Abstract
3D cryo-electron microscopy reconstruction methods are uniquely able to reveal structures of many important macromolecules and macromolecular complexes. EMDataBank.org, a joint effort of the Protein Data Bank in Europe (PDBe), the Research Collaboratory for Structural Bioinformatics (RCSB), and the National Center for Macromolecular Imaging (NCMI), is a “one-stop shop” resource for global deposition and retrieval of cryoEM map, model and associated metadata. The resource unifies public access to the two major EM Structural Data archives: EM Data Bank (EMDB) and Protein Data Bank (PDB), and facilitates use of EM structural data of macromolecules and macromolecular complexes by the wider scientific community.
Introduction
Structural biology of macromolecules has become an indispensable branch of molecular biology. Researchers use the results from structural studies to explain the functions and mechanisms of biological processes at the molecular level, leading to more targeted experiments to explore structure and function. Many key biological processes are carried out by large macromolecular complexes, including signal transduction, genome replication, transcription, translation, chaperonin-assisted protein folding, viral infection, and motility. It is becoming increasingly feasible to determine three-dimensional structures of these complexes in different functional or chemical states using cryo electron microscopy (cryoEM).
Specimens for cryoEM studies come in many forms and shapes, e.g. two- or three-dimensional crystals (Gonen et al., 2005; Henderson et al., 1990; Schmid et al., 2004), one-dimensional filaments or tubular crystals possessing helical symmetry (Unwin, 2005; Wang et al., 2006), and individual particles with or without symmetry (Gabashvili et al., 2000; Olson et al., 1990; Zhou et al., 2001). CryoEM is also being applied to large samples consisting of irregular ensembles of complexes and cells using tomographic reconstruction methods (Baumeister, 2004; Murphy and Jensen, 2007). Other chapters in this volume describe preparation and image reconstruction methods for the different specimen types including 2D crystals, helical arrays, single particles, and unique structures.
At present, cryoEM researchers are rapidly producing a large body of knowledge regarding the 3D structural arrangements of components within large macromolecular complexes, within subcellular assemblies, and even within whole cells, based on map volumes with resolution limits ranging from 80 Å to 2 Å. Interpretation varies according to the map resolution, available tools, and additional knowledge of the system and/or its components and may involve either segmentation, rigid body fitting of atomic coordinates determined using X-ray crystallography or NMR, or ab initio model building.
Public access to cryoEM map volumes and their associated fitted model interpretations permits independent assessment and interpretation of structural results and stimulates development of new tools for visualization, fitting, and validation. The EM Data Bank (EMDB) is the major repository for 3D map volumes solved using electron microscopy (Tagari et al., 2002), while the Protein Data Bank (PDB) collects atomic coordinates fitted into EM map volumes (Dutta et al., 2009). The Unified Data Resource for CryoEM (EMDataBank.org) was created in order to unify data deposition, processing and retrieval of maps and fitted models. This chapter provides an overview of the EM structural data archives and the unified resource, including historical context, current content and use, and future prospects.
EM Structural Data Archives
Maps
The EMDB was established at the European Bioinformatics Institute (EBI) in Hinxton, UK and began operations in 2002. It was initially supported by two European Union-funded projects, the Integration of Information about Macromolecular Structure project (IIMS) and the 3DEM Network of Excellence (3DEM NoE). An IIMS-sponsored workshop was held in Nov 2002 that focused on data exchange, harvesting, deposition issues, and presentation of EM data to non-specialists. Guidelines and release policies were set for the newly founded EMDB, and the workshop established the database as a resource for the international community, with an announcement published in Structure (Fuller, 2003), followed by an editorial in Nature Structural Biology (2003). The workshop concluded with a strong endorsement of EM map volume deposition and linkage of EMDB with other archival databases in biomedical research.
Working closely with IIMS project partners, leading European electron microscopy laboratories and PDB partners, an initial data model was produced for electron microscopy derived maps. A web-based deposition system, EMDEP, was developed to handle data capture (Henrick et al., 2003). EMDEP validates data via an interactive depositor-driven operation, and it relies on the knowledge and expertise of the experimenters for the complete and accurate description of the structural experiment and its results. The captured metadata, e.g., sample description, specimen preparation, imaging, reconstruction, fitting details are stored in a “header” file, and the deposited map is converted to a common format for redistribution. A database query tool, EMSEARCH, was also designed and implemented to enable web-based searches.
By the time the IIMS project was completed in December 2003, the EMDB had become an operational public database with 65 map volumes deposited by major EM laboratories in Europe and the USA. At this time the PDB began to see a significant increase in EM-related coordinate depositions, in many cases models that were fitted into maps deposited to EMDB.
Models
The PDB archive was established in 1971 as a public repository for X-ray crystal structures of biological macromolecules (Bernstein et al., 1977), and is presently maintained by the global organization world-wide PDB (wwPDB, Berman et al., 2003), a consortium consisting of the RCSB PDB (www.pdb.org), the Protein Data Bank in Europe (PDBe) at EBI (www.ebi.ac.uk/pdbe), the Protein Data Bank Japan (PDBj, www.pdbj.org), and the Biological Magnetic Resonance Bank (bmrb.wisc.edu). The number of structures in PDB has grown from the initial seven to over 65,000 entries. Over time, the PDB began to collect coordinates of structures determined by methods other than X-ray crystallography. In the 1980's, coordinates and restraint data determined from NMR methods began to be included in the PDB and these now represent about 15% of the archive. In the 1990's, model coordinates for structures determined using EM began to be archived, beginning with models for bacteriorhodopsin (Henderson et al., 1990), and the RecA hexamer (Yu and Egelman, 1997). EM structures account currently for less than 0.5% of all PDB entries, but the rate of deposition is increasing more rapidly than for any other experimental method.
EM Dictionary Development
Two workshops held in 2004 invited the EM community to participate in development of an improved data model for describing cryoEM experiments, and also set in motion efforts to unify deposition and access to EM maps and models.
The 2004 3DEM NoE workshop at EBI reviewed tools and software practices used in the field of cryoEM, examined data items and data models required to fully describe EM experiments, and allocated tasks to different groups to develop the required standards. The workshop also defined goals for further development of the database including providing archiving capabilities for cryo-electron tomography, providing cross-referencing between EMDB maps and PDB coordinate models and converting common map formats in a lossless manner.
The second 2004 workshop held at the Research Collaboratory for Structural Bioinformatics (RCSB) at Rutgers, USA and co-sponsored by the National Center for Macromolecular Imaging (NCMI) aimed to develop a global community consensus on data items needed for deposition of 3D map volumes and fitted atomic models derived from cryoEM studies. In addition to discussion of desired improvements in the areas of visualization, data mining and data integration, a unanimous recommendation of workshop attendees was the need to develop a “one-stop shop” for deposition of EM map and model data in order to eliminate the duplication of effort involved in creating separate depositions to EMDB and PDB.
Based on recommendations gathered at the 2004 workshops, a revised and expanded EM dictionary was created in a three-way collaboration between EBI, RCSB, and NCMI with broad community input and was presented in 2005 at the 3DEM Gordon Research Conference in New Hampshire as well as a 3D-EM Developers workshop held in the UK. Recommended conventions for exchange of cryoEM data were published (Heymann et al., 2005), and a standards task force was created to gather information on the different cryoEM map and image conventions and formats to facilitate conversion. In June 2005, a notice posted to the 3DEM community email bulletin board (3dem.ucsd.edu) announced the intention of EBI and RCSB to jointly collaborate towards further development of 3DEM database services.
Unified Resource
Following the recommendations of the EM community to create a one-stop shop for EM, the Unified Data Resource for CryoEM (EMDataBank.org) was established in 2007 with funding from the National Institutes of Health/NIGMS as a joint effort of EBI, RCSB, and NCMI. The resource is creating a global deposition and retrieval network for cryoEM map, model and associated metadata, as well as a portal for software tools for standardized map format conversion, map, segmentation and model assessment, visualization, and data integration. The first goal of this three-way collaboration was completed in early 2008 when RCSB joined EBI as a second EMDB deposition and retrieval site. Joint EMDB (map) and PDB (model) deposition systems were developed and put into operation at both EBI and RCSB in early 2009, and web-based 3D visualization tools have been integrated into EMDB atlas pages. Efforts to improve uniformity and usability of the EM structural data in both EMDB and PDB databases are ongoing, and additional services for data harvesting and evaluation are planned.
Deposition and Content
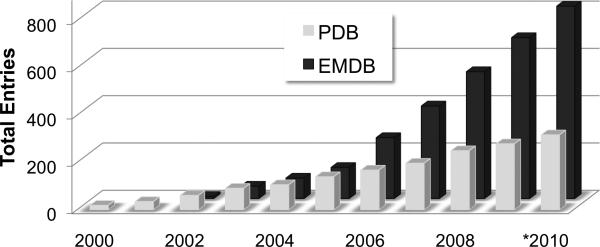
The EMDB currently holds more than 800 map volume entries while PDB holds more than 300 entries of coordinates fitted into EM map volumes (Figure 1). Map volume and fitted model deposition rates are on the rise, currently ~150 and ~40 per year, respectively, with roughly 40% of all published EM structures being captured in the databases. As the importance of EM-derived structural information continues to increase, it is anticipated that more journals and funding agencies will require deposition.
Figure 1.
EM entries in EMDB and PDB, cumulative by year (*2010 data is through May).
Each EMDB entry holds a single map volume plus associated experimental metadata; each associated PDB entry holds the fitted coordinate models and associated experimental metadata plus primary sequence information for each polymer. The metadata information that is shared by both databases is automatically transferred during the joint deposition process and includes:
Sample description, including components and stoichiometry
Sample and specimen preparation, including buffer, grid, vitrification, staining
Imaging experiment, including microscope type, microscope settings, detector
Particle selection, image correction, reconstruction method, resolution, resolution method
Coordinate fitting description
The correspondences between maps and associated fitted coordinate models are maintained in both archives. EMDB entries can optionally hold associated masks, structure factors, and/or layerline data; PDB entries can also hold structure factors. The underlying dictionaries for the two databases have direct translations and are regularly being updated to reflect changes in experimental apparatus and methods. For readers interested in depositing EM structural data, some guidelines are provided in the last section of this chapter.
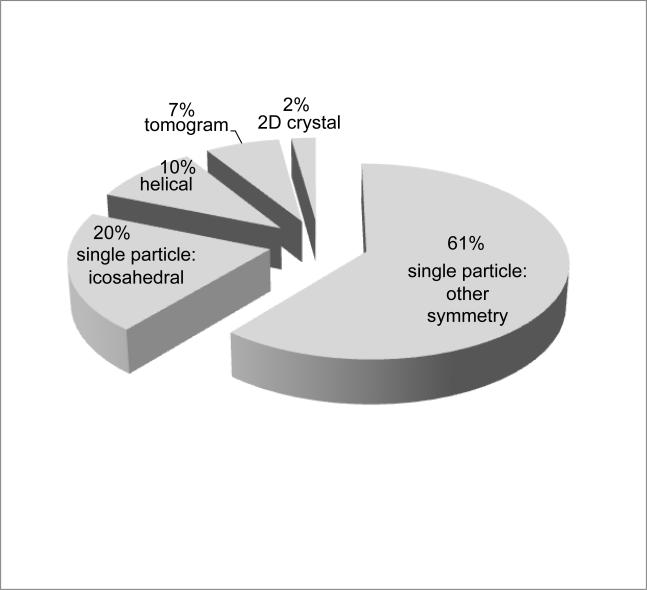
The map archive includes several different types of maps generated by electron microscopy imaging (Figure 2). The majority of entries are single particle reconstructions (81%), which represent ensemble averages of thousands of individual imaged particles, often with additional symmetry averaging. The largest class of single particle specimens represented are the viruses (20% of all holdings), the majority of which are icosahedrally averaged (Chiu and Rixon, 2002; Huiskonen and Butcher, 2007; Lee and Johnson, 2003). Virus entries typically represent distinct states of maturation, or complexes with antibodies or receptors.
Figure 2.
Current distribution of EM map entry types.
The second largest class of single particle specimens represented are the ribosomes (15%). Ribosome entries define key structural conformations encountered in translating messenger RNA into protein, or elucidate structural variations across diverse species (Frank, 2009; Mitra and Frank, 2006). Other single particle specimens represented include macromolecular machines involved in protein folding, protein degradation, energy metabolism, cell cycle processes, DNA replication, DNA repair, RNA transcription, and RNA splicing.
The map archive also holds densities for 2D crystals and for helical arrays, including intracellular filaments and microtubules, flagella, and helical crystals. There are also several tomographic maps of unique structures as well as maps that represent 3D averages of aligned tomograms. Diverse specimens currently held of this type include flagellar motors, insect flight muscle tissue, and desmosomes. A gallery of representative map volumes is presented in Figure 3.
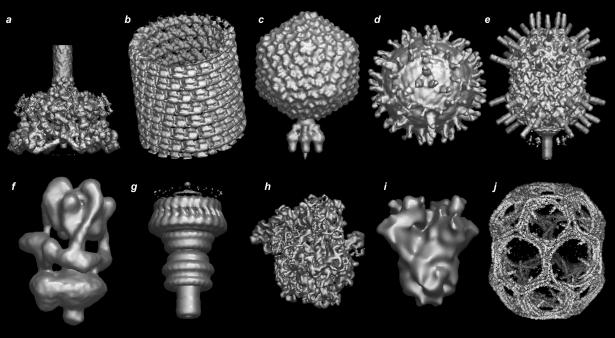
Figure 3.
Image gallery of representative EM map entries (arbitrary scales). Top row from left to right: (a) EMD-1048: bacteriophage T4 baseplate (Kostyuchenko et al., 2003); (b) EMD-1129: GDP-tubulin (Wang and Nogales, 2005); (c) EMD-1222: bacteriophage P22 (Chang et al., 2006); (d) EMD-1234: West Nile Virus decorated with neutralizing antibody Fab (Kaufmann et al., 2006); (e) EMD-1265: bacteriophage phi29 (Xiang et al., 2006). Bottom row from left to right: (f) EMD-1590: vacuolar ATPase motor (Muench et al., 2009); (g) EMD-1617: Shigella flexneri T3SS needle complex (Hodgkinson et al., 2009); (h) EMD-5036: E. coli 70S ribosome complex (Villa et al., 2009); (i) EMD-5114: BK potassium channel (Wang and Sigworth, 2009); (j) EMD-5119: clathrin lattice (Fotin et al., 2004). Images were created using AstexViewer.
Coordinates of EM entries are obtained using a variety of modelling methods including manual docking, rigid body fitting, homology modelling, and computational refinement algorithms. EM entries in the PDB are classified either under electron microscopy or electron crystallography as the experimental method. For structures with regular point or helical symmetry, coordinates are given for the asymmetric unit along with a set of transformation matrices to build the biological assembly (Lawson et al., 2008).
Access
Access to EM structural data and related services is through the EMDataBank.org web site. The EMDB and PDB archives are updated weekly on Wednesdays at 00:00 GMT. EMDB is distributed on two ftp mirrors supported by the two EMDataBank.org distribution partners in the UK and the USA, while PDB is distributed on ftp mirrors supported by each of the wwPDB partners.
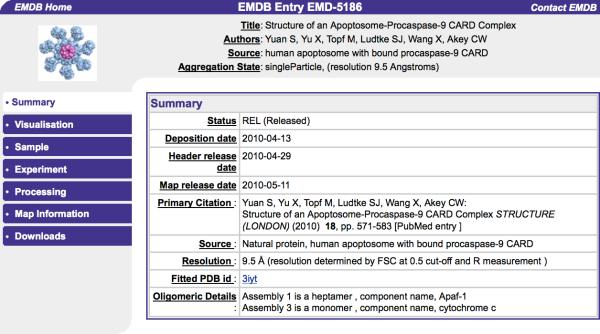
The EMSEARCH web service is also maintained and updated weekly at both distribution sites. EMSEARCH enables browsing and searching of EMDB metadata uploaded into a relational database. Simple searches can be performed based on author name, title, sample name, citation abstract word, aggregation type, resolution, and release date range. Search summaries link to atlas pages for each entry, which include summary, visualization, sample, experiment, processing, map information, and download pages (Figure 4).
Figure 4.
Example EMDB atlas page. [note to editor: color reproduction is not required for understanding]
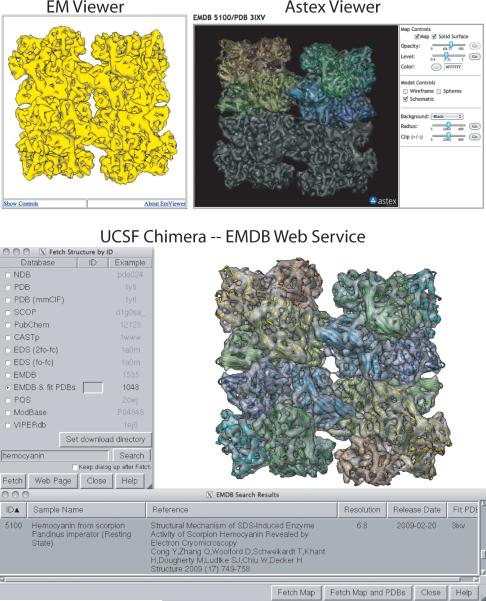
The atlas visualization page provides several ways to view EM maps. In addition to a static 2D image provided by the deposition author, it is also possible to launch two different Java-based 3D viewers (Figure 5, left and right top panels). EMViewer, developed by Powei Feng and Joe Warren at Rice University in collaboration with NCMI, provides a simple, single isosurface representation of a map at a predetermined contour level. Clicking on the “Launch EMViewer” button will bring up a simple 3D representation of the map that can be manipulated by mouse drag and click actions. AstexViewer, a molecular graphics program originally developed to display crystallographic data (Hartshorn, 2002) was recently re-released under an open source license and has been adapted by EMDataBank.org for display of EM maps and associated PDB coordinate models. To improve web download speed and minimize memory requirements, a compact map format is used (BRIX), and larger maps are also down-sampled by a factor of 2-5. Current capabilities include ability to control map contour level, opacity, color, solid vs. mesh surface rendering, and concurrent display of a PDB coordinate entry. Viewing large maps may require increasing Java Applet Runtime memory allocation.
Figure 5.
Tools for 3D visualization of EM structural data. EM Viewer (top left) and Astex Viewer (top right) are Java-based 3D viewers that can be launched from the EMDB atlas “Visualization” pages. EM Viewer employs a compact single contour-level mesh representation for lightweight map viewing; Astex Viewer enables exploration of maps and their fitted models with adjustable map contour level. The EMDB web service implemented in UCSF Chimera (bottom panel) expedites search of EMDB and subsequent map + model download. Scorpion hemocyanin (Cong et al., 2009, EMD-5100 and PDB id 3ixv) is the example shown in each of the panels. [note to editor: this figure requires color reproduction]
Map volumes are distributed in standard CCP4/MRC format and can be viewed with locally installed software such as UCSF Chimera (Pettersen et al., 2004), Pymol (Delano, 2002), VMD (Hsin et al., 2008) Coot (Emsley and Cowtan, 2004) or other graphics programs, enabling investigation with a more extensive set of tools. Links for map download are available on atlas download and map information pages. Recent distributions of UCSF Chimera enable direct downloads of EMDB maps plus associated fitted PDB models via simple queries (e.g. author name, title) to the EMSEARCH relational database through the EMDataBank.org beta-web service, which is a self-contained programming interface based on the SOAP protocol (Figure 5, bottom panel).
EM Navigator at PDBj (emnavi.protein.osaka-u.ac.jp) is an additional resource for browsing, viewing, and downloading maps and fitted coordinate models from the EMDB and PDB databases. Each of the wwPDB partners (wwpdb.org) has web interfaces to database representations of the PDB archive with advanced searching and browsing capabilities. ViperDB (Natarajan et al., 2005), a database specifically for icosahedral viruses, also holds some EM maps and related coordinates.
Examples of Use
A major goal of the EMDataBank.org unified data resource is to archive EM-derived structural information in a way that will enable further research. The impact of availability of structural data for smaller proteins has already been amply demonstrated by the success of the PDB, which is accessed globally by thousands of individuals every day. Listed here are a few examples of how archived EM data facilitates subsequent scientific exploration by other investigators.
Building up molecular pictures of large macromolecular assemblies
The availability of a coordinate model representation of a macromolecular complex leads directly to a dramatic increase in our fundamental understanding of biological function. Atomic coordinates permit 3D mapping of a wide variety of data including sites of mutations leading to disease, modification sites, antibody recognition sites, amino-acid sequence variability, and electrostatic properties. But in many cases, high-resolution structural models are not available for every component within a cryoEM map at the time it is first interpreted. By preserving map and model information together in a freely accessible database, new structural information can be incorporated as it becomes available. Examples include (1) reinterpretation of ribosome stalk regions in multiple EMDB-archived maps after the crystal structure of the L7/L12 complex was determined (Diaconu et al., 2005), and (2) progression towards a complete fitted coordinate model for bacteriophage T4 EM reconstructions based on fitting of components as they have become available (Aksyuk et al., 2009a; Aksyuk et al., 2009b).
Structural basis for interpreting data
Structural knowledge can be crucial for interpreting biochemical and biophysical data. For instance, dengue and West Nile virus cryoEM structures (Kuhn et al., 2002; Mukhopadhyay et al., 2003) have led the way to mapping sites of glycosylation (Hanna et al., 2005), amino-acid sequence variability (Modis et al., 2005), neutralizing antibody binding (Nybakken et al., 2005), design of vaccines (Ledizet et al., 2005), and a structural basis for understanding membrane fusion (Modis et al., 2004). Recently, multiple EMDB map entries were examined in light of biochemical data designed to distinguish between two distinct models for the hexameric subunit arrangement of two closely related disaggregating proteins, ClpB and Hsp10 (Wendler and Saibil, 2010).
Crystal structure phasing
CryoEM maps can be used to initiate crystallographic phasing. Isomorphous replacement phasing methods that are routinely used in X-ray crystallography are technically difficult to apply to large macromolecular complexes. A low-resolution cryoEM map of the complex under study thus provides a valuable complementary source of phase information. For high-symmetry structures, such as icosahedral viruses, application of robust computational averaging and extension algorithms to initial cryoEM-derived phases is often sufficient to complete the structure determination without additional phase information. Examples of crystal structures phased with cryoEM envelopes include proteases (Bosch et al., 2001; Wang et al., 1998), ribosomes (Ban et al., 1998; Cate et al., 1999; Thygesen et al., 1996), and icosahedral viruses (Dokland et al., 1997; Dokland et al., 1998; Grimes et al., 1998; Helgstrand et al., 2003; Prasad et al., 1999; Reinisch et al., 2000; Wynne et al., 1999). Additional case studies are described by Xiong (2008), and use of a stain EM reconstruction for phasing is described by Trapani et al. (2010). Routine archiving of EM-derived maps facilitates the use of this method by the crystallography community.
Comparative studies
By capturing the coordinates, maps, and metadata and making them available through searchable databases, it is possible to easily compare structures, select structures for further analysis using a variety of criteria, perform experimental design for new analyses, and design new algorithms to improve the state of the art methodology. For example, observation of publicly available maps and coordinates led to the conclusion that the double-stranded DNA tailed phages and herpesvirus have a similar fold in their major capsid proteins though there is little sequence similarity among them (Baker et al., 2005). In a second example, a flexible fitting approach applied to 43 EMDB map volumes of bacterial 70S ribosome in various functional states revealed global conformational differences between the EM structures involving large-scale ratchet-like deformations (Matsumoto and Ishida, 2009).
Software development
Availability of EM map volumes in the EMDB facilitates development and validation of software for map viewing, analysis, manipulation, coordinate model fitting and validation. Development of algorithms for fitting of EM maps with atomic coordinates is a particularly active area. For low to medium-resolution studies in which the source coordinates are likely to be derived from crystallographic or NMR studies, algorithms being explored include rigid-body fitting (Wriggers, 2010), normal mode analysis (Tama et al., 2004), spatial interpolation (Rusu et al., 2008), conformational sampling under low-resolution restraints (Schroder et al., 2007), molecular dynamics flexible fitting (Trabuco et al., 2009), and simulated annealing approaches (Tan et al., 2008; Topf et al., 2008). For medium to high-resolution studies where de novo model building becomes possible, methods are being developed for skeletonization and secondary structure element detection (Baker et al., 2007) and incorporation of structure prediction from primary sequence (DiMaio et al., 2009). Additional examples of algorithm development facilitated by public availability of EMDB maps include investigations of map denoising (Jiang et al., 2003), B-factor sharpening (Fernandez et al., 2008), map resolution determination (Sousa and Grigorieff, 2007), and automated segmentation (Baker et al., 2006; Pintilie et al., 2010). The UCSF Chimera team has made particularly effective use of the EMDB resource for development and testing of a large, versatile set of tools for manipulating volume data (Goddard et al., 2007; Pettersen et al., 2004).
Future Prospects
3D cryo-electron microscopy reconstruction methods are uniquely able to reveal structural aspects of many important macromolecules and macromolecular complexes, and for this reason the field is in a period of rapid expansion and development. Based on current growth of EM entries and publication of EM structures, the total number of structures of large biological assemblies contributed by EM is anticipated to approach 10,000 by the year 2020.
In the near future, deposition and archiving of EM structural data will be integrated in a common tool that is being developed by the wwPDB partners to handle depositions from all structural biology methods. In addition, as the field matures, validation tools and criteria for assessment of map and fitted coordinate models will play an important role in providing guidance to users of cryoEM derived structural data. To this end, a validation task force is being assembled by the unified data resource partners (NCMI, RCSB and EBI) to develop recommendations as to how best to assess the quality of both maps and models that have been obtained from cryoEM data. The recommendations will form the basis for a validation suite that will be used by EMDB and PDB.
Guide to Deposition of EM Structural Data
This section provides an overview of joint map + fitted coordinate model deposition to the EM DataBank (EMDB) and Protein Data Bank (PDB). To prepare for deposition, the following items should be available:
Map File. One map file may be uploaded per EMDB entry. If an experiment yields more than one 3D map volume, the maps must be deposited in separate entries. Map formats accepted for upload include SPIDER, MRC, and CCP4; maps can be compressed (tar, zip, gzip). The uploaded map will be converted to EMDB redistribution format (currently CCP4/MRC).
Model File. Atomic models in PDB or mmCIF format must be in the same coordinate frame as the map. Models may include species homologs or homology models, and/or may contain either full-residues or alpha-carbon/phosphate backbone traces. For structures with regular point or helical symmetry, coordinates for one asymmetric unit may be deposited along with transformation matrices to build the assembly (Lawson et al., 2008). Detailed information about coordinate formats, annotation and processing policies can be found at wwpdb.org/docs.html.
Experiment Info. Basic information about the experiment, including a description of the assembly, specimen preparation, microscope imaging, image processing and reconstruction, and coordinate fitting will be requested during deposition.
External Database IDs. In addition, external database ids such as GO (geneontology.org), INTERPRO (www.ebi.ac.uk/interpro), Virus ICTVdB (www.ncbi.nlm.nih.gov/ICTVdb), PDB (coordinates used for fitting), and PUBMED (if published) will be requested.
Map Deposition using EMDEP
EM structural data can be submitted to deposition sites at PDBe (UK) or RCSB-PDB (USA). Go to emdatabank.org/deposit.html to select the deposition site. To initiate an EMDEP session, click on “Start Session.” Select “new deposition”, or select “based on previous submission” if the experiment is related to a prior deposition.
The map deposition is created page by page. When the relevant information on the page is entered and saved, the symbol for the page on the left hand menu changes from a red arrow to a green circle. Before submission, it is possible to go back and change answers on completed pages. Help text is available for every data item by clicking on the item.
Many pages contain sections for entering metadata information about the experiment that can be duplicated. For instance, two “sample component” sections should be completed for each unique component in an Fab:virus complex assembly, one section for the Fab, and a second section for the virus. Multiple imaging sessions/microscopes can be defined.
Atomic coordinates from existing PDB entries (e.g., X-ray or NMR structures) used in fitting of the map are specified by the depositor on the “fitting” page, which can also be duplicated as needed. In contrast, fitted atomic coordinates representing the depositor's molecular interpretation of the EM map are deposited to the PDB following the map deposition (see below), and the resulting PDB id is associated with the map entry by the database curator.
Fitted Model Deposition
After completing map deposition using EMDEP at either site, a link will appear on the EMDEP left hand menu to initiate deposition of one or more models to PDB with automatic transfer of experimental metadata (sample description, microscope type, etc). At the PDBe site, the link opens an AutoDep session; at the RCSB-PDB site, the link opens an EM-Adit session. Sequence information must be provided for each protein or nucleic acid entity should include the entire sequence of the imaged material, including any mutations or expression tags.
Accession IDs
After completion of map and model depositions, accession IDs are assigned by the EMDB and PDB, respectively. The accession ids are associated with each other by the two databases. All provided ids should be included in the primary publication describing the EM structure.
Acknowledgements
Many current and past members of the EMDataBank.org team have made significant contributions to the development of the Unified Data Resource for CryoEM including Kim Henrick, Wah Chiu, Helen Berman, Gerard Kleywegt, Richard Newman, John Westbrook, Glen van Ginkel, Batsal Devkota, Matt Baker, Tom Oldfield, Christoph Best, Gaurav Sahni, Raul Sala, Chunxiao Bi, Powei Feng, Joe Warren, Matt Dougherty, Steve Ludtke, and Ian Rees. The Resource is funded by National Institutes of Health GM079429 to Baylor College of Medicine, Rutgers University, and the European Bioinformatics Institute.
Footnotes
Book Chapter for Methods in Enzymology, Volume on CryoEM, Grant Jensen, editor
References
- Aksyuk AA, et al. The tail sheath structure of bacteriophage T4: a molecular machine for infecting bacteria. EMBO J. 2009a;28:821–9. doi: 10.1038/emboj.2009.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksyuk AA, et al. The structure of gene product 6 of bacteriophage T4, the hinge-pin of the baseplate. Structure. 2009b;17:800–8. doi: 10.1016/j.str.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Baker ML, et al. Common ancestry of herpesviruses and tailed DNA bacteriophages. J Virol. 2005;79:14967–70. doi: 10.1128/JVI.79.23.14967-14970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker ML, et al. Identification of secondary structure elements in intermediate-resolution density maps. Structure. 2007;15:7–19. doi: 10.1016/j.str.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker ML, et al. Automated segmentation of molecular subunits in electron cryomicroscopy density maps. J Struct Biol. 2006 doi: 10.1016/j.jsb.2006.05.013. in press. [DOI] [PubMed] [Google Scholar]
- Ban N, et al. A 9 A resolution X-ray crystallographic map of the large ribosomal subunit. Cell. 1998;93:1105–15. doi: 10.1016/s0092-8674(00)81455-5. [DOI] [PubMed] [Google Scholar]
- Baumeister W. Mapping molecular landscapes inside cells. Biol Chem. 2004;385:865–72. doi: 10.1515/BC.2004.113. [DOI] [PubMed] [Google Scholar]
- Berman HM, et al. Announcing the worldwide Protein Data Bank. Nat Struct Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- Bernstein FC, et al. Protein Data Bank: a computer-based archival file for macromolecular structures. J. Mol. Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Bosch J, et al. Purification, crystallization, and preliminary X-ray diffraction analysis of the Tricorn protease hexamer from Thermoplasma acidophilum. J Struct Biol. 2001;134:83–7. doi: 10.1006/jsbi.2001.4360. [DOI] [PubMed] [Google Scholar]
- Cate JH, et al. X-ray crystal structures of 70S ribosome functional complexes. Science. 1999;285:2095–104. doi: 10.1126/science.285.5436.2095. [DOI] [PubMed] [Google Scholar]
- Chang J, et al. Cryo-EM asymmetric reconstruction of bacteriophage P22 reveals organization of its DNA packaging and infecting machinery. Structure. 2006;14:1073–82. doi: 10.1016/j.str.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Chiu W, Rixon FJ. High resolution structural studies of complex icosahedral viruses: a brief overview. Virus Res. 2002;82:9–17. doi: 10.1016/s0168-1702(01)00381-1. [DOI] [PubMed] [Google Scholar]
- Cong Y, et al. Structural mechanism of SDS-induced enzyme activity of scorpion hemocyanin revealed by electron cryomicroscopy. Structure. 2009;17:749–58. doi: 10.1016/j.str.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delano WL. The Pymol Molecular Graphics System. Delano Scientific; Palo Alto: 2002. [Google Scholar]
- Diaconu M, et al. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell. 2005;121:991–1004. doi: 10.1016/j.cell.2005.04.015. [DOI] [PubMed] [Google Scholar]
- DiMaio F, et al. Refinement of protein structures into low-resolution density maps using rosetta. J Mol Biol. 2009;392:181–90. doi: 10.1016/j.jmb.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokland T, et al. Structure of a viral procapsid with molecular scaffolding. Nature. 1997;389:308–13. doi: 10.1038/38537. [DOI] [PubMed] [Google Scholar]
- Dokland T, et al. Structure determination of the phiX174 closed procapsid. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):878–90. doi: 10.1107/s0907444998002467. [DOI] [PubMed] [Google Scholar]
- Dutta S, et al. Data deposition and annotation at the worldwide protein data bank. Mol Biotechnol. 2009;42:1–13. doi: 10.1007/s12033-008-9127-7. [DOI] [PubMed] [Google Scholar]
- Editorial, A database for 'em Nat Struct Biol. 2003;10:313. doi: 10.1038/nsb0503-313. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fernandez JJ, et al. Sharpening high resolution information in single particle electron cryomicroscopy. J Struct Biol. 2008;164:170–5. doi: 10.1016/j.jsb.2008.05.010. [DOI] [PubMed] [Google Scholar]
- Fotin A, et al. Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature. 2004;432:573–9. doi: 10.1038/nature03079. [DOI] [PubMed] [Google Scholar]
- Frank J. Single-particle reconstruction of biological macromolecules in electron microscopy--30 years. Q Rev Biophys. 2009;42:139–58. doi: 10.1017/S0033583509990059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller SD. Depositing electron microscopy maps. Structure (Camb) 2003;11:11–2. doi: 10.1016/s0969-2126(02)00942-5. [DOI] [PubMed] [Google Scholar]
- Gabashvili IS, et al. Solution structure of the E. coli 70S ribosome at 11.5 A resolution. Cell. 2000;100:537–49. doi: 10.1016/s0092-8674(00)80690-x. [DOI] [PubMed] [Google Scholar]
- Goddard TD, et al. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157:281–7. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Gonen T, et al. Lipid-protein interactions in double-layered two-dimensional AQP0 crystals. Nature. 2005;438:633–8. doi: 10.1038/nature04321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes JM, et al. The atomic structure of the bluetongue virus core. Nature. 1998;395:470–8. doi: 10.1038/26694. [DOI] [PubMed] [Google Scholar]
- Hanna SL, et al. N-linked glycosylation of west nile virus envelope proteins influences particle assembly and infectivity. Journal of Virology. 2005;79:13262–13274. doi: 10.1128/JVI.79.21.13262-13274.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartshorn MJ. AstexViewer: a visualisation aid for structure-based drug design. J Comput Aided Mol Des. 2002;16:871–81. doi: 10.1023/a:1023813504011. [DOI] [PubMed] [Google Scholar]
- Helgstrand C, et al. The refined structure of a protein catenane: the HK97 bacteriophage capsid at 3.44 A resolution. J Mol Biol. 2003;334:885–99. doi: 10.1016/j.jmb.2003.09.035. [DOI] [PubMed] [Google Scholar]
- Henderson R, et al. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J Mol Biol. 1990;213:899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- Henrick K, et al. EMDep: a web-based system for the deposition and validation of high-resolution electron microscopy macromolecular structural information. J Struct Biol. 2003;144:228–37. doi: 10.1016/j.jsb.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Heymann JB, et al. Common conventions for interchange and archiving of three-dimensional electron microscopy information in structural biology. J Struct Biol. 2005;151:196–207. doi: 10.1016/j.jsb.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Hodgkinson JL, et al. Three-dimensional reconstruction of the Shigella T3SS transmembrane regions reveals 12-fold symmetry and novel features throughout. Nat Struct Mol Biol. 2009;16:477–85. doi: 10.1038/nsmb.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin J, et al. Using VMD: an introductory tutorial. Curr Protoc Bioinformatics. 2008 doi: 10.1002/0471250953.bi0507s24. Chapter 5, Unit 5 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huiskonen JT, Butcher SJ. Membrane-containing viruses with icosahedrally symmetric capsids. Curr Opin Struct Biol. 2007;17:229–36. doi: 10.1016/j.sbi.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Jiang W, et al. Applications of bilateral denoising filter in biological electron microscopy. J Struc Biol. 2003;144:114–122. doi: 10.1016/j.jsb.2003.09.028. [DOI] [PubMed] [Google Scholar]
- Kaufmann B, et al. West Nile virus in complex with the Fab fragment of a neutralizing monoclonal antibody. Proc Natl Acad Sci U S A. 2006;103:12400–4. doi: 10.1073/pnas.0603488103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostyuchenko VA, et al. Three-dimensional structure of bacteriophage T4 baseplate. Nat Struct Biol. 2003;10:688–93. doi: 10.1038/nsb970. [DOI] [PubMed] [Google Scholar]
- Kuhn RJ, et al. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108:717–25. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson CL, et al. Representation of viruses in the remediated PDB archive. Acta Crystallographica Section D-Biological Crystallography. 2008;64:874–882. doi: 10.1107/S0907444908017393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledizet M, et al. A recombinant envelope protein vaccine against West Nile virus. Vaccine. 2005;23:3915–3924. doi: 10.1016/j.vaccine.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Lee KK, Johnson JE. Complementary approaches to structure determination of icosahedral viruses. Curr Opin Struct Biol. 2003;13:558–69. doi: 10.1016/j.sbi.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Matsumoto A, Ishida H. Global conformational changes of ribosome observed by normal mode fitting for 3D Cryo-EM structures. Structure. 2009;17:1605–13. doi: 10.1016/j.str.2009.09.017. [DOI] [PubMed] [Google Scholar]
- Mitra K, Frank J. Ribosome dynamics: insights from atomic structure modeling into cryo-electron microscopy maps. Annu Rev Biophys Biomol Struct. 2006;35:299–317. doi: 10.1146/annurev.biophys.35.040405.101950. [DOI] [PubMed] [Google Scholar]
- Modis Y, et al. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427:313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- Modis Y, et al. Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. Journal of Virology. 2005;79:1223–1231. doi: 10.1128/JVI.79.2.1223-1231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muench SP, et al. Cryo-electron microscopy of the vacuolar ATPase motor reveals its mechanical and regulatory complexity. J Mol Biol. 2009;386:989–99. doi: 10.1016/j.jmb.2009.01.014. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, et al. Structure of West Nile virus. Science. 2003;302:248. doi: 10.1126/science.1089316. [DOI] [PubMed] [Google Scholar]
- Murphy GE, Jensen GJ. Electron cryotomography. Biotechniques. 2007;43:413, 415, 417. doi: 10.2144/000112568. passim. [DOI] [PubMed] [Google Scholar]
- Natarajan P, et al. Exploring icosahedral virus structures with VIPER. Nat Rev Microbiol. 2005;3:809–17. doi: 10.1038/nrmicro1283. [DOI] [PubMed] [Google Scholar]
- Nybakken GE, et al. Structural basis of West Nile virus neutralization by a therapeutic antibody. Nature. 2005;437:764–768. doi: 10.1038/nature03956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson NH, et al. The three-dimensional structure of frozen-hydrated Nudaurelia capensis beta virus, a T = 4 insect virus. J Struct Biol. 1990;105:111–22. doi: 10.1016/1047-8477(90)90105-l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pintilie GD, et al. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J Struct Biol. 2010 doi: 10.1016/j.jsb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad BV, et al. X-ray crystallographic structure of the Norwalk virus capsid. Science. 1999;286:287–90. doi: 10.1126/science.286.5438.287. [DOI] [PubMed] [Google Scholar]
- Reinisch KM, et al. Structure of the reovirus core at 3.6 A resolution. Nature. 2000;404:960–7. doi: 10.1038/35010041. [DOI] [PubMed] [Google Scholar]
- Rusu M, et al. Biomolecular pleiomorphism probed by spatial interpolation of coarse models. Bioinformatics. 2008;24:2460–6. doi: 10.1093/bioinformatics/btn461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid MF, et al. Structure of the acrosomal bundle. Nature. 2004;431:104–7. doi: 10.1038/nature02881. [DOI] [PubMed] [Google Scholar]
- Schroder GF, et al. Combining efficient conformational sampling with a deformable elastic network model facilitates structure refinement at low resolution. Structure. 2007;15:1630–41. doi: 10.1016/j.str.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa D, Grigorieff N. Ab initio resolution measurement for single particle structures. J Struct Biol. 2007;157:201–10. doi: 10.1016/j.jsb.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Tagari M, et al. New electron microscopy database and deposition system. Trends Biochem Sci. 2002;27:589. doi: 10.1016/s0968-0004(02)02176-x. [DOI] [PubMed] [Google Scholar]
- Tama F, et al. Normal mode based flexible fitting of high-resolution structure into low-resolution experimental data from cryo-EM. J Struct Biol. 2004;147:315–26. doi: 10.1016/j.jsb.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Tan RK, et al. YUP.SCX: coaxing atomic models into medium resolution electron density maps. J Struct Biol. 2008;163:163–74. doi: 10.1016/j.jsb.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thygesen J, et al. The suitability of multi-metal clusters for phasing in crystallography of large macromolecular assemblies. Structure. 1996;4:513–8. doi: 10.1016/s0969-2126(96)00057-3. [DOI] [PubMed] [Google Scholar]
- Topf M, et al. Protein structure fitting and refinement guided by cryo-EM density. Structure. 2008;16:295–307. doi: 10.1016/j.str.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabuco LG, et al. Molecular dynamics flexible fitting: a practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods. 2009;49:174–80. doi: 10.1016/j.ymeth.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapani S, et al. Macromolecular crystal data phased by negative-stained electron-microscopy reconstructions. Acta Cryst D. 2010;66:514–521. doi: 10.1107/S0907444910002763. [DOI] [PubMed] [Google Scholar]
- Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4A resolution. J Mol Biol. 2005;346:967–89. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Villa E, et al. Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis. Proc Natl Acad Sci U S A. 2009;106:1063–8. doi: 10.1073/pnas.0811370106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HW, Nogales E. Nucleotide-dependent bending flexibility of tubulin regulates microtubule assembly. Nature. 2005;435:911–5. doi: 10.1038/nature03606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, et al. Crystal structure determination of Escherichia coli ClpP starting from an EM-derived mask. J Struct Biol. 1998;124:151–63. doi: 10.1006/jsbi.1998.4058. [DOI] [PubMed] [Google Scholar]
- Wang L, Sigworth FJ. Structure of the BK potassium channel in a lipid membrane from electron cryomicroscopy. Nature. 2009;461:292–5. doi: 10.1038/nature08291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YA, et al. The structure of a filamentous bacteriophage. J Mol Biol. 2006;361:209–15. doi: 10.1016/j.jmb.2006.06.027. [DOI] [PubMed] [Google Scholar]
- Wendler P, Saibil HR. Cryo electron microscopy structures of Hsp100 proteins: crowbars in or out? Biochem Cell Biol. 2010;88:89–96. doi: 10.1139/o09-164. [DOI] [PubMed] [Google Scholar]
- Wriggers W. Using Situs for the integration of multi-resolution structures. Biophys Rev. 2010;2:21–27. doi: 10.1007/s12551-009-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynne SA, et al. The crystal structure of the human hepatitis B virus capsid. Mol Cell. 1999;3:771–80. doi: 10.1016/s1097-2765(01)80009-5. [DOI] [PubMed] [Google Scholar]
- Xiang Y, et al. Structural changes of bacteriophage phi29 upon DNA packaging and release. EMBO J. 2006;25:5229–39. doi: 10.1038/sj.emboj.7601386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y. From electron microscopy to X-ray crystallography: molecular-replacement case studies. Acta Crystallogr D Biol Crystallogr. 2008;64:76–82. doi: 10.1107/S090744490705398X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Egelman EH. The RecA hexamer is a structural homologue of ring helicases. Nat Struct Biol. 1997;4:101–4. doi: 10.1038/nsb0297-101. [DOI] [PubMed] [Google Scholar]
- Zhou ZH, et al. Electron cryomicroscopy and bioinformatics suggest protein fold models for rice dwarf virus. Nat Struct Biol. 2001;8:868–73. doi: 10.1038/nsb1001-868. [DOI] [PubMed] [Google Scholar]