Abstract
Background
Proteins that selectively transport water across the membranes of cells are recognized as important in the normal functioning of the body systems of vertebrates. There are 13 known mammalian aquaporins (AQP0 to AQP12), some of which have been shown to have unexpected cellular roles beyond transmembrane water transport. The availability of non-mammalian vertebrate animal models has the potential to provide insight into the emergence of diverse function in the aquaporins. The domesticated chicken (Gallus gallus) is the premier avian model for biological research; however, only a limited number of studies have compared chicken and mammalian aquaporins. The identification of aquaporins that share functional motifs or are expressed in the same tissues in human and chicken could allow the further functional analyses of homologous aquaporins in both species. We hypothesize that integrative analyses of protein sequences and body site expression of human, mouse, rat and chicken aquaporins has the potential to yield novel biological hypotheses about the unexpected cellular roles of aquaporins beyond transmembrane water transport.
Results
A total of 76 aquaporin transcript models derived from 47 aquaporin genes were obtained for human, mouse, rat and chicken. Eleven body sites (brain, connective tissue, head, heart, liver, muscle, ovary, pancreas, small intestine, spleen and testis) were identified in which there is suggested expression of at least one mammalian and one chicken aquaporin. This study demonstrates that modern on-line analysis tools, a novel matrix integration technique, and the availability of the chicken genome for comparative genomics and expression analysis enables hypothesis generation in several important areas including: (i) alternative transcription and speciation effects on the conservation of functional motifs in vertebrate aquaporins; (ii) the emergence of basolateral targeting in mammalian species; (iii) the potential of the cysteine-rich AQP11 as a possible target in the pathophysiology of neurodegenerative disorders such as autism that involve Purkinje cells; and (iv) possible impairment of function of pancreas-expressed AQP12 during pancreatotropic necrosis in avian influenza virus infection.
Conclusion
The investigation of aquaporin function in chicken and mammalian species has the potential to accelerate the discovery of novel knowledge of aquaporins in both avian and mammalian species.
Background
Proteins that selectively transport water across the membranes of cells are recognized as important in normal functioning of the body systems of vertebrates. These homologous proteins are collectively referred to as aquaporins and include a subset called aquaglyceroporins that are able to transport glycerol, urea and other small solutes in addition to water [1]. There are 13 known mammalian aquaporins (AQP0 to AQP12). These aquaporins vary in tissue and developmental expression across mammalian species and unexpected cellular roles for the aquaporins beyond transmembrane water transport have been identified [2]. However, the cellular and molecular strategies for these roles are not completely understood. The water-only aquaporins are AQP0, AQP1, AQP2, AQP4, AQP5, AQP6 and AQP8, while aquaglyceroporins are AQP3, AQP7, AQP9 and AQP10 [1]. The transport specificities and roles of AQP11 and AQP12 in health and disease are not completely described [1,3-5]. Aquaporins typically have six transmembrane regions and five loops (A to E) with two characteristic Asparagine-Proline-Alanine (NPA) motifs in loops B and E (Figure 1) [6]. Based on the number of citations in PubMed, avian aquaporins have been poorly investigated compared to those of human, mouse and rat [7].
Figure 1.
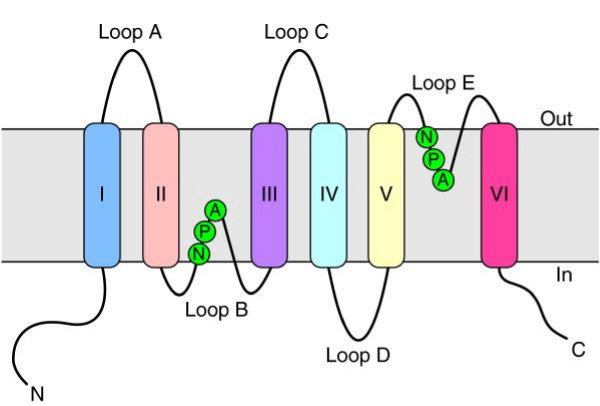
Topology of an aquaporin protein within the membrane. The protein consists of six transmembrane helices (I-VI) connected by five loops (A-E) and includes two internal tandem repeats (I-III and IV-VI, respectively). Loops B and E, containing the conserved NPA motifs (in the single-letter amino-acid code), form short α helices that fold back into the membrane from opposite sides. C, carboxyl terminus; N, amino terminus. Figure reproduced from [6].
The domesticated chicken Gallus gallus is the premier avian model for biological research [8-10]. Furthermore, the publication of the chicken genome and availability of large-scale gene expression data presents new opportunities to compare the expression of avian and mammalian aquaporin genes. Mammalian aquaporins function in diverse biological processes including development of lens, inner ear, teeth and oral facial tissues; maintenance of sperm motility; synthesis and secretion of milk; and urine concentration. In addition, malfunction of mammalian aquaporins have been implicated in diverse disease processes such as altered fat metabolism, brain edema, cancer, congenital cataract, dry mouth, dry skin, infertility, hearing loss, nephrogenic diabetes, renal failure, and wet lung syndrome [11]. This broad range of pathologies linked to altered aquaporin expression has further supported the potential of aquaporins as drug targets for water-transport related disorders [12].
Apart from transmembrane/epithelial water transport, recently suggested unexpected cellular roles of mammalian aquaporins include cell migration, cell volume regulation, mitochondria metabolism, neural signal transduction, renal glycerol clearance and vesicular swelling [11,13]. The co-expression of several members of the aquaporin gene family in the same tissue such as small intestine [14] makes it difficult to determine their individual role apart from trans-epithelial water transport. It is not clear whether alternative transcription/splicing of an aquaporin gene to produce more than one isoform contributes to these unexpected cellular roles. Mice with deficient or mutated aquaporin are currently used as models to study novel cellular roles of human aquaporins [11]. There is an increasing need to have alternative non-mammalian models for aquaporin function in humans [14]. The chick optic tectum has been used to investigate the role of AQP4 in blood brain barrier development [15]. Furthermore, in chick lens development, AQP0 interacts with lens-fiber gap junctions during lens development [16].
There are a limited numbers of reports that have compared the body site expression of chicken and mammalian aquaporins. Comparison of nucleotide sequences of chicken AQP2, AQP4 and AQP5 to their rat and human orthologs has revealed an overall identity of 75–90% and similarity in tissue distribution [7]. AQP9 has not been shown to be expressed in mammalian kidney, but recently found expressed in young chicken kidney [17]. In addition, water-deprivation in early development of rodents and chicken results in upregulation of the kidney AQP2 [17,18]. The distribution of AQP4 in the circumventricular organs of chicken and rat brains have been compared [19]. Taken together, the identification of aquaporins that share sequence similarity or are expressed in identical tissues in human and chicken could allow the further functional analyses of aquaporins in avian species.
The objectives of the investigation reported in this article were to (1) determine the impact of alternative transcription and speciation on functional motifs of aquaporin gene transcript models predicted from the genomes of human, mouse, rat and chicken; and (2) determine body sites common to human, mouse, rat and chicken with suggested aquaporin expression. We hypothesize that integrative analyses of protein sequences and body site expression of human, mouse, rat and chicken aquaporins has the potential to yield novel biological hypotheses about the unexpected cellular roles of aquaporins beyond transmembrane water transport. Our high-throughput comparative sequence analysis revealed the potential impact of alternative transcription and speciation on the conservation of functional motifs in human, mouse, rat and chicken aquaporins. Furthermore, by using controlled vocabulary of terms describing body sites in the UniGene database, an integrated view of suggested tissue expression of aquaporins for these four organisms was generated.
The integrative analyses of protein sequences and tissue expression profiles presented in this article provides novel insights into the potential function of cysteine-rich AQP11 in the pathophysiology of autism and evidence for involvement of pancreas-expressed AQP12 in the pathology of highly pathogenic avian influenza virus infections. The investigation of aquaporin function in chicken could accelerate the discovery of novel knowledge on human aquaporins especially during early development.
Results
Comparison of predicted aquaporin transcripts from human, mouse, rat and chicken
In order to determine the impact of alternative transcription as well as speciation on the conservation of functional motifs in aquaporins, we first compiled the aquaporin types with entries in the Entrez Gene [20] and Ensembl [21] genomic resources (Table 1) for human, mouse, rat and chicken. This dataset provided the basis for comparative sequence analysis of the transcripts predicted in the Ensembl genome resource. Multiple sequence alignment was performed on each aquaporin type in which at least one mammalian and one chicken transcript were available. A total of 76 sequences were retrieved from the Ensembl. Multiple sequence alignments for each aquaporin type are available as Additional File 1. A summary of the number of transcripts examined for 9 aquaporin types and key observations from sequence alignment are presented in Table 2. The impact of alternative transcription and speciation of these vertebrate aquaporins on the conservation of the two water-transport motifs is summarized in Table 3. We further describe below the results obtained for AQP3, AQP4, AQP11 and AQP12.
Table 1.
Aquaporin entries in Entrez Gene, UniGene, and Ensembl genomic resources
| AQP | Human | Mouse | Rat | Chicken | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Entrez Gene | Unigene | Ensembl* | Entrez Gene | Unigene | Ensembl* | Entrez Gene | Unigene | Ensembl* | Entrez Gene | Unigene | Ensembl* | |
| AQP 0 | 4284 | Hs.574026 | 135517 | 17339 | Mm.31625 | 25389 | 25480 | Rn.23532 | 3132 | 374124 | Gga.67 | - |
| AQP 1 | 358 | Hs.76152 | 106125 | 11826 | Mm.18625 | 4655 | 25240 | Rn.1618 | 11648 | 420384 | Gga.2680 | 5209 |
| AQP 2 | 359 | Hs.130730 | 167580 | 11827 | Mm.20206 | 23013 | 25386 | Rn.90076 | 297 | - | - | - |
| AQP 3 | 360 | Hs.234642 | 165272 | 11828 | Mm.34043 | 28435 | 65133 | Rn.11109 | 9797 | 426894 | Gga.23800 | 2452 |
| AQP 4 | 361 | Hs.315369 | 171885 | 11829 | Mm.250786 | 24411 | 25293 | Rn.90091 | 16043 | 421088 | Gga.11374 | 15128 |
| AQP 5 | 362 | Hs.298023 | 161798 | 11830 | Mm.45580 | 44217 | 25241 | Rn.10066 | 17685 | 431305 | Gga.6412 | 2720 |
| AQP 6 | 363 | Hs.54505 | 86159 | 11831 | Mm.202309 | 43144 | 29170 | Rn.48667 | 296 | - | - | 10260 |
| AQP 7 | 364 | Hs.455323 | 165269 | 11832 | Mm.8728 | 28427 | 29171 | Rn.11111 | 9686 | 426892 | Gga.21944 | 18534 |
| AQP 8 | 343 | Hs.176658 | 103375 | 11833 | Mm.273175 | 30762 | 29172 | Rn.6315 | 14652 | 416566 | Gga.6178 | 5988 |
| AQP 9 | 366 | Hs.104624 | 103569 | 64008 | Mm.335570 | 32204 | 65054 | Rn.30018 | 15949 | 415402 | Gga.12485 | 4261 |
| AQP 10 | 89872 | Hs.259048 | 143595 | 435743 | - | - | - | - | - | - | - | - |
| AQP 11 | 282679 | Hs.503345 | 178301 | 66333 | Mm.29756 | 42797 | 286758 | Rn.20144 | 13358 | 426725 | Gga.8876 | 1614 |
| AQP 12 | 375318 | Hs.437167 | 184945 | 208760 | Mm.235537 | 45091 | 367316 | Rn.20532 | 4452 | 424861 | Gga.19694 | 6436 |
*Ensembl Identifiers obtained had a total of 11 numerals with the appropriate number of zeros as padding. Furthermore, the identifiers were preceded by ENSG, ENSMUSG, ENSRNOG, and ENSGAL are for human, mouse, rat and chicken genes respectively. The isoform of Human AQP12 used for analysis was Aquaporin 12B.
Table 2.
Aquaporin isoforms examined for each aquaporin type and key observations from comparative sequence analysis*
| AQP Type | Organism | Observations from sequence alignment | |||
|---|---|---|---|---|---|
| Human | Mouse | Rat | Chicken | ||
| AQP1 | 2 | 1 | 1 | 1 | Human transcript ENST00000265298 lacked the first 94 amino acids of ENST00000311813 which is present in the AQP1 from mouse, rat and chicken. This N-terminal region of the sequence contained the first NPA motif. However, the second NPA motif starting at position 192 in ENST00000311813 is conserved in all the 5 protein sequences |
| AQP3 | 3 | 1 | 1 | 1 | Human transcript ENST00000379492 lacked the first 34 amino acids present in the other isoforms. |
| AQP4 | 3 | 2 | 2 | 2 | All the organisms had multiple transcripts. |
| AQP5 | 1 | 2 | 2 | 3 | The first NPA motif was observed in all the protein sequences. Two chicken transcripts ENSGALT00000041244 and ENSGALT00000041246 and two rodent transcripts ENSMUST00000088200 and ENSRNOT00000040874 produced isoforms that lacked the second NPA motif. |
| AQP7 | 4 | 3 | 1 | 1 | All nine protein sequences of AQP7 contained atypical water transport NPA motifs. The first motif is NAA while the second motif was NPS. |
| AQP8 | 1 | 2 | 1 | 1 | The first water-transport motif for chicken AQP8 was NPV |
| AQP9 | 1 | 4 | 1 | 2 | Except for mouse transcript ENSMUST00000113569, all other AQP9 protein sequences had the second NPA motif. |
| AQP11 | 1 | 2 | 2 | 1 | The first water transporting motif was atypical (NPC) in all the isoforms examined. Only three protein isoforms from transcripts ENSRNOT00000018091, ENSMUST00000084986 and ENST00000313578 contained the second NPA motif. |
| AQP12 | 2 | 1 | 1 | 1 | Human transcript ENST00000373309 lacked the first NPA-like motif (NPT) present in the other sequences. |
*Additional File 1 contains the multiple sequence alignment for AQP0 to AQP12. Ensembl Identifiers preceded by ENST, ENSMUST, ENSRNOT, and ENSGALT are for human, mouse, rat and chicken transcripts respectively.
Table 3.
Prioritize aquaporin isoforms for investigation for roles other than water-transport
| Aquaporin | Organism | Ensembl ID | Missing NPA or NPA-like motif |
|---|---|---|---|
| AQP1 | Human | ENST00000265298 | First |
| AQP3 | Human | ENST00000343952 | Second |
| AQP3 | Human | ENST00000379492 | Second |
| AQP4 | Human | ENST00000339532 | First |
| AQP4 | Human | ENST00000383170 | Second |
| AQP5 | Mouse | ENSMUST00000088200 | Second |
| AQP5 | Rat | ENSRNOT00000040874 | Second |
| AQP5 | Chicken | ENSGALT00000041244 | Second |
| AQP5 | Chicken | ENSGALT00000041246 | Second |
| AQP9 | Mouse | ENSMUST00000113569 | Second |
| AQP12 | Human | ENST00000373309 | First |
Three human transcripts were retrieved for AQP3 and one each for mouse, rat and chicken from the Ensembl (Figure 2). The protein isoform from human AQP3 transcript ENST00000379492 lacked the first 34 amino acids present in the other isoforms. This region contained the YRLL motif known to be important for basolateral sorting in epithelial cells [22-24]. The YRLL motif was replaced by a NKLV motif in the only chicken isoform obtained from transcript ENSGALT00000003868. The first NPA motif was conserved in all the AQP3 isoforms while the second NPA motif was absent in two human AQP3 isoform sequences (ENST00000343952 and ENST00000379492). The impact of alternative transcription and speciation is illustrated by protein features predicted for the three human AQP3 transcripts and one chicken AQP transcript (Figure 3). Note the difference in number of transmembrane helices predicted for the human transcripts. Interestingly, the chicken transcript lacks a prediction for signal peptide. This observation led us to further analyze the Ensembl generated pairwise sequence alignments of the chicken AQP3 with other vertebrate AQP3 showing 1-to-1 ortholog prediction. The AQP3 (ENSOANG00000014661) of the platypus (Ornithorhynchus anatinus) had a YKLL motif aligned to the NKLV motif of the chicken sequence (Figure 4).
Figure 2.
ClustalW multiple sequence alignment of AQP3 protein isoforms predicted by the Ensembl software system. Position 19 to 22 contains the YRLL motif for basolateral targeting. This motif is replaced in chicken AQP3 with NKLV.
Figure 3.

Impact of alternative transcription and speciation on protein features of AQP3 isoforms. There are differences in the predicted protein features of AQP3 isoforms obtained from human and chicken gene loci. In the chicken homolog, no signal peptide cleavage predicted. Images were obtained from Ensembl gene information pages. HGNC is abbreviation for Human Genome Nomenclature Committee.
Figure 4.
Multiple sequence alignment of AQP3 sequences from selected vertebrates. The platypus (Ornithorhynchus anatinus), a beaked mammal whose females lay eggs, had an YKLL motif aligned to the NKLV motif of the chicken sequence.
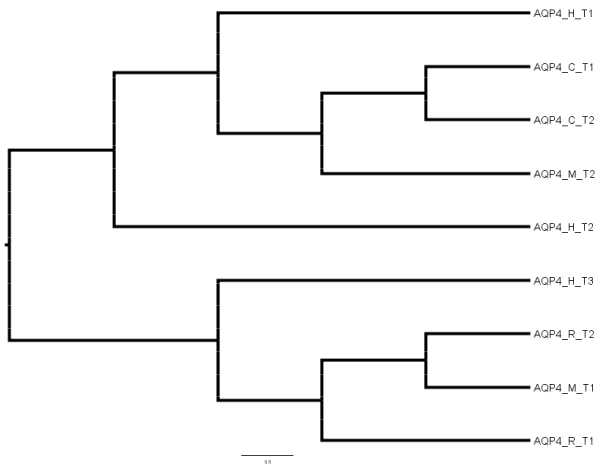
AQP4 was the only aquaporin type in which the four organisms had at least two transcripts. Multiple sequence alignment of the amino acid sequences from the 9 transcripts and construction of phylogenetic tree revealed two classes of transcripts (Figure 5). The two rat transcripts (ENSRNOT00000048109 and ENSRNOT00000021961) clustered with a human transcript (ENST00000383170) and a mouse transcript (ENSMUST00000079081). In the case of the remaining 5 AQP4 transcripts, the chicken and mouse protein sequences were clustered with human transcript ENST00000339532.
Figure 5.
Phylogenetic tree of AQP4 protein sequences from human (H), mouse (M), rat (R) and chicken (C). The symbols and their corresponding Ensembl transcripts in brackets are: AQP4_C_T1 (ENSGALT00000024413); AQP4_C_T2 (ENSGALT00000036809); AQP4_M_T1 (ENSMUST00000079081); AQP4_M_T2 (ENSMUST00000115856); AQP4_R_T1 (ENSRNOT00000021961); AQP4_R_T2 (ENSRNOT00000048109); AQP4_H_T1 (ENST00000339532); AQP4_H_T2 (ENST00000383168); AQP4_H_T3 (ENST00000383170).
In the case of AQP11, the first water transporting motif was atypical (NPC) in all the isoforms examined. Only three protein isoforms from transcripts ENSRNOT00000018091, ENSMUST00000084986 and ENST00000313578 contained the second NPA motif. AQP11 sequences had a high number of cysteine residues compared to other aquaporin types including a triplet CCC present in the N-terminal of protein sequences produced by transcripts ENSMUST00000055379, ENSMUST00000084986 and ENST00000313578. The two rat sequences had a CYC motif while chicken had a CAC motif. Cysteine residues are known to interact with sulfhydryl-reactive metals such as mercury, cadmium, lead, and arsenic [25]. A total of five transcripts for AQP12 were retrieved for the four species. The protein sequence for one of the human transcripts (ENST00000373309) lacked the first NPA-like motif (NPT) present in the other sequences. All contained the second NPA motif.
Body site expression profiles for human, mouse, rat and chicken aquaporins
A total of 44 UniGene aquaporin entries from human (13), mouse (12), rat (10) and chicken (9) were identified has having suggested expression in at least one of 57 body sites based on Expressed Sequence Tags (EST) counts (Table 4). A total of 51 binary signatures described the expression of aquaporins from the organisms compared. The visualized matrix of signatures is presented in Figure 6. Furthermore, 11 body sites (brain, connective tissue, head, heart, liver, muscle, ovary, pancreas, small intestine, spleen and testis) were identified in which both chicken and mammalian aquaporins were expressed. The brain had the highest count for aquaporin expression. All four organisms expressed AQP1, AQP4 and AQP11 in the brain. There was evidence of expression of AQP12 in the intestine, pancreas, stomach, and tongue as well as expression in the pancreas for all species except rat (Figure 6, Additional file 2). We mapped chicken aquaporins to body sites in order to prioritize them for further functional analysis (Table 5).
Table 4.
Controlled UniGene body site terms associated with expressed aquaporins from human, mouse, rat and chicken
| Body Site | AQP Count | Body Site | AQP Count | Body Site | AQP Count | Body Site | AQP Count |
|---|---|---|---|---|---|---|---|
| Adipose tissue | 5 | Extraembryonic tissue | 3 | Muscle | 14 | Spleen | 6 |
| Adrenal gland | 4 | Eye | 19 | Nasopharynx | 3 | Stomach | 10 |
| Ascites | 1 | Fertilized ovum | 1 | Nerve | 2 | Testis | 15 |
| Bladder | 6 | Head | 3 | Ovary | 10 | Thymus | 8 |
| Blood | 3 | Heart | 10 | Pancreas | 11 | Thyroid | 6 |
| Bone | 4 | Inner ear | 3 | Parathyroid | 2 | Tongue | 3 |
| Bone marrow | 5 | Intestine | 13 | Pharynx | 2 | Trachea | 2 |
| Brain | 23 | Joint | 3 | Pineal gland | 2 | Umbilical cord | 2 |
| Cervix | 3 | Kidney | 21 | Pituitary gland | 2 | Uterus | 9 |
| Colon | 2 | Larynx | 4 | Placenta | 8 | Vagina | 2 |
| Connective tissue | 12 | Liver | 15 | Prostate | 10 | Vascular | 3 |
| Dorsal root ganglion | 1 | Lung | 14 | Salivary gland | 4 | Vibrissa | 1 |
| Embryonic tissue | 12 | Lymph node | 1 | Skin | 8 | ||
| Epididymis | 1 | Mammary gland | 10 | Small intestine | 2 | ||
| Esophagus | 2 | Mouth | 2 | Spinal cord | 2 |
Figure 6.
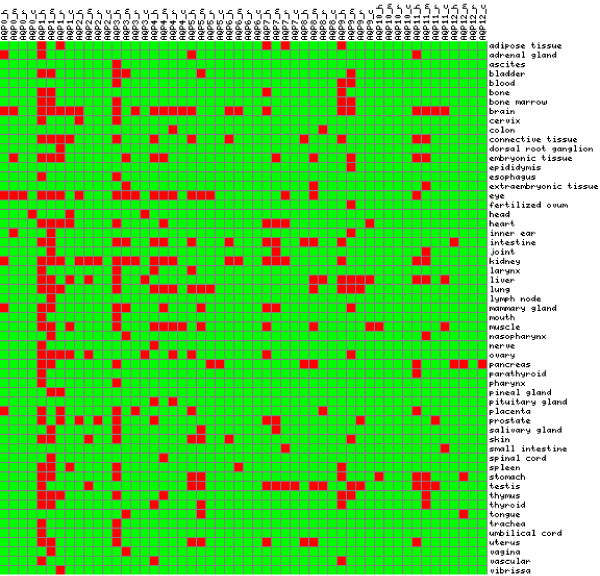
Visualization of comparison of suggested body site expression of UniGene data for human (h), mouse (m), rat (r), and chicken (c) aquaporins. Each aquaporin type is represented by 4 boxes corresponding to the four-digit binary number. Red box indicate presence of expression (1) while green box indicate absence of expression (0).
Table 5.
Body sites and corresponding expressed chicken aquaporins that could be used as models for understanding mammalian aquaporins
| Aquaporin Type | Chicken UniGene ID | Body Site |
|---|---|---|
| AQP0 | Gga.67 | Head |
| AQP1 | Gga.2680 | Brain, Connective Tissue, Head, Heart, Liver, Muscle, Ovary, Spleen |
| AQP3 | Gga.23800 | Head, Liver, Ovary |
| AQP4 | Gga.11374 | Brain, Muscle |
| AQP5 | Gga.6412 | Pancreas |
| AQP7 | Gga.21944 | Testis |
| AQP9 | Gga.12485 | Heart, Liver, Muscle |
| AQP11 | Gga.8876 | Brain, Liver, Muscle, Small Intestine |
| AQP12 | Gga.19694 | Pancreas |
Discussion
Aquaporin cell surface proteins are emerging as important proteins beyond water transport [2,11,13,26]. However, the molecular, cellular and evolutionary basis for these novel roles are not completely known. Our high-throughput comparative sequence analysis revealed the impact of alternative transcription and speciation on the conservation of functional motifs in human, mouse, rat and chicken aquaporins. Furthermore, by using a controlled vocabulary of terms describing body sites in the UniGene database, an integrated view of suggested tissue expression of aquaporins in these four organisms was generated (Figure 6). We were unable to retrieve information from the Ensembl database on chicken AQP2 and AQP0 (Entrez Gene ID: 374124) although chicken AQP0 (MIP) has been studied in the context of early development of lens fiber and its association with gap junction proteins [16]. The predicted chicken AQP6 was annotated as AQP2-like. The annotation of AQP0, AQP2 and AQP6 from the chicken genome warrants further investigation.
Alternate splicing of mRNA as a means of generating protein diversity can occur by exon skipping [27]. All the 76 aquaporins transcripts analyzed were multi-exon transcripts and could be subject to alternative splicing with impact on protein function. For the nine aquaporin types compared, our sequence alignments uncovered aquaporin isoforms that lacked one of the two water-transport motifs (Table 3). We hypothesize that isoforms lacking one of the two NPA or NPA-like motifs characteristic will have impaired or abolished water transport function. They are also candidates for investigating roles other than water function. Another mechanism for variation that can impact function of aquaporins is single nucleotide polymorphisms (SNPs). Previous work has demonstrated that novel genetic variants of AQP4 resulting from single nucleotide polymorphisms (SNP) showed reduced water permeability [28]. We observed from the Ensembl database (Release 50 July 2008) that a total of 19 reference SNPs were mapped to the following chicken aquaporins: AQP1, AQP7, AQP8, AQP9 and AQP12. As the chicken genome become further characterized for polymorphisms, it may become possible to gain insights into impact of SNP variation on aquaporin function and organism phenotype.
Most proteins that target the plasma membrane contain signals within their cytoplasmic termini that permit their recruitment into endocytic vesicles, which in turn facilitates their selective compartmentalization in the apical or basolateral membranes selectively [23]. We were interested in identifying avian protein isoforms in which the functional motifs were different from mammalian aquaporin isoforms. We have previously investigated the compartmentalization of AQP3 and AQP10 in the human intestine in which we observed that the basolateral sorting motif "YRLL" is present in AQP3 but absent in AQP10 [22]. Based on the UniGene suggested expression profiles there was no suggestion for the expression of chicken AQP3 in the intestine. Furthermore, the signal peptide for targeting was not predicted for chicken AQP3 in the Ensembl resource (Figure 3). Tissue expression for this chicken AQP3 was suggested for head, liver, and ovary. Comparative sequence analysis provided evidence that chicken AQP3 lacks the YRLL motif (NKLV was observed) suggesting that it may not be targeted to the basolateral membrane of the intestine. The multiple sequence alignment of the AQP3 protein sequences from the four organisms revealed that the amino acids immediately before and after the YRLL and NKLV motifs were conserved (Figure 2).
According to the ProTeus (PROtein TErminUS) tool [29], the NKLV protein signature is a short linear significant signature in termini of proteins with a corresponding Gene Ontology Cellular Component of "extracellular". Based on alignments available at the Ensembl resource [21] for AQP3 from fish (Danio rerio, Oryzias latipes, Takifugu rubripes, and Tetraodon nigroviridis) and an amphibian (Xenopus tropicalis), the ancestral motifs of the sorting motifs found in human may be NKLL or NMLM (Figure 4). Thus, the presence of the amino acid tyrosine (Y) in this sorting tetrapeptide in the platypus suggests the point in the evolution of AQP3 where the asparagine residue was replaced by the critical tyrosine required for sorting function [30]. The genome of O. anatinus has been proposed has useful for informing human genome sequence and critical link to understanding the differences between avian and mammalian genomes [31,32]. These observations provide novel evolutionarily insights into the localization signals encoded in the termini of AQP3.
We have used a binary encoding integration strategy to gain a comprehensive view of suggested tissue expression of aquaporins for four organisms. Higher-order patterns in a binary vector space that encodes the presence (1) or absence (0) of feature of interest is an approach for integrating genome-wide numerical datasets [33-35]. Several advantages offered by the binary integration of high-throughput gene expression data include computational efficiency and noise resilience [35]. Our matrix revealed that AQP4, in addition to AQP1 and AQP11 were expressed in the brain of all the four organisms. These three aquaporin types are candidates for comparative experimental investigation of aquaporin in brain function. In addition, the analysis delivered a set of body sites in which there was evidence of expression of at least one mammalian aquaporin and chicken aquaporin.
The functions of AQP11 and AQP12 in vertebrate physiology are not completely understood [1,3]. We observed that AQP11 had the highest number of cysteine residues compared to other aquaporins. AQP11 is expressed in the Purkinje cells of the brain cerebellar [1], a site that have been implicated in the pathophysiology of autism [36,37]. Exposure of chick embryo to the environmental metal pollutant methylmercury led to reduction in the number of Purkinje cells [38]. Furthermore, there were adverse post-natal behavioral, morphological and biochemical consequences. Mercury ions are known to regulate aquaporin function by interacting with cysteine residues [39,40]. Interestingly, the first pore forming motif of AQP11 found in Loop B has a motif of NPC instead of the NPA. However, the second motif is NPA consistent with other AQPs. AQP11 is the only human aquaporin with the tri-cysteine (CCC) motif. These observations led us to hypothesize that AQP11 may be a target in the pathophysiology of neurodegenerative disorders like autism. We are currently studying the specificity and affinity of a range of cations to further understand the interaction of cations with aquaporins in the function of the central nervous system. With the availability of the chicken genome, the chick embryo has the potential to serve as an important model for the study of the development of neurodegenerative disorders [38,41].
In the case of AQP12, there was suggested expression in the pancreas of human, mouse and chicken (Table 4) with chicken having the highest expression level. AQP12 is localized intracellularly in the pancreatic acinar cells, the site for synthesis of digestive enzymes [3]. Histopathological studies have confirmed that the highly pathogenic H5N1 virus that causes avian influenza in chicken and other avian species causes multifocal necrosis in the pancreatic acinar cells suggesting that the effect of avian influenza on the function of AQP12 warrants further investigation.
Conclusion
This study demonstrates that the chicken genome combined with a comprehensive controlled vocabulary-facilitated integration of UniGene suggested body site expression can drive generation of hypotheses related to the function of avian and mammalian aquaporins. We describe the generation of hypotheses related to (i) the impact of alternative transcription and speciation on the conservation of functional motifs in human, mouse, rat and chicken aquaporins; (ii) identification of the emergence of basolateral targeting in mammalian species (iii) possibility of cysteine-rich AQP11 as target in the pathophysiology of neurodegenerative disorders; and (iv) impact on the function of pancreas-expressed AQP12 during pancreatotropic necrosis associated with avian influenza virus infection.
Methods
Comparison of predicted aquaporin transcripts from human, mouse, rat and chicken
Predictions of genes encoding aquaporins from human, mouse, rat and chicken genomes were obtained from the Ensembl project (Release 50 July 2008) [21] and the Entrez Gene database at the National Center for Biotechnology Information (NCBI) [20]. In both databases, the human aquaporin information was used as the starting point to extract predicted homologous proteins in the other three species. The amino acid sequences encoding predicted transcripts associated with aquaporin genes in which both an Ensembl and Entrez Gene record exist were further compared. We sought to determine the conservation of functional motifs among predicted transcripts from orthologous genes. Therefore, the amino acid sequence diversity encoded by aquaporin transcripts was determined by multiple sequence alignment of orthologs using ClustalW. In order to visualize the relationship of selected sequences, neighbor-joining tree bootstrapped using 1,000 random samples of sites from the alignment was constructed using the ClustalW software at the DNA Data Bank of Japan [42]. FigTree software version 1.12 [43] was used to view the phylogenetic trees.
UniGene expression profiles for human, mouse, rat and chicken aquaporins
Each NCBI UniGene Cluster contains set of transcript sequences that appear to come from the same transcription locus (gene or expressed pseudogene) as well as other information including expression profile in body sites and developmental stages [20]. The UniGene cluster identifier for each of the aquaporins was extracted from the Entrez Gene record and verified manually. In instances where more than one UniGene entry was associated the Entrez Gene, the unambiguous UniGene entry was selected. The value of the Transcript per million (TPM) for each body site was programmatically extracted from the UniGene Expression Profile Viewer page.
There was need to identify chicken and mammalian aquaporins that were expressed in the same body site based on controlled vocabulary term used in UniGene. Therefore, a 4-digit binary signature was constructed to encode the presence or absence of species aquaporin expression. Thus a matrix consisting of 52-digit binary signature and the number of body sites was constructed and visualized using matrix2png [44].
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
RDI, RVR and HHPC conceived and designed the study. RDI wrote scripts to retrieve compare and integrate datasets and drafted the manuscript. RVR carried out sequence alignment and motif analysis and helped to draft the manuscript. CDJ assembled the aquaporin datasets from multiple organisms, evaluated binary matrix integration and helped to draft the manuscript. TOO developed scripts and computational pipelines to facilitate literature extraction, search and visualization of binary matrices; and helped to draft the manuscript. HHPC helped to interpret the study results, coordinated the study and helped to draft the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Multiple sequence alignment of aquaporins isoforms from human, mouse, rat and chicken. ClustalW multiple sequence alignment of aquaporin types from human, mouse, rat and chicken. Identifiers preceded by ENST, ENSMUST, ENSRNOT, and ENSGALT are for human, mouse, rat and chicken transcripts respectively.
UniGene Transcripts per million and spot intensity of human, mouse and chicken AQP12 expressed in pancreas.
Contributor Information
Raphael D Isokpehi, Email: raphael.isokpehi@jsums.edu.
Rajendram V Rajnarayanan, Email: rrajnarayanan@tougaloo.edu.
Cynthia D Jeffries, Email: centjeffries@yahoo.com.
Tolulola O Oyeleye, Email: oyeleyetolulola@yahoo.com.
Hari HP Cohly, Email: hari.cohly@jsums.edu.
Acknowledgements
Mississippi NSF-EPSCoR "Innovations through Computational Sciences" Award (EPS-0556308); Mississippi Computational Biology Consortium Seed Grant Program; Research Centers in Minority Institutions (RCMI) – Center for Environmental Health at Jackson State University (NIH-NCRR 2G12RR013459); Bioinformatics in Biodefense Career Development Program (DHS-2007-ST-104-000007) and NIH RIMI Grant 1P20MD002725-01 to Tougaloo College. Disclaimer: The views and conclusions contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the funding agencies.
This article has been published as part of BMC Genomics Volume 10 Supplement 2, 2009: Proceedings of the Avian Genomics Conference and Gene Ontology Annotation Workshop. The full contents of the supplement are available online at http://www.biomedcentral.com/1471-2164/10?issue=S2
References
- Gorelick DA, Praetorius J, Tsunenari T, Nielsen S, Agre P. Aquaporin-11: a channel protein lacking apparent transport function expressed in brain. BMC Biochem. 2006;7:14. doi: 10.1186/1471-2091-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos MC, Saadoun S, Verkman AS. Aquaporins and cell migration. Pflugers Arch. 2008;456:693–700. doi: 10.1007/s00424-007-0357-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Rai T, Kuwahara M, Ko SB, Uchida S, Sasaki S. et al. Identification of a novel aquaporin, AQP12, expressed in pancreatic acinar cells. Biochem Biophys Res Commun. 2005;330:832–838. doi: 10.1016/j.bbrc.2005.03.046. [DOI] [PubMed] [Google Scholar]
- Okada S, Misaka T, Tanaka Y, Matsumoto I, Ishibashi K, Sasaki S, Aquaporin-11 knockout mice and polycystic kidney disease animals share a common mechanism of cyst formation. FASEB J. 2008. [DOI] [PubMed]
- Tchekneva EE, Khuchua Z, Davis LS, Kadkina V, Dunn SR, Bachman S. et al. Single Amino Acid Substitution in Aquaporin 11 Causes Renal Failure. J Am Soc Nephrol. 2008;19:1955–1964. doi: 10.1681/ASN.2008030296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse E, Uehlein N, Kaldenhoff R. The aquaporins. Genome Biol. 2006;7:206. doi: 10.1186/gb-2006-7-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Lorca R, Munoz-Cabello AM, Toledo-Aral JJ, Ilundain AA, Echevarria M. Aquaporins in chicken: localization of ck-AQP5 along the small and large intestine. Comp Biochem Physiol A Mol Integr Physiol. 2006;143:269–277. doi: 10.1016/j.cbpa.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Buza TJ, McCarthy FM, Burgess SC. Experimental-confirmation and functional-annotation of predicted proteins in the chicken genome. BMC Genomics. 2007;8:425. doi: 10.1186/1471-2164-8-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogburn LA, Porter TE, Duclos MJ, Simon J, Burgess SC, Zhu JJ. et al. Functional genomics of the chicken – a model organism. Poult Sci. 2007;86:2059–2094. doi: 10.1093/ps/86.10.2059. [DOI] [PubMed] [Google Scholar]
- Kohonen P, Nera KP, Lassila O. Avian model for B-cell immunology – new genomes and phylotranscriptomics. Scand J Immunol. 2007;66:113–121. doi: 10.1111/j.1365-3083.2007.01973.x. [DOI] [PubMed] [Google Scholar]
- Verkman AS. Dissecting the roles of aquaporins in renal pathophysiology using transgenic mice. Semin Nephrol. 2008;28:217–226. doi: 10.1016/j.semnephrol.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigeri A, Nicchia GP, Svelto M. Aquaporins as targets for drug discovery. Curr Pharm Des. 2007;13:2421–2427. doi: 10.2174/138161207781368738. [DOI] [PubMed] [Google Scholar]
- Verkman AS. More than just water channels: unexpected cellular roles of aquaporins. J Cell Sci. 2005;118:3225–3232. doi: 10.1242/jcs.02519. [DOI] [PubMed] [Google Scholar]
- Li H, Kamiie J, Morishita Y, Yoshida Y, Yaoita E, Ishibashi K. et al. Expression and localization of two isoforms of AQP10 in human small intestine. Biol Cell. 2005;97:823–829. doi: 10.1042/BC20040091. [DOI] [PubMed] [Google Scholar]
- Nicchia GP, Nico B, Camassa LM, Mola MG, Loh N, Dermietzel R. et al. The role of aquaporin-4 in the blood-brain barrier development and integrity: studies in animal and cell culture models. Neuroscience. 2004;129:935–945. doi: 10.1016/j.neuroscience.2004.07.055. [DOI] [PubMed] [Google Scholar]
- Yu XS, Yin X, Lafer EM, Jiang JX. Developmental regulation of the direct interaction between the intracellular loop of connexin 45.6 and the C terminus of major intrinsic protein (aquaporin-0) J Biol Chem. 2005;280:22081–22090. doi: 10.1074/jbc.M414377200. [DOI] [PubMed] [Google Scholar]
- Sugiura K, Aste N, Fujii M, Shimada K, Saito N. Effect of hyperosmotic stimulation on aquaporins gene expression in chick kidney. Comp Biochem Physiol A Mol Integr Physiol. 2008;151:173–179. doi: 10.1016/j.cbpa.2008.06.014. [DOI] [PubMed] [Google Scholar]
- Bonilla-Felix M, Jiang W. Aquaporin-2 in the immature rat: expression, regulation, and trafficking. J Am Soc Nephrol. 1997;8:1502–1509. doi: 10.1681/ASN.V8101502. [DOI] [PubMed] [Google Scholar]
- Goren O, Adorjan I, Kalman M. Heterogeneous occurrence of aquaporin-4 in the ependyma and in the circumventricular organs in rat and chicken. Anat Embryol (Berl) 2006;211:155–172. doi: 10.1007/s00429-005-0067-8. [DOI] [PubMed] [Google Scholar]
- Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V. et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36:D13–D21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Suarez XM, Schuster MK. Using the Ensembl genome server to browse genomic sequence data. Curr Protoc Bioinformatics. 2007;Chapter 1(Unit) doi: 10.1002/0471250953.bi0115s16. [DOI] [PubMed] [Google Scholar]
- Cohly HH, Isokpehi R, Rajnarayanan RV. Compartmentalization of aquaporins in the human intestine. Int J Environ Res Public Health. 2008;5:115–119. doi: 10.3390/ijerph5020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey PJ, Meise KS, Coffey RJ. Basolateral sorting of transforming growth factor-alpha precursor in polarized epithelial cells: characterization of cytoplasmic domain determinants. Exp Cell Res. 2003;285:159–174. doi: 10.1016/S0014-4827(03)00035-1. [DOI] [PubMed] [Google Scholar]
- Marks MS, Woodruff L, Ohno H, Bonifacino JS. Protein targeting by tyrosine- and di-leucine-based signals: evidence for distinct saturable components. J Cell Biol. 1996;135:341–354. doi: 10.1083/jcb.135.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quig D. Cysteine metabolism and metal toxicity. Altern Med Rev. 1998;3:262–270. [PubMed] [Google Scholar]
- Verkman AS. Mammalian aquaporins: diverse physiological roles and potential clinical significance. Expert Rev Mol Med. 2008;10:e13. doi: 10.1017/S1462399408000690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Goren A, Ast G. Alternative splicing: current perspectives. Bioessays. 2008;30:38–47. doi: 10.1002/bies.20692. [DOI] [PubMed] [Google Scholar]
- Sorani MD, Zador Z, Hurowitz E, Yan D, Giacomini KM, Manley GT. Novel variants in human Aquaporin-4 reduce cellular water permeability. Hum Mol Genet. 2008;17:2379–2389. doi: 10.1093/hmg/ddn138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahir I, Linial M. ProTeus: identifying signatures in protein termini. Nucleic Acids Res. 2005;33:W277–W280. doi: 10.1093/nar/gki371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Traub LM. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- Grutzner F, Deakin J, Rens W, El-Mogharbel N, Marshall Graves JA. The monotreme genome: a patchwork of reptile, mammal and unique features? Comp Biochem Physiol A Mol Integr Physiol. 2003;136:867–881. doi: 10.1016/j.cbpb.2003.09.014. [DOI] [PubMed] [Google Scholar]
- Warren WC, Hillier LW, Marshall Graves JA, Birney E, Ponting CP, Grutzner F. et al. Genome analysis of the platypus reveals unique signatures of evolution. Nature. 2008;453:175–183. doi: 10.1038/nature06936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazko G, Coleman M, Mushegian A. Similarity searches in genome-wide numerical data sets. Biol Direct. 2006;1:13. doi: 10.1186/1745-6150-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazko G, Gordon A, Mushegian A. The choice of optimal distance measure in genome-wide datasets. Bioinformatics. 2005;21(Suppl 3):iii3–11. doi: 10.1093/bioinformatics/bti1201. [DOI] [PubMed] [Google Scholar]
- Shmulevich I, Zhang W. Binary analysis and optimization-based normalization of gene expression data. Bioinformatics. 2002;18:555–565. doi: 10.1093/bioinformatics/18.4.555. [DOI] [PubMed] [Google Scholar]
- Cohly HH, Panja A. Immunological findings in autism. Int Rev Neurobiol. 2005;71:317–341. doi: 10.1016/S0074-7742(05)71013-8. [DOI] [PubMed] [Google Scholar]
- Kern JK, Jones AM. Evidence of toxicity, oxidative stress, and neuronal insult in autism. J Toxicol Environ Health B Crit Rev. 2006;9:485–499. doi: 10.1080/10937400600882079. [DOI] [PubMed] [Google Scholar]
- Carvalho MC, Nazari EM, Farina M, Muller YM. Behavioral, Morphological and Biochemical Changes after in ovo Exposure to Methylmercury in Chicks. Toxicol Sci. 2008;106:180–185. doi: 10.1093/toxsci/kfn158. [DOI] [PubMed] [Google Scholar]
- Kuwahara M, Gu Y, Ishibashi K, Marumo F, Sasaki S. Mercury-sensitive residues and pore site in AQP3 water channel. Biochemistry. 1997;36:13973–13978. doi: 10.1021/bi9711442. [DOI] [PubMed] [Google Scholar]
- Savage DF, Stroud RM. Structural basis of aquaporin inhibition by mercury. J Mol Biol. 2007;368:607–617. doi: 10.1016/j.jmb.2007.02.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller YM, Rivero LB, Carvalho MC, Kobus K, Farina M, Nazari EM. Behavioral impairments related to lead-induced developmental neurotoxicity in chicks. Arch Toxicol. 2008;82:445–451. doi: 10.1007/s00204-007-0266-6. [DOI] [PubMed] [Google Scholar]
- CLUSTALW WWW System. http://clustalw.ddbj.nig.ac.jp/top-e.html
- FigTree software version 1.12. http://tree.bio.ed.ac.uk/software/figtree/
- Pavlidis P, Noble WS. Matrix2png: a utility for visualizing matrix data. Bioinformatics. 2003;19:295–296. doi: 10.1093/bioinformatics/19.2.295. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multiple sequence alignment of aquaporins isoforms from human, mouse, rat and chicken. ClustalW multiple sequence alignment of aquaporin types from human, mouse, rat and chicken. Identifiers preceded by ENST, ENSMUST, ENSRNOT, and ENSGALT are for human, mouse, rat and chicken transcripts respectively.
UniGene Transcripts per million and spot intensity of human, mouse and chicken AQP12 expressed in pancreas.