Abstract
Structural information on complex biological RNA molecules can be exploited to design tectoRNAs or artificial modular RNA units that can self-assemble through tertiary interactions thereby forming nanoscale RNA objects. The selective interactions of hairpin tetraloops with their receptors can be used to mediate tectoRNA assembly. Here we report on the modulation of the specificity and the strength of tectoRNA assembly (in the nanomolar to micromolar range) by variation of the length of the RNA subunits, the nature of their interacting motifs and the degree of flexibility of linker regions incorporated into the molecules. The association is also dependent on the concentration of magnesium. Monitoring of tectoRNA assembly by lead(II) cleavage protection indicates that some degree of structural flexibility is required for optimal binding. With tectoRNAs one can compare the binding affinities of different tertiary motifs and quantify the strength of individual interactions. Furthermore, in analogy to the synthons used in organic chemistry to synthesize more complex organic compounds, tectoRNAs form the basic assembly units for constructing complex RNA structures on the nanometer scale. Thus, tectoRNA provides a means for constructing molecular scaffoldings that organize functional modules in three-dimensional space for a wide range of applications.
INTRODUCTION
The use of DNA as a medium to construct a variety of three-dimensional (3D) nanoscale objects of defined topology has been extensively developed by the work of Seeman (1,2). For example, branched DNA molecular units assembled through the formation of Watson–Crick base pairings followed by covalent ligation, have been used to create geometrical nanoscale objects (3,4) as well as two-dimensional (2D) quasi-crystalline arrays (5,6). To date, DNA nanotechnology has essentially exploited self-assembly by Watson–Crick base complementarity (6–8) and, to a lesser extent, formation of guanine quartet structures (9,10). A switchable nano-molecular device based on the conversion of a stretch of right-handed B-DNA duplex into a left-handed Z-DNA duplex was recently constructed, opening the way to controllable DNA nanostructures under appropriate conditions (11).
In contrast to DNA, which is often regarded as a rather simple and monotonous helical structure, natural RNA molecules exhibit a breathtaking range of complex 3D structures that give rise to diverse functions (12). Perhaps the most stunning and sophisticated machine based on RNA is the ribosome (13–17). Interestingly, complex RNA molecules can be analyzed as hierarchically organized modular objects (18). Despite the fact that RNA offers a wider structural diversity than DNA, it is only recently that the growing body of structural information has been applied to create artificial nanoscale RNA objects. We have recently shown that specific, non-Watson–Crick RNA tertiary interactions can be exploited to construct ‘tectoRNA’ molecular units, defined as RNA molecules capable of self-assembly to form nanoscale structures (19). The use of such type of tertiary interactions allows one to control and modulate the assembly process by manipulating cation concentration (e.g. Mg2+), and employing modularly designed ‘selector’ RNA molecules. For the self-assembly of one-dimensional arrays, we designed a basic modular unit that comprises a 4-way junction with an interacting module on each helical arm. The interacting module is either a GAAA loop or a specific GAAA loop receptor (20). Each tectoRNA was shown to interact with two other tectoRNAs via the formation of four loop–receptor interactions, two with each partner molecule. Although RNA nano-objects containing more than 16 units could be obtained, it was clear, at that point, that more knowledge was required to control the size and shape of the nanoscale objects generated through tectoRNA self-assembly.
For instance, to construct directional, ordered assemblies, in which the tectoRNA units assemble with a specific orientation and in a specific linear order, two or more specific interaction motifs are needed. Moreover, to vary and control the size and diameter of the linear assemblies, the distance between two interaction motifs should be varied. In the present work we have tested the effects on the binding affinity of different tectoRNA molecules of varying the length, helical twist and flexibility of the linker region that separates the interacting motifs in each module. We demonstrate that several of these tectoRNAs can associate at sub-micromolar concentrations, suggesting that a wide range of assembly units can be used for RNA nano-construction. Moreover, this work shows that tectoRNAs can be used as scaffoldings to determine binding affinities of different RNA–RNA interactions.
MATERIALS AND METHODS
Design of the RNA scaffoldings
The sequence of each RNA molecule was chosen to minimize the occurrence of alternative secondary structures. Sequences were checked by submitting them to the RNA folding program Mfold (version 3.0, http://bioinfo.math.rpi.edu/~zukerm/rna/mfold-3.0.html) which predicts the thermodynamically favored secondary structure of a given RNA sequence, for comparison with the desired secondary structure. The three major RNA scaffoldings presented in this paper were manually modeled in three dimensions using MANIP (21) and the 3D views were generated with DRAWNA (22). The 3D models of the dimeric forms of each scaffolding, were assembled by manipulating two identical loop–receptor motifs, extracted from the structure of the P4–P6 domain of the group I intron (NDB file URX053) (23) to allow the insertion of A-form helices of 10, 21 or 32 bp.
DNA template and RNA synthesis
RNA molecules were prepared in vitro by run-off transcription of DNA templates using T7 RNA polymerase. DNA templates for in vitro transcription were generated by amplifying a synthetic DNA molecule (moleculeN.mat), coding for the antisense sequence of the desired RNA molecule, with a forward primer (moleculeN.fwd) of 36 nt containing the T7 RNA polymerase promoter, and a reverse primer of 20 nt (moleculeN.rev). Sequences of oligonucleotides used are given in Table S1 (see Supplementary Material available at NAR Online). The forward and reverse primers were designed to hybridize to the template sequence (moleculeN.mat) with Tm ~52°C. Typical PCR reactions were carried out in a total volume of 200 µl and contained 2 mM MgCl2, 50 mM KCl, 10 mM Tris–HCl pH 8.0, 0.5% NP-40, 1 mg/ml gelatin, 0.5 mM of each dNTP, 2 nM of moleculeN.mat, and 1 µM moleculeN.fwd and moleculeN.rev. The reactions were calibrated to produce 150 pmol of DNA template after 15 cycles (94°C, 1 min 15 s; 52°C, 1 min 15 s; 72°C, 1 min 15 s). After purification of the PCR products using the QiaQuick PCR purification kit (Qiagen), 25 pmol of each DNA template were incubated for 3 h at 37°C with T7 RNA polymerase (10 U/µl) in a buffer containing 15 mM MgCl2, 2 mM spermidine, 50 mM Tris 7.5, 2.5 mM of each NTP, 10 mM DTT, 0.01 µg/µl inorganic pyrophosphatase and 0.8 U/µl RNasin. After the reaction, the DNA templates were degraded by incubating with FPLC pure RQ1 DNase (0.3 U/µl) for 20 min at 37°C. The RNA product was purified by denaturing polyacrylamide gel electrophoresis (PAGE) (10–15% acrylamide, 8 M urea). After elution overnight at room temperature, in 300 mM NaCl, 10 mM Tris pH 7.5, 0.5 mM EDTA, the RNA was ethanol precipitated, rinsed twice with 85% ethanol, dried and dissolved in water. Transcription reactions typically yielded 400–500 pmol of RNA.
RNA labeling
Each RNA transcript (10–20 pmol) was 3′ end-labeled with [32P]pCp in a reaction volume of 10 µl, using T4 RNA ligase (4 U/µl) and 5′-[32P]pCp (5 µl; 3000 Ci/mmol) for 12 h at 4°C, in a buffer containing 50 mM Tris pH 7.8, 10 mM MgCl2, 20 mM DTE, 10 µg/ml BSA, 0.1 mM ATP and 10% DMSO. After addition of 10 µl of 8 M urea/blue (0.01% bromophenol blue, 0.01% xylene cyanol), the labeled material was purified on denaturing polyacrylamide gels (10–15% acrylamide, 8 M urea).
Dissociation constant (Kd) determination by native gel electrophoresis
RNA samples containing a fixed amount (0.1 nM) of 3′-end [32P]pCp-labeled RNA and sufficient unlabeled RNA to give the indicated concentrations were heated in water at 90°C for 1 min, immediately cooled on ice for 2 min and then allowed to dimerize for 15–30 min at 30°C in 89 mM Tris–borate pH 8.3, 15 mM Mg(OAc)2 and 5% glycerol. We have checked that under these conditions the equilibrium between monomeric and dimeric forms is reached after 15 min for the parent molecules (1 and 2 in Fig. 1). Dimerization was allowed to occur for 30 min for all the other molecules tested. For analysis, 10 µl of the RNA sample was combined with 1 µl of gel-loading buffer (same buffer with 0.01% bromophenol blue, 0.01% xylene cyanol) and run at 4°C on 9% (30:1) non-denaturing polyacrylamide gels. The gel and running buffers were identical to those used for dimerization except that glycerol was not included. Monomer and dimer bands were quantified on a BioImager BAS2000 (Fuji) and the dimer formation was correlated with RNA concentration. Kds were determined as the concentration at which half the RNA molecules are dimerized.
Figure 1.
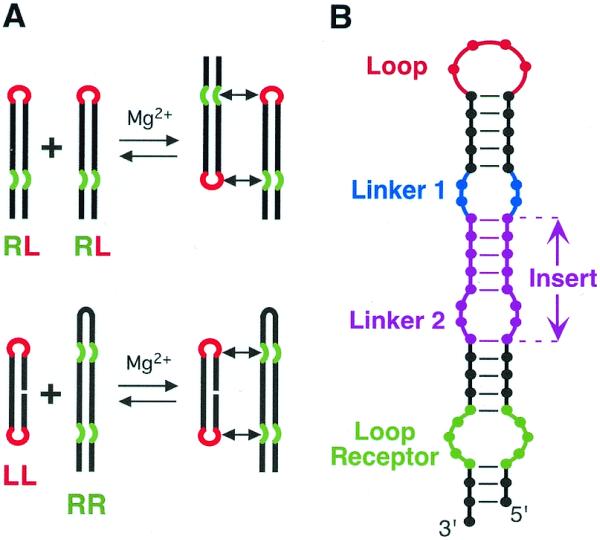
TectoRNAs employing two loop–receptor motifs and their modes of assembly. (A) Two modes of assembly used in this study. (B) Schematic of RNA assembly unit showing elements varied for this study. The tetraloop (L) is shown in red, the tetraloop receptor (R) in green, the linker (or hinge) in blue and the insert [comprising a helix and a second linker (or hinge)] in magenta (refer also to the 2D and 3D models in Figs 2 and 3).
Lead (Pb2+)-induced cleavage
RNA samples at various concentrations (including a fixed amount, 1–2 nM, of 3′-end [32P]pCp-labeled RNA) were treated as indicated above, except that the dimerization buffer comprised 15 mM Mg(OAc)2 and 25 mM HEPES pH 7.5. After addition of 1 mg of crude tRNA, Pb2+ cleavage was induced by adding 8 or 16 mM Pb(OAc)2 and stopped after 5 min by adding 50 mM EDTA followed by ethanol precipitation. RNA fragments were separated on denaturing polyacrylamide gels (15% acrylamide, 8 M urea).
RESULTS
Design of tectoRNAs capable of self-assembly
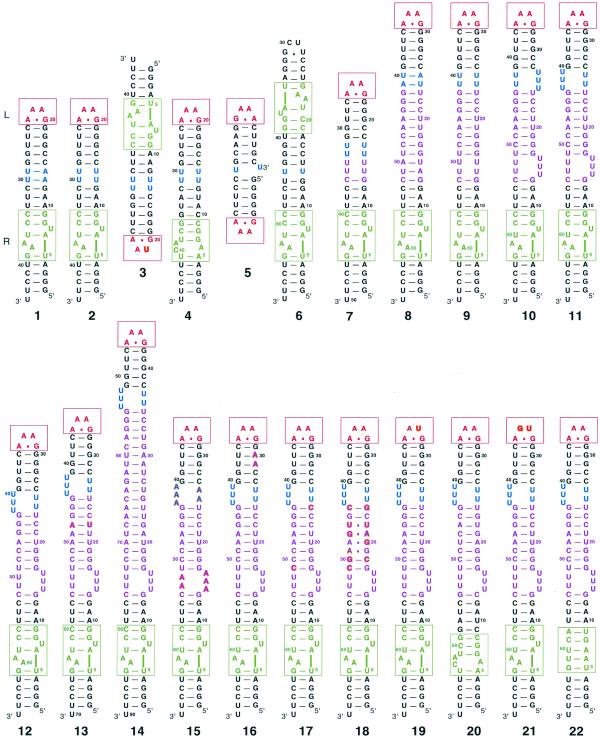
We have previously defined two modes of tectoRNA assembly (19) (Fig. 1A). The first mode implies assembly between molecules each with a tetraloop and a loop receptor whereas the second implies the recognition of a molecule with two tetraloops by another with two receptors. The schematic of the tectoRNA scaffolding used for the design of almost all the molecules from this work is shown in Figure 1B. Besides the structural features of the tetraloop–receptor interaction that impose specific constraints on the design, the distance and structural nature of linkers and insert regions that separate the two interacting modules are also expected to dramatically influence tectoRNA self-assembly (Fig. 1). Sequences and secondary structures of all constructs are shown in Figure 2, and 3D models of selected complexes in Figure 3. The structural features and Kd of tectoRNA complexes are summarized in Table 1.
Figure 2.
Secondary structures of all molecules reported (see Table 1). All molecules in the second row are derived from molecule 11. The bases that differ from molecule 11 are shown in bold. A red box is drawn around the tetraloop and a green box around the tetraloop receptor. The color code is the same as Figure 1. Molecules 3 and 4 as well as 5 and 6 are drawn to show their mode of interaction. All molecules were synthesized as described in the Materials and Methods.
Figure 3.
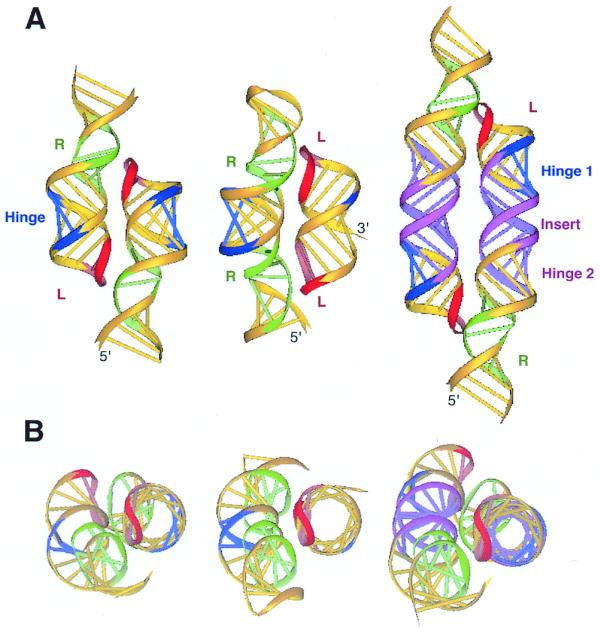
3D models of representative tectoRNA units. (A) Dimers formed by molecules 1 (left), 5 interacting with 6 (center), and 8 (right). (B) The respective top views are shown. The color code is the same as in Figures 1 and 2. L, tetraloop; R, tetraloop receptor.
Table 1. Kd of tectoRNA complexes.
| Molecule
number |
Loops/receptors |
Insert
length (bp) |
Nature of
linker (hinge) |
Substitutions |
Dimerization
in conc. range <15 µM |
Kd value (nM) | Comments |
| 1 | RL-GAAA | 0 | (all W.C. pairs) | – | Yes | 4.3 ± 0.4 | |
| 2 | RL-GAAA | 0 | U2/U2 | – | Yes | 4.2 ± 0.8 | |
| 3 | L-GUAA | 0 | U2/U2 | – | No | – | |
| 4 | R-GUAA | 0 | U2/U2 | – | No | – | |
| 3 + 4 | L-GUAA + R-GUAA | Yes | 150 ± 21 | Kinetic equilibrium | |||
| 2 + 3 | L-GUAA + RL-GAAA | Yes | 4600 ± 1400 | Kinetic equilibrium | |||
| 2 + 4 | R-GUAA + RL-GAAA | No | – | ||||
| 5 | LL-GAAA | +1 | U/Nick | – | No | – | |
| 6 | RR-GAAA | –1 | U/U | – | No | ||
| 5 + 6 | LL-GAAA + RR-GAAA | Yes | 210 ± 85 | Kinetic equilibrium | |||
| 7 | RL-GAAA | +3 | GU4/U4C | – | No | – | |
| 8 | RL-GAAA | +11 | (all W.C. pairs) | – | Yes | 100 ± 3 | Kinetic equilibrium |
| 9 | RL-GAAA | +11 | U2-U2/U2-U2 | – | Yes | 500 ± 260 | Kinetic equilibrium |
| 10 | RL-GAAA | +11 | U3-U3/U2-U2 | – | Yes | 68 ± 28 | |
| 11 | RL-GAAA | +11 | U3-U2/U3-U2 | – | Yes | 60 ± 14 | |
| 12 | RL-GAAA | +10 | U3-U2/U3-U2 | – | Yes | 540 ± 65 | Kinetic equilibrium |
| 13 | RL-GAAA | +12 | U3-U2/U3-U2 | – | Yes | 68 ± 23 | |
| 14 | RL-GAAA | +22 | U3-U2/U3-U2 | No | – | ||
| 15 | RL-GAAA | +11 | A3-A2/A3-A2 | – | Yes | 130 ± 15 | Kinetic equilibrium |
| 16 | RL-GAAA | +11 | U3-U2/U3-U2 | G29A and G30A | Yes | 70 | Kinetic equilibrium |
| 17 | RL-GAAA | +11 | U3-U2/U3-U2 | U24C and U51C | Yes | 70 | |
| 18 | RL-GAAA | +11 | U3-U2/U3-U2 | G18C, U20G, U21A, C22U, U24G, G44C, G46U, A47G, C49A, C50G, U51C | Yes | 26 ± 4 | |
| 19 | L-GUAA | +11 | U3-U2/U3-U2 | No | – | ||
| 20 | R-GUAA | +11 | U3-U2/U3-U2 | No | – | ||
| L-GUAA + R-GUAA | Yes | 3500 | Kinetic equilibrium | ||||
| 21 | L-GUGA | +11 | U3-U2/U3-U2 | No | – | ||
| 22 | R-GUGA | +11 | U3-U2/U3-U2 | No | – | ||
| 21 + 22 | L-GUGA + R-GUGA | No | – |
The structural features of each tectoRNA are summarized in columns 2–5, including the nature of the tetraloop and the specificity of the tetraloop receptor (column 2), the length of the linker insert (column 3), the nature of the linker (column 4) and base substitutions in the molecule (column 5). Kd were measured at 15 mM Mg(OAc)2, by quantitating bands from native gel electrophoresis as described in the Materials and Methods and are given in column 7. Kd values with standard deviations are averages of parameters measured from three independent experiments. For some constructs, the mobility of the dimer band increased as RNA concentration decreased, eventually merging with the monomer band. In these cases Kd was estimated from the mobility of the RNA dimer. This behavior was attributed to kinetic equilibrium between monomer and dimer, as indicated in column 8.
In a previous work (19), we have taken advantage of the X-ray structure of the tertiary interaction formed by the GAAA tetraloop and its ‘11-nucleotide motif’ receptor (23), to build by computer modeling the smallest tectoRNA scaffolding able to self-dimerize upon formation of two interaction motifs. The best representative was molecule 1 for which a regular A-form double helix of 10 Watson–Crick base pairs between the GAAA loop and the GAAA loop receptor was found to be optimal for self-dimerization (see Figs 2 and 3). Dimerization of molecule 1 was dependent upon magnesium and occurred with high affinity [Kd = 4.3 nM at 15 mM Mg(OAc)2], as determined by PAGE mobility shift analysis [Fig. 4; see also fig. 2 from (19)]. The cooperative self-association upon magnesium was found to require the binding of two magnesium ions per dimer (19). Molecule 2 was then generated to determine whether the binding affinity could be increased by the introduction of structural flexibility within the tectoRNA. To do so, we changed the two adjacent U–A base pairs within linker 1 to U–U mismatches known to be less stable than classical Watson–Crick base pairs (24). Uridines can bulge out of the helix or can base pair with each other in a number of ways (25). Nevertheless, according to the crystallographic structure of tandem UU base pairs within an RNA dodecamer, the tandem U–U mismatches within the tectoRNA context was expected to form U·U wobble pairs, altering the course of the helical axis by 11–12° (26). As previously reported, molecule 2 dimerized with an affinity that was indistinguishable from that of molecule 1, even though Pb2+ cleavage experiments showed the hinge region (linker 1) to be more flexible in molecule 2 (19).
Figure 4.
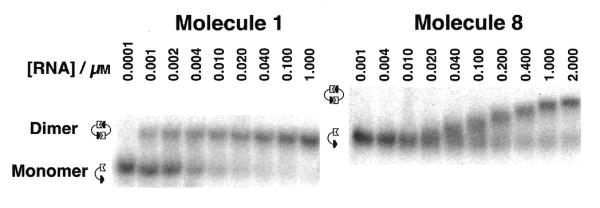
Autoradiograms of native gels used to characterize the dimerization of tectoRNAs. Experiments were carried out in the presence of 15 mM Mg(OAc)2, as described in the Materials and Methods. The behavior of molecule 1 (left) shows two distinct bands, one for the monomer and one for the dimer. By contrast, molecule 8 (right) is typical of molecules showing fast exchange kinetics. The mobility of the dimer band of 8 varies continuously with concentration, in the concentration range where significant populations of each form exist in solution, indicating a dynamic equilibrium between monomer and dimer forms. Because of their different size, molecules 1 and 8 have different migration mobility on native polyacrylamide gels.
Given that no difference in Kd was observed for molecules 1 and 2, we decided to use molecule 2 for further experiments to test the ability of this scaffolding to accommodate other interaction motifs. We reasoned that the slightly greater flexibility of 2 could allow for small structural adjustments to accommodate different motifs in their optimal orientations. Thus, we synthesized molecules 3 and 4 in which either the loop or the receptor motif was changed. In molecule 3 (‘L-GUAA’, Table 1), GUAA replaces the GAAA loop, whereas in molecule 4 (‘R-GUAA’), the GAAA receptor is replaced by a GUAA-specific receptor motif found by in vitro selection (27). As expected, neither molecule dimerized, but 3 and 4 did associate with Kd = 150 ± 21 nM. Molecule 3 (L-GUAA) also associated with molecule 2, although the binding affinity was considerably weaker, but still significant (Kd = 4600 nM). These results proved the generality of our tectoRNA design that allows one to swap motifs with little change in scaffolding structure. Moreover, the differences in binding affinity observed with these constructs agreed with previous work on ribozymes, that indicated that the GAAA loop–receptor interaction is stronger than the GUAA interaction (27).
The flexibility of the scaffolding was increased even more in molecules 5 and 6 (Fig. 2). These were modeled by taking the molecule 2 dimer and swapping the loop and receptor motifs, thus creating molecule 5, with two GAAA loops and molecule 6, with two GAAA receptors (Fig. 3). As predicted from modeling, 5 and 6 associated, although neither was capable of self-dimerization. However, the Kd of the 5 + 6 complex was found to be higher than that of the 2 dimer or even of the 3 + 4 complex (Table 1). This experiment showed that two RNA modules linked by a single-strand could still bind cooperatively, although the affinity was less than when the two modules were more rigidly pre-organized for binding.
Self-assembly of tectoRNAs of greater length
According to 3D models (Fig. 3), the helical distance separating the two interacting motifs and the resulting helical twist are critical for the association. Indeed, when molecule 7 was constructed by adding an insert (Fig. 1B) comprising a GC base pair and two more Us to each strand of the linker in molecule 2 (Fig. 2), it showed no detectable dimerization in the range of RNA concentrations studied (up to 15 µM). Within the context of molecule 7, the extra nucleotides prevented the cooperative association of both tertiary interaction motifs required for RNA complexation, probably because the loop was not in phase with its receptor.
Computer modeling indicated that the optimal length for a regular A-form helical insert would be 11 ± 1 nt (Fig. 3). In fact, when we constructed molecule 8 by inserting an 11 bp insert, we obtained a molecule that dimerized with relatively high affinity (Kd = 100 nM) compared with molecules 1 and 2 (Fig. 4). To improve self-association with the longer tectoRNA design, we constructed molecules 9, 10 and 11 to test the effect of substituting the Watson–Crick base pairs within linkers 1 and 2 by U-rich internal loops (see Fig. 2). As for the shorter tectoRNA design, we reasoned that these loops might act as hinges to increase the flexibility of the tectoRNA to favor a better induced fit for the loop receptor interaction. Molecule 9, which had two symmetrical U2/U2 linkers, dimerized with the lowest affinity (Table 1). This result was somewhat surprising as the introduction of symmetrical tandem UUs had no measurable consequence on the association of the shorter molecule 2. Nevertheless, according to the tandem UUs crystallographic structure, the overall bend introduced by the two tandem UUs could have been detrimental to the tertiary interactions induced fit of molecule 9. By contrast, molecules 10 and 11 that incorporated two asymmetric U3/U2 linkers, dimerized with comparable affinities, greater than that of the molecule 8 (see Table 1). While the Kds of 10 and 11 were significantly lower than that of 8 (∼65 nM versus 100 nM), they were, however, still higher than for molecules 1 or 2 (4 nM). As U3/U2 asymmetric internal loops are less stable than U2/U2 symmetric ones (28), this result is consistent with the idea that structural flexibility within the structure could tune the longer tectoRNA to associate better.
As exemplified in Figure 4, we observed two types of behavior for tectoRNA dimerization. For those molecules that dimerized with a Kd <70 nM, two distinct bands corresponding to the monomer and dimer forms were observed in the concentration range for total RNA in which both forms are significantly populated. Kd could be readily calculated from the integrated intensities of these bands. For molecules which dimerized with Kd >70 nM, the mobility of the dimer band varied continuously in the equivalent concentration range, reflecting a dynamic equilibrium. In that case, Kd could be estimated from the concentration at which the mobility was intermediate between that of the monomer and dimer forms, obtained from limiting concentrations.
Monitoring tectoRNA self-assembly by Pb2+-induced cleavage
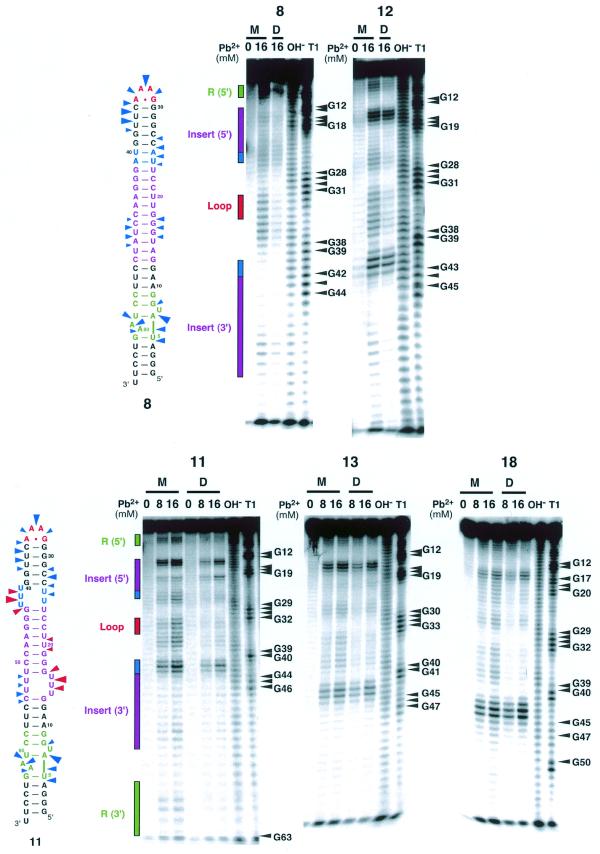
Pb2+ cleavage experiments were carried out to confirm that the longer molecules do in fact bind in the same manner as molecules 1 and 2, i.e. using the tertiary interactions. Pb2+ generally does not bind to or cleave the phosphodiester backbone in regular A-form helical regions of RNA molecules, but rather cleaves RNA backbone regions that are single-stranded, kinked or generally more flexible (29). However, Pb2+ may substitute sometimes at specific magnesium-binding sites (30). Figure 5 shows Pb2+ probing data for molecules 8, 11, 12, 13 and 18. Radiolabeled RNA molecules were reacted with Pb2+ (8 or 16 mM) in the monomer or in the dimer state in the presence of magnesium ions. As shown in Figure 5, for each molecule tested, Pb2+-cleavage of the monomer occurred primarily in the U-rich linker regions, in the GAAA hairpin loop and in the receptor motif. Also, for all molecules, dimerization led to protections in the GAAA loop and in the receptor motif, whereas the U-rich linkers remained reactive in the dimers. It has been reported that U-rich internal loops substituting for J5/5a within the P4–P6 domain of the Tetrahymena ribozyme were essentially unstructured and acted as flexible hinges (31). The comparison of Pb2+ cleavage within molecules 8, 9, 10 and 11 was also consistent with the idea that U-rich linkers, especially asymmetric ones, were acting as flexible hinges within the long tectoRNA context. One cannot rule out that U-rich linkers could specifically bind Pb2+. However, this hypothesis is unlikely considering X-ray structural studies (26), which did not show any divalent cations bound at the level of tandem UUs. Interestingly, the same protections towards Pb2+ cleavage occurred regardless of the Kd values, which range from 60 (molecule 11) to 540 nM (molecule 12). This result suggests that all the molecules tested assemble into similar dimeric structures despite their structural differences occurring at some distances from the loop receptor interactions. Thus, the difference in Kd seems to reflect the ease at which induced fit can occur for each of these molecules.
Figure 5.
Lead(II)-induced cleavage patterns for molecules 8, 11, 12, 13 and 18 in their monomeric and dimeric states. Monomer lanes are indicated by M and dimer lanes by D. To maintain the RNA in the monomer state, RNA concentrations were set to 1 nM. To achieve dimerization, the RNA concentration was adjusted to 5 µM for molecules 8 and 12, and 1 µM for 11, 13 and 18, well above the Kd in all cases. Lanes labeled OH– and T1 correspond to alkaline treatment and digestion with RNase T1, respectively. Lead(II)-induced cleavage was performed as described in the Materials and Methods. Phosphates that are cleaved in the monomeric RNA but are mostly protected in the dimeric RNA are indicated with blue arrows on the secondary structures for 8 and 11. Sites that are cleaved in both monomeric and dimeric forms are shown with red arrows. The size of the arrows is roughly proportional to the extent of cleavage in the monomer.
Tuning tectoRNAs self-assembly with minimal structural changes
The insert in molecule 11 was changed by ±1 bp to produce molecules 12 and 13. A large decrease in dimerization affinity was observed for 12, which is one base pair shorter. No measurable effect was found for molecule 13, indicating that the optimal geometric arrangement of the two tertiary motifs probably lies between molecules 11 (11 bp insert) and 13 (12 bp insert). The insert was extended by an additional helical turn to give molecule 14. This, however, did not dimerize in the range of concentrations studied (up to 15 µM).
Molecule 15 was constructed as a further test of the effect of linker flexibility on binding. It is derived from molecule 11 by replacing the asymmetric U3/U2 linkers with A3/A2. This is, in fact, the J4/5 motif from the Group I intron (NDB file URX053) which forms two tandem A·A trans Hoogsteen/Shallow-groove (‘sheared’) pairs with the fifth adenosine unpaired but inserted into the helix (23). Despite being less stable than a tandem AAs symmetrical internal loop (32), this motif adopts a specific structure that is poorly cleaved by Pb2+ (data not shown). Thus, this motif is expected to be structurally more rigid than the U3/U2 linker. Molecule 15 dimerized with less affinity than the more flexible molecule 11 from which it was derived and with slightly less affinity than the rigid molecule 8 (which has a linker consisting entirely of Watson–Crick base pairs). This result provides further evidence of the importance of flexibility in the longer scaffolding for optimal formation of tertiary interactions.
Molecules 16 and 17 incorporated more subtle variations into molecule 11, and in fact displayed binding affinities that were indistinguishable from 11. These molecules replace G·U wobble base pairs at the base of the GAAA hairpin loop (16), or adjacent to the U-rich linkers (17), with G–C pairs. This indicates that the perturbations to regular A-type helices introduced by G·U wobble pairs are not as great in the context of the tectoRNA.
Molecule 18 was derived from 11 by replacing four Watson–Crick pairs of the insert with four non-canonical base pairs comprising a recurrent internal loop motif found in 16S and 23S rRNA. Sequence analysis indicates that this motif is related to bacterial loop E of 5S rRNA (33) and probably comprises three ‘sheared’ (i.e., trans Hoogsteen/Shallow-groove) A·G pairs. An example of this motif occurs in the crystal structure (NDB file UR0009) of domain IV of Escherichia coli 4.5S RNA (34). The crystal structure reveals a greater twist for this motif compared with a comparable stretch of canonical base pairs. Remarkably, molecule 18 was found to dimerize with the highest affinity of any of the long constructs (26 nM), approaching that of the short constructs. As the loop E motif introduces an additional helical twist of half a base pair with respect to a regular helix of the same length (34), the overall helical twist of the molecule 18 insert is just between that of molecules 11 and 13. Thus, this result points to the importance of helical twist in optimizing tertiary motifs and the role of tracts of non-canonical pairs (symmetric loops) in modulating helical twist in natural RNA molecules.
We also experimented with motif substitution within the long constructs. Molecules 19 and 20 are analogs to 3 and 4 in the short series (Fig. 2). In molecule 19 the GAAA loop is replaced by GUAA while in 20 the GAAA receptor is replaced by the GUAA receptor. Neither 19 nor 20 dimerized, but the combination 19 + 20 did associate; the Kd was ~3500 nM. Replacing the GAAA loop with GUGA also resulted in a molecule that was incapable of dimerization. The receptor for GUGA is the shallow groove of a canonical helix with the sequence 5′-GA-3′/3′-CU-5′ (35). Molecule 22 was constructed with this receptor properly positioned. Association of 21 and 22 was not, however, observed at concentrations up to 15 µM. This tertiary interaction is known, in fact, to be weaker than that of GUAA for its receptor (27).
TectoRNAs as a tool for characterizing thermodynamically tertiary interactions
Special care must be taken when the energetic contribution of a specific tertiary interaction to the stability of RNA is estimated. The affinities measured are highly dependent on the structural context of the tertiary interaction within the RNA. Thus, as far as the contribution of a specific tertiary interaction to the overall stability of an RNA molecule is concerned, it is unlikely that any absolute thermodynamic data relative to a tertiary interaction will be of general interest. Nevertheless, by comparing the affinities of different specific tertiary interactions within a similar structural context, it is possible to extract meaningful information. The Kd for a single interacting GAAA loop receptor motif was estimated to be 65 µM within the scaffolding of molecule 2. The simple substitution of the second adenine in the tetraloop by a uracil increased the Kd by three orders of magnitude, indicating that at least 4 kcal/mol of binding were lost in the tertiary interaction. The Kd for tertiary interaction occurring between the GUAA tetraloop and its receptor was found to be 2.3 mM, indicating that the strength of this interacting motif is 150 times weaker than that of the GAAA loop receptor motif.
The ratio of the Kds for the 3 + 4 and the analogous 19 + 20 complexes is 3500/150 = 23. Within experimental error this equals the ratio of the Kds for the dimerization of 1 and of 11 (60/4.3 = 14). This indicates that the GUAA loop receptor motif within the long constructs introduces a degree of destabilization comparable with the one in the short constructs, independent of the second interacting motif. This suggests that the degree of destabilization due to a change within one of the two interacting motifs is independent of the tectoRNA structural context. This might not be the case for all RNA structures, however.
DISCUSSION
Recent work on the folding dynamics of complex, biologically active RNA molecules indicate that folding occurs in a hierarchical manner over a large range of time scales (36,37). Long-range tertiary interactions are responsible for critical steps to stabilize the final, fully compacted, biologically active structures of RNA molecules. Estimates of the thermodynamic parameters for these interactions are invaluable for modeling the dynamics of RNA folding and interpreting kinetic data, as well as predicting 3D structure. The present work provides one way to construct RNA scaffoldings that allow one to readily obtain thermodynamic parameters for RNA tertiary interactions. As such interactions are generally too weak to allow one to study them individually, the scaffoldings described here rely on cooperative, bimolecular RNA association using two motifs. This work indicates, however, that careful attention must be paid when designing the scaffoldings to optimize the simultaneous binding of both motifs. In addition to adjusting the helical length, one can increase the flexibility of the linkers. Another approach is to introduce helical motifs comprising non-canonical base pairs to tune the overall helical twist. Used in combination, the two approaches can result in dramatic changes in the binding affinities.
To our knowledge, the tectoRNAs described here are the first RNA molecules shown to self-dimerize by taking advantage of only non-Watson–Crick tertiary interactions. Although loop–receptor interactions participate to the assembly of large RNA molecules into their active structure (see for example 20,35,38), RNA dimerization occurs via classical Watson–Crick loop–loop interactions in nature (see for example 39–42). This could be explained by the greater strength of Watson–Crick pairings as well as their possible greater robustness to single point mutations. Indeed, a Kd in the nanomolar range can quite easily be reached by a single Watson–Crick pairing interaction (43), which is not the case for the loop–receptor interaction. Non-canonical base pairs can however contribute significantly to the stability of the Watson–Crick pairing (43).
Besides being tools for thermodynamic analysis, these tectoRNAs and their derivatives have several other interesting uses. In the emerging field of nanobiotechnology (44), they can potentially serve as molecular units for the controlled construction of RNA nano-objects of desired 3D structures. We have already used the first generation RNA unit to build one-dimensional self-assembling RNA nano-objects (19). This can be extended by the use of RNA units of greater length. For instance, by fusing two RNA units from the second generation or combining RNA units from the first generation to RNA units from the second, it is possible to modulate the width of the RNA one-dimensional array. One can also imagine creating modular units that self-assemble in a particular order by suitable permutation of interacting motifs.
Secondly, these tectoRNAs can be used as scaffolding to present various RNA structural motifs or RNA binding sites for proteins or organic molecules, in order to favor their study by NMR or their crystallization for X-ray analysis. We are presently carrying out crystallization trials that show that some of these molecules are indeed able to crystallize readily under standard conditions (J.Ng, personal communication). Moreover, tectoRNA can be employed to act as scaffolding for the selection of novel tertiary interactions.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Jessica Yingling for her help at the early stage of this work. This work was carried out in Strasbourg, France, and N.B.L. acknowledges support by grants from the NIH (1R15 GM55898) and the NIH Fogarty Institute (1-F06-TW02251-01). L.J. and E.W. thank the CNES for support (grant 96/CNES/0247).
References
- 1.Seeman N.C. (1998) DNA nanotechnology: novel DNA constructions. Annu. Rev. Biophys. Biomol. Struct., 27, 225–248. [DOI] [PubMed] [Google Scholar]
- 2.Seeman N.C. (1999) DNA engineering and its application to nanotechnology. Trends Biotechnol., 17, 437–443. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y. and Seeman,N.C. (1994) The construction of a DNA octahedron. J. Am. Chem. Soc., 116, 1661–1669. [Google Scholar]
- 4.Chen J.H. and Seeman,N.C. (1991) Synthesis from DNA of a molecule with the connectivity of a cube. Nature, 350, 631–633. [DOI] [PubMed] [Google Scholar]
- 5.Winfree E., Liu,F., Wenzler,L.A. and Seeman,N.C. (1998) Design and self-assembly of two-dimensional DNA crystals. Nature, 394, 539–544. [DOI] [PubMed] [Google Scholar]
- 6.Mao C., Sun,W. and Seeman,N.C. (1999) Designed Two-Dimensional DNA Holliday Junction Arrays Visualized by Atomic Force Microscopy. J. Am. Chem. Soc., 121, 5437–5443. [Google Scholar]
- 7.Scheffler M., Dorenbeck,A., Jordan,S., Wüstefeld,M. and von Kiedrowski,G. (1999) Self-Assembly of Trisoligonucleotidyls: The Case for Nano-Acetylene and Nano-Cyclobutadiene. Angew. Chem. Int. Ed. Engl., 38, 3311–3315. [PubMed] [Google Scholar]
- 8.Protozanova E. and Macgregor,R.B.J. (1996) Frayed Wires: A thermally Stable Form of DNA with Two Distinct Structural Domains. Biochemistry, 35, 16638–16645. [DOI] [PubMed] [Google Scholar]
- 9.Venczel E.A. and Sen,D. (1996) Synapsable DNA. J. Mol. Biol., 257, 219–224. [DOI] [PubMed] [Google Scholar]
- 10.Fahlman R.P. and Sen,D. (1999) ‘Synapsable’ DNA Double Helices: Self-Selective Modules for Assembling DNA Superstructures. J. Am. Chem. Soc., 121, 11079–11085. [Google Scholar]
- 11.Mao C., Sun,W., Shen,Z. and Seeman,N.C. (1999) A nanomechanical device based on the B-Z transition of DNA. Nature, 397, 144–146. [DOI] [PubMed] [Google Scholar]
- 12.Batey R.T., Rambo,R.P. and Doudna,J.A. (1999) Tertiary Motifs in RNA Structure and Folding. Angew. Chem. Int. Ed. Engl., 38, 2326–2343. [DOI] [PubMed] [Google Scholar]
- 13.Westhof E. and Leontis,N. (2000) Atomic Glimpses on a Billion-Year-Old Molecular Machine. Angew. Chem. Int. Ed. Engl., 39, 1587–1591. [DOI] [PubMed] [Google Scholar]
- 14.Frank J. (2000) The ribosome–a macromolecular machine par excellence. Chem. Biol., 7, R133–R141. [DOI] [PubMed] [Google Scholar]
- 15.Ban N., Nissen,P., Hansen,J., Moore,P.B. and Steitz,T.A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science, 289, 905–920. [DOI] [PubMed] [Google Scholar]
- 16.Schluenzen F., Tocilj,A., Zarivach,R., Harms,J., Gluehmann,M., Janell,D., Bashan,A., Bartels,H., Agmon,I., Franceschi,F. and Yonath,A. (2000) Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell, 102, 615–623. [DOI] [PubMed] [Google Scholar]
- 17.Wimberly B.T., Brodersen,D.E., Clemons,W.M.,Jr, Morgan-Warren,R.J., Carter,A.P., Vonrhein,C., Hartsch,T. and Ramakrishnan,V. (2000) Structure of the 30S ribosomal subunit. Nature, 407, 327–339. [DOI] [PubMed] [Google Scholar]
- 18.Westhof E., Masquida,B. and Jaeger,L. (1996) RNA tectonics: towards RNA design. Fold Des., 1, R78–R88. [DOI] [PubMed] [Google Scholar]
- 19.Jaeger L. and Leontis,N.B. (2000) Tecto-RNA: One-dimensional Self-assembly through Tertiary Interactions. Angew. Chem. Int. Ed. Engl., 14, 2521–2524. [DOI] [PubMed] [Google Scholar]
- 20.Costa M. and Michel,F. (1995) Frequent use of the same tertiary motif by self-folding RNAs. EMBO J., 14, 1276–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Massire C. and Westhof,E. (1999) MANIP: an interactive tool for modelling RNA. J. Mol. Graph. Model., 16, 197–205. [DOI] [PubMed] [Google Scholar]
- 22.Massire C., Gaspin,C. and Westhof,E. (1994) DRAWNA: a program for drawing schematic views of nucleic acids. J. Mol. Graph., 12, 201–206. [DOI] [PubMed] [Google Scholar]
- 23.Cate J.H., Gooding,A.R., Podell,E., Zhou,K., Golden,B.L., Kundrot,C.E., Cech,T.R. and Doudna,J.A. (1996) Crystal structure of a group I ribozyme domain: principles of RNA packing. Science, 273, 1678–1685. [DOI] [PubMed] [Google Scholar]
- 24.Wu M., McDowell,J.A. and Turner,D.H. (1995) A periodic table of symmetric tandem mismatches in RNA. Biochemistry, 34, 3204–3211. [DOI] [PubMed] [Google Scholar]
- 25.Leontis N.B. and Westhof,E. (1998) Conserved geometrical base-pairing patterns in RNA. Q. Rev. Biophys., 31, 399–455. [DOI] [PubMed] [Google Scholar]
- 26.Lietzke S.E., Barnes,C.L., Berglund,J.A. and Kundrot,C.E. (1996) The structure of an RNA dodecamer shows how tandem U-U base pairs increase the range of stable RNA strctures and the diversity of recognition sites. Structure, 4, 917–930. [DOI] [PubMed] [Google Scholar]
- 27.Costa M. and Michel,F. (1997) Rules for RNA recognition of GNRA tetraloops deduced by in vitro selection: Comparison with in vivo evolution. EMBO J., 16, 3289–3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schroeder S.J. and Turner,D.H. (2000) Factors affecting the thermodynamic stability of small asymmetric internal loops in RNA. Biochemistry, 39, 9257–9274. [DOI] [PubMed] [Google Scholar]
- 29.Gornicki P., Baudin,F., Romby,P., Wiewiorowski,M., Kryzosiak,W., Ebel,J.P., Ehresmann,C. and Ehresmann,B. (1989) Use of lead(II) to probe the structure of large RNAs. Conformation of the 3′ terminal domain of E. coli 16S rRNA and its involvement in building the tRNA binding sites. J. Biomol. Struct. Dyn., 6, 971–984. [DOI] [PubMed] [Google Scholar]
- 30.Moine H., Ehresmann,B., Ehresmann,C. and Romby,P. (1998) In Simons,R.W. and Grunberg-Manago,M. (eds), RNA Structure and Function. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 77–116.
- 31.Szewczak A.A. and Cech,T.R. (1997) An RNA internal loop acts as a hinge to facilitate ribozyme folding and catalysis. RNA, 3, 838–849. [PMC free article] [PubMed] [Google Scholar]
- 32.Peritz A.E., Kierzek,R., Sugimoto,N. and Turner,D.H. (1991) Thermodynamic study of internal loops in oligoribonucleotides: symmetric loops are more stable than asymmetric loops. Biochemistry, 30, 6428–6436. [DOI] [PubMed] [Google Scholar]
- 33.Leontis N.B. and Westhof,E. (1998) The 5S rRNA loop E: chemical probing and phylogenetic data versus crystal structure. RNA, 4, 1134–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jovine L., Hainzl,T., Oubridge,C., Scott,W.G., Li,J., Sixma,T.K., Wonacott,A., Skarzynski,T. and Nagai,K. (2000) Crystal structure of the ffh and EF-G binding sites in the conserved domain IV of escherichia coli 4.5S RNA. Structure Fold Des ., 8, 527–540. [DOI] [PubMed] [Google Scholar]
- 35.Jaeger L., Michel,F. and Westhof,E. (1994) Involvement of a GNRA tetraloop in long-range RNA tertiary interactions. J. Mol. Biol., 236, 1271–1276. [DOI] [PubMed] [Google Scholar]
- 36.Brion P. and Westhof,E. (1997) Hierarchy and dynamics of RNA folding. Annu. Rev. Biophys. Biomol. Struct., 26, 113–137. [DOI] [PubMed] [Google Scholar]
- 37.Woodson S.A. (2000) Recent insights on RNA folding mechanisms from catalytic RNA. Cell Mol. Life Sci., 57, 796–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Massire C., Jaeger,L. and Westhof,E. (1998) Derivation of the three-dimensional architecture of bacterial ribonuclease P RNAs from comparative sequence analysis. J. Mol. Biol., 279, 773–793. [DOI] [PubMed] [Google Scholar]
- 39.Tomizawa J. (1984) Control of ColE1 plasmid replication: the process of binding of RNA I to the primer transcript. Cell, 38, 861–870. [DOI] [PubMed] [Google Scholar]
- 40.Wagner E.G. and Simons,R.W. (1994) Antisense RNA control in bacteria, phages and plasmids. Annu. Rev. Microbiol., 48, 713–742. [DOI] [PubMed] [Google Scholar]
- 41.Paillart J.C., Marquet,R., Skripkin,E., Ehresmann,C. and Ehresmann,B. (1996) Dimerization of retroviral genomic RNAs: structural and functional implications. Biochimie, 78, 639–653. [DOI] [PubMed] [Google Scholar]
- 42.Ferrandon D., Koch,I., Westhof,E. and Nusslein-Volhard,C. (1997) RNA-RNA interaction is required for the formation of specific bicoid mRNA 3′ UTR-STAUFEN ribonucleoprotein particles. EMBO J., 16, 1751–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lodmell J.S., Ehresmann,C., Ehresmann,B. and Marquet,R. (2000) Convergence of natural and artificial evolution on the RNA loop-loop interaction: The HIV-1 dimerization initiation site. RNA, 6, 1267–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lowe C.R. (2000) Nanobiotechnology: the fabrication and applications of chemical and biological nanostructures. Curr. Opin. Struct. Biol., 10, 428–434. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.