Abstract
Background
Gene silencing due to aberrant DNA methylation is a frequent event in hepatocellular carcinoma (HCC) and also in hepatocellular adenoma (HCA). However, very little is known about epigenetic defects in fibrolamellar carcinoma (FLC), a rare variant of hepatocellular carcinoma that displays distinct clinical and morphological features.
Methodology/Principal Findings
We analyzed the methylation status of the APC, CDH1, cyclinD2, GSTπ1, hsa-mir-9-1, hsa-mir-9-2, and RASSF1A gene in a series of 15 FLC and paired normal liver tissue specimens by quantitative high-resolution pyrosequencing. Results were compared with common HCC arising in non-cirrhotic liver (n = 10). Frequent aberrant hypermethylation was found for the cyclinD2 (19%) and the RASSF1A (38%) gene as well as for the microRNA genes mir-9-1 (13%) and mir-9-2 (33%). In contrast to common HCC the APC and CDH1 (E-cadherin) genes were found devoid of any DNA methylation in FLC, whereas the GSTπ1 gene showed comparable DNA methylation in tumor and surrounding tissue at a moderate level. Changes in global DNA methylation level were measured by analyzing methylation status of the highly repetitive LINE-1 sequences. No evidence of global hypomethylation could be found in FLCs, whereas HCCs without cirrhosis showed a significant reduction in global methylation level as described previously.
Conclusions
FLCs display frequent and distinct gene-specific hypermethylation in the absence of significant global hypomethylation indicating that these two epigenetic aberrations are induced by different pathways and that full-blown malignancy can develop in the absence of global loss of DNA methylation. Only quantitative DNA methylation detection methodology was able to identify these differences.
Introduction
Inactivation of tumor suppressor genes by aberrant methylation of cytosine residues in the promoter region is an important molecular alteration contributing to the development and progression of malignant tumors [1]. It can already be found in pre-malignant lesions and in-situ carcinomas indicating that this epigenetic alteration is an early event in carcinogenesis [2]. In colonic carcinoma acquired genetic and epigenetic defects complement one another in the process of malignant transformation [3]. The diagnostic and prognostic potential of altered DNA methylation patterns is currently being unraveled [4].
Aberrant DNA methylation is a well described phenomenon in common hepatocellular carcinoma [5] and also in hepatocellular adenoma [6]. However, very little is known about epigenetic defects in fibrolamellar carcinoma (FLC), a rare variant of hepatocellular carcinoma that displays unique clinical and morphologicall features [7], [8]. FLC occurs in the absence of chronic liver disease in children and young adults and is characterized by large eosinophilic tumor cells and abundant deposition of collagen between tumor cells ( Figure 1 ). The few existing studies of genetic defects in FLCs indicate that chromosomal instability is a rare event in FLCs and that mutations frequently found in common HCC (e.g., in the TP53 or the CTNNB1 gene) occur at a much lower frequency if at all [9], [10]. We performed epigenetic profiling of a series of FLCs (n = 15) in comparison to common HCC arising in non-cirrhotic livers. For this purpose the global methylation level as well as gene-specific hypermethylation at 7 loci was assessed using quantitative pyrosequencing methodology.
Figure 1. Representative histology of FLC and common HCC without cirrhosis.
Representative histology of fibrolamellar carcinoma (A) and common hepatocellular carcinoma in non-cirrhotic liver (B). FLCs show large eosinpphilic tumor cells containing cytoplasmic globuli (arrow). There are abundant collagenous bands (arrowheads) separating nests of tumor cells. HCC of common type show solid nests and trabecules of smaller and paler cells without formation of collagenous bands (HE stained, original magnification A): 200×, B): 100×).
Results
Selection of patients and genes under study
Upon review 15 cases of FLC were identified from the archives of the Institute of Pathology, Medizinische Hochschule Hannover. 10 cases of common HCC arising in non-cirrhotic livers were used as a control group ( Figure 1 ). Patient age, sex and tumor stage are summarized in Table 1 .
Table 1. Overview of patients.
| FLC cases | |||||||
| no. | sex | age | UICC-classification | Vascular invasion | AFIP Grade | HBV | HCV |
| 1 | male | 26 | pT4, pN1 | neg. | n/a | ||
| 2 | male | 20 | pT3, pN1 | present | neg. | neg. | |
| 3 | male | 20 | pT4, pN1, pM1 | present | neg. | neg. | |
| 4 | male | 39 | pT1, pN0 | absent | n/a | n/a | |
| 5 | male | 19 | pT2, pNx | present | neg. | neg. | |
| 6 | male | 24 | pT3, pN1, pM1 | present | neg. | neg. | |
| 7 | female | 15 | pT4, pN1 | neg. | neg. | ||
| 8 | female | 32 | pT1, pNx | absent | neg. | neg. | |
| 9 | male | 19 | pT3, pN1 | present | neg. | neg. | |
| 10 | female | 28 | pT1, pNx | absent | n/a | n/a | |
| 11 | female | 13 | pT3, pN0, pM1 | present | neg. | neg. | |
| 12 | male | 28 | pT3, pN1, pM1 | present | neg. | neg. | |
| 13 | female | 36 | pT1, pN0 | absent | HBV+ | neg. | |
| 14 | female | 13 | pT1, pNx | absent | neg. | neg. | |
| 15 | male | 22 | pT1, pN0 | absent | neg. | neg. | |
The methylation status of the following loci was analyzed in a series of 15 FLC samples and the surrounding normal appearing tissue employing quantitative high-resolution pyrosequencing technology: APC, CDH1, cyclinD2, ESR1, GSTπ1, LINE-1, MINT31, hsa-mir-9-1, hsa-mir-9-2, RASSF1A, SFRP1, SOCS-1. With the exception of the microRNA genes these loci are reported to be frequently hypermethylated in common hepatocellular carcinoma in more than one study (see [5] and references therein). Common HCC was chosen as a reference because HCC and FLC are regarded as to arise not only in the same organ but also in the very same cell type. Aberrant hypermethylation of microRNA genes hsa-mir-9-1 and hsa-mir-9-2 in liver tumors has been recently discovered in our group (Albat and Lehmann, in preparation). Gen-specific hypermethylation very often takes place in the context of a generalized hypomethylation ([11] and the methylation status of the highly repetitive LINE-1 sequence is a suitable surrogate marker for assessing this global loss of methylation [12]. Therefore, methylation analysis of LINE-1 was included.
Since fibrolamellar carcinoma is a rare subtype of liver carcinoma [7], several specimens were quite old (up to 20 years) and collected under non-standardized conditions. Therefore, yield and quality of genomic DNA extracted from the paraffin blocks was highly variable within this series and for several loci only a subset of samples gave reproducible results. For these reasons, the further analysis and discussion focus on the following loci: APC, CDH1, cyclinD2, GSTπ1, LINE-1, hsa-mir-9-1, hsa-mir-9-2, and RASSF1A.
For these 8 loci high-quality pyrosequencing data could be obtained for all specimens with only very few exceptions: The results of 399 out of 400 measurements (99.8%) are summarized in Figures 2 and 3 .
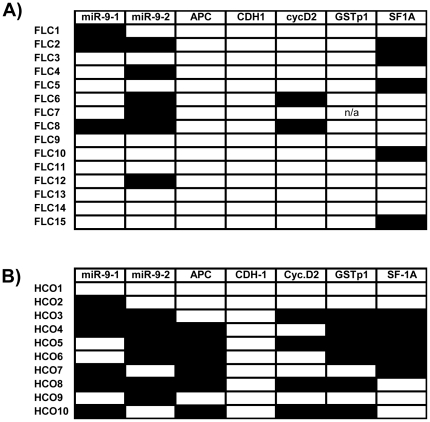
Figure 2. Summary of methylation data for FLC and HCC without cirrhosis.
Frequent aberrant hypermethylation in FLC is obvious. A black box indicates “hypermethylated” according to the stringent threshold definition (mean of the control group plus 2× STD, see text for details).
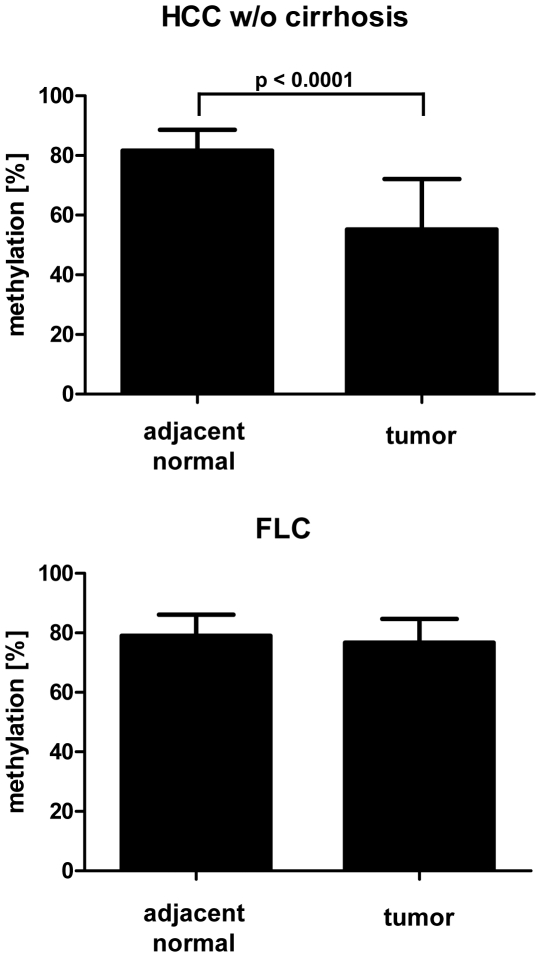
Figure 3. Global methylation level in FLC and HCC without cirrhosis.
The Methylation level of LINE-1 sequences was measured quantitatively using pyrosequencing. The Methylation level of these repetitive elements reflects very well the overall methylation level of the genome [12].
Definition of “hypermethylation”
DNA methylation levels for all genes under study displayed a quite high variation in the non-tumorous adjacent tissue. The range of variation ranged from 6 percentage points for the CDH1 (E-cadherin) gene (2–8%) to up to 49% percentage points for GSTπ1 gene (7–56%). Therefore, two different definitions for scoring a tumor sample as “hypermethylated” were applied:
a) “hypermethylated” is defined by levels in excess of two standard deviations above the mean of the control group of adjacent non-tumorous liver tissue (“mean of control group +2× standard deviation”, [6]).
b) “hypermethylated” is defined by levels of methylation 50% higher than in the corresponding non-neoplastic liver tissue from the same patient. In order to avoid over-interpretation of data due to background fluctuations (e.g., comparing 6% in the tumor fraction with 3.5% in the adjacent normal tissue fraction) only those samples were scored “hypermethylated” in which the methylation level in the tumor fraction were above 10%. Definition a) is much more stringent, because the above mentioned high variation in the control group of normal appearing adjacent non-tumorous specimens causes a high standard deviation and thereby a high threshold value. For this reason, this threshold setting may conceal significant differences between normal and tumor for individual cases. For example, methylation level of the cyclinD2 gene in the tumor fraction of FLC 15 is 18.8%, in the surrounding normal tissue only 5.7%. Both values are below the threshold defined following definition a) (27.8% for cyclinD2 in FLC) but are clearly indicating increased DNA methylation in the tumor tissue (with statistical significance).
For a detailed comparison of the results using both definitions see Figure S1.
Hypermethylation of cyclinD2 and RASSF1A in FLC
Aberrant hypermethylation of the cyclinD2 and the RASSF1A gene has been described for common hepatocellular carcinoma arising in cirrhotic liver tissue [13] as well as in many other carcinomas, including breast, gastric, and colon carcinoma [14].
In fibrolamellar carcinoma these two genes are also frequent targets of aberrant hypermethylation. Both genes were found to be hypermethylated in 2/15 (13.3%) and 5/15 (33.3%) of FLC cases, respectively. There were no statistical significant differences in comparison to the control group of common HCC (cyclinD2: 4/10, RASSF1A: 6/10, p = 0.18 and 0.24, respectively, Fisher's exact test, two-sided).
Hypermethylation of microRNA genes hsa-mir-9-1 and hsa-mir-9-2 in FLC
Aberrant hypermethylation of microRNA genes has been reported for several human malignancies, including colon cancer [15], breast cancer [16], acute lymphoblastic leukemia [17], and chronic myelogenous leukemia [18]. So far, only two studies reported microRNA gene hypermethylation in liver tumors: according to Datta et al., 2 out of 4 HCC specimens tested showed hypermethylation of the hsa-mir-1 gene [19]. In a more comprehensive study, Furuta et al. could demonstrate aberrant methylation of microRNA genes miR-124, miR-203, and miR-375 in a series of 41 common hepatocellular carcinoma cases [20]. Our own systematic study of a range of liver tumors including fibrolamellar carcinoma identified several frequently hypermethylated microRNA genes (Albat and Lehmann, in preparation). In FLC, hsa-mir-9-1 is hypermethylated in 3/15 cases (20%), in HCC without cirrhosis in 6/10 cases (60%). Hsa-mir-9-2 is hypermethylated in 6/15 cases (40%) in FLC and in 6/10 cases (60%) in HCC without cirrhosis. For both microRNA genes there are no significant differences in hypermethylation between FLC and HCC without cirrhosis (Fisher's exact test, two-sided, p = 0.087 and 0.43, respectively) and the hypermethylation in both entities is quite frequent (20 to 60%).
Absence of APC, CDH1, and GSTπ1 gene hypermethylation in FLC
All specimens from patients with FLC (tumor and adjacent normal tissue) showed only little or no methylation in the APC and the CDH1 promoter not qualifying for hypermethylation status according to the definitions outlined above.
On the contrary, 3 samples showed a clear reduction of the methylation levels of the APC gene in FLC compared to adjacent non-neoplastic tissue from the same patient.
The GSTπ1 gene showed variable methylation levels in the FLC specimens (15–50%) but to a very similar extent in nearly all samples of surrounding normal liver tissue (10–55%). Therefore, following the stringent threshold defined above, no FLC sample qualifies as “hypermethylated”. Applying the cases-specific definition of hypermethylation (“50% more than in the paired normal tissue”, see above), 4/17 (23.5%) qualify as “hypermethylated” in the GSTπ1 gene.
Since the APC gene and the GSTπ1 gene were found to be hypermethylated in 60% (6/10 cases each) of common HCC differences between FLC and HCC groups were significantly different (p = 0.0009 for both genes, Fisher's exact test, two-sided).
Global methylation level in FLC
Global loss of methylation is well described for many solid tumors and also for hepatocellular carcinoma [21]. Therefore, the methylation level of LINE-1 sequences, highly repetitive DNA elements scattered throughout the human genome, was assessed using quantitative pyrosequencing. The methylation level of these elements correlates very well with the global methylation level of the genome under study and therefore can serve as surrogate marker for the overall methylation level [12]. In comparison to adjacent non-neoplastic liver tissue, no significant demethylation of LINE-1 sequences was found in FLCs, indicating the absence of widespread hypomethylation in this liver tumor ( Figure 3 ). In contrast, in common HCC from non-cirrhotic liver samples a significant decrease of LINE-1 methylation in comparison to adjacent non-neoplastic liver tissue was identified (p<0.0001, Figure 3 ). The LINE-1 methylation level in the non-neoplastic liver tissue from both patient groups (FLC and HCC) did not show any differences (81.6+/−7% versus 79.1+/−7%, p = 0.26, M-Whitney-U, two-tailed).
Cluster analysis of methylation data
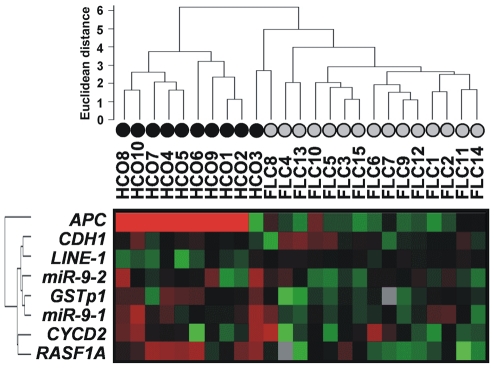
Unsupervised clustering of all quantitative log-transformed methylation data revealed a very good separation of the FLC samples from the HCC samples ( Figure 4 ). Within these two patient groups the separation between tumor and adjacent non-neoplastic tissue is less pronounced (see Figure S2). The clustering shown in Figure 4 underlines the distinct methylation profile of FLC in comparison to common HCC arising in non-cirrhotic liver already apparent from Figure 2 .
Figure 4. Cluster analysis of methylation data for FLC and common HCC without cirrhosis.
The quantitative methylation data were log-transformed and translated into a color code.
Discussion
This is the first study that uses high-resolution quantitative methodology for comparison of gene-specific and global DNA methylation patterns of a large series of FLCs (n = 15) with those in common HCC arising in non-cirrhotic liver. Interesting differences between these two entities were revealed.
FLCs frequently harbor gene-specific hypermethylation in the absence of global hypomethylation. For several loci the frequency of aberrant hypermethylation is indistinguishable from conventional hepatocellular carcinoma arising in the non-cirrhotic liver, whereas for the APC and the CDH1 gene statistically significant differences in hypermethylation could be found.
These data demonstrate that global reduction in DNA methylation and gene-specific hypermethylation appear to represent independent events during tumor evolution of FLCs. These observations are in accordance with studies focusing on common hepatocellular carcinoma [22] and also other epithelial tumors (e.g. prostate carcinoma [23] or urothelial carcinoma [24]), which also show that global and gene-specific hypermethylation are independent events. The data also demonstrate that full-blown malignancy (i.e., carcinoma) can develop in the absence of global hypomethylation.
Our results are also in line with the data from Kim et al. [25], who showed that in common HCC global hypomethylation can take place independent from cirrhosis.
The less distinct separation of FLC from adjacent tissue and HCC from adjacent tissue, respectively, supports the concept of “field cancerization” [26], which involves epigenetic field defects also in the non-neoplastic tissue adjacent to full-blown malignancy.
One FLC case was HBV positive (no. 13) and one common HCC case was HCV positive (no. 10). But in both cases careful reanalysis of all methylation data did not reveal any peculiarity in comparison to the hepatitis-negative cases (see also Figure S1).
The results presented in this study are partly at variance with another study of DNA methylation in a series of 5 FLCs describing low levels of methylation in all FLCs without any difference in comparison to adjacent non-neoplastic liver tissues [27]. We found unequivocal and frequent hypermethylation of several loci (e.g., hsa-mir-9-1, hsa-mir-9-2, cyclinD2, RASSF1A) as well as total absence of hypermethylation of the CDH1 (E-cadherin) gene. By contrast, the CDH1 gene is reported by Vivekanandan and Torbenson to be methylated in 4/4 normal liver samples and 5/5 FLC and 4/3 FLC metastases. In part, these discrepancies can be explained by selection of genes. microRNA genes and the APC gene were not included by Vivekanandan and Torbenson. However, the most likely reason is difference in detection methodology: Vivekanandan and Torbenson used exclusively conventional qualitative methylation-specific PCR (MSP). From Figure 1 in their publication it can be deduced that they scored any sample showing a PCR product using M-primers as “methylated”, regardless of band intensity and ratio between M- and U-band.
This explains for example the occurrence of 100% methylation of the CDH1 gene in normal liver samples and the inability to detect differences in methylation levels between tumor specimens and adjacent non-neoplastic liver tissue. Employing the very same MSP primers for the analysis of all FLC specimens a weak “M-band” of variable intensity relative to the corresponding “U-band” was observed for 7 tumor and 6 adjacent tissue samples (see Figure S3).
Several groups (including our own) have demonstrated the importance of employing quantitative methods in studying aberrations in DNA methylation [6], [28], [29]. A mere qualitative analysis is not able to identify highly significant differences between tumor and surrounding tissue as well as between different tumor types.
Conclusions
The results presented demonstrate the presence of frequent aberrant DNA methylation in fibrolamellar carcinoma. However, these differences were only discernible employing quantitative methodology. In comparison to common HCC arising in non-cirrhotic liver clear differences exist, separating these morphologically and clinically distinct entities also on the epigenetic level. The aberrant hypermethylation in FLC specimens takes place in the absence of a global loss of methylation, indicating that these two epigenetic aberrations are well separated phenomena and that full-blown malignancy can develop in the absence of global hypomethylation.
Materials and Methods
Patient material
Cases of FLC (n = 15) and HCC of non-cirrhotic liver (n = 10) from the period from 1988 to 2007 were retrieved from the archive of the Institute of Pathology, Medizinische Hochschule Hannover, Germany and analyzed anonymously. The local Ethics committee (“Ethik-Kommission der Medizinischen Hochschule Hannover”, head: Prof. Dr. Tröger) exempted this study from review because all specimens under study were retrieved anonymously and retrospectively (left-over samples from diagnostic procedures) and waived the need for consent due to the fact the samples received were anonymous. Age, sex, and TNM stage of the tumors were extracted from the histological reports (see Table 1 ). Clinical follow up data were not available. Cases were independently reviewed by two diagnostic histopathologists (PF, FP). Areas of tumor and non-tumor tissue were marked and DNA was isolated from unstained serial sections using these marked slides as guidance for manual microdissection.
DNA extraction and bisulfite treatment
Genomic DNA was isolated from formalin-fixed paraffin-embedded specimens using proteinase K-digest over night (50 mM Tris pH 8,1; 1 mM EDTA; 0,5% Tween 20; 10 µg/ml proteinase K), followed by exhaustive organic extraction and ethanol precipitation. Subsequently, DNA samples were treated with sodium bisulfite using the EZ DNA Methylation Kit™ (Zymo Research, HiSS Diagnostics, Freiburg, Germany) following the manufacturer's instructions and finally eluted in 40 µL elution buffer.
Generation of the PCR-products for methylation analysis
PCR products were generated in a 25 µL reaction volume with 400 nmol/L of forward, 40 nmol/L reverse and 400 nmol/L universal biotinylated primers, 200 µmol/L of each dNTP, 1.5 mmol/L or 2.5 mmol/L MgCl2 (see Table 2 for all primer sequences and reaction conditions), 1× Platinum-Taq reaction buffer and 1.25 units PlatinumTaq™ (Invitrogen, Karlsruhe, Germany). PCR conditions were 95°C for 5 minutes, followed by 45 cycles with denaturation at 95°C for 30 seconds, annealing at 55°C or 60°C (see Table 1) for 45 seconds, and elongation at 72°C for 30 seconds finished with 1 cycle final elongation at 72°C for 5 minutes. The reverse primer is tagged by a sequence recognized by the universal primer. Therefore, a single (expansive) biotinylated primer can be used for all different gene-specific assays [30].
Table 2. Primer sequences.
| Gene | Forward primer | MgCl2 [mM] | TAnn [°C] | Size (bp) |
| mir-9-1 | f: GGG AAA TGG GGTATT AGA AAT TTTr: [GGG ACA CCG CTG ATC GTT TA] CAA CAA CAA AAA CCT CAA ACA CPyro: TTT TTG GGT TTG GAT | 1,5 | 60 | 140 |
| mir-9-2 | f: GGA AGA GAT GTT GAT TGA GAA AAr: [GGG ACA CCG CTG ATC GTT TA] TAA TCA ACC AAC TAC CCC ACPyro: GGG ATT GTT GTA ATG TTG | 1,5 | 60 | 114 |
| APC | f: GGA GAG AGA AGT AGT TGT GTA ATT Tr: [GGG ACA CCG CTG ATC GTT TA]A CTA CAC CAA TAC AAC CAC ATA TCPyro: TTA GGG TGT TTT TTA TTT T | 2,5 | 55 | 123 |
| CDH1 | f: AGA TTT TAG TAA TTT TAG GTT AGA GGr: [GGG ACA CCG CTG ATC GTT TA]C TAA TTA ACT AAA AAT TCA CCT ACCPyro a: ATT TTA GGT TAG AGG GTT ATPyro b: TTT GGG GAG GGG TT | 1,5 | 55 | 134 |
| Cyclin-D2 | f: GTA TTT TTT GTA AAG ATA GTT TTG ATTr: [GGG ACA CCG CTG ATC GTT TA] CCA AAC TTT CTC CCT AAA AACPyro: ATA GTT TTG ATT TAA GGA TG | 1,5 | 55 | 117 |
| ESR1 | f: GGY GAG GTG TAT TTG GAT AGT AGr: [GGG ACA CCG CTG ATC GTT TA]C TAT TAA ATA AAA AAA AAC CCC CPyro: GTA TTT GGA TAG TAG TAA GTT | 2,5 | 55 | 208 |
| GSTπ1 | f: GGG GAG GGA TTA TTT TTA TAA Gr: [GGG ACA CCG CTG ATC GTT TA]A ATT AAC CCC ATA CTA AAA ACT CTPyro: GGA TTA TTT TTA TAA GGT | 2,5 | 55 | 173 |
| LINE-1 | n/a (Qiagen) | 1,5 | 50 | n/a |
| MINT31 | f: GTT TAG GGG TGA TGG TTT TAGr: [GGG ACA CCG CTG ATC GTT TA]A AAC ACT TCC CCA ACA TC TACPyro: GTG GTG ATG GAG GTT AT | 1,5 | 55 | 188 |
| RASSF1A | f: AGT TTG GAT TTT GGG GGA GGr: 5′-Biotin-CAA CTC AAT AAA CTC AAA CTC CCCPyro: GGG TTY GTT TTG TGG TTT | 1,5 | 60 | 136 |
| SFRP1 | f: TTG GGG ATT GYG TTT TTT GTTr: [GGG ACA CCG CTG ATC GTT TA] ACT CTA CRC CCT ATT CTC CPyro: GAG GTT TTT GGA AGT TTG | 1,5 | 55 | 108 |
| SOCS1-a | f: GTG AAG ATG GTT TYG GGA TTTr: [GGG ACA CCG CTG ATC GTT TA]C AAC RAA ACC CCC AAC ATA CPyro: TTY GAG TTG TTG GAG TAT TA | 1,5 | 55 | 150 |
| SOCS1-b | f: GTT TTT AGY GTG AAG ATG GTT Tr: [GGG ACA CCG CTG ATC GTT TA] CTA ACR AAA CAA CTC CTA CAA CPyro: GTT TTT ATT TGG ATG GTA G | 1,5 | 55 | 221 |
| univ-bio | 5′-Biotin-[GGG ACA CCG CTG ATC GTT TA] |
Y = Pyrimidine (C/T).
R = Purine (A/G).
f = forward primer.
r = reverse primer.
Pyro = pyrosequencing primer.
Sequences in square brackets resemble universal tag for biotinylated primer.
Methylation analysis using Pyrosequencing
PCR products (5–20 µL) were added to a mix consisting of 3 µL Streptavidin Sepharose HP™ (Amersham Biosciences, Freiburg, Germany) and 37 µL binding buffer (Qiagen, Hilden, Germany) and mixed at 1200 rpm for 5 minutes at room temperature.
Using the Vacuum Prep Tool™ (Qiagen, Hilden, Germany), single-stranded PCR products were prepared following the manufacturer's instructions. The sepharose beads with the single stranded templates attached were released into a PSQ 96 Plate Low™ (Qiagen, Hilden, Germany) containing a mix of 12 µL annealing buffer (Qiagen, Hilden, Germany) and 500 nmmol/L of the corresponding sequencing primer (see Table 2 ). Pyrosequencing™ reactions were performed in a PyroMark MD System (Qiagen, Hilden, Germany) according to the manufacturer's instructions using the PyroGold SQA™ Reagent Kit (Qiagen, Hilden, Germany). CpG site quantification was performed using the new methylation Software Pyro Q-CpG™.
Criteria for Pyrogram™ selection were as follows: sufficient peak height of >25 units for a single nucleotide (arbitrary units for light emission calculated by the software), sharp symmetric peaks without any irregularities or side-peaks, and a wide reading length with a high reliability until the end of the sequence. Furthermore, the absence of any significant signals at the positions where a bisulfite treatment control was included or where control nucleotides were dispensed to check for unspecific background signals.
Methylation analysis using conventional Methylation-specific PCR
For conventional qualitative methylation specific PCR the primer pairs described by Vivekanandan and Torbenson were used [27]. 20–50 ng bisulfite treated DNA were amplified using 0.5 units of Taq polymerase (Platinium Tag, Invitrogen, Karlsruhe Germany) in the presence of 200 µM dNTPs, 1.5 mM MgCl2 and 10 pmol of each primer in the reaction buffer provided by the manufacturer in a final volume of 25 µl. After an initial denaturation of 2 min at 95°C, 40 cycles consisting of 30 sec at 95°C, 30 sec at 65°C and 40 sec at 72°C followed.
The PCR products were resolved on a 6% PAA gel and visualized employing ethodium bromide staining.
Statistical analysis
Statistical differences were calculated using the Mann-Whitney-U test. All calculations were performed using the software package GraphPad Prism (version 5.01 for Windows, La Jolla, CA, USA). p<0.05 were considered statistically significant.
For cluster analyses the quantitative methylation data from all samples were log-transformed and uploaded into the statistical package BRB array tool (version 3.5.0-Patch_2) [31]. Hierarchical clustering was then performed applying Euclidean distance as a dissimilarity metric and the complete linkage clustering method.
Supporting Information
Comparison of the two different definitions of “hypermethylated” (see “Results”) if applied to all methylation measurements performed in this study.
(0.26 MB TIF)
Separate clustering of methylation data for FLC and adjacent non-neoplastic tissue (A) and common HCC from non-cirrhotic liver and adjacent non-neoplastic tissue (B).
(1.37 MB TIF)
MSP results for three tumor/adjacent tissue sample pairs using the primers described by Vivekanandan and Torbenson. FLC7 shows a weak “M-band” in the adjacent tissue, FLC8 in the tumor specimen and FLC4 in both fractions. Altogether 7 tumor and 6 adjacent tissue specimens displayed an “M-band” of variable intensity relative to the corresponding “U-band”.
(0.07 MB TIF)
Acknowledgments
The authors would like to thank Ms Britta Hasemeier for indispensable help in preparing the figures.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by SFB/TRR77-“Liver cancer” (Project B1) of the Deutsche Forschungsgemeinschaft (DFG). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol. 2005;2(Suppl 1):S4–11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 2.Chan AO, Rashid A. CpG island methylation in precursors of gastrointestinal malignancies. Curr Mol Med. 2006;6:401–408. doi: 10.2174/156652406777435417. [DOI] [PubMed] [Google Scholar]
- 3.Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135:1079–1099. doi: 10.1053/j.gastro.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 5.Huang J. Current progress in epigenetic research for hepatocarcinomagenesis. Sci China C Life Sci. 2009;52:31–42. doi: 10.1007/s11427-009-0014-7. [DOI] [PubMed] [Google Scholar]
- 6.Lehmann U, Berg-Ribbe I, Wingen LU, Brakensiek K, Becker T. Distinct methylation patterns of benign and malignant liver tumors revealed by quantitative methylation profiling. Clin Cancer Res. 2005;11:3654–3660. doi: 10.1158/1078-0432.CCR-04-2462. [DOI] [PubMed] [Google Scholar]
- 7.Liu S, Chan KW, Wang B, Qiao L. Fibrolamellar hepatocellular carcinoma. Am J Gastroenterol. 2009;104:2617–2624. doi: 10.1038/ajg.2009.440. [DOI] [PubMed] [Google Scholar]
- 8.Torbenson M. Review of the clinicopathologic features of fibrolamellar carcinoma. Adv Anat Pathol. 2007;14:217–223. doi: 10.1097/PAP.0b013e3180504913. [DOI] [PubMed] [Google Scholar]
- 9.Wilkens L, Bredt M, Flemming P, Kubicka S, Klempnauer J, et al. Cytogenetic aberrations in primary and recurrent fibrolamellar hepatocellular carcinoma detected by comparative genomic hybridization. Am J Clin Pathol. 2000;114:867–874. doi: 10.1309/BMTT-JBPD-D13H-1UVD. [DOI] [PubMed] [Google Scholar]
- 10.Kakar S, Chen X, Ho C, Burgart LJ, Sahai V, et al. Chromosomal changes in fibrolamellar hepatocellular carcinoma detected by array comparative genomic hybridization. Mod Pathol. 2009;22:134–141. doi: 10.1038/modpathol.2008.178. [DOI] [PubMed] [Google Scholar]
- 11.Ehrlich M. Cancer-linked DNA hypomethylation and its relationship to hypermethylation. Curr Top Microbiol Immunol. 2006;310:251–274. doi: 10.1007/3-540-31181-5_12. [DOI] [PubMed] [Google Scholar]
- 12.Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, et al. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schagdarsurengin U, Wilkens L, Steinemann D, Flemming P, Kreipe HH, et al. Frequent epigenetic inactivation of the RASSF1A gene in hepatocellular carcinoma. Oncogene. 2003;22:1866–1871. doi: 10.1038/sj.onc.1206338. [DOI] [PubMed] [Google Scholar]
- 14.Pfeifer GP, Yoon JH, Liu L, Tommasi S, Wilczynski SP, et al. Methylation of the RASSF1A gene in human cancers. Biol Chem. 2002;383:907–914. doi: 10.1515/BC.2002.097. [DOI] [PubMed] [Google Scholar]
- 15.Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- 16.Lehmann U, Hasemeier B, Christgen M, Muller M, Romermann D, et al. Epigenetic inactivation of microRNA gene hsa-mir-9-1 in human breast cancer. J Pathol. 2008;214:17–24. doi: 10.1002/path.2251. [DOI] [PubMed] [Google Scholar]
- 17.Agirre X, Vilas-Zornoza A, Jimenez-Velasco A, Martin-Subero JI, Cordeu L, et al. Epigenetic silencing of the tumor suppressor microRNA Hsa-miR-124a regulates CDK6 expression and confers a poor prognosis in acute lymphoblastic leukemia. Cancer Res. 2009;69:4443–4453. doi: 10.1158/0008-5472.CAN-08-4025. [DOI] [PubMed] [Google Scholar]
- 18.Bueno MJ, Perez de Castro I, Gomez de Cedron M, Santos J, Calin GA, et al. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell. 2008;13:496–506. doi: 10.1016/j.ccr.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 19.Datta J, Kutay H, Nasser MW, Nuovo GJ, Wang B, et al. Methylation mediated silencing of MicroRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res. 68:5049–5058. doi: 10.1158/0008-5472.CAN-07-6655. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Furuta M, Kozaki KI, Tanaka S, Arii S, Imoto I, et al. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2010;31:766–776. doi: 10.1093/carcin/bgp250. [DOI] [PubMed] [Google Scholar]
- 21.De Smet C, Loriot A. DNA hypomethylation in cancer: Epigenetic scars of a neoplastic journey. Epigenetics. 2010;5(3) doi: 10.4161/epi.5.3.11447. [DOI] [PubMed] [Google Scholar]
- 22.Lee HS, Kim BH, Cho NY, Yoo EJ, Choi M, et al. Prognostic implications of and relationship between CpG island hypermethylation and repetitive DNA hypomethylation in hepatocellular carcinoma. Clin Cancer Res. 2009;15:812–820. doi: 10.1158/1078-0432.CCR-08-0266. [DOI] [PubMed] [Google Scholar]
- 23.Cho NY, Kim JH, Moon KC, Kang GH. Genomic hypomethylation and CpG island hypermethylation in prostatic intraepithelial neoplasm. Virchows Arch. 2009;454:17–23. doi: 10.1007/s00428-008-0706-6. [DOI] [PubMed] [Google Scholar]
- 24.Park SY, Yoo EJ, Cho NY, Kim N, Kang GH. Comparison of CpG island hypermethylation and repetitive DNA hypomethylation in premalignant stages of gastric cancer, stratified for Helicobacter pylori infection. J Pathol. 2009;219:410–416. doi: 10.1002/path.2596. [DOI] [PubMed] [Google Scholar]
- 25.Kim MJ, White-Cross JA, Shen L, Issa JP, Rashid A. Hypomethylation of long interspersed nuclear element-1 in hepatocellular carcinomas. Mod Pathol. 2009;22:442–449. doi: 10.1038/modpathol.2008.203. [DOI] [PubMed] [Google Scholar]
- 26.Dakubo GD, Jakupciak JP, Birch-Machin MA, Parr RL. Clinical implications and utility of field cancerization. Cancer Cell Int. 2007;7:2. doi: 10.1186/1475-2867-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vivekanandan P, Torbenson M. Epigenetic instability is rare in fibrolamellar carcinomas but common in viral-associated hepatocellular carcinomas. Mod Pathol. 2008;21:670–675. doi: 10.1038/modpathol.2008.32. [DOI] [PubMed] [Google Scholar]
- 28.Brakensiek K, Langer F, Kreipe H, Lehmann U. Low level of DAP-kinase DNA methylation in myelodysplastic syndrome. Blood. 2004;104:1586–1587. doi: 10.1182/blood-2004-03-0898. [DOI] [PubMed] [Google Scholar]
- 29.Lee ES, Issa JP, Roberts DB, Williams MD, Weber RS, et al. Quantitative promoter hypermethylation analysis of cancer-related genes in salivary gland carcinomas: comparison with methylation-specific PCR technique and clinical significance. Clin Cancer Res. 2008;14:2664–2672. doi: 10.1158/1078-0432.CCR-07-1232. [DOI] [PubMed] [Google Scholar]
- 30.Colella S, Shen L, Baggerly KA, Issa JP, Krahe R. Sensitive and quantitative universal Pyrosequencing methylation analysis of CpG sites. Biotechniques. 2003;35:146–150. doi: 10.2144/03351md01. [DOI] [PubMed] [Google Scholar]
- 31.Simon R, Lam A, Li MC, Ngan M, Menenzes S, et al. Analysis of Gene Expression Data Using BRB-Array Tools. Cancer Inform. 2007;3:11–17. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of the two different definitions of “hypermethylated” (see “Results”) if applied to all methylation measurements performed in this study.
(0.26 MB TIF)
Separate clustering of methylation data for FLC and adjacent non-neoplastic tissue (A) and common HCC from non-cirrhotic liver and adjacent non-neoplastic tissue (B).
(1.37 MB TIF)
MSP results for three tumor/adjacent tissue sample pairs using the primers described by Vivekanandan and Torbenson. FLC7 shows a weak “M-band” in the adjacent tissue, FLC8 in the tumor specimen and FLC4 in both fractions. Altogether 7 tumor and 6 adjacent tissue specimens displayed an “M-band” of variable intensity relative to the corresponding “U-band”.
(0.07 MB TIF)