Djinovic-Carugo et al., 2002). In these proteins, spectrin-domains appear as repeated units that connect other actin-binding domains and regulate properties of the cytoskeletal structures. In PRC1, this domain appears to have evolved to mediate microtubule binding, an entirely different role for this protein fold. Further, the spectrin fold is unrelated to the other protein folds (e.g Cap/Gly motifs and calponin-homology domains) that are known to mediate interactions with the microtubule lattice. It has been proposed that some protein surfaces are evolutionarily selected to act as a versatile protein interaction platform. This is best exemplified in the Fc domain of IgG, which is structurally adapted to interact with several different protein scaffolds (DeLano et al., 2000). Similarly, it appears that the microtubule surface has evolved to bind a large variety of unrelated structural motifs and thereby accommodate diverse MAPs that can carry out a wide range of functions (Amos and Schlieper, 2005).
Microtubule binding by the spectrin domain is augmented by a Lys/Arg rich unstructured domain in PRC1. Synergistic microtubule binding by structured and unstructured domains have also been seen in other MAPs such as Ndc80 (Guimaraes et al., 2008; Miller et al., 2008), suggesting that this feature may be a frequent adaptation in MAPs. There are also at least two other functions ascribed to these unstructured microtubule binding domains. First, these unstructured domains are often sites of phospho-regulation (Holt et al., 2009). Interestingly, Cdk1 phosphorylation sites in PRC1 map to the unstructured Lys/Arg domain (Zhu et al., 2006). Our data suggest that these phosphorylations would directly attenuate microtubule affinity by reducing the net positive charge of the domain. Dephosphorylation of these residues would activate PRC1's microtubule binding at anaphase, as would be needed for PRC1's functions during the final stages of cell division. Second, as has been suggested for Ndc80, these domains are proposed to allow a mode of attachment which does not significantly resist microtubule movement. We find that PRC1 does not strongly oppose microtubule sliding by kinesin-5, indicating that moderate binding affinities and diffusive microtubule interactions of PRC1 can permit microtubule movement while maintaining attachment. This result is also consistent with the reported diffusion constant of Ase1 which indicates that >100 crosslinker molecules would be needed to generate a resistive force greater than 1.5 pN, which is in the range of the force required to inhibit kinesin-5 movement (Kapitein et al., 2008a; Korneev et al., 2007; Valentine and Block, 2009). This would be relevant in vivo, when microtubule bundling by Ase1 and sliding by kinesin-5 are both required for spindle elongation in anaphase B (Khmelinskii et al., 2009).
Crossbridges between two PRC1-crosslinked microtubules are formed by PRC1's dimerization domain. These are seen to project at an angle of 70° relative to the microtubule lattice. Binding at a fixed angle relative to the microtubule lattice has also been reported for Ndc80 (Wilson-Kubalek et al., 2008) but the implications of defined projection angles for any microtubule binding protein is thus far unknown. The crossbridge also determines the inter-microtubule spacing between two microtubules. This inter-microtubule distance could also affect the ability of motor proteins to bind and slide PRC1 crosslinked microtubules, providing an additional mechanism for activating or deactivating specific motors at these structures. For example, the reported length of kinesin-5 motor is approximately 95 nm, which is ~ 2-fold greater than the 37 nm inter-microtubule spacing of PRC1 crosslinked microtubules (Kashina et al., 1996). Hence, the reduction in the velocity of microtubule sliding by kinesin-5 seen in our experiments at high PRC1 concentrations could result from the inability of kinesin-5 to bind properly and efficiently walk along both microtubules with dense PRC1 crosslinks. Such modulation of motor activity through control of inter-filament spacing has been proposed to affect the magnitude of active forces generated in striated muscles during muscle contraction (Millman, 1998).
Based on our results, we propose a structural model for how polarity-specific crosslinking is mediated by PRC1 (Figure 7A, B). The ability of a microtubule associated protein to distinguish parallel and anti-parallel filaments relies on two factors. First, PRC1 molecules must decode filament polarity when bound to one microtubule. Our results show that the spectrin domain is the most ordered region in a PRC1-microtubule complex and uses a well-defined surface for microtubule binding. This suggests that this domain makes contacts with the microtubule lattice that decode filament orientation. Second, the microtubule polarity needs to be transmitted across the linker to the second spectrin domain in the PRC1 homodimer. Our data show that the dimerization domain in PRC1 has a single conformation when crosslinking two microtubules. The structural rigidity of this domain is likely to be responsible for PRC1's selectivity for anti-parallel microtubules. Though specificity can be achieved by oriented binding of the spectrin domain and rigidity in the linker domain, the weak microtubule binding affinity of the spectrin domain alone does not explain the extensive filament bundling induced by PRC1. To increase binding and consequently crosslinking efficiency, PRC1 uses an unstructured positively charged domain for maintaining long-lived associations with microtubules. Additionally, the flexibility in the linker domain of PRC1, which appears to have more than one conformation when bound to one microtubule, may allow for an initial contact with a second microtubule that could have a wide-range of orientations. Relative to a highly rigid structure, such flexibility could increase the crosslinking efficiency of PRC1 molecules. These features would enable PRC1 to stay associated with the first microtubule encountered, explore its length by 1-D diffusion, and thereby increase the probability of capturing a second filament for establishing anti-parallel linkages between two microtubules.
FIGURE 7.
PRC1 is a compliant, microtubule-overlap tracking protein that tunes structural rigidity to specifically crosslink two anti-parallel microtubules. (A) A model for how PRC1 can align microtubules into anti-parallel arrays. The spectrin domain in PRC1 can make oriented contacts with the microtubule to decode filament polarity. The unstructured domain acts to enhance the binding affinity, while allowing diffusion along microtubules. The dimerization domain is not entirely rigid on a single microtubule but adopts a specific conformation when crosslinking two microtubules. (B) The rigidity and flexibility of the different domains in PRC1 can facilitate sorting of randomly oriented microtubules into anti-parallel arrays and allow PRC1 to function as a selective `mark' for microtubule overlap regions.
Many cellular processes require recognition of a “mark” at precise locations. Post-translational modifications such as ubiquitination and methylation are some of the common marking mechanisms for proteins and DNA in cells. In the microtubule cytoskeleton, the +TIP-proteins track and identify microtubule growing microtubule plus-ends (Akhmanova and Steinmetz, 2008). Similarly, by recognizing specific microtubule geometries and forming compliant crosslinks, the MAP65 proteins can mark anti-parallel microtubule overlap during the self-organization of dynamic cytoskeletal networks.
EXPERIMENTAL PROCEDURES
Crystallization of PRC1-S
PRC1-S (341–466) was concentrated to 50 mg/ml in motility buffer (80 mM PIPES (pH 6.8), 1 mM MgCl2, 1 mM EGTA) and 150 mM KCl and crystallized by hanging drop vapor diffusion at 4°C using an equal volume of protein sample and crystallization solution consisting of 0.1 M CHES (pH 9.5) and 30–35% PEG 3350. Crystals (needles of dimension 25 × 25 × 300 μm) appeared between 1 – 3 weeks. Microseeding was used to obtain crystals of the selenomethionyl derivative of the construct. Crystals were frozen in liquid nitrogen after a 5 min soak in a solution comprising of the crystallization solution with 5–10 % v/v glycerol. Both native and SAD datasets were collected along single needles at NE-CAT beamline 24ID-E at APS at the peak selenium wavelength 0.97918 Å and processed as described in the supplement.
Cryo-EM and image analysis
Microtubules were polymerized as previously described (Wilson-Kubalek et al., 2008). Microtubules (0.166 mg/ml in motility buffer) were applied to plasma cleaned C-flat grids and incubated with PRC1-NΔC (0.53 mg/ml) or PRC1-SC' (0.9 mg/ml). The sample was then vitrified in liquid ethane, using a manual plunger. The data sets were collected using a GATAN cryoholder and a FEI Tecnai F20 transmission electron microscope equipped with a Gatan Ultrascan 4000×4000 pixel CCD camera. Images of the decorated microtubules were recorded in low-dose conditions (<10 e/Å2) at a magnification of ~ 29,000 and a nominal defocus range from 1.5 to 2.0 μm. For image analysis of the PRC1-microtubule complexes, PRC1-decorated 15-protofilament microtubules were selected from CCD images. 3D reconstructions were calculated using Phoelix, essentially as described elsewhere (Whittaker et al., 1995).
3D EM reconstructions
Surface representations of side- and top- views of the cryo-EM reconstructions (Figure 4 and S4) were produced using the Chimera software package (Pettersen et al., 2004). To interpret the PRC1 densities protruding from the microtubule density, we manually docked the crystal structure of the spectrin domain into the EM density reconstructions using Chimera.
Cryotomography
Tilt-series images were acquired using the Serial EM software package (Mastronarde, 2005) on a Tecnai F20 microscope through a range of +/− 50° at 1.5° increments. The specimen was subjected to a total dose of ~100 electrons. The tomographic image reconstruction was performed using the IMOD software package (Kremer et al., 1996).
Fluorescence microscopy
All experiments were performed on an instrument described previously (Kapitein et al., 2008b). GFP was imaged with TIRF illumination from a 491-nm laser source (Cobalt Calypso 50; Solamere Technology) and X-rhodamine was imaged with wide-field illumination. For all experiments, oxygen scavenging mix (OS) comprising of 25 mM glucose, 40 mg/ml glucose oxidase, 35 mg/ml catalase and 0.5% beta-mercaptoethanol was included in the final buffer.
Kinesin-5/PRC1 sliding experiments were performed as described previously with some modifications (Kapitein et al., 2008b). Briefly, X-rhodamine-labeled biotinylated taxol-stabilized microtubules were immobilized on the coverslip by first coating the surface with biotinylated BSA followed by streptavidin. After a brief incubation with casein to block non-specific binding to the surface, GFP-PRC1-FL was added and allowed to bind the immobilized microtubules. This was followed by addition of X-rhodamine-labeled microtubules which generated a `sandwich' with two microtubules crosslinked by PRC1. After washing the solution microtubules, the final assay mix comprising of GFP-PRC1-FL and/or kinesin-5 in motility buffer supplemented with 80 mM KCl, 2 mM MgCl2, 1 mg/mL k-casein, 1 mM MgATP, 20 μM taxol and OS mix was added to the flow chamber. TIRF and wide-field images were acquired near simultaneously with 0.2 s exposure, 0.1 s−1 frame rate and EM gain set to 100 and 200 for TIRF and wide-field respectively. Each pair of moving microtubule was analyzed by kymographs using MetaMorph (MDS Analytical Technologies). Sliding velocities were calculated from the slopes of the diagonal streaks generated by the movement of one speckled microtubule over an immobilized microtubule. In a few rare events (< 5 %), we observed an abrupt ~2-fold change in microtubule movement velocity in the kymographs. Based on our analyses of kinesin-5 sliding alone (without PRC1), we believe this is due to kinesin-5 switching between walking on both microtubules it crosslinks to walking on only one of the two microtubules. This change in motility mode leads to a 2-fold reduction in relative filament sliding. In these cases, both velocities were measured and are included in the histograms shown.
HIGHLIGHTS.
Human MAP65, PRC1 uses structured and unstructured microtubule binding domains
PRC1 is flexible on one microtubule but rigid when crosslinking two filaments
PRC1 crosslinks do not substantially resist kinesin-5 driven filament sliding
Compliant PRC1 crosslinks can dynamically track antiparallel microtubule overlap
Supplementary Material
ACKNOWLEDGMENTS
We thank K. Rajashankar (Argonne Advanced Photon Source) and Deena Oren (Rockefeller University Structural Biology Resource Center (SBRC)) for technical assistance. T.M.K. is grateful to the NIH (GM65933 and GM65933-S1) for support. R.S. is a recipient of the Rockefeller Women and Science postdoctoral fellowship. R.A.M. acknowledges support from the NIH (GM52468). We also acknowledge the National Resource for Automated Molecular Microscopy (NIH RR17573), the SBRC (NIH 1S10RR022321-01), and the Keck Facility at Yale University (NIH 1S10RR023748-01) for instrument use.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers Coordinates have been deposited with PDB IDs 3NRX (native crystal structure), 3NRY (SelenoMet crystal structure) and EMDB accession code EMD-5205 (EM map)
REFERENCES
- Akhmanova A, Steinmetz MO. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol. 2008;9:309–322. doi: 10.1038/nrm2369. [DOI] [PubMed] [Google Scholar]
- Amos LA, Schlieper D. Microtubules and maps. Adv Protein Chem. 2005;71:257–298. doi: 10.1016/S0065-3233(04)71007-4. [DOI] [PubMed] [Google Scholar]
- Carter AP, Garbarino JE, Wilson-Kubalek EM, Shipley WE, Cho C, Milligan RA, Vale RD, Gibbons IR. Structure and functional role of dynein's microtubule-binding domain. Science. 2008;322:1691–1695. doi: 10.1126/science.1164424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J, Jensen CG, Jensen LC, Bush M, Lloyd CW. The 65-kDa carrot microtubule-associated protein forms regularly arranged filamentous cross-bridges between microtubules. Proc Natl Acad Sci U S A. 1999;96:14931–14936. doi: 10.1073/pnas.96.26.14931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Dos Reis G, Maiolica A, Polka J, De Luca JG, De Wulf P, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL, Ultsch MH, de Vos AM, Wells JA. Convergent solutions to binding at a protein-protein interface. Science. 2000;287:1279–1283. doi: 10.1126/science.287.5456.1279. [DOI] [PubMed] [Google Scholar]
- Djinovic-Carugo K, Gautel M, Ylanne J, Young P. The spectrin repeat: a structural platform for cytoskeletal protein assemblies. FEBS Lett. 2002;513:119–123. doi: 10.1016/s0014-5793(01)03304-x. [DOI] [PubMed] [Google Scholar]
- Gaillard J, Neumann E, Van Damme D, Stoppin-Mellet V, Ebel C, Barbier E, Geelen D, Vantard M. Two microtubule-associated proteins of Arabidopsis MAP65s promote antiparallel microtubule bundling. Mol Biol Cell. 2008;19:4534–4544. doi: 10.1091/mbc.E08-04-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M. The 3Ms of central spindle assembly: microtubules, motors and MAPs. Nat Rev Mol Cell Biol. 2009;10:9–20. doi: 10.1038/nrm2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes GJ, Dong Y, McEwen BF, Deluca JG. Kinetochoremicrotubule attachment relies on the disordered N-terminal tail domain of Hec1. Curr Biol. 2008;18:1778–1784. doi: 10.1016/j.cub.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentrich C, Surrey T. Microtubule organization by the antagonistic mitotic motors kinesin-5 and kinesin-14. J Cell Biol. 2010;189:465–480. doi: 10.1083/jcb.200910125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt LJ, Tuch BB, Villen J, Johnson AD, Gygi SP, Morgan DO. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson ME, Loughlin R, Loiodice I, Fu C, Brunner D, Nedelec FJ, Tran PT. Crosslinkers and motors organize dynamic microtubules to form stable bipolar arrays in fission yeast. Cell. 2007;128:357–368. doi: 10.1016/j.cell.2006.12.030. [DOI] [PubMed] [Google Scholar]
- Jiang W, Jimenez G, Wells NJ, Hope TJ, Wahl GM, Hunter T, Fukunaga R. PRC1: a human mitotic spindle-associated CDK substrate protein required for cytokinesis. Mol Cell. 1998;2:877–885. doi: 10.1016/s1097-2765(00)80302-0. [DOI] [PubMed] [Google Scholar]
- Kapitein LC, Janson ME, van den Wildenberg SM, Hoogenraad CC, Schmidt CF, Peterman EJ. Microtubule-driven multimerization recruits ase1p onto overlapping microtubules. Curr Biol. 2008a;18:1713–1717. doi: 10.1016/j.cub.2008.09.046. [DOI] [PubMed] [Google Scholar]
- Kapitein LC, Kwok BH, Weinger JS, Schmidt CF, Kapoor TM, Peterman EJ. Microtubule cross-linking triggers the directional motility of kinesin-5. J Cell Biol. 2008b;182:421–428. doi: 10.1083/jcb.200801145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashina AS, Baskin RJ, Cole DG, Wedaman KP, Saxton WM, Scholey JM. A bipolar kinesin. Nature. 1996;379:270–272. doi: 10.1038/379270a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskii A, Roostalu J, Roque H, Antony C, Schiebel E. Phosphorylation-dependent protein interactions at the spindle midzone mediate cell cycle regulation of spindle elongation. Dev Cell. 2009;17:244–256. doi: 10.1016/j.devcel.2009.06.011. [DOI] [PubMed] [Google Scholar]
- Korneev MJ, Lakamper S, Schmidt CF. Load-dependent release limits the processive stepping of the tetrameric Eg5 motor. Eur Biophys J. 2007;36:675–681. doi: 10.1007/s00249-007-0134-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Loiodice I, Staub J, Setty TG, Nguyen NP, Paoletti A, Tran PT. Ase1p organizes antiparallel microtubule arrays during interphase and mitosis in fission yeast. Mol Biol Cell. 2005;16:1756–1768. doi: 10.1091/mbc.E04-10-0899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AL, Compton DA. Structural and regulatory roles of nonmotor spindle proteins. Curr Opin Cell Biol. 2008;20:101–106. doi: 10.1016/j.ceb.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Miller SA, Johnson ML, Stukenberg PT. Kinetochore attachments require an interaction between unstructured tails on microtubules and Ndc80(Hec1) Curr Biol. 2008;18:1785–1791. doi: 10.1016/j.cub.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millman BM. The filament lattice of striated muscle. Physiol Rev. 1998;78:359–391. doi: 10.1152/physrev.1998.78.2.359. [DOI] [PubMed] [Google Scholar]
- Neef R, Gruneberg U, Kopajtich R, Li X, Nigg EA, Sillje H, Barr FA. Choice of Plk1 docking partners during mitosis and cytokinesis is controlled by the activation state of Cdk1. Nat Cell Biol. 2007;9:436–444. doi: 10.1038/ncb1557. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Ribbeck K, Groen AC, Santarella R, Bohnsack MT, Raemaekers T, Kocher T, Gentzel M, Gorlich D, Wilm M, Carmeliet G, et al. NuSAP, a mitotic RanGTP target that stabilizes and cross-links microtubules. Mol Biol Cell. 2006;17:2646–2660. doi: 10.1091/mbc.E05-12-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasabe M, Machida Y. MAP65: a bridge linking a MAP kinase to microtubule turnover. Curr Opin Plant Biol. 2006;9:563–570. doi: 10.1016/j.pbi.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Schuyler SC, Liu JY, Pellman D. The molecular function of Ase1p: evidence for a MAP-dependent midzone-specific spindle matrix. Microtubule-associated proteins. J Cell Biol. 2003;160:517–528. doi: 10.1083/jcb.200210021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slep KC, Vale RD. Structural basis of microtubule plus end tracking by XMAP215, CLIP-170, and EB1. Mol Cell. 2007;27:976–991. doi: 10.1016/j.molcel.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thadani R, Ling YC, Oliferenko S. The fission yeast TACC protein Mia1p stabilizes microtubule arrays by length-independent crosslinking. Curr Biol. 2009;19:1861–1868. doi: 10.1016/j.cub.2009.09.063. [DOI] [PubMed] [Google Scholar]
- Valentine MT, Block SM. Force and premature binding of ADP can regulate the processivity of individual Eg5 dimers. Biophys J. 2009;97:1671–1677. doi: 10.1016/j.bpj.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat Struct Mol Biol. 2007;14:54–59. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- Whittaker M, Carragher BO, Milligan RA. PHOELIX: a package for semi-automated helical reconstruction. Ultramicroscopy. 1995;58:245–259. doi: 10.1016/0304-3991(95)00057-8. [DOI] [PubMed] [Google Scholar]
- Wilson-Kubalek EM, Cheeseman IM, Yoshioka C, Desai A, Milligan RA. Orientation and structure of the Ndc80 complex on the microtubule lattice. J Cell Biol. 2008;182:1055–1061. doi: 10.1083/jcb.200804170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Sato M, Fujita A, Yamamoto M, Toda T. The roles of fission yeast ase1 in mitotic cell division, meiotic nuclear oscillation, and cytokinesis checkpoint signaling. Mol Biol Cell. 2005;16:1378–1395. doi: 10.1091/mbc.E04-10-0859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C. NuMA: a nuclear protein involved in mitotic centrosome function. Microsc Res Tech. 2000;49:467–477. doi: 10.1002/(SICI)1097-0029(20000601)49:5<467::AID-JEMT9>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Zhu C, Lau E, Schwarzenbacher R, Bossy-Wetzel E, Jiang W. Spatiotemporal control of spindle midzone formation by PRC1 in human cells. Proc Natl Acad Sci U S A. 2006;103:6196–6201. doi: 10.1073/pnas.0506926103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.