Abstract
Background
Centromere identity is determined epigenetically by deposition of CenH3, a centromere-specific histone H3 variant that dictates kinetochore assembly. The molecular basis of the contribution of CenH3 to centromere/kinetochore functions is, however, incompletely understood, as its interactions with the rest of centromere/kinetochore components remain largely uncharacterised at the molecular/structural level.
Principal Findings
Here, we report on the contribution of Drosophila CenH3CID to recruitment of BubR1, a conserved kinetochore protein that is a core component of the spindle attachment checkpoint (SAC). This interaction is mediated by the N-terminal domain of CenH3CID (NCenH3CID), as tethering NCenH3CID to an ectopic reporter construct results in BubR1 recruitment and BubR1-dependent silencing of the reporter gene. Here, we also show that this interaction depends on a short arginine (R)-rich motif and that, most remarkably, it appears to be evolutionarily conserved, as tethering constructs carrying the highly divergent NCenH3 of budding yeast and human also induce silencing of the reporter. Interestingly, though NCenH3 shows an exceedingly low degree of conservation, the presence of R-rich motives is a common feature of NCenH3 from distant species. Finally, our results also indicate that two other conserved sequence motives within NCenH3CID might also be involved in interactions with kinetochore components.
Conclusions
These results unveil an unexpected contribution of the hypervariable N-domain of CenH3 to recruitment of kinetochore components, identifying simple R-rich motives within it as evolutionary conserved structural determinants involved in BubR1 recruitment.
Introduction
Centromere function ensures accurate chromosome segregation during mitosis and meiosis, as the centromere dictates assembly of the kinetochore that, in turn, regulates the spindle attachment checkpoint (SAC), which delays anaphase onset until all chromosomes are correctly attached in a bipolar fashion to the mitotic spindle. Centromere identity is regulated epigenetically by deposition of the centromere-specific histone H3 variant CenH3 that, being exclusively found at centromeres, constitutes the structural and functional foundation for kinetochore assembly and function [1], [2], [3], [4], [5], [6], [7], [8]. CenH3 is essential for viability, being required for centromeric localisation of all centromere/kinetochore proteins analysed to date.
Little is known, however, about the actual molecular/structural basis of the contribution(s) of CenH3 to kinetochore assembly and function, as its interactions with the rest of centromere/kinetochore proteins is just beginning to be understood. In this context, it was recently reported that CENP-N and CENP-C, which are components of the constitutive centromere associated network (CCAN) in vertebrates [9], [10], directly interact with human CenH3CENP-A-containing nucleosomes in vitro [11], [12]. These interactions involve the C-terminal and centromere targeting (CATD) domains of CenH3CENP-A, respectively. Here, we report on the contribution of the N-terminal domain of Drosophila CenH3CID (NCenH3CID) to recruitment of BubR1, an evolutionarily conserved kinetochore protein that is a core component of the spindle attachment checkpoint (SAC) [13], [14], [15]. This interaction is mediated by a simple arginine (R)-rich motif within the hypervariable NCenH3CID domain. Our results also suggest that this interaction is likely conserved in the highly divergent NCenH3 of budding yeast and humans. Most remarkably, though NCenH3 is poorly conserved through evolution [16], [17], the presence of R-rich motives is a common feature of NCenH3 from distant species, including budding yeast and humans [2]. In Drosophila, NCenH3CID contains two other conserved motives that might also mediate interactions with kinetochore components. Altogether, these results indicate that conserved sequence motives within the hypervariable NCenH3 domain mediate centromere/kinetochore interactions.
Results and Discussion
Targeting NCenH3CID to an ectopic white-reporter construct silences reporter expression
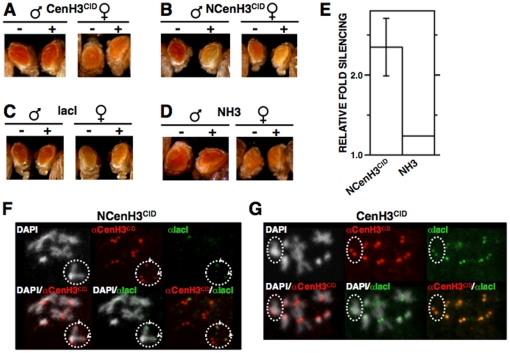
To analyse the contribution of CenH3CID to the regulation of centromere/kinetochore interactions, we performed ectopic targeting experiments using Drosophila transgenic lines carrying a white-reporter transgene that contains multiple binding sites for the bacterial lacI repressor at the regulatory region, about 500 bp upstream from the reporter gene. In these experiments, lines S9.2 and 157.1 were used, which contain 46 and 256 lacI-repeats inserted on the third- and X-chromosome, respectively [18]. We anticipated that, if resulting in recruitment of kinetochore proteins, tethering of fused CenH3CID-lacI proteins would interfere with expression of the white-reporter gene, which is easily monitored by analysing changes in eye pigmentation. As shown in Figure 1A, in line S9.2, expression of a CenH3CID-lacI fusion does not significantly affect white expression. In contrast, expression of an NCenH3CID-lacI fused protein, which carries only the N-terminal domain of CenH3CID, significantly silences reporter expression (Figure 1B and 1E). This effect is specific of NCenH3CID, as no silencing is observed in flies expressing the lacI-DNA-binding domain alone (Figure 1C) [18], [19], or, more important, an NH3-lacI construct (Figure 1D and 1E), carrying the N-terminal domain of canonical histone H3. Similar results were obtained when expression was carried out in line 157.1 (Figure S1A, D and E), though, in this case, the observed effects are weaker since expression of the white-reporter is low in control flies expressing no fused proteins [18], [19]. Immunolocalisation experiments showed that CenH3CID-lacI fails to localise to the reporter construct but, on the contrary, incorporates to all centromeres (Figures 1G and S2A), indicating that, in CenH3CID-lacI, the histone-fold domain (HFD) of CenH3CID predominates over the lacI-DNA-binding domain, so that CenH3CID-lacI incorporates into nucleosomes, like endogenous CenH3CID does, being specifically deposited at centromeres. On the other hand, NCenH3CID-lacI exclusively localises to the ectopic reporter sites (Figure 1F and S2B). From these results, we conclude that lack of silencing observed in the case of CenH3CID-lacI is actually the consequence of its failure to localise to the ectopic reporter sites.
Figure 1. Tethering NCenH3CID to a white-reporter silences reporter expression.
(A–D) The eye phenotype of S9.2 flies expressing the indicated lacI-fused proteins (+) is compared to that of siblings where no fused protein is expressed (−). Results are presented for both female and male individuals. (E) Quantitative analysis is presented for lines expressing NCenH3CID-lacI and NH3CID-lacI constructs. Relative fold silencing is expressed as the ratio between OD480 of control S9.2 lines expressing no fused protein and that of lines expressing the indicated constructs. For NCenH3CID-lacI, results correspond to the average of three independent lines. For NH3CID-lacI, results are presented for a single representative line. (F and G) NCenH3CID-lacI, but not CenH3CID-lacI, bind to the ectopic white-reporter construct. Fused proteins were expressed in 157.1 flies, where the white-reporter is inserted at a distal position on the X-chromosome, and localisation was determined in mitotic chromosomes by immunostaining with αlacI (green) and αCenH3CID (red), which also detects endogenous CenH3CID at centromeres. Dotted circles indicate X-chromosomes. Arrows indicate co-localisation of αCenH3CID and αlacI signals at ectopic sites on the X-chromosome, reflecting binding of NCenH3CID-lacI to the reporter. DNA was stained with DAPI. See Figure S4 for a description of the constructs.
Tethering NCenH3CID-lacI results in ectopic BubR1 recruitment
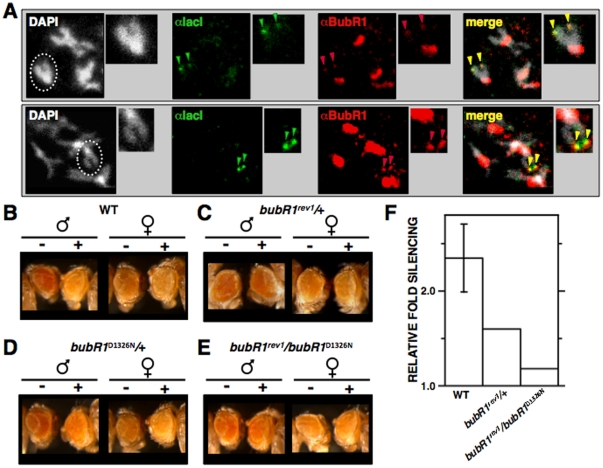
Next, we addressed whether ectopic targeting of NCenH3CID-lacI actually results in recruitment of kinetochore proteins. For this purpose, immunolocalisation experiments using antibodies against several Drosophila kinetochore proteins were performed (Figures 2 and 3). In mitotic chromosomes, tethering NCenH3CID-lacI to the reporter construct results in recruitment of BubR1, as distinct ectopic αBubR1 signals are detected on the X-chromosome in approximately 15% of chromosomes (N = 60; p<0.001) (Figure 2A). These αBubR1 signals overlap with αlacI signals, which reflect binding of NCenH3CID-lacI to the lacI-repeats and, therefore, mark the position corresponding to the reporter. Ectopic αBubR1 signals are weak compared to those observed at the kinetochore, indicating that ectopic BubR1 recruitment is less efficient than at the kinetochore, which might simply reflect the limited number of NCenH3CID-lacI molecules that can be targeted to the reporter, a maximum of 256 copies. It is also possible that additional factors are involved in stabilising BubR1 at the kinetochore. Actually, as discussed below (see “General considerations and implications”), recruitment and maintenance of BubR1 at kinetochores might involve different mechanisms. In good agreement with these results, silencing of the reporter depends on BubR1, as it is suppressed by bubR1 mutations (Figure 2B–F). In these experiments, bubR1rev1 and bubR1D1326N mutants were used, which correspond to a deletion and a point-mutation at the catalytic kinase-domain, respectively [20], [21]. In heterozygous bubR1rev1/+ flies, silencing induced by NCenH3CID-lacI is strongly suppressed (Figure 2C and 2F), when compared to control wild-type flies (Figure 2B and 2F). This dominant suppressor effect is observed in approximately 50% of the off-spring (N = 75), the rest showing only slight or no suppression. bubR1rev1 mutation is homozygous lethal, so that silencing induced by NCenH3CID-lacI could not be analysed in homozygous bubR1rev1/bubR1rev1 flies. However, suppression is enhanced in trans-heterozygous bubR1rev1/bubR1D1326N, where the complete off-spring (N = 55) shows strong suppression (Figure 2E and 2F). On the other hand, heterozygous bubR1D1326N/+ flies show only slight suppression (Figure 2D).
Figure 2. NCenH3CID mediates BubR1 recruitment.
(A) Localisation of NCenH3CID-lacI and BubR1 was determined in mitotic chromosomes from 157.1 flies by immunostaining with αlacI (green) and αBubR1 (red). Dotted circles indicate X-chromosomes. Arrows indicate co-localisation of αlacI and αBubR1 signals at ectopic sites on the X-chromosome, reflecting recruitment of BubR1 by NCenH3CID-lacI. Enlarged images are shown on the right of each panel for easier visualisation. DNA was stained with DAPI. Two independent examples are presented. On the bottom, pictures were recorded to a higher intensity to better visualise the ectopic αBubR1 signals observed on the X-chromosome. (B–E) Silencing induced by NCenH3CID-lacI depends on BubR1. The eye phenotype of S9.2 flies expressing NCenH3CID-lacI (+) or not (−) is presented in the indicated genetic backgrounds. Results are presented for both female and male individuals. (F) Quantitative analysis is presented for flies expressing NCenH3CID-lacI in the indicated genetic backgrounds. Relative fold silencing is expressed as the ratio between OD480 of control S9.2 flies expressing no fused protein and that of flies expressing NCenH3CID-lacI in the indicated genetic backgrounds. For wild-type, results correspond to the average of three independent lines. For mutants, results are presented for a single representative line.
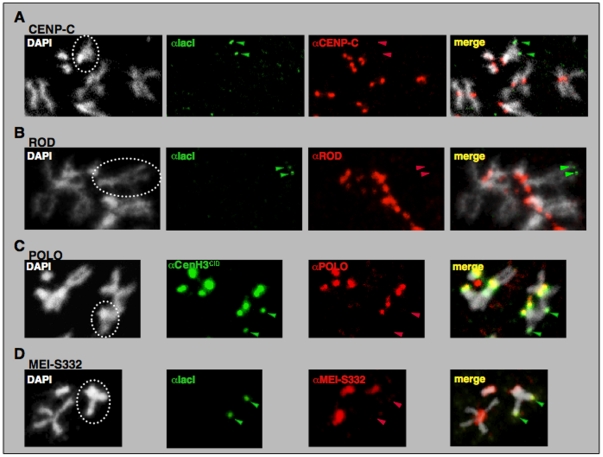
Figure 3. Targeting of NCenH3CID-lacI to the reporter construct in 157.1 flies does not results in ectopic recruitment of CENP-C.
(A), ROD (B), POLO (C) or MEI-S332 (D). Localisation of NCenH3CID-lacI (green) and CENP-C, ROD, POLO or MEI-S332 (red) was determined in mitotic chromosomes by immunostaining with the indicated specific antibodies. Dotted circles indicate X-chromosomes. Arrows indicate ectopic αlacI or αCenH3CID signals on the X-chromosome, which reflect binding of NCenH3CID-lacI to the reporter construct.
Results described above indicate that, in Drosophila, NCenH3CID is involved in recruitment of BubR1, an evolutionarily conserved kinetochore protein, which is a core component of the spindle attachment checkpoint (SAC). Current models for SAC function suggest that unattached kinetochores recruit SAC components, such as BubR1, to generate a diffusible signal that delays anaphase onset. Therefore, ectopic recruitment of BubR1 could reflect formation of a functional ectopic kinetochore. This possibility, however, is highly unlikely since targeting NCenH3CID does not result in recruitment of other essential kinetochore components, including CENP-C [22], ROD [23], [24], POLO [25], and MEI-S332/Sgo [26], [27] (Figure 3). Also in agreement with this hypothesis, ectopic targeting of NCenH3CID does not induce any detectable proliferation defects (not shown), which is contrary to what would be expected if resulting in efficient formation of an ectopic kinetochore [28]. Altogether, these results argue against formation of a functional kinetochore, indicating that NCenH3CID is not sufficient by itself to support kinetochore assembly. Actually, it was recently reported that, in vertebrates, both the C-terminal and central CATD domains of CenH3CENP-A mediate interactions with two essential CCAN-components, CENP-C and CENP-N [11], [12], being, therefore, required for full kinetochore assembly.
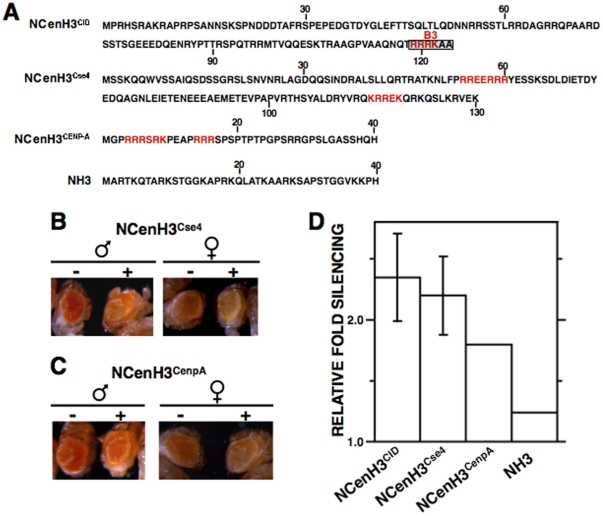
Silencing induced by CenH3CID depends on a simple arginine-rich sequence motif
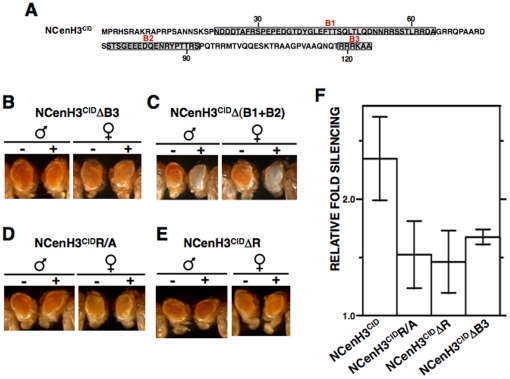
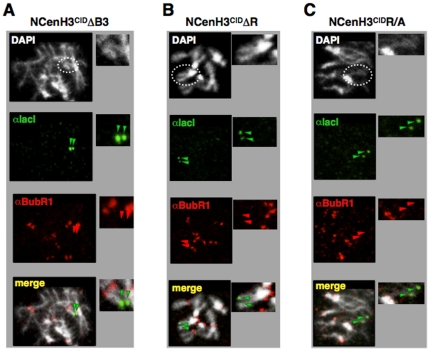
Next, we asked about the molecular basis of the contribution of NCenH3CID to BubR1 recruitment. Within the Drosophila genus, NCenH3CID shows significant variability. However, sequence comparison of NCenH3CID from a broad group of Drosophila species [17], allowed identification of three sequence motives (B1, B2 and B3) (Figure 4A) that, being evolutionarily conserved over 25 million years, were good candidates to mediate recruitment of BubR1. To test this possibility, we performed deletion analyses, where the contribution of each conserved motif to silencing of the white-reporter was determined in ectopic targeting experiments. As shown in Figure 4, motif B3 has a major contribution, as its deletion strongly impairs silencing (Figure 4B and 4F) and is capable by itself to induce robust silencing of the reporter (Figure 4C). Moreover, deletion of motives B1 or B2 has no detectable effect on reporter silencing (Figure S3B and S3C). Motif B3 corresponds to a rather simple sequence, 119RRRKAA124, showing a peculiar enrichment in arginine (R) residues. As a matter of fact, silencing induced by NCenH3CID is strongly impaired when R-residues within B3 are replaced by alanine (A) (Figure 4D and 4F) or deleted (Figure 4E and 4F), and, concomitant to the lack of silencing, no ectopic αBubR1 signals are detected in these cases (Figure 5). Altogether, these results identify R-residues within B3 as involved in BubR1 recruitment.
Figure 4. Silencing induced by NCenH3CID depends on a conserved simple arginine (R)-rich motif.
(A) Amino acid sequence of the N-terminal domain of D. melanogaster CenH3CID. Sequence motives (B1, B2 and B3) that are conserved amongst distant Drosophila species are indicated. (B–E) The eye phenotype of S9.2 flies expressing the indicated NCenH3CID-lacI deletions (+) is compared to that of siblings where no fused protein is expressed (−). Results are presented for both female and male individuals. (F) Quantitative analysis is presented for lines expressing the indicated constructs. Relative fold silencing is expressed as the ratio between OD480 of control S9.2 lines expressing no fused protein and that of lines expressing each construct. Results correspond to the average of four (NCenH3CIDR/A), three (NCenH3CID), and two (NCenH3CIDΔR and NCenH3CIDΔB3) independent lines. See Figure S4 for a description of the constructs.
Figure 5. R-residues within B3 motif mediate recruitment of BubR1.
Co-localisation of BubR1 with NCenH3CIDΔB3-lacI (A), NCenH3CIDΔR-lacI (B) and NCenH3CIDR/A-lacI (C) was determined in mitotic chromosomes from 157.1 flies by immunostaining with αlacI (green) and αBubR1 (red). Dotted circles indicate X-chromosomes. Arrows indicate the position of the reporter construct on the X-chromosome. Enlarged images are shown on the right of each panel for easier visualisation. DNA was stained with DAPI.
Highly divergent NCenH3 of budding yeast and human also silence reporter expression
In comparison to canonical histone H3, CenH3 is much less well conserved [16], [17]. Homology, that at the histone-fold domain ranges from 40% to 60% identity, is, however, insignificant for NCenH3 that, showing strong variability both in size (ranging from 20 to 200 aa) and sequence, cannot be aligned across different eukaryotic lineages. Therefore, in this context, we asked whether the effects described above are restricted to Drosophila CenH3CID. To address this question, lacI-fusions carrying NCenH3 from Saccharomyces cerevisiae (NCenH3Cse4) or humans (NCenH3CenpA) were expressed in Drosophila S9.2 reporter flies and the extent to which they induce silencing of the reporter determined. Both constructs induce silencing (Figure 6B–D), though it is less robust than that observed for NCenH3CID, being significant only in a fraction of lines analysed. In the case of NCenH3Cse4, 75% (N = 4) of lines show strong silencing similar to that observed for NCenH3CID (Figure 6B and 6D). On the other hand, 50% (N = 4) of lines expressing NCenH3CenpA-lacI show significant silencing, which is slightly weaker than that induced by NCenH3CID-lacI (Figure 6C and 6D). Similar results were obtained when expression was performed in 157.1 flies (Figure S1B and C). These results strongly suggest that the contribution of NCenH3 to recruitment of BubR1 is conserved through evolution, from S. cerevisiae to humans, which is in contrast to its low degree of conservation. It must be noted, however, that a common feature of NCenH3 from distant species is its enrichment in R-residues in comparison to canonical histone H3. As a matter of fact, R-rich motives similar to motif B3 that mediates BubR1 recruitment in Drosophila, are present at NCenH3 from most species, including those analysed here, but absent in canonical histone H3 [2] (Figure 6A).
Figure 6. Expression of fused proteins carrying highly divergent NCenH3 of budding yeast (NCenH3Cse4) and human (NCenH3CENP-A) also silence reporter expression.
(A) Amino acid sequence of the N-terminal domain of D. melanogaster CenH3CID, S. cerevisiae CenH3Cse4, human CenH3CENP-A, and canonical histone H3 (NH3). Motif B3 that is responsible for silencing induced by D. melanogaster NCenH3CID is indicated. R-rich motives (basic sequences containing ≥40% R) are highlighted in red. (B and C) The eye phenotype of S9.2 flies expressing the indicated lacI-fused proteins (+) is compared to that of siblings where no fused protein is expressed (−). Results are presented for both female and male individuals. (F) Quantitative analysis is presented for lines expressing the indicated constructs. Relative fold silencing is expressed as the ratio between OD480 of control S9.2 lines expressing no fused protein and that of lines expressing each construct. For NCenH3CID-lacI and NCenH3Cse4-lacI, results correspond to the average of three independent lines. For NCenH3CENP-A-lacI, where only 50% of the lines show significant silencing, and NH3-lacI, results are presented for a single representative line. See Figure S4 for a description of the constructs.
General considerations and implications
Current models for kinetochore assembly and function suggest that presence of CenH3 at the centromere results in a specialised chromatin structure, which provides a physical foundation to build the kinetochore. CenH3-kinetochore interactions remain, however, incompletely understood at the molecular level. Results reported here identify simple R-rich motives within the hypervariable NCenH3CID as evolutionary conserved structural determinants involved in BubR1 recruitment. Whether the contribution of NCenH3CID to BubR1 recruitment is direct or mediated by additional unidentified factor(s) is uncertain, as GST-pull down assays failed to detect any direct physical interaction between BubR1 and NCenH3CID in vitro or upon co-transfection into cultured S2-cells (not shown).
These results, which are based on ectopic targeting experiments, are likely relevant in the context of the endogenous locus. BubR1 recruitment is detected in early prometaphase, when kinetochores are bound by only few or no microtubules [29]. Our results suggest that BubR1 recruitment does not require full kinetochore assembly, as ectopic targeting of NCenH3CID, which does not result in formation of a functional kinetochore, is capable of recruiting BubR1. Several observations support this hypothesis. On one hand, in chicken DT40 cells, strong reduction of CenH3CENP-A to levels that severely impair centromeric localisation of several CCAN-components (CENP-C, -H, and -I), as well as some outer kinetochore proteins (Nuf2/Hec1, Mad2, and CENP-E), shows only a moderate effect on the initial recruitment of BubR1 in early prometaphase [30]. Furthermore, in Drosophila, cid mutants, that fail to assemble the kinetochore, show a BubR1-dependent early mitotic delay [31]. Altogether, these observations suggest a BubR1-CenH3 interaction occurring early in mitosis, prior to full kinetochore assembly. Whether this interaction is mediated by NCenH3 remains, however, to be determined. On the other hand, association of BubR1 to metaphase kinetochores appears to depend strongly on kinetochore assembly, as it is strongly destabilised in CenH3CENP-A-depleted DT40 cells [30], suggesting that initial recruitment and maintenance of BubR1 at kinetochores involve different mechanisms and, perhaps, fulfil different functions. Actually, BubR1 is known to play multiple roles during mitosis [32].
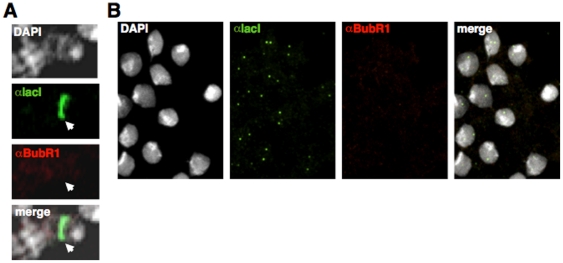
NCenH3CID-BubR1 interaction appears to be regulated during cell-cycle progression, as no ectopic αBubR1 signals are detected on polytene chromosomes, which constitute a special type of interphase chromatin, or in interphase nuclei from larval neuroblasts (Figure 7), indicating that, like at the kinetochore, ectopic recruitment of BubR1 by NCenH3CID is constrained to mitosis. In contrast, our results show that NCenH3CID-mediated silencing of the white-reporter depends on BubR1, indicating that BubR1 is required to repress reporter expression at interphase. BubR1 recruitment at mitosis might stabilise binding of factor(s) required for repression at interphase. It is also possible that BubR1 facilitates chromatin modification, as recent results show that Bub1, a closely related SAC-kinase that plays partially redundant functions, regulates H2AS121-phosphorylation in fission yeast, which lacks BubR1 [33].
Figure 7. Ectopic targeting of NCenH3CID does not induce BubR1 recruitment in interphase.
Localisation of NCenH3CID-lacI and BubR1 was determined in polytene chromosomes (A) and in interphase cells from brain squashes (B) by immunostaining with αlacI (green) and αBubR1 (red). αlacI signals indicate binding of NCenH3CID-lacI to the reporter construct. DNA was stained with DAPI.
In addition to the B3 R-rich motif, Drosophila NCenH3CID contains two other evolutionarily conserved regions (motives B1 and B2) (Figure 4A), which might also mediate centromere/kinetochore interactions. Support for this hypothesis comes from the observation that constructs containing only motif B1 or B2 retain some silencing competence (Figure S3D and S3E). However, in comparison to motif B3, the contribution of motives B1 and B2 to silencing is only minor, as by themselves induce much weaker silencing (Figure S3D and S3E), and their deletion does not significantly affect silencing (Figure S3B and S3C, and Figure 4C). Whether silencing induced by motives B1 and B2 also reflects interaction with kinetochore proteins remains, however, to be determined.
Altogether, these observations favour the hypothesis that some kinetochore proteins bind CenH3-chromatin through the recognition of specific sequence motives within the hypervariable NCenH3 domain, unveiling its essential contribution to CenH3 functionality. Results obtained in S. cerevisiae support this hypothesis, as NCenH3Cse4 is essential for viability and, moreover, interacts genetically with components of COMA, a kinetochore complex that is functionally related to CCAN and mediates protein-protein interactions with other centromere/kinetochore proteins, including the essential CBF3 complex [34], [35].
Materials and Methods
Fly stocks
For targeting experiments, CenH3CID, NCenH3CID, NH3, NCenH3Cse4 and NCenH3CenpA were fused at N-terminus of the lacI-DNA-binding domain (see Figure S4 for a description of the constructs), using plasmid lacIST-Topo-TA and cloned into pNHT4 plasmid [36], where expression is driven by the hsp70-promoter. For deletion-analyses, NCenH3CID-lacIST-Topo-TA was used as template and the various deletions cloned to vector pNHT4 (Figure S4). Transgenic lines were generated in +;+/+;ry506/ry506 flies by standard procedures. All constructs were tested for expression and their ability to target the reporter construct determined in polytene chromosomes (Figure S5). bubR1rev1 and bubR1D1326N mutants are described elsewhere [20], [21]. Lines S9.2 or 157.1 are also described elsewhere [18].
Tethering experiments
For tethering experiments, heterozygous flies carrying the indicated lacI-constructs were crossed to homozygous S9.2 or 157.1 reporter flies. Crosses were, then, subjected to daily heat-shock treatment for 45 min at 37°C and the eye phenotype of flies expressing the corresponding lacI-fused protein compared to that of siblings, of the same sex and age, expressing no fused protein. For each lacI-construct, at least four independent lines were analysed. When silencing induced by NCenH3CID-lacI was analysed in bubR1rev1 and bubR1D1326N mutant backgrounds, heterozygous NCenH3CID-lacI/+ males were crossed to bubR1rev1/+ or bubR1D1326N/+ reporter S9.2 females. When silencing was analysed in trans-heterozygous bubR1rev1/bubR1D1326N flies, bubR1D1326N/+ reporter S9.2 females were crossed to bubR1rev1/+ males carrying NCenH3CID-lacI. For quantitative analyses, eye pigment was extracted with 30% acid-ethanol (pH = 2) according to [37] and OD480 determined in a Nanodrop 1000/3.7. Extraction was performed from 20 heads obtained from male individuals. Relative fold silencing was then expressed as the ratio between OD480 of control S9.2 line expressing no fused protein and that of lines expressing the corresponding constructs.
Immunostaining experiments
For immunostaining experiments, homozygous flies carrying the indicated lacI-constructs were crossed to homozygous 157.1 reporter flies and crosses were subjected to daily heat-shock treatment for 45 min at 37°C to the third-instar larvae stage. After the last heat-shock treatment, larvae were left to recover at 25°C for 2 h prior to dissection. Brains were, then, incubated in 0.5 mg/ml colcemid in PBS for 1.5 h before fixation in 3.7% formaldehyde. Neuroblasts squashes and immunostainings were performed as described elsewhere [19], [38] using rabbit polyclonal αCenH3CID (1∶500), rabbit polyclonal αBubR1 (Rb666) (1∶1000) [39], mouse monoclonal αPolo (mab294) (1∶100) [40], rabbit αCenpC polyclonal antibody (1∶5000) [22], rabbit αRod polyclonal antibody (1∶200) [41], guinea pig αMEI-S332 polyclonal antibody (1∶1000) [27] and mouse monoclonal αlacI (clone 9A5, Upstate) (1∶150). For visualization, slides were mounted in Mowiol (Calbiochem-Novabiochem) containing 0.2 ng/ml DAPI (Sigma) and visualized by confocal microscopy (Leica TCS SP2-AOBS).
Supporting Information
Tethering NCenH3-lacI to the ectopic white-reporter of 157.1 flies induces silencing of the reporter gene. (A–E) The eye phenotype of flies expressing the indicated fused proteins (+) is compared to that of siblings where no fused protein is expressed (−). Results are presented only for male individuals.
(0.46 MB PDF)
In interphase nuclei, CenH3CID-lacI incorporates to centromeres. CenH3CID-lacI (A) and NCenH3CID-lacI (B) were expressed in 157.1 flies carrying an ectopic white reporter construct inserted at a distal position on the X-chromosome. Localisation of the fused proteins was determined in interphase cells from brain squashes of third instar larvae by immunostaining with αCenH3CID (red) and αlacI (green). In cells expressing CenH3CID-lacI (A), all αCenH3CID signals co-localise with αlacI, indicating incorporation of CenH3CID-lacI to centromeres. In contrast, in cells expressing NCenH3CID-lacI (B), co-localisation is restricted to two-spots (indicated by the arrows), reflecting binding of the fused protein to the ectopic reporter construct.
(0.09 MB PDF)
Motives B1 and B2 retain silencing competence. (A) Amino acid sequence of the N-terminal domain of D. melanogaster CenH3CID. Conserved sequence motives (B1, B2 and B3) are indicated. (B–E) The eye phenotype of S9.2 flies expressing the indicated NCenH3CID-lacI deletions (+) is compared to that of siblings where no fused protein is expressed (−). Results are presented for both female and male individuals. See Figure S4 for a description of the constructs.
(1.45 MB PDF)
Constructs used in these experiments. Schematic representation of fused proteins used in these experiments. The position of motives B1, B2 and B3 is indicated. Numbers correspond to amino acid positions on the corresponding sequences. DNA-binding domain of lacI is indicated in red.
(0.02 MB PDF)
Constructs used in these experiments target the reporter construct in polytene chromosomes. Localisation of the indicated constructs was determined in polytene chromosomes by immunostaining with αlacI (green). DNA was stained with DAPI.
(0.15 MB PDF)
Acknowledgments
We are thankful to Drs R.E. Karess, C.F. Lehner, T.L. Orr-Weaver, C.E. Sunkel and L.L. Wallrath for materials. We are also most thankful to Esther Fuentes for technical support.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: SMG acknowledges receipt of a FPU fellowship from MCINN. This work was financed by grants from “Ministerio de Ciencia e Innovación” (BFU2009-07111; CSD2006-00049) and “Comissió Interdepartamental de Recerca i Innovació Tecnològica” (2009SGR1023). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Malik HS, Henikoff S. Major evolutionary transitions in centromere complexity. Cell. 2009;138:1067–1082. doi: 10.1016/j.cell.2009.08.036. [DOI] [PubMed] [Google Scholar]
- 2.Torras-Llort M, Moreno-Moreno O, Azorín F. Focus on the centre: the role of chromatin on the regulation of centromere identity and function. EMBO J. 2009;28:2337–2348. doi: 10.1038/emboj.2009.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bloom K. Centromere dynamics. Curr Opin Genet Dev. 2007;17:1–6. doi: 10.1016/j.gde.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 4.Ekwall K. Epigenetic control of centromere behavior. Annu Rev Genet. 2007;41:63–81. doi: 10.1146/annurev.genet.41.110306.130127. [DOI] [PubMed] [Google Scholar]
- 5.Morris CA, Moazed D. Centromere assembly and propagation. Cell. 2007;128:647–650. doi: 10.1016/j.cell.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 6.Sullivan KF. A solid foundation: functional specialization of centromeric chromatin. Curr Opin Gen Dev. 2001;11:182–188. doi: 10.1016/s0959-437x(00)00177-5. [DOI] [PubMed] [Google Scholar]
- 7.Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nature Rev Genet. 2008;9:923–937. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith MM. Centromeres and variant histones: what, where, when and why? Curr Opin Cell Biol. 2002;14:279–285. doi: 10.1016/s0955-0674(02)00331-9. [DOI] [PubMed] [Google Scholar]
- 9.Foltz DR, Jansen LET, Black BE, Bailey AO, Yates JR, III, et al. The human CENP-A centromeric nucleosome-associated complex. Nature Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 10.Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, et al. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nature Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- 11.Carroll CW, Milks KJ, Straight AF. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J Cell Biol. 2010;189:1143–1155. doi: 10.1083/jcb.201001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carroll CW, Silva MCC, Godek KM, Jansen LET, Straight AF. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nature Cell Biol. 2009;11:896–902. doi: 10.1038/ncb1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lew DJ, Burke DJ. The spindle assembly and spindle position checkpoints. Annu Rev Genet. 2003;37:251–282. doi: 10.1146/annurev.genet.37.042203.120656. [DOI] [PubMed] [Google Scholar]
- 14.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nature Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 15.Straight AF. Checkpoint proteins and kinetochores. Curr Biol. 1997;7:R613–R616. doi: 10.1016/s0960-9822(06)00315-0. [DOI] [PubMed] [Google Scholar]
- 16.Malik HS, Henikoff S. Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics. 2001;157:1293–1298. doi: 10.1093/genetics/157.3.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malik HS, Vermaak D, Henikoff S. Recurrent evolution of DNA-binding motifs in the Drosophila centromeric histone. Proc Natl Acad Sci USA. 2002;99:1449–1454. doi: 10.1073/pnas.032664299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Danzer JR, Alvarez P, Belmont AS, Wallrath LL. Effects of tethering HP1 to euchromatic regions of the Drosophila genome. Development. 2003;130:1817–1824. doi: 10.1242/dev.00405. [DOI] [PubMed] [Google Scholar]
- 19.Font-Burgada J, Rossell D, Auer H, Azorín F. Drosophila HP1c isoform interacts with the zinc-finger proteins WOC and Relative-of-WOC to regulate gene expression. Genes Dev. 2008;22:3007–3023. doi: 10.1101/gad.481408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Basu J, Bousbaa H, Logarinho E, Li Z, Williams BC, et al. Mutations in the essential spindle checkpoint gene bub1 cause chromosome missegregation and fail to block apoptosis in Drosophila. J Cell Biol. 1999;146:13–28. doi: 10.1083/jcb.146.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malmanche N, Owen S, Gegick S, Steffensen S, Tomkiel JE, et al. Drosophila BubR1 is essential for meiotic sister-chromatid cohesion and maintenance of synaptonemal complex. Curr Biol. 2007;17:1–9. doi: 10.1016/j.cub.2007.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heeger S, Leismann O, Schittenhelm R, Schraidt O, Heidmann S, et al. Genetic interactions of separase regulatory subunits reveal the diverged Drosophila Cenp-C homolog. Genes Dev. 2005;19:2041–2053. doi: 10.1101/gad.347805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Basto R, Gomes R, Karess RE. Rough deal and Zw10 are required for metaphase checkpoint in Drosophila. Nature Cell Biol. 2000;2:939–943. doi: 10.1038/35046592. [DOI] [PubMed] [Google Scholar]
- 24.Williams BC, Li Z, Liu S, Williams EV, Leung G, et al. Zwilch, a new component, of the ZW10/ROD complex required for kinetochore functions. Mol Biol Cell. 2003;14:1379–1391. doi: 10.1091/mbc.E02-09-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Logarinho E, Sunkel C. The Drosophila POLO kinase localises to multiple compartments of the mitotic apparatus and is required for phosphorylation of MPM2 reactive epitopes. J Cell Sci. 1998;111:2897–2909. doi: 10.1242/jcs.111.19.2897. [DOI] [PubMed] [Google Scholar]
- 26.Blower MD, Karpen GH. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nature Cell Biol. 2001;3:730–739. doi: 10.1038/35087045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang TT-L, Bickel SE, Young LM, Orr-Weaver TL. Maintenance of sister-chromatid cohesion at the centromere by the Drosophila MEI-S332 protein. Genes Dev. 1998;12:3843–3856. doi: 10.1101/gad.12.24.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heun P, Erhardt S, Blower MD, Weiss S, Skora AD, et al. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev Cell. 2006;10:303–315. doi: 10.1016/j.devcel.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffman DB, Pearson CG, Yen TJ, Howell BJ, Salmon ED. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol Biol Cell. 2001;12:1995–2009. doi: 10.1091/mbc.12.7.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Régnier V, Vagnarelli P, Fukagawa T, Zerjal T, Burns E, et al. CENP-A is required for accurate chromosome segregation and sustained kinetochore association of BubR1. Mol Cell Biol. 2005;25:3967–3981. doi: 10.1128/MCB.25.10.3967-3981.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blower MD, Daigle T, Kaufman T, Karpen G. Drosophila CENP-A mutations cause a BubR1-dependent early mitotic delay without normal localization of kinetochore components. PLoS Genet. 2006;2:e110. doi: 10.1371/journal.pgen.0020110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rahmani Z, Gagou ME, Lefebvre C, Emre D, Karess RE. Separating the spindle, checkpoint, and timer functions of BubR1. J Cell Biol. 2009;187:597–605. doi: 10.1083/jcb.200905026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawashima SA, Yamagishi Y, Honda T, Ishiguro K-i, Watanabe Y. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing Shugoshin. Science. 2010;327:172–177. doi: 10.1126/science.1180189. [DOI] [PubMed] [Google Scholar]
- 34.Chen Y, Baker RE, Keith KC, Harris K, Stoler S, et al. The N terminus of centromere H3-like protein Cse4p performs an essential function distinct from that of the histone fold domain. Mol Cell Biol. 2000;20:7037–7048. doi: 10.1128/mcb.20.18.7037-7048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keith KC, Baker RE, Chen Y, Harris K, Stoler S, et al. Analysis of primary structural determinants that distinguish the centromere-specific functions of histone variant Cse4p from histone H3. Mol Cell Biol. 1999;19:6130–6139. doi: 10.1128/mcb.19.9.6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagao T, Leuzinger S, Acampora D, Simeone A, Finkelstein R, et al. Developmental rescue of Drosophila cephalic defects by the human Otx genes. Proc Natl Acad Sci USA. 1998;95:3737–3742. doi: 10.1073/pnas.95.7.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ephrussi B, Herold JL. Studies of eye pigments of Drosophila. I. Methods of extraction and quantitative estimation of the pigment components. Genetics. 1944:148–175. doi: 10.1093/genetics/29.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gatti M, Bonaccorsi S, Pimpinelli S. Looking at Drosophila mitotic chromosomes. Methods Cell Biol. 1994;44:371–391. doi: 10.1016/s0091-679x(08)60924-3. [DOI] [PubMed] [Google Scholar]
- 39.Logarinho E, Bousbaa H, Dias JM, Lopes C, Amorim I, et al. Different spindle checkpoint proteins monitor microtubule attachment and tension at kinetochores in Drosophila cells. J Cell Sci. 2004;117:1757–1771. doi: 10.1242/jcs.01033. [DOI] [PubMed] [Google Scholar]
- 40.Llamazares S, Moreira A, Tavares A, Girdham C, Spruce BA, et al. Polo encodes a protein kinase homolog required for mitosis in Drosophila. Genes Dev. 1991;5:2153–2165. doi: 10.1101/gad.5.12a.2153. [DOI] [PubMed] [Google Scholar]
- 41.Scaërou F, Aguilera I, Saunders R, Kane N, Blottière L, et al. The rough deal protein is a new kinetochore component required for accurate chromosome segregation in Drosophila. J Cell Sci. 1999;112:3757–3768. doi: 10.1242/jcs.112.21.3757. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tethering NCenH3-lacI to the ectopic white-reporter of 157.1 flies induces silencing of the reporter gene. (A–E) The eye phenotype of flies expressing the indicated fused proteins (+) is compared to that of siblings where no fused protein is expressed (−). Results are presented only for male individuals.
(0.46 MB PDF)
In interphase nuclei, CenH3CID-lacI incorporates to centromeres. CenH3CID-lacI (A) and NCenH3CID-lacI (B) were expressed in 157.1 flies carrying an ectopic white reporter construct inserted at a distal position on the X-chromosome. Localisation of the fused proteins was determined in interphase cells from brain squashes of third instar larvae by immunostaining with αCenH3CID (red) and αlacI (green). In cells expressing CenH3CID-lacI (A), all αCenH3CID signals co-localise with αlacI, indicating incorporation of CenH3CID-lacI to centromeres. In contrast, in cells expressing NCenH3CID-lacI (B), co-localisation is restricted to two-spots (indicated by the arrows), reflecting binding of the fused protein to the ectopic reporter construct.
(0.09 MB PDF)
Motives B1 and B2 retain silencing competence. (A) Amino acid sequence of the N-terminal domain of D. melanogaster CenH3CID. Conserved sequence motives (B1, B2 and B3) are indicated. (B–E) The eye phenotype of S9.2 flies expressing the indicated NCenH3CID-lacI deletions (+) is compared to that of siblings where no fused protein is expressed (−). Results are presented for both female and male individuals. See Figure S4 for a description of the constructs.
(1.45 MB PDF)
Constructs used in these experiments. Schematic representation of fused proteins used in these experiments. The position of motives B1, B2 and B3 is indicated. Numbers correspond to amino acid positions on the corresponding sequences. DNA-binding domain of lacI is indicated in red.
(0.02 MB PDF)
Constructs used in these experiments target the reporter construct in polytene chromosomes. Localisation of the indicated constructs was determined in polytene chromosomes by immunostaining with αlacI (green). DNA was stained with DAPI.
(0.15 MB PDF)