Abstract
The blood-brain barrier (BBB) maintains brain homeostasis and limits the entry of toxins and pathogens into the brain. Despite its importance, little is known about the molecular mechanisms regulating the development and function of this crucial barrier. In this study we have developed methods to highly purify and gene profile endothelial cells from different tissues, and by comparing the transcriptional profile of brain endothelial cells with those purified from the liver and lung, we have generated a comprehensive resource of transcripts that are enriched in the BBB forming endothelial cells of the brain. Through this comparison we have identified novel tight junction proteins, transporters, metabolic enzymes, signaling components, and unknown transcripts whose expression is enriched in central nervous system (CNS) endothelial cells. This analysis has identified that RXRalpha signaling cascade is specifically enriched at the BBB, implicating this pathway in regulating this vital barrier. This dataset provides a resource for understanding CNS endothelial cells and their interaction with neural and hematogenous cells.
Introduction
The vasculature of the CNS forms an endothelial barrier, not found in other tissues, that limits the flow of molecules and ions between the blood and the brain. This BBB is crucial for maintaining brain homeostasis and for limiting the penetration of toxins and pathogens into the brain [1]. The importance of the BBB is highlighted by the severe pathology of diseases in which it is disrupted, including stroke, edema, brain traumas and multiple sclerosis [2]. The BBB also poses an obstacle for the treatment of neurological disorders as it can greatly limit drug delivery into the brain [3].
Many of the properties of the BBB are manifested in the endothelial cells which make up the walls of the blood vessels. Endothelial cells in the brain differ from endothelial cells in other tissues in that they are held together by high electrical resistance tight junctions and contain few transcytotic vesicles, limiting the paracellular and transcellular flow of molecules from the blood into the brain [4]. In addition, CNS endothelial cells express a variety of transporters, both to provide the CNS with specific nutrients, and also to efflux potential toxins from the CNS. Whereas the endothelial cells form the barrier, transplantation studies have demonstrated that these properties are not intrinsic to the endothelial cells but signaled by the brain microenvironment [5]. Evidence suggests that the BBB is induced and regulated by its close association with astrocytes, pericytes and other neural cells, however the molecular nature of these interactions remains a mystery [6], [7], [8].
Several studies have taken different approaches to understand the genomics of the BBB, including microarray analysis, subtractive hybridization and serial analysis of gene expression [9], [10], [11]. These studies have provided information about the gene expression of CNS vessels but had two limitations. First, because these studies used whole vessel fractions, which contain many different cell types, the transcriptomes of the individual cellular components, the endothelial cells and the pericytes, are not yet known. Second, these studies compared the gene expression of brain vessels with whole tissue fractions instead of purified peripheral vessels. Paramount to understanding the function of the BBB is a comparison of purified endothelial cells from brain with purified endothelial cells from peripheral tissues.
Here we utilize fluorescence-activated cell sorting (FACS) to highly purify endothelial cells from the brain, liver and lung of Tie2GFP transgenic mice [12]. Using Affymetrix microarrays to gene profile these purified cell populations, we have identified a set of genes whose expression is highly enriched at the BBB. These genes include tight junction molecules, transporters, signaling molecules, and other molecule classes. This analysis has implicated Wnt and RXRalpha signaling cascades in regulating this crucial barrier.
Materials and Methods
Ethics Statement
All experiments were approved by Stanford University IACUC committee, approved protocol #10726.
FACS purification of GFP+ cells
Homozygous Tie2GFP mice (strain 003658) were obtained from Jackson labs and bred to maintain homozygosity. Cell suspensions were prepared from cerebral cortex, liver or lung based on procedures previously described [13], [14]. For brain samples, the cerebral cortex was dissected away from the forebrain, then the meninges were peeled off with fine forceps. For liver samples, peripheral regions of each lobe were utilized to avoid the hepatic portal vein ensuring the tissue vasculature consisted primarily of capillaries. Whole lung lobes were utilized. Each tissue was diced with a scalpel, and enzymatically dissociated with a papain solution (40 U/ml Worthington-3126) containing L-cysteine (0.4 mg/ml, Sigma C 7477) and DNase (125 U/ml, Worthington LS002007) for 1.5 hours at 35°C, prior to mechanical trituration in a solution containing ovomucoid (2 mg/ml, Roche 109878), DNase (125 U/ml) and BSA (1 mg/ml, Sigma A8806), to yield a cell suspension, which was recovered by centrifugation. Cell suspensions were re-suspended in FACS buffer (DPBS, 0.02%BSA with propidium iodide), and endothelial cells were FACS purified based on GFP fluorescence utilizing a FACS Vantage SE sorter (Becton Dickinson) and CellQuest software. For each tissue, background GFP fluorescence was determined by FACS analysis of cell suspensions from wild type FvB mice (Charles River), and dead cells were eliminated by high propidium iodide fluorescence. Forward scatter and side scatter analysis were also used as gates to limit the sorting to single live cells. For additional pericyte/VSM depletion, cell suspensions were stained with a goat anti-mouse PDGFRbeta primary antibody (R & D systems), and a donkey anti-goat secondary antibody labeled with APC. For these experiments, background APC fluorescence was gated with cell suspensions incubated with secondary, but not primary antibody. In each case, two rounds of sorting were performed for maximal purity, based on reanalysis. Cortical GFP− cells were isolated from Tie2GFP+ mice with gating based on background GFP fluorescence from wild type FvB mice.
GeneChip Analysis
Purification of RNA, generation of biotinylated cRNA, subsequent hybridization to Affymetrix Mouse Genome 430 2.0 Arrays and raw image analysis with Affymetrix GCOS 1.3 software was performed as previously described [14]. Three biological replicates were used for each analysis of adult cells, whereas five biological replicates were utilized for postnatal samples. Each biological replicate represents a purification from different mice performed on different days. The Significance Analysis of Microarrays (SAM) method was used to determine genes that were significantly different between cell populations. For this analysis, samples were grouped into 6 groups: brain parenchyma (GFP−), brain vascular (GFP+), brain endothelial (GFP+PDGFRbeta−), postnatal brain GFP+, liver endothelial and lung endothelial. Probe sets were included in this statistical analysis if they were defined as being called present and having an intensity level greater than 200 in at least two-thirds of the samples in at least one group. The significance thresholds were set for a false discovery rate of less than 1% with a fold-change of greater than 2.0×. For each data analysis presented in this paper, fold-enrichment was calculated as expression value of one data set divided by the second. In each table presented, probe sets were included only if gene expression was ≥200 in the enriched data set. In comparisons of brain endothelial to lung and liver endothelial samples, or comparisons of adult and postnatal brain samples, GFP+ samples for each tissue were compared, and CNS pericyte/VSM enriched transcripts (brain GFP+/GFP+PDGFRbeta−>2) were excluded. To identify signaling and metabolic pathways enriched in specific data sets, Ingenuity Pathway Analysis software core analysis (Ingenuity systems) was utilized to determine the statistical significance of enrichment for the pathways within a given data set.
In situ Hybridization
In situ hybridization on P20 fresh frozen mouse brain and liver sections was performed as described [15] with a few modifications, omitting the treatment with Proteinase K. The antisense mRNA probes were generated using the DIG RNA Labeling Kit (Roche Applied Science, Indianapolis, IN). Ten genes were analyzed by in situ hybridization on P20 fresh frozen mouse brain and liver sections: Pecam (SmaI, T7), Cldn5 (XhoI, T3), Apcdd1 (HindIII, T7), Abcb1a (EcoRV, T7), Slco1c1 (XhoI, T3), Slco1a4 (XhoI, T3), Slc22a8 (XhoI, T3), Slc7a5 (SalI, T3), Itih5 (SalI, T3), Foxq1 (EcoRV, T7). Apcdd1 cDNA was amplified by RT-PCR using total RNA isolated from FACS isolated Tie2GFP P60 brain endothelial cells. The other full length or partial cDNAs were obtained as mouse ESTs from Open Biosystems (Hunstville Al).
Results
Purification of CNS Vascular Cells from Tie2GFP Mice
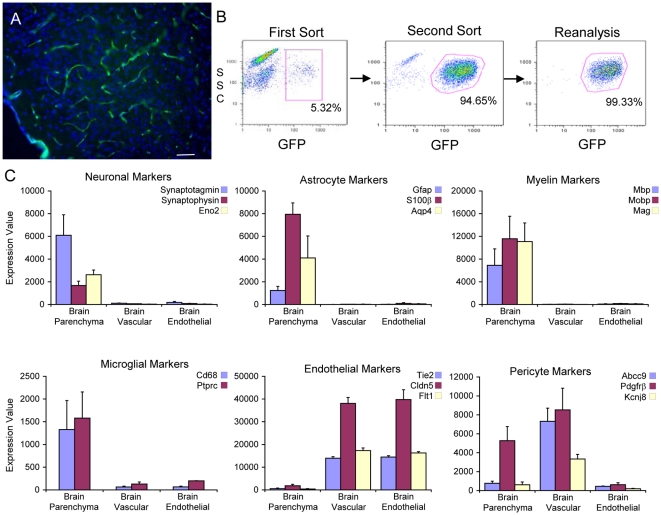
To purify cells from the vasculature of different tissues we took advantage of Tie2GFP transgenic mice available from Jackson labs. These mice express the green fluoresecent protein (GFP) under the pan-endothelial Tie2 promotor, and GFP fluorescence can be visualized in the vasculature of the brain, liver, lung, spleen, kidney, muscle and other tissue cryosections (Figure 1a)[12]. When single cell suspensions of adult cerebral cortex were analyzed by FACS, approximately 5% of the cells were GFP+, a figure which matches the proportion of vascular cells in the brain (Figure 1b)[12]. Using a double FACS procedure to ensure the highest purity (Figure 1b) we purified three different cell populations: GFP− (parenchymal), GFP+ (vascular cell), and GFP+PDGFRbeta− (endothelial cell). For the latter cell population an anti-PDGFRbeta depletion step was utilized to deplete pericytes/vascular smooth muscle cells (VSMs) from the sample.
Figure 1. Purification of endothelial cells from the cerebral cortex of Tie2GFP mice.
A) GFP is expressed in the vasculature of Tie2GFP transgenic mice. GFP (green) can be visualized in a vascular pattern in tissue cryosections of cerebral cortex of an adult Tie2GFP transgenic mouse. Nuclei were stained with DAPI (blue). Bar, 50 microns. B) FACS purification of GFP+ cells from the cerebral cortex of Tie2GFP mice. Single cell cortical suspensions were sorted for GFP fluorescence twice, isolating a GFP+ cell population that is greater than 99.3% pure. Y-axis represents side scatter co-efficient, X-axis represents GFP fluorescence, numerical values represent percent of total cells sorted that are within gated region. C) Purity of FACS sorted cells. GeneChip transcriptional profiling of GFP− (parenchyma), GFP+ (vascular) and GFP+PDGFRbeta− (endothelial) fractions from the cerebral cortex of Tie2GFP mice. Neuronal, astrocyte, oligodendrocyte, microglial, endothelial and pericyte markers were analyzed to detect the purity of sample for each FACS sorted sample. The GFP+ sample contains a mixture of endothelial cells and pericytes, such that an additional PDGFRbeta negative selection is needed to generate a pure endothelial cell population. Error Bars represent standard error of the mean (SEM).
The purity of these cell populations was ascertained by using Affymetrix microarrays to compare the relative mRNA levels of specific markers for neurons (synaptotagmin I, synaptophysin, neuronal enolase), astrocytes (GFAP, Aq4, s100beta), oligodendrocyte lineage cells (MBP, MAG, MOBP), microglial cells (cd68, Ptprc), endothelial cells (Tie2, claudin 5, and Flt1) and pericytes/VSMs (PDGFRbeta, Abcc9, and Kcnj8) (Figure 1c). As expected, the neuronal and glial markers were all present in the brain parenchyma sample and low to absent in the vascular and endothelial cell samples, indicating these samples did not contain neural cells. The vascular cell sample contained endothelial and pericyte/VSM cell markers indicating it was a mixture of these two cell populations, whereas the endothelial cell sample contained endothelial cell markers but not pericyte/VSM markers. This separation allowed us to purify endothelial cells and identify the endothelial cell transcriptome. Furthermore, comparing the vascular and endothelial cell data sets we are able to identify many genes that are highly enriched to pericytes/VSM. A master data table of all data sets described can be found in Table S1.
In Table S2, we list the enriched CNS endothelial genes, defined as those GeneChip probe sets significantly enriched greater than two-fold in the endothelial:parenchymal fractions (GFP+ PDGFRbeta−:GFP−). We further present a list of the pericyte/VSM enriched genes, as defined as enriched greater than two-fold in the Vascular:Endothelial (GFP+:GFP+PDGFRbeta−) fractions (Table S3). In Table 1 we list the most endothelial and pericyte enriched probe sets which provide many new endothelial and pericyte/VSM specific markers within the brain.
Table 1. Most enriched CNS endothelial and pericyte transcripts.
| A.Probe Set ID | Gene Symbol | Brain Endothelial/Parenchyma | B.Probe Set ID | Gene Symbol | Vascular/Endothelial |
| 1421078_at | Tcf23 | 96.1 | 1423341_at | Cspg4 | 40.9 |
| 1438558_x_at | Foxq1 | 71.9 | 1418773_at | Fads3 | 38.8 |
| 1422905_s_at | Fmo2 | 62.1 | 1455098_a_at | Vtn | 32.7 |
| 1460371_at | Hspa12b | 62.1 | 1416286_at | Rgs4 | 27.8 |
| 1438227_at | She | 59.4 | 1437902_s_at | Rarres2 | 27.1 |
| 1436939_at | Unc45b | 55.4 | 1459713_s_at | Tmem16a | 25.8 |
| 1419467_at | Clec14a | 52.4 | 1425103_at | Ace2 | 24.6 |
| 1448117_at | Kitl | 52.3 | 1433525_at | Ednra | 24.3 |
| 1447933_at | Kif26a | 52.0 | 1426285_at | Lama2 | 22.2 |
| 1438325_at | Evi1 | 51.3 | 1434141_at | Gucy1a3 | 22.0 |
| 1422966_a_at | Tfrc | 48.3 | 1417439_at | Cd248 | 21.9 |
| 1416564_at | Sox7 | 48.0 | 1448649_at | Enpep | 21.6 |
| 1443832_s_at | Sdpr | 47.9 | 1417148_at | Pdgfrb | 20.1 |
| 1438532_at | Hmcn1 | 47.7 | 1438658_a_at | Edg3 | 20.0 |
| 1449184_at | Pglyrp1 | 47.6 | 1436411_at | Atp13a5 | 20.0 |
| 1452352_at | Ctla2b | 44.8 | 1438751_at | Slc30a10 | 19.6 |
| 1435701_at | Cyyr1 | 44.8 | 1427221_at | Xtrp3s1 | 18.9 |
| 1436419_a_at | 1700097N02Rik | 44.2 | 1448421_s_at | Aspn | 18.2 |
| 1449328_at | Ly75 | 42.0 | 1418142_at | Kcnj8 | 17.7 |
| 1428345_at | Ppapdc2 | 42.0 | 1436203_a_at | 1110059G02Rik | 16.4 |
| 1442698_at | --- | 41.1 | 1435751_at | Abcc9 | 16.3 |
| 1440926_at | Flt1 | 40.8 | 1424186_at | Ccdc80 | 15.4 |
| 1456061_at | Gimap8 | 40.7 | 1452474_a_at | Art3 | 15.2 |
| 1452013_at | Atp10a | 40.6 | 1424902_at | Plxdc1 | 15.0 |
| 1418059_at | Eltd1 | 39.6 | 1424733_at | P2ry14 | 14.6 |
| 1438587_at | 4933417M04Rik | 39.3 | 1455050_at | E130203B14Rik | 14.3 |
| 1441907_s_at | Cd93 | 38.9 | 1423477_at | Zic1 | 13.3 |
| 1440225_at | Gpr116 | 38.8 | 1448918_at | Slco3a1 | 13.1 |
| 1451500_at | Ushbp1 | 38.7 | 1448132_at | Slc19a1 | 12.8 |
| 1456152_at | Paqr5 | 38.6 | 1437165_a_at | Pcolce | 12.8 |
| 1447584_s_at | Myct1 | 37.8 | 1448123_s_at | Tgfbi | 12.5 |
| 1459888_at | LOC545261 | 37.6 | 1455271_at | LOC620695 | 12.5 |
| 1422028_a_at | Ets1 | 37.0 | 1417011_at | Sdc2 | 12.4 |
| 1457867_at | Sgpp2 | 36.9 | 1434893_at | Atp1a2 | 12.4 |
| 1417616_at | St6galnac2 | 36.8 | 1440879_at | Abca9 | 11.9 |
| 1418979_at | Akr1c14 | 36.5 | 1430700_a_at | Pla2g7 | 11.7 |
| 1420459_at | Dscr6 | 36.4 | 1424099_at | 2310016C16Rik | 11.7 |
| 1438027_at | 5830443L24Rik | 35.8 | 1422058_at | Nodal | 11.7 |
| 1457141_at | Aqp11 | 35.7 | 1417534_at | Itgb5 | 11.6 |
| 1416832_at | Slc39a8 | 35.6 | 1419663_at | Ogn | 10.1 |
| 1429177_x_at | Sox17 | 35.3 | 1422545_at | Tbx2 | 10.1 |
| 1456768_a_at | Mmrn2 | 35.0 | 1420872_at | Gucy1b3 | 10.0 |
| 1444181_at | Gimap5 | 35.0 | 1435741_at | Pde8b | 9.9 |
| 1419758_at | Abcb1a | 34.7 | 1454966_at | Itga8 | 9.8 |
| 1426914_at | Marveld2 | 34.5 | 1449860_at | Higd1b | 9.4 |
| 1432176_a_at | Eng | 34.3 | 1416271_at | Perp | 8.8 |
| 1439221_s_at | Cd40 | 34.2 | 1420688_a_at | Sgce | 8.7 |
| 1448862_at | Icam2 | 33.6 | 1430136_at | Grm3 | 8.2 |
| 1448471_a_at | Ctla2a | 33.3 | 1435805_at | Lin7a | 8.0 |
| 1435418_at | Slc22a8 | 33.0 | 1418157_at | Nr2f1 | 7.6 |
A) Most enriched CNS endothelial transcripts. List of the 50 most enriched endothelial cell transcripts defined as the greatest ratio of brain endothelial cell (GFP+ PDGFRbeta−): brain parenchyma (GFP−) values analyzed by affymetrix GeneChips.
B) Most enriched CNS pericyte transcripts. List of the 50 most enriched pericyte transcripts defined as the greatest ratio of brain vascular (GFP+): brain endothelial (GFP+ PDGFRbeta−) values analyzed by Affymetrix GeneChips.
Identification of the Blood-Brain Barrier Transcriptome
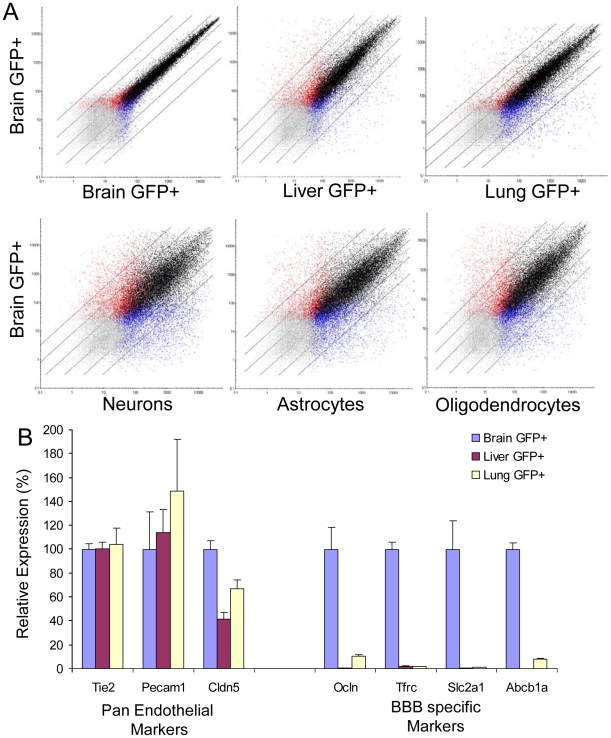
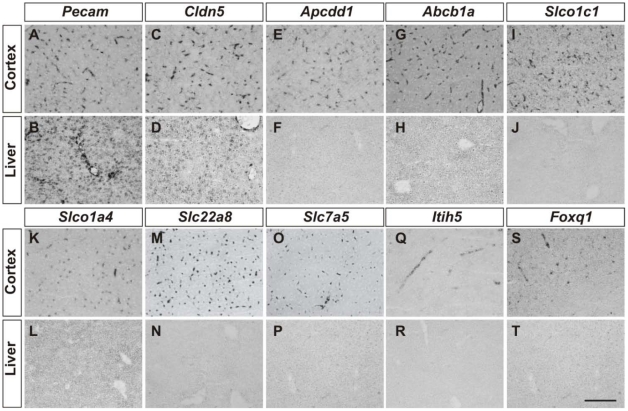
In order to identify blood-brain barrier enriched transcripts, we next utilized GeneChip analysis to compare the gene expression in GFP+ cells isolated from the brain, liver and lung of adult Tie2GFP mice. In each case, we used a double FACS purification procedure in order to isolate GFP+ cells for maximal purity. Comparison of the gene profiles of brain and liver, or brain and lung, revealed that the GFP+ cell populations in these tissues were very similar. This is graphically represented by a dot plot, in which each dot represents a probe set on the Affymetrix GeneChip, and their position relative to the logarithmic X and Y axes represents expression levels in the different tissues analyzed (Figure 2a). In these analyses, the vast majority of genes lie near the diagonal axis, indicating that their expression levels are not more than two-fold different between the samples compared. There are, however, outliers, genes which are expressed at vastly different levels in the vasculature of the different tissues. These differences were not identified between two biological replicates of brain GFP+ cells (Figure 2a). Furthermore, far greater variation was observed between the transcriptional profile of brain endothelial cells and those for neurons and glia (Figure 2a, data for neurons and glia generated by Cahoy et al 2008). The genes which are expressed at high levels in the brain vasculature, but low or absent from the liver and lung, are BBB-enriched genes. For BBB-enriched genes we identify genes that are enriched in the Brain GFP+ (vascular) cells, compared to liver GFP+ cells and lung GFP+ cells, and eliminate any pericyte enriched genes by comparing Brain GFP+PDGFRbeta− endothelial cells with Brain GFP+ vascular cells. Preliminary validation of this data set comes from analyzing characterized endothelial genes. For instance, known pan-endothelial genes, including Tie2, PECAM1 and claudin 5, are expressed at similar levels in all tissue data sets, whereas established BBB-specific genes glut-1, p-glycoprotein, occludin and the transferrin receptor, are expressed at significantly higher levels in the brain vasculature than the liver or lung (Figure 2b). To further validate our data set we used in situ hybridization to analyze the expression pattern of the several top brain endothelial transcripts enriched compared with liver (Itih5, Apcdd1, Abcb1a and Slco1c1) and lung endothelial cells (Slc22a8, Slc7a5, Slco1a4 and Foxq1) (Figure 3). In each case, we observed expression in vascular cells in the brain but not liver. Interestingly, the pattern observed was different between the probe sets tested. For instance, Itih5 and Foxq1 appeared to be expressed sporadically and specifically in larger vessels, whereas Apcdd1, Slco1a4 and Slc22a8 appear to be expressed in smaller vessels. Indeed, hetereogeneity between the different segments of the CNS vasculature has been observed[16], and specific transcripts may be differentially expressed between arteries, arterioles, capillaries, venules and veins.
Figure 2. Comparison of transcriptional profiles of brain endothelial cells with liver and lung endothelial cells.
A) Affymetrix GeneChips were utilized to compare gene expression in GFP+ cells isolated from cerebral cortex, liver and lung of Tie2GFP transgenic mice. The data is represented as a dot plot on a logarithmic scale, where each point reflects a probe set on the GeneChip. Black dots indicate probe sets identified as present in both samples, red dots indicate probe sets identified as present in the brain but not peripheral sample, blue dots indicate probe sets present in the peripheral but not brain sample, and grey dots represent probe sets identified as absent in both samples. Two biological replicates of the brain GFP+ samples were compared to identify variation between replicates. In addition, brain GFP+ cells were compared with profiles generated for neurons and glia by Cahoy et al (2008). Black diagonal lines represent cutoffs for two-fold, four-fold and eight-fold differences. B) Validation of transcriptional profiling by comparison of GeneChip expression with known CNS endothelial markers. Expression values are given relative to the brain GFP+ sample. Pan endothelial transcripts are expressed in all GFP+ samples, whereas BBB transcripts were enriched in the Brain GFP+ sample. Error bars represent SEM.
Figure 3. Selective expression of blood-brain barrier-enriched genes in CNS endothelial cells.
Expression of Pecam (A, B), Cldn5 (C, D), Apcdd1 (E, F), Abcb1a1 (G, H), Slco1c1 (I, J), Slco1a4 (K, L), Slc22a8 (M, N), Slc7a5 (O, P), Itih5 (Q, R) and Foxq1 (S, T) mRNA in the brain and liver of P20 mouse pups. The top four brain endothelial transcripts enriched compared with liver (Itih5, Apcdd1, Abcb1a and Slco1c1) and lung endothelial cells (Slc22a8, Slc7a5, Slco1a4 and Foxq1) were analyzed, excluding transcripts also expressed in the brain parenchyma (brain GFP+:brain GFP−<2), and those expressed at <10000 to ensure that the signal would be above the detection limit of the in situ hybridization. Note that Pecam1 and Cldn5 mRNAs are expressed in the endothelial cells in both brain and liver. However, the transcripts for the blood-brain barrier enriched genes are only present in brain but not liver endothelial cells. Bar, 200 microns.
From this analysis, we conclude that we have generated a comprehensive data set representing the blood-brain barrier transcriptome. In Table 2 we list the most BBB enriched genes: those with the greatest difference between expression levels in the brain GFP+ cells versus liver and lung GFP+ cells, with parenchymal and pericyte/VSM transcripts excluded from these lists. All BBB enriched genes are listed in Table S6. In addition to identifying BBB enriched transcripts, we can also use this data set to identify genes which are enriched in the liver and lung endothelial cells compared with brain endothelial cells (Table S7). These genes might be important for regulating the permeability of non-CNS vessels, or imparting specific functions of non-CNS endothelial cells. In the following sections we present functional subsets of BBB enriched genes including tight junction proteins, transporters, signaling molecules and metabolic pathways that together constitute the blood-brain barrier.
Table 2. Most BBB enriched transcripts.
| A.Probe Set ID | Gene Symbol | Brain/Liver | Brain/Lung | B.Probe Set ID | Gene Symbol | Brain/Liver | Brain/Lung |
| 1441946_at | Itih5 | 3124.8 | 213.3 | 1418706_at | Slc38a3 | 85.5 | 821.1 |
| 1449070_x_at | Apcdd1 | 3087.4 | 75.2 | 1435418_at | Slc22a8 | 774.6 | 764.3 |
| 1419758_at | Abcb1a | 1848.7 | 12.9 | 1418326_at | Slc7a5 | 15.8 | 734.2 |
| 1423343_at | Slco1c1 | 1745.2 | 310.2 | 1420405_at | Slco1a4 | 847.4 | 450.2 |
| 1437967_at | Titin | 948.1 | 63.9 | 1423424_at | Zic3 | 438.5 | 441.2 |
| 1448147_at | Tnfrsf19 | 908.6 | 18.9 | 1438558_x_at | Foxq1 | 317.0 | 404.8 |
| 1420405_at | Slco1a4 | 847.4 | 450.2 | 1423343_at | Slco1c1 | 1745.2 | 310.2 |
| 1447787_x_at | Gja7 | 825.5 | 3.8 | 1428223_at | Mfsd2 | 777.8 | 224.9 |
| 1428223_at | Mfsd2 | 777.8 | 224.9 | 1441946_at | Itih5 | 3124.8 | 213.3 |
| 1435418_at | Slc22a8 | 774.6 | 764.3 | 1426215_at | Ddc | 142.0 | 171.7 |
| 1418094_s_at | Car4 | 772.1 | 2.9 | 1434734_at | E130016E03Rik | 168.2 | 167.1 |
| 1419700_a_at | Prom1 | 589.5 | 69.8 | 1453072_at | Gpr160 | 10.8 | 165.4 |
| 1429154_at | Slc35f2 | 552.5 | 69.2 | 1415802_at | Slc16a1 | 59.3 | 148.3 |
| 1447272_s_at | Atp10a | 527.2 | 42.8 | 1442347_at | Lrp8 | 387.2 | 145.4 |
| 1429348_at | Sema3c | 511.0 | 1.3 | 1449184_at | Pglyrp1 | 371.8 | 116.0 |
| 1448557_at | 1200015N20Rik | 481.7 | 10.9 | 1454734_at | Lef1 | 89.5 | 108.6 |
| 1428663_at | 5133401H06Rik | 472.0 | 4.8 | 1436417_at | Slc19a3 | 249.7 | 105.8 |
| 1425779_a_at | Tbx1 | 469.4 | 25.2 | 1426399_at | Vwa1 | 202.8 | 103.8 |
| 1423424_at | Zic3 | 438.5 | 441.2 | 1418383_at | Apcdd1 | 448.3 | 95.6 |
| 1457867_at | Sgpp2 | 429.8 | 9.3 | 1460459_at | Paqr5 | 72.3 | 80.2 |
| 1460465_at | A930038C07Rik | 419.3 | 9.8 | 1439609_at | --- | 116.6 | 70.8 |
| 1423608_at | Itm2a | 415.0 | 25.3 | 1419700_a_at | Prom1 | 589.5 | 69.8 |
| 1451255_at | MGI:1927471 | 412.6 | 41.0 | 1451047_at | Itm2a | 266.5 | 69.5 |
| 1427157_at | E030025D05Rik | 390.4 | 0.8 | 1429154_at | Slc35f2 | 552.5 | 69.2 |
| 1442347_at | Lrp8 | 387.2 | 145.4 | 1437967_at | Titin | 948.1 | 63.9 |
| 1416301_a_at | Ebf1 | 380.1 | 11.8 | 1423858_a_at | Hmgcs2 | 63.9 | 62.7 |
| 1425415_a_at | Slc1a1 | 372.5 | 12.5 | 1417061_at | Slc40a1 | 1.1 | 59.2 |
| 1449184_at | Pglyrp1 | 371.8 | 116.0 | 1452661_at | Tfrc | 48.1 | 55.4 |
| 1439505_at | Clic5 | 357.9 | 0.9 | 1435123_at | C030014M07Rik | 52.6 | 52.2 |
| 1426431_at | Jag2 | 325.6 | 4.4 | 1424549_at | Degs2 | 156.4 | 50.0 |
| 1438558_x_at | Foxq1 | 317.0 | 404.8 | 1456140_at | Zic5 | 61.5 | 48.7 |
| 1448117_at | Kitl | 261.9 | 1.2 | 1426599_a_at | Slc2a1 | 234.7 | 48.5 |
| 1448873_at | Ocln | 255.5 | 9.8 | 1417714_x_at | Hba-a1 | 5.2 | 47.7 |
| 1438452_at | Nebl | 253.1 | 4.7 | 1427931_s_at | Pdxk | 31.0 | 43.7 |
| 1437360_at | Pcdh19 | 249.8 | 19.4 | 1447272_s_at | Atp10a | 527.2 | 42.8 |
| 1436417_at | Slc19a3 | 249.7 | 105.8 | 1451255_at | Lsr | 412.6 | 41.0 |
| 1454622_at | Slc38a5 | 237.6 | 13.5 | 1435026_at | Spock2 | 180.3 | 37.7 |
| 1426599_a_at | Slc2a1 | 234.7 | 48.5 | 1456147_at | st8sia6 | 95.7 | 36.9 |
| 1433933_s_at | Slco2b1 | 226.7 | 18.5 | 1426082_a_at | Slc16a4 | 94.0 | 34.0 |
| 1418412_at | Tpd52l1 | 216.4 | 25.6 | 1417022_at | Slc7a3 | 38.6 | 33.0 |
| 1454795_at | Cobll1 | 216.0 | 5.9 | 1435465_at | Kbtbd11 | 129.9 | 30.5 |
| 1426399_at | Vwa1 | 202.8 | 103.8 | 1456616_a_at | Bsg | 24.7 | 30.1 |
| 1452968_at | Cthrc1 | 200.9 | 7.2 | 1452202_at | Pde2a | 0.7 | 30.1 |
| 1459903_at | Sema7a | 194.3 | 4.2 | 1417184_s_at | Hbb-b | 3.0 | 29.0 |
| 1418979_at | 9030611N15Rik | 182.3 | 9.9 | 1442698_at | --- | 5.7 | 28.5 |
| 1435026_at | Spock2 | 180.3 | 37.7 | 1437268_at | Lancl3 | 53.2 | 28.0 |
| 1434734_at | Rad54b | 168.2 | 167.1 | 1418412_at | Tpd52l1 | 216.4 | 25.6 |
| 1437479_x_at | Tbx3 | 163.6 | 1.8 | 1429206_at | Rhobtb1 | 2.7 | 25.4 |
| 1424549_at | Degs2 | 156.4 | 50.0 | 1451675_a_at | Alas2 | 6.1 | 25.4 |
| 1426215_at | Ddc | 142.0 | 171.7 | 1451857_a_at | 5730593N15Rik | 31.2 | 25.3 |
The 50 most enriched transcripts in brain GFP+ cells compared with (A) liver, or (B) lung. Fold-enrichment given as a ratio of brain GFP+: peripheral GFP+ microarray expression values. To highlight BBB enriched transcripts, only probe sets ≥ two-fold enriched in brain endothelial cells compared with brain parenchyma (GFP-) are listed, and pericyte genes are excluded.
Identification of Novel BBB Enriched Tight Junction Molecules
Tight junctions (TJs) are crucial components of the blood-brain barrier [17], but many of the key molecules that constitute these junctions are not yet known. In Table 3, we list the tight junction transcripts expressed by brain endothelial cells, and their relative expression levels in liver and lung endothelial cells. Among the claudin gene family, claudin 5 and claudin 12 are expressed by endothelial cells in all tissues, as described in the literature [18], [19]. Claudin 1, 3, and 11 have also been suggested to be expressed at the BBB [11], [20], [21], however, we did not see a definitive signal for any of these molecules. We also did not see expression of these molecules in CNS vessels by mRNA in situ hybridization and immunofluoresence imaging (data not shown), suggesting that claudin 5 and 12 are the key claudins expressed at the BBB. Whereas these endothelial claudins are expressed by endothelial cells in all tissues analyzed, we found that other tight junction transmembrane molecules including occludin, marveld2 (tricellulin), and Jam4 are highly enriched in brain endothelial cells. The specific expression of occludin at the BBB has been well documented [22], however, the enrichment of Jam4 and marveld2 in CNS endothelial cells is a novel finding. Marveld2 functions to seal tight junctions at points where three cells meet [23], [24]. The expression of this molecule by brain endothelial cells suggests that sealing tripartite adhesions may be critical for forming a high electrical resistance barrier. Endothelial cells of BBB capillaries form TJs with adjacent endothelial cells, as well as intra-cellular junctions as they fold over on to themselves forming the vessel tube. Thus, tripartite adhesions at the BBB could be the interface between intercellular tight junction strands and intracellular tight junction strands.
Table 3. BBB enriched tight junction molecules.
| Probe set | Gene Symbol | Brain GFP+ | Liver GFP+ | Lung GFP+ |
| Transmembrane Molecules | ||||
| 1417839_at | Cldn5 | 38079.1 | 15781.7 | 25539.9 |
| 1433781_a_at | Cldn12 | 1728.6 | 1554.1 | 2732.0 |
| 1448873_at | Ocln | 10303.7 | 40.3 | 1054.4 |
| 1451407_at | Jam4 | 1053.1 | 21.9 | 219.6 |
| 1436568_at | Jam2 | 11440.3 | 11372.0 | 3897.8 |
| 1426914_at | Marveld2 | 2877.8 | 72.7 | 554.4 |
| Cytoplasmic Adaptors | ||||
| 1450985_a_at | Tjp2 | 9089.6 | 3809.5 | 9019.6 |
| 1417749_a_at | Tjp1 | 25536.7 | 14771.9 | 16378.3 |
| 1426691_at | Tjap1 | 509.3 | 588.0 | 572.8 |
| 1455179_at | Mpp7 | 787.1 | 195.1 | 436.3 |
| 1434954_at | Mpp5 | 1197.4 | 1166.8 | 884.7 |
| 1450919_at | Mpp1 | 1438.7 | 3789.9 | 1506.7 |
| 1435461_at | Magi3 | 3487.4 | 1785.9 | 2131.1 |
| 1452309_at | Cgnl1 | 8039.7 | 430.1 | 1562.4 |
| 1415691_at | Dlg1 | 3574.6 | 2188.6 | 3425.2 |
| 1426465_at | Dlgap4 | 773.3 | 1219.4 | 1842.6 |
| 1427064_a_at | Scrib | 551.2 | 605.5 | 625.0 |
| 1420851_at | Pard6g | 1176.7 | 426.6 | 4731.0 |
| 1434775_at | Pard3 | 2325.2 | 62.8 | 1465.7 |
| 1450937_at | Lin7c | 2913.0 | 2055.6 | 2396.9 |
| 1424595_at | F11r | 4332.0 | 5220.9 | 4775.5 |
| 1460356_at | Esam1 | 21470.6 | 16937.8 | 15387.4 |
| 1433956_at | Cdh5 | 2893.8 | 4790.3 | 6622.2 |
| 1420985_at | Ash1l | 2011.1 | 1239.5 | 2094.6 |
| 1426623_a_at | Arhgap17 | 845.4 | 1530.9 | 891.0 |
| 1452387_a_at | Amotl2 | 2714.8 | 5359.0 | 2375.6 |
| 1428785_at | Amotl1 | 1989.5 | 6058.1 | 2486.1 |
| 1454890_at | Amot | 5943.5 | 3708.8 | 835.5 |
| 1433676_at | Wnk1 | 15543.9 | 10052.6 | 12021.5 |
Transcripts defined by MGI as tight junction molecules, or related family members, expressed in brain GFP+ cells with an expression value ≥500 by Affymetrix GeneChip analysis. Transcripts highlighted in red are enriched at least 3-fold over liver endothelial cells.
Several intracellular accessory tight junction molecules are also expressed in all endothelial cells. These include ZO-1, ZO-2, Mpdz, MPP1, MPP5, MPP6, MPP7, Dlgh1, Dlgh5, Dlgap4, lglh and Scrib. These proteins may play integral roles in linking the tight junction to the actin cytoskeleton as well as regulating the function of the junctions. However, they are expressed by all endothelial cells, and therefore their presence alone does not define the BBB. In our data set we have also identified several interesting cytoplasmic tight junction molecules as being highly enriched at the BBB, including cingulin-like 1 (jacop) and pard3. Cingulin-like 1 is also of great interest as it is homologous to cingulin, which links tight junctions to the actin cytoskeleton [25]. Perhaps the difference between peripheral and CNS endothelial cells is not the expression of the structural claudins, but in how claudins are regulated by the cytoskeleton. The pard family molecules are highly conserved throughout evolution from worms to flies to mammals and are involved in the formation of asymmetry in cells during development [26], [27]. This is very interesting as a defining feature of blood-brain barrier forming endothelial cells is that the tight junctions form highly asymmetric cells, and that different transporters are localized specifically to the luminal or abluminal surface [28].
In each case, the BBB enriched TJ molecules identified, whether occludin, marveld2, jam4, cingulin-like 1, or pard3, all have far greater expression levels in the brain endothelial cells than liver endothelial cells, with lung endothelial cells expressing at intermediate levels. This pattern follows the known level of permeability of the vessels in each tissue. BBB vessels form a tight impermeable barrier, liver sinusoidal capillaries are among the leakiest vessels in the body, whereas lung vessels have an intermediate permeability [29], [30]. The fact that the expression levels of enriched tight junction molecules follows this pattern suggests that we may be able to identify novel molecules involved in barrier function by examining our data set for probe sets whose expression level increases in the liver GFP+ to lung GFP+ to brain GFP+ microarray analyses. These quantitative profiles should be highly useful for identifying candidate genes that may regulate the differential permeability observed in blood vessels of various tissues throughout the body. With this in mind, novel molecules that follow a similar pattern may also be important for BBB formation. A number of these molecules are known to associate with cellular junctions in other tissues including Sorbs1, Sorbs2, Enah, Neo1 and Lims2, and thus may be important for BBB formation [31], [32], [33], [34], [35]. In addition we may be able to identify novel transmembrane molecules involved in structural adhesions. Intriguing candidates are Itm2a, Apcdd1, Tmeff1, Tmem23, Ly75, Ly6c, cd34, Pcdh19 and Extl3. Also, several Golgi protein glycosylation enzymes, including St6galnac2, St8sia4 and st8sia6, are identified using this analysis, suggesting that processing of proteins may be important in regulating BBB formation.
Elucidation of the Set of BBB-Enriched Transporters
Another key aspect of the blood-brain barrier is the expression of transporters by CNS endothelial cells. Whereas many transporters have already been identified as highly enriched at the BBB, a genome wide analysis has not been performed before. In Table 4 we present a list of transporters expressed by the CNS vasculature. We divide these into four categories: BBB enriched transporters which are enriched in CNS endothelial cells compared with peripheral endothelial cells; pan-endothelial transporters identified in all endothelial cells analyzed; pericyte enriched transporters (Table 4); and ubiquitous transporters expressed by all cells analyzed (data not shown). Elucidating the substrates of these transporters may provide a greater understanding of how endothelial cells participate in the metabolic division of labor within the CNS. Furthermore, these transporters may be utilized to target specific molecules for delivery into the brain for therapeutic purposes.
Table 4. Transporters of the BBB.
| A.Probe set ID | Gene Symbol | Brain GFP+ | Liver GFP+ | Lung GFP+ | B.Probe set ID | Gene Symbol | Brain GFP+ | Liver GFP+ | Lung GFP+ |
| 1433933_s_at | Slco2b1 | 7798.5 | 34.4 | 420.7 | 1439368_a_at | Slc9a3r2 | 11220.0 | 5884.7 | 9141.2 |
| 1423343_at | Slco1c1 | 30482.2 | 17.5 | 98.3 | 1438559_x_at | Slc44a2 | 7412.4 | 30206.2 | 17458.8 |
| 1420405_at | Slco1a4 | 38443.9 | 45.4 | 85.4 | 1433645_at | Slc44a1 | 8029.7 | 5969.1 | 5828.9 |
| 1418326_at | Slc7a5 | 13681.2 | 866.3 | 18.6 | 1425364_a_at | Slc3a2 | 18715.9 | 8790.5 | 12701.4 |
| 1417022_at | Slc7a3 | 2336.3 | 60.5 | 70.8 | 1416832_at | Slc39a8 | 11166.0 | 16155.8 | 1380.6 |
| 1454991_at | Slc7a1 | 31330.8 | 696.7 | 2873.3 | 1429593_at | Slc38a2 | 3477.9 | 2701.2 | 3444.1 |
| 1437149_at | Slc6a6 | 21228.3 | 1608.7 | 6912.5 | 1452059_at | Slc35f5 | 1790.0 | 1341.4 | 2176.9 |
| 1436137_at | Slc6a17 | 243.6 | 114.6 | 98.1 | 1453300_at | Slc35d2 | 1199.2 | 912.8 | 421.2 |
| 1451486_at | Slc46a3 | 2002.6 | 343.5 | 392.6 | 1433898_at | Slc25a30 | 1114.3 | 1358.5 | 556.7 |
| 1447227_at | Slc40a1 | 953.7 | 116.1 | 22.8 | 1424735_at | Slc25a25 | 677.4 | 578.3 | 766.9 |
| 1433751_at | Slc39a10 | 36219.1 | 1987.1 | 3932.0 | 1452717_at | Slc25a24 | 3275.4 | 2221.4 | 2520.1 |
| 1454622_at | Slc38a5 | 5337.6 | 22.5 | 396.4 | 1450395_at | Slc22a5 | 2034.3 | 1478.3 | 1606.7 |
| 1418706_at | Slc38a3 | 8758.2 | 102.4 | 10.7 | 1429726_at | Slc16a9 | 6134.7 | 164.5 | 4913.5 |
| 1429154_at | Slc35f2 | 3223.2 | 5.8 | 46.6 | 1418257_at | Slc12a7 | 1271.8 | 1519.8 | 2671.3 |
| 1433750_at | Slc31a1 | 3655.1 | 1091.9 | 953.6 | 1451227_a_at | Slc10a3 | 1095.1 | 1157.3 | 620.5 |
| 1436164_at | Slc30a1 | 23687.8 | 3827.4 | 4884.1 | 1418774_a_at | Atp7a | 2948.3 | 2002.4 | 3669.4 |
| 1434773_a_at | Slc2a1 | 41376.6 | 267.8 | 1419.6 | 1442145_at | Atp13a3 | 534.7 | 250.3 | 2057.8 |
| 1424211_at | Slc25a33 | 936.5 | 332.8 | 188.8 | 1455083_at | Atp11c | 4660.2 | 3234.7 | 3048.5 |
| 1453149_at | Slc25a32 | 1131.7 | 368.3 | 253.8 | 1451388_a_at | Atp11b | 5863.6 | 1539.2 | 2495.8 |
| 1423109_s_at | Slc25a20 | 1082.6 | 408.0 | 515.9 | 1436544_at | Atp10d | 3373.8 | 1319.2 | 2658.9 |
| 1435418_at | Slc22a8 | 9605.2 | 12.4 | 12.6 | |||||
| 1448299_at | Slc1a1 | 8321.1 | 61.9 | 521.2 | |||||
| 1436417_at | Slc19a3 | 9686.6 | 38.8 | 91.6 | |||||
| 1426082_a_at | Slc16a4 | 1724.0 | 18.3 | 50.8 | |||||
| 1418445_at | Slc16a2 | 3708.5 | 689.0 | 232.1 | |||||
| 1415802_at | Slc16a1 | 32259.1 | 544.3 | 217.5 | |||||
| 1447851_x_at | Atp10a | 16691.2 | 52.4 | 676.0 | |||||
| 1422906_at | Abcg2 | 24521.4 | 2038.1 | 4007.5 | |||||
| 1443870_at | Abcc4 | 17102.3 | 2475.8 | 872.6 | |||||
| 1419759_at | Abcb1a | 42011.6 | 129.7 | 3931.4 |
A) BBB enriched transporters. Transporters from the Slc, Slco, ATP and Abc gene families with enriched expression in brain endothelial cells. Transporters identified in Table S6, all of which have at least a two-fold greater expression value in brain GFP+ than liver and lung GFP+ cells, pericyte genes excluded.
B) Endothelial transporters. Transporters expressed in multiple endothelial samples. These transporters were identified as endothelial specific in Table S2, but were not significantly enriched in brain GFP+ cells compared to either the liver GFP+ or lung GFP+ samples.
C) Pericyte enriched transporters. A list of pericyte enriched transporters from Table S3, defined as those transcripts whose GeneChip expression values have a brain vascular:endothelial ratio of ≥2.
Identification of Highly BBB-Enriched Signaling and Metabolic Pathways
The molecular signals that regulate the formation and function of the BBB remain largely unknown. We therefore took advantage of Ingenuity Pathway Analysis (IPA) software to analyze the CNS endothelial enriched transcripts to identify signaling pathways that are enriched at the BBB or in peripheral endothelial cells (Table 5a,c). We identified several pathways as being significantly associated with brain endothelial cells, including the Wnt/beta-catenin signaling pathway[36]. This pathway has been demonstrated to be specifically activated in CNS endothelial cells, and is important for regulating CNS-specific angiogenesis and inducing BBB properties including the expression of glut-1 and tight junction proteins[36], [37], [38]. The identification of the BBB inducing Wnt pathway validates this database as a method to identify pathways that may be important for BBB formation.
Table 5. Signaling cascades and metabolic pathways enriched at the BBB and peripheral endothelial cells.
| A.BBB enriched signaling pathway | P-value | B.BBB enriched metabolic pathway | P-value |
| LPS/IL-1 Mediated Inhibition of RXR Function | 1.20E-05 | Glycosphingolipid Biosynthesis - Neolactoseries | 0.004 |
| Serotonin Receptor Signaling | 0.001 | Glycine, Serine and Threonine Metabolism | 0.008 |
| Clathrin-mediated Endocytosis Signaling | 0.003 | Phenylalanine Metabolism | 0.010 |
| Dopamine Receptor Signaling | 0.008 | Nitrogen Metabolism | 0.025 |
| PXR/RXR Activation | 0.025 | Glycolysis/Gluconeogenesis | 0.027 |
| Regulation of IL-2 Expression in Activated and Anergic T Lymphocytes | 0.039 | Synthesis and Degradation of Ketone Bodies | 0.031 |
| Role of Wnt/GSK-3beta Signaling in the Pathogenesis of Influenza | 0.039 | Histidine Metabolism | 0.035 |
| Wnt/beta-catenin Signaling | 0.040 | Methionine Metabolism | 0.035 |
| Type II Diabetes Mellitus Signaling | 0.049 | ||
A–B) BBB enriched pathways. Ingenuity Pathway Analysis (IPA) software was utilized to identify signaling cascades (A) and metabolic pathways (B) that are significantly enriched at the BBB. For this analysis BBB enriched transcripts identified in Table S6 were utilized. Pathways in which p-value <0.05 are displayed.
C–D) Peripheral vascular enriched pathways. IPA software was utilized to identify signaling cascades (C) and metabolic pathways (D) that are significantly enriched in peripheral vessels compared to brain vessels. For this analysis peripheral enriched transcripts identified in Table S7 were utilized. Pathways in which p-value <0.05 are displayed.
The “LPS/IL-1 mediated inhibition of retinoic x receptor (RXR) function” pathway was enriched with the highest confidence level in the CNS endothelial cells compared with liver and lung endothelial cells. The core of this pathway is a transcriptional cassette activated by the nuclear receptor RXRalpha, which can also be inhibited by a cytoplasmic kinase cascade initiated by LPS, IL-1 or TNFalpha signals. Many transcripts whose expression is turned on by RXRalpha are specifically expressed by CNS endothelial cells. These transcripts include Oatp2, Mrp4, Mdr1, Aldh, Pltp, Cpt, Hmgcs and Alas1, some of which are BBB enriched transporters presented in Table 4. Remarkably, the “LPS/IL-1 mediated inhibition of RXR function” pathway is also enriched in peripheral endothelial cells compared with brain endothelial cells. However, the transcripts enriched in peripheral endothelial cells encode molecules which inhibit RXRalpha function, including secreted molecules, membrane receptors and cytoplasmic signaling components (Lbp, IL-1, Cd14, Myd6b, Nfp). This suggests that a fundamental difference between CNS and peripheral endothelial cells is that RXRalpha mediated transcription is activated in CNS endothelial cells and inhibited in peripheral endothelial cells. In addition we have identified that neurotransmitter signaling, regulation of IL-2, and other pathways are enriched in brain endothelial cells. These findings strongly suggest in addition to Wnt signaling, RXRalpha, and other signaling may impart BBB specific properties to CNS endothelial cells.
IPA software was also used to identify metabolic pathways enriched at the BBB or in peripheral endothelial cells (Table 5b,d). Glycolysis/gluconeogenesis and amino acid metabolism are enriched at the BBB, suggesting that brain endothelial cells may be intimately involved in the production of energy metabolites and amino acids for neurons, a function that has been thought to be uniquely served by astrocytes.
Prediction of Novel Cell-Cell Signaling Interactions Between Vascular and Parenchymal Cell Types
From the endothelial and pericyte transcriptome data sets, we next generated predictions about the molecular nature of the cell-cell interactions by which endothelial cells and pericytes communicate. These interactions may be critical for vascular contraction, remodeling, differentiation, and permeability. In fact of the top 50 pericyte enriched genes, 40 are predicted by David Bionformatics or Mouse Genome Informatics (MGI) to encode transmembrane or secreted proteins, and thus are potentially important for endothelial-pericyte signaling. Using this analysis we identified previously known endothelial-pericyte signals, thus validating this approach. For instance, endothelial secreted PDGF-BB regulates the proliferation and survival of CNS pericytes through PDGFRbeta as evidenced by the complete lack of CNS pericytes in PDGFB or PDGFRbeta deficient mice [39], [40]. In our data set, PDGFB is enriched in endothelial cells whereas PDGFRbeta is specifically expressed by pericytes. Other potential endothelial-to-pericyte signals include endothelial-derived Jagged2 or Dll4 activation of pericyte-expressed Notch3, or endothelial-derived sema3c, sema3g, sema4c and sema7a signaling to pericyte-expressed plexin B2, Plxdc1 and Plxdc2. Putative pericyte-to-endothelial signals include pericyte derived TGFbeta superfamily members bmp5 and nodal, and endothelial cell expressed activin receptors (Acvr1, Acvrl1) and bmp receptors (Bmpr2). In addition we identified angiotensin I converting enzyme 2 (Ace2) as highly expressed by pericytes, and thus likely to be involved in generating ligands for agtrl1 receptors made by endothelial cells. These ligand-receptor pairs are thus candidates to play important roles in brain angiogenesis and BBB development.
We further looked for novel vascular-parenchymal cell interactions. Recent work suggests that neural-vascular cellular interactions are important for the development of the nervous system. Comparing the vascular enriched genes with those expressed by neurons and glia [14] can lead to the discovery of signaling pathways important for the development of the CNS. Astrocytes possess long processes whose endfeet ensheath blood vessels. This close association has been implicated in regulating angiogenesis, vascular contraction and blood-brain barrier permeability [41]. Among astrocyte enriched genes defined by Cahoy et al (2008), 798 of 2618 are predicted by DAVID bioinformatics to be membrane or secreted. Of these, many have putative receptors expressed by endothelial cells. For instance putative astrocytes/endothelial interactions may be mediated through ApoE/Lrp8; BMP7/ActrIIa; AGT/Agtrl1; Wnt7a,Wnt7b/Fzd4,Fzd6; ang1/tie-2, and VEGFa/Flt1, Flt, Kdr. Blood vessels also interact with neural stem cells in the developing and adult nervous system, providing a “vascular niche” that maintains stem cells, promotes their division, and expands neurogenesis [42]. Along these lines, we identified a high level of expression of potential oligodendrocyte precursor cell (OPC) mitogens within endothelial cells including cxcl12/sdf1, kitl, csf1, and PDGFB, with cognate receptors being highly expressed by OPCs, respectively cmkor1/cxcr7, c-kit, csf1r, and PDGFRalpha [43], [44], [45], [46].
Developmental Changes in BBB Gene Expression
In order to understand developmental changes that regulate angiogenesis and formation of the BBB, we used FACS to purify and then profile vascular cells (GFP+) from the cerebral cortex of Tie2GFP mice of different ages. We compared cells purified from postnatal (P2–8) mice and adult (p60–70) mice. In Table S4 and Table S5 we list the CNS vascular transcripts significantly up-regulated and down-regulated during development from postnatal to adult mice, and in Table 6 we list the signaling cascades and metabolic pathways identified by IPA that significantly change during development. Interestingly, Interferon, Prolactin, Growth Hormone, and circadian rhythm signaling are significantly up-regulated during development suggesting these pathways may regulate the maturation of the vasculature. On the other hand, integrin signaling, protein ubiquitination and hypoxia signaling are all down-regulated during development suggesting that these pathways may be involved in regulating angiogenesis.
Table 6. Developmentally regulated brain endothelial signaling cascades and metabolic pathways.
| A.Up-regulated Signaling Pathway | P-value | C.Down-regulated Signaling Pathway | P-value |
| Interferon Signaling | 1.60E-04 | Mitochondrial Dysfunction | 4.50E-05 |
| Prolactin Signaling | 3.90E-04 | Airway Pathology in Chronic Obstructive Pulmonary Disease | 0.004 |
| Growth Hormone Signaling | 0.001 | RAN Signaling | 0.004 |
| Circadian Rhythm Signaling | 0.001 | Parkinson's Signaling | 0.006 |
| Synaptic Long Term Potentiation | 0.003 | Sphingosine-1-phosphate Signaling | 0.006 |
| Phospholipase C Signaling | 0.005 | Molecular Mechanisms of Cancer | 0.007 |
| IL-3 Signaling | 0.007 | Endothelin-1 Signaling | 0.007 |
| Cholecystokinin/Gastrin-mediated Signaling | 0.007 | Integrin Signaling | 0.010 |
| Factors Promoting Cardiogenesis in Vertebrates | 0.008 | Apoptosis Signaling | 0.011 |
| Renin-Angiotensin Signaling | 0.009 | Regulation of eIF4 and p70S6K Signaling | 0.017 |
| Molecular Mechanisms of Cancer | 0.009 | p70S6K Signaling | 0.017 |
| NRF2-mediated Oxidative Stress Response | 0.010 | Cell Cycle Regulation by BTG Family Proteins | 0.018 |
| GNRH Signaling | 0.011 | mTOR Signaling | 0.018 |
| JAK/Stat Signaling | 0.011 | Protein Ubiquitination Pathway | 0.019 |
| IGF-1 Signaling | 0.012 | Cytotoxic T Lymphocyte-mediated Apoptosis of Target Cells | 0.029 |
| Thrombopoietin Signaling | 0.017 | Glioma Invasiveness Signaling | 0.030 |
| Huntington's Disease Signaling | 0.017 | Hypoxia Signaling in the Cardiovascular System | 0.032 |
| Hypoxia Signaling in the Cardiovascular System | 0.021 | IL-8 Signaling | 0.032 |
| Role of Macrophages, Fibroblasts and Endothelial Cells in Arthritis | 0.025 | Production of NO and Reactive Oxygen Species in Macrophages | 0.035 |
| FLT3 Signaling in Hematopoietic Progenitor Cells | 0.026 | Death Receptor Signaling | 0.042 |
| BMP signaling pathway | 0.026 | Induction of Apoptosis by HIV1 | 0.045 |
| EGF Signaling | 0.029 | Fcγ Receptor-mediated Phagocytosis in Macrophages | 0.049 |
| p53 Signaling | 0.030 | HMGB1 Signaling | 0.049 |
| Glioblastoma Multiforme Signaling | 0.034 | ||
| Cardiomyocyte Differentiation via BMP Receptors | 0.035 | ||
| Endoplasmic Reticulum Stress Pathway | 0.035 | ||
| VDR/RXR Activation | 0.036 | ||
| Corticotropin Releasing Hormone Signaling | 0.036 | ||
| Retinoic acid Mediated Apoptosis Signaling | 0.039 | ||
| Human Embryonic Stem Cell Pluripotency | 0.041 | ||
| p38 MAPK Signaling | 0.041 | ||
| Prostate Cancer Signaling | 0.044 | ||
| Ephrin Receptor Signaling | 0.046 | ||
| Melanocyte Development and Pigmentation Signaling | 0.046 | ||
| Aryl Hydrocarbon Receptor Signaling | 0.046 | ||
| Oncostatin M Signaling | 0.048 | ||
| CNTF Signaling | 0.048 | ||
| Activation of IRF by Cytosolic Pattern Recognition Receptors | 0.048 | ||
| FGF Signaling | 0.049 |
A–D) IPA software analysis of developmentally regulated (2-fold up or down) transcripts, identifies signaling cascades (A,C) and metabolic pathways (B,D) that are up-regulated (A,B) or down regulated (C,D) during development. For this analysis developmentally regulated pathways identified in Table S4 (up-regulated) and Table S5 (down-regulated) were utilized. Pathways with p-value<0.05 are listed.
Discussion
Identification of the Blood-Brain Barrier Transcriptome
In this paper we provide a new resource for understanding gene expression in the developing and adult CNS vasculature. Previous gene profiling studies of brain vessels had several limitations [9], [10], [11], [47] including use of whole vessel fractions, which contain many different cell types, and the lack of elucidation of peripheral endothelial profiles for comparison. We have combined FACS purification with GeneChip analysis to generate a transcriptional profile of highly purified endothelial cells from the CNS, and through comparison with liver and lung endothelial cells we have generated a comprehensive list of transcripts enriched at the BBB. These transcripts encode tight junction molecules, transporters, metabolic enzymes, signaling cascades, and proteins of unknown function, that together form the highly specialized interface between the blood and the brain. This represents a novel resource for three reasons. First, we separate CNS endothelial cells from pericytes/VSM, and thus generate a transcriptional profile of endothelial cells, the key cells that form the BBB. Second, we compare brain endothelial cells with peripheral endothelial cells isolated from the liver and lung, identifying transcripts which are specific to barrier forming brain endothelial cells. Finally, we have isolated cells from different ages and thus can identify developmental changes in the BBB. The BBB transcriptome database should be highly useful for understanding the molecular control of CNS angiogenesis, BBB permeability, and the cell-cell interactions in the developing CNS.
Identification of Candidates Transcripts that Control Vascular Permeability
A hallmark of the BBB is that CNS endothelial cells are held together by high electrical resistance tight junctions [17], [48]. Of the best characterized tight junction molecules expressed at the BBB, claudin 5, occludin, ZO-1 and ZO-2, only occludin is specifically expressed at the blood-brain barrier and not by non-barrier forming vessels [18], [22]. The fact that occludin deficient mice still form a functional BBB suggests that the expression of this molecule does not impart BBB function [49]. It therefore remains a mystery as to what causes CNS endothelial cells to form these high electrical resistance tight junctions. In this study we have identified several tight junction molecules whose expression is enriched at the BBB, including marveld2, cingulin-like-1, and pard3, that may play a crucial role in the formation of BBB tight junctions. Our data sets will also be useful to identify novel molecules not previously known to be associated with tight junction proteins, which may be important for BBB integrity. In addition to the highly specialized vasculature of the blood-brain barrier, there is great heterogeneity in the structure and function of vessels in all tissues [29], [30]. By analyzing the relative levels of transcripts expressed by the endothelial cells in different tissues we can generate hypotheses as to the relevance of different genes for the tissue specific functions of each vessel, including permeability, leukocyte trafficking, hemostasis, angiogenesis, immunity, and vasomotor tone.
Signaling Pathways Enriched at the BBB
Transplantation experiments have established that the brain environment signals brain endothelial cells to form the BBB [5], but the identity of these signals are largely unknown. In particular, we have identified transcripts from a number of cascades that are enriched in the vasculature of the CNS including Wnt/beta-catenin, RXRalpha and other signaling pathways. The identification of the Wnt pathway, a known BBB inducing signaling pathway, validates this database as a method to identify pathways that may be important for BBB formation.
RXRalpha is a nuclear receptor which activates the transcription of a series of molecules. The activity of RXRalpha can be inhibited by a cytoplasmic kinase cascade initiated by LPS, IL-1 or TNFalpha signals [50]. Remarkably, genes in the inhibitory cassette of this pathway, including Lbp, IL-1, Cd14, Myd6b, Nfp are enriched in peripheral endothelial cells, whereas transcripts downstream of RXRalpha are enriched in CNS endothelial cells. Therefore, a fundamental difference between CNS and peripheral endothelial cells is the expression of the RXRalpha transcriptional cassette in CNS endothelial cells, and its suppression in peripheral endothelial cells. RXRalpha is a retinoic acid nuclear receptor which raises the question: Does retinoic acid (RA) regulate the BBB? This seems possible as addition of RA to endothelial cell lines in vitro increases the expression of BBB specific molecules Pgp and gamma-glutamyl transpeptidase [51], [52]. This matches our result that many of the RXRalpha regulated transcripts are the BBB enriched transporters, including Oatp2, Mdr1 and Mrp4. Interestingly, this pathway is inhibited by immune system modulating cytokines (IL-1) and bacterial LPS. Perhaps infection and inflammation alter the levels of BBB specific molecules through regulation of RXRalpha activity.
Targeting Tumor Angiogenesis
In addition to identifying novel barrier forming molecules, our data also identifies transcripts that may be important for angiogenesis and/or vascular pathfinding. This is evident as we have identified integrins and semaphorins, both of which have been implicated in the formation of new vessels [53], [54], [55]. Identifying such molecules may be important for developing treatments for brain tumors, as formation of new vessels is a critical step in tumor progression. Disruption of tumor angiogenesis, without harming mature vessels, is a powerful technique for limiting tumor growth [56], [57]. To accomplish this, it is key to identify potential targets found on angiogenic vessels, that are absent from mature vessels or parenchymal tissue. In this resource we have identified transcripts whose expression level changes from developmental stages where robust angiogenesis is occurring, to adult animals where angiogenesis is largely completed. Potential targets include prion protein doublet (prnd), madcam1, tachykinin receptor 3, Slc7a8, and Cxcr4. Each of these transcripts is enriched in postnatal brain endothelial cells as compared with adult brain, liver and lung endothelial cells, and adult CNS parenchyma.
In summary, we have generated a comprehensive data set describing the transcriptome of the BBB. This will provide a valuable resource for understanding the development and function of this crucial barrier, as well as its role modulating CNS function.
Supporting Information
Complete Excel spreadsheet. The Excel spreadsheet containing the comprehensive list of probe sets from the mouse 430-2 GeneChip, with expression values for adult brain GFP−, adult brain GFP+, adult brain GFP+PDGFRbeta−, liver GFP+, lung GFP+, and postnatal brain GFP+ samples. Each expression value is the average of at least three biological replicates.
(9.96 MB XLS)
Endothelial specific genes. A data set containing probe sets from the mouse 430–2 GeneChip statistically enriched in CNS endothelial cells compared with CNS parenchyma as identified by probe sets with GFP+PDGFRbeta−:GFP− >2 with expression value in GFP+PDGFRbeta−>200. SAM algorithms were utilized to identify significance.
(0.87 MB XLS)
Pericyte specific genes. A data set containing probe sets from the mouse 430–2 GeneChip statistically enriched in CNS pericytes compared with CNS endothelial cells as identified by probe sets with GFP+:GFP+PDGFRbeta−>2 with expression value in GFP+>200. SAM algorithms were utilized to identify significance.
(0.10 MB XLS)
Developmentally up-regulated CNS vascular genes. A data set containing probe sets from the mouse 430–2 GeneChip statistically up-regulated in CNS vessels during development as identified by probe sets with adult GFP+:postnatal>2 with expression value in adult GFP+>200. Pericyte genes were eliminated from this dataset. SAM algorithms were utilized to identify significance.
(0.43 MB XLS)
Developmentally down-regulated CNS vascular genes. A data set containing probe sets from the mouse 430–2 GeneChip statistically down-regulated in CNS vessels during development as identified by probe sets with adult GFP+:postnatal<2 with expression value in postnatal GFP+>200. Pericyte genes were eliminated from this dataset. SAM algorithms were utilized to identify significance.
(0.40 MB XLS)
BBB enriched genes. A data set containing probe sets from the mouse 430–2 GeneChip statistically enriched in CNS endothelial cells compared with those from the CNS parenchyma as identified by probe sets with BrainGFP+:LiverGFP+>2 and BrainGFP+:LungGFP+>2 with expression value in BrainGFP+>200. Additionally separate work sheets are presented for data sets containing BrainGFP+:LiverGFP+>2 with expression value in BrainGFP+>200, and BrainGFP+:LungGFP+>2 with expression value in BrainGFP+>200. Pericyte genes were eliminated from each dataset. SAM algorithms were utilized to identify significance.
(1.01 MB XLS)
Peripheral endothelial enriched genes. A data set containing probe sets from the mouse 430–2 GeneChip statistically enriched in peripheral endothelial cells compared with those from the CNS as identified by probe sets with LiverGFP+:BrainGFP+ >2 and LungGFP+:BrainGFP+ >2 with expression value in LiverGFP+>200 and LungGFP+>200. SAM algorithms were utilized to identify significance.
(0.28 MB XLS)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the National Institute of Neurological Disease (R01 NS045621; B.A.B.), http://www.ninds.nih.gov/; the Myelin Repair Foundation (B.A.B, R.D.), http://www.myelinrepair.org/; the National Multiple Sclerosis Society RG3936A7 (B.A.B.), http://www.nationalmssociety.org/index.aspx; and the American Heart Association Graduate Fellowship (R.D.), http://www.americanheart.org/presenter.jhtml?identifier=1200000. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Zlokovic BV. The blood-brain barrier in health and chronic neurodegenerative disorders. Neuron. 2008;57:178–201. doi: 10.1016/j.neuron.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Ballabh P, Braun A, Nedergaard M. The blood-brain barrier: an overview: structure, regulation, and clinical implications. Neurobiol Dis. 2004;16:1–13. doi: 10.1016/j.nbd.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 3.Pardridge WM. Blood-brain barrier delivery. Drug Discov Today. 2007;12:54–61. doi: 10.1016/j.drudis.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 4.Rubin LL, Staddon JM. The cell biology of the blood-brain barrier. Annu Rev Neurosci. 1999;22:11–28. doi: 10.1146/annurev.neuro.22.1.11. [DOI] [PubMed] [Google Scholar]
- 5.Stewart PA, Wiley MJ. Developing nervous tissue induces formation of blood-brain barrier characteristics in invading endothelial cells: a study using quail--chick transplantation chimeras. Dev Biol. 1981;84:183–192. doi: 10.1016/0012-1606(81)90382-1. [DOI] [PubMed] [Google Scholar]
- 6.Janzer RC, Raff MC. Astrocytes induce blood-brain barrier properties in endothelial cells. Nature. 1987;325:253–257. doi: 10.1038/325253a0. [DOI] [PubMed] [Google Scholar]
- 7.Weidenfeller C, Svendsen CN, Shusta EV. Differentiating embryonic neural progenitor cells induce blood-brain barrier properties. J Neurochem. 2007;101:555–565. doi: 10.1111/j.1471-4159.2006.04394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rubin LL, Hall DE, Porter S, Barbu K, Cannon C, et al. A cell culture model of the blood-brain barrier. J Cell Biol. 1991;115:1725–1735. doi: 10.1083/jcb.115.6.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li JY, Boado RJ, Pardridge WM. Blood-brain barrier genomics. J Cereb Blood Flow Metab. 2001;21:61–68. doi: 10.1097/00004647-200101000-00008. [DOI] [PubMed] [Google Scholar]
- 10.Li JY, Boado RJ, Pardridge WM. Rat blood-brain barrier genomics. II. J Cereb Blood Flow Metab. 2002;22:1319–1326. doi: 10.1097/01.WCB.0000040944.89393.0f. [DOI] [PubMed] [Google Scholar]
- 11.Enerson BE, Drewes LR. The rat blood-brain barrier transcriptome. J Cereb Blood Flow Metab. 2006;26:959–973. doi: 10.1038/sj.jcbfm.9600249. [DOI] [PubMed] [Google Scholar]
- 12.Motoike T, Loughna S, Perens E, Roman BL, Liao W, et al. Universal GFP reporter for the study of vascular development. Genesis. 2000;28:75–81. doi: 10.1002/1526-968x(200010)28:2<75::aid-gene50>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 13.Huettner JE, Baughman RW. Primary culture of identified neurons from the visual cortex of postnatal rats. J Neurosci. 1986;6:3044–3060. doi: 10.1523/JNEUROSCI.06-10-03044.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaeren-Wiemers N, Gerfin-Moser A. A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: in situ hybridization using digoxigenin-labelled cRNA probes. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- 16.Macdonald JA, Murugesan N, Pachter JS. Endothelial cell heterogeneity of blood-brain barrier gene expression along the cerebral microvasculature. J Neurosci Res. 2009;88:1457–1474. doi: 10.1002/jnr.22316. [DOI] [PubMed] [Google Scholar]
- 17.Gloor SM, Wachtel M, Bolliger MF, Ishihara H, Landmann R, et al. Molecular and cellular permeability control at the blood-brain barrier. Brain Res Brain Res Rev. 2001;36:258–264. doi: 10.1016/s0165-0173(01)00102-3. [DOI] [PubMed] [Google Scholar]
- 18.Morita K, Sasaki H, Furuse M, Tsukita S. Endothelial claudin: claudin-5/TMVCF constitutes tight junction strands in endothelial cells. J Cell Biol. 1999;147:185–194. doi: 10.1083/jcb.147.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nitta T, Hata M, Gotoh S, Seo Y, Sasaki H, et al. Size-selective loosening of the blood-brain barrier in claudin-5-deficient mice. J Cell Biol. 2003;161:653–660. doi: 10.1083/jcb.200302070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee SW, Kim WJ, Choi YK, Song HS, Son MJ, et al. SSeCKS regulates angiogenesis and tight junction formation in blood-brain barrier. Nat Med. 2003;9:900–906. doi: 10.1038/nm889. [DOI] [PubMed] [Google Scholar]
- 21.Hawkins BT, Abbruscato TJ, Egleton RD, Brown RC, Huber JD, et al. Nicotine increases in vivo blood-brain barrier permeability and alters cerebral microvascular tight junction protein distribution. Brain Res. 2004;1027:48–58. doi: 10.1016/j.brainres.2004.08.043. [DOI] [PubMed] [Google Scholar]
- 22.Hirase T, Staddon JM, Saitou M, Ando-Akatsuka Y, Itoh M, et al. Occludin as a possible determinant of tight junction permeability in endothelial cells. J Cell Sci. 1997;110 ( Pt 14):1603–1613. doi: 10.1242/jcs.110.14.1603. [DOI] [PubMed] [Google Scholar]
- 23.Riazuddin S, Ahmed ZM, Fanning AS, Lagziel A, Kitajiri S, et al. Tricellulin is a tight-junction protein necessary for hearing. Am J Hum Genet. 2006;79:1040–1051. doi: 10.1086/510022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ikenouchi J, Furuse M, Furuse K, Sasaki H, Tsukita S. Tricellulin constitutes a novel barrier at tricellular contacts of epithelial cells. J Cell Biol. 2005;171:939–945. doi: 10.1083/jcb.200510043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohnishi H, Nakahara T, Furuse K, Sasaki H, Tsukita S, et al. JACOP, a novel plaque protein localizing at the apical junctional complex with sequence similarity to cingulin. J Biol Chem. 2004;279:46014–46022. doi: 10.1074/jbc.M402616200. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki A, Ohno S. The PAR-aPKC system: lessons in polarity. J Cell Sci. 2006;119:979–987. doi: 10.1242/jcs.02898. [DOI] [PubMed] [Google Scholar]
- 27.Macara IG. Par proteins: partners in polarization. Curr Biol. 2004;14:R160–162. [PubMed] [Google Scholar]
- 28.Cornford EM, Hyman S. Localization of brain endothelial luminal and abluminal transporters with immunogold electron microscopy. NeuroRx. 2005;2:27–43. doi: 10.1602/neurorx.2.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aird WC. Phenotypic heterogeneity of the endothelium: II. Representative vascular beds. Circ Res. 2007;100:174–190. doi: 10.1161/01.RES.0000255690.03436.ae. [DOI] [PubMed] [Google Scholar]
- 30.Aird WC. Phenotypic heterogeneity of the endothelium: I. Structure, function, and mechanisms. Circ Res. 2007;100:158–173. doi: 10.1161/01.RES.0000255691.76142.4a. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, Chen K, Guo L, Wu C. Characterization of PINCH-2, a new focal adhesion protein that regulates the PINCH-1-ILK interaction, cell spreading, and migration. J Biol Chem. 2002;277:38328–38338. doi: 10.1074/jbc.M205576200. [DOI] [PubMed] [Google Scholar]
- 32.Zhang M, Liu J, Cheng A, Deyoung SM, Chen X, et al. CAP interacts with cytoskeletal proteins and regulates adhesion-mediated ERK activation and motility. Embo J. 2006;25:5284–5293. doi: 10.1038/sj.emboj.7601406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Srinivasan K, Strickland P, Valdes A, Shin GC, Hinck L. Netrin-1/neogenin interaction stabilizes multipotent progenitor cap cells during mammary gland morphogenesis. Dev Cell. 2003;4:371–382. doi: 10.1016/s1534-5807(03)00054-6. [DOI] [PubMed] [Google Scholar]
- 34.Hoffman LM, Jensen CC, Kloeker S, Wang CL, Yoshigi M, et al. Genetic ablation of zyxin causes Mena/VASP mislocalization, increased motility, and deficits in actin remodeling. J Cell Biol. 2006;172:771–782. doi: 10.1083/jcb.200512115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kioka N, Ueda K, Amachi T. Vinexin, CAP/ponsin, ArgBP2: a novel adaptor protein family regulating cytoskeletal organization and signal transduction. Cell Struct Funct. 2002;27:1–7. doi: 10.1247/csf.27.1. [DOI] [PubMed] [Google Scholar]
- 36.Daneman R, Agalliu D, Zhou L, Kuhnert F, Kuo CJ, et al. Wnt/beta-catenin signaling is required for CNS, but not non-CNS, angiogenesis. Proc Natl Acad Sci U S A. 2009;106:641–646. doi: 10.1073/pnas.0805165106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liebner S, Corada M, Bangsow T, Babbage J, Taddei A, et al. Wnt/beta-catenin signaling controls development of the blood-brain barrier. J Cell Biol. 2008;183:409–417. doi: 10.1083/jcb.200806024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stenman JM, Rajagopal J, Carroll TJ, Ishibashi M, McMahon J, et al. Canonical Wnt signaling regulates organ-specific assembly and differentiation of CNS vasculature. Science. 2008;322:1247–1250. doi: 10.1126/science.1164594. [DOI] [PubMed] [Google Scholar]
- 39.Lindahl P, Hellstrom M, Kalen M, Betsholtz C. Endothelial-perivascular cell signaling in vascular development: lessons from knockout mice. Curr Opin Lipidol. 1998;9:407–411. doi: 10.1097/00041433-199810000-00004. [DOI] [PubMed] [Google Scholar]
- 40.Lindahl P, Johansson BR, Leveen P, Betsholtz C. Pericyte loss and microaneurysm formation in PDGF-B-deficient mice. Science. 1997;277:242–245. doi: 10.1126/science.277.5323.242. [DOI] [PubMed] [Google Scholar]
- 41.Abbott NJ, Ronnback L, Hansson E. Astrocyte-endothelial interactions at the blood-brain barrier. Nat Rev Neurosci. 2006;7:41–53. doi: 10.1038/nrn1824. [DOI] [PubMed] [Google Scholar]
- 42.Shen Q, Goderie SK, Jin L, Karanth N, Sun Y, et al. Endothelial cells stimulate self-renewal and expand neurogenesis of neural stem cells. Science. 2004;304:1338–1340. doi: 10.1126/science.1095505. [DOI] [PubMed] [Google Scholar]
- 43.Maysami S, Nguyen D, Zobel F, Pitz C, Heine S, et al. Modulation of rat oligodendrocyte precursor cells by the chemokine CXCL12. Neuroreport. 2006;17:1187–1190. doi: 10.1097/01.wnr.0000227985.92551.9a. [DOI] [PubMed] [Google Scholar]
- 44.Ida JA, Jr, Dubois-Dalcq M, McKinnon RD. Expression of the receptor tyrosine kinase c-kit in oligodendrocyte progenitor cells. J Neurosci Res. 1993;36:596–606. doi: 10.1002/jnr.490360512. [DOI] [PubMed] [Google Scholar]
- 45.Pringle NP, Mudhar HS, Collarini EJ, Richardson WD. PDGF receptors in the rat CNS: during late neurogenesis, PDGF alpha-receptor expression appears to be restricted to glial cells of the oligodendrocyte lineage. Development. 1992;115:535–551. doi: 10.1242/dev.115.2.535. [DOI] [PubMed] [Google Scholar]
- 46.Sawada M, Itoh Y, Suzumura A, Marunouchi T. Expression of cytokine receptors in cultured neuronal and glial cells. Neurosci Lett. 1993;160:131–134. doi: 10.1016/0304-3940(93)90396-3. [DOI] [PubMed] [Google Scholar]
- 47.Calabria AR, Shusta EV. Blood-brain barrier genomics and proteomics: elucidating phenotype, identifying disease targets and enabling brain drug delivery. Drug Discov Today. 2006;11:792–799. doi: 10.1016/j.drudis.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 48.Ueno M. Molecular anatomy of the brain endothelial barrier: an overview of the distributional features. Curr Med Chem. 2007;14:1199–1206. doi: 10.2174/092986707780597943. [DOI] [PubMed] [Google Scholar]
- 49.Saitou M, Furuse M, Sasaki H, Schulzke JD, Fromm M, et al. Complex phenotype of mice lacking occludin, a component of tight junction strands. Mol Biol Cell. 2000;11:4131–4142. doi: 10.1091/mbc.11.12.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y, Moser AH, Shigenaga JK, Grunfeld C, Feingold KR. Downregulation of liver X receptor-alpha in mouse kidney and HK-2 proximal tubular cells by LPS and cytokines. J Lipid Res. 2005;46:2377–2387. doi: 10.1194/jlr.M500134-JLR200. [DOI] [PubMed] [Google Scholar]
- 51.El Hafny B, Chappey O, Piciotti M, Debray M, Boval B, et al. Modulation of P-glycoprotein activity by glial factors and retinoic acid in an immortalized rat brain microvessel endothelial cell line. Neurosci Lett. 1997;236:107–111. doi: 10.1016/s0304-3940(97)00679-4. [DOI] [PubMed] [Google Scholar]
- 52.Lechardeur D, Schwartz B, Paulin D, Scherman D. Induction of blood-brain barrier differentiation in a rat brain-derived endothelial cell line. Exp Cell Res. 1995;220:161–170. doi: 10.1006/excr.1995.1302. [DOI] [PubMed] [Google Scholar]
- 53.Proctor JM, Zang K, Wang D, Wang R, Reichardt LF. Vascular development of the brain requires beta8 integrin expression in the neuroepithelium. J Neurosci. 2005;25:9940–9948. doi: 10.1523/JNEUROSCI.3467-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gu C, Rodriguez ER, Reimert DV, Shu T, Fritzsch B, et al. Neuropilin-1 conveys semaphorin and VEGF signaling during neural and cardiovascular development. Dev Cell. 2003;5:45–57. doi: 10.1016/s1534-5807(03)00169-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gu C, Yoshida Y, Livet J, Reimert DV, Mann F, et al. Semaphorin 3E and plexin-D1 control vascular pattern independently of neuropilins. Science. 2005;307:265–268. doi: 10.1126/science.1105416. [DOI] [PubMed] [Google Scholar]
- 56.Bertolini F, Shaked Y, Mancuso P, Kerbel RS. The multifaceted circulating endothelial cell in cancer: towards marker and target identification. Nat Rev Cancer. 2006;6:835–845. doi: 10.1038/nrc1971. [DOI] [PubMed] [Google Scholar]
- 57.Noonan DM, Benelli R, Albini A. Angiogenesis and cancer prevention: a vision. Recent Results Cancer Res. 2007;174:219–224. doi: 10.1007/978-3-540-37696-5_19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complete Excel spreadsheet. The Excel spreadsheet containing the comprehensive list of probe sets from the mouse 430-2 GeneChip, with expression values for adult brain GFP−, adult brain GFP+, adult brain GFP+PDGFRbeta−, liver GFP+, lung GFP+, and postnatal brain GFP+ samples. Each expression value is the average of at least three biological replicates.
(9.96 MB XLS)
Endothelial specific genes. A data set containing probe sets from the mouse 430–2 GeneChip statistically enriched in CNS endothelial cells compared with CNS parenchyma as identified by probe sets with GFP+PDGFRbeta−:GFP− >2 with expression value in GFP+PDGFRbeta−>200. SAM algorithms were utilized to identify significance.
(0.87 MB XLS)
Pericyte specific genes. A data set containing probe sets from the mouse 430–2 GeneChip statistically enriched in CNS pericytes compared with CNS endothelial cells as identified by probe sets with GFP+:GFP+PDGFRbeta−>2 with expression value in GFP+>200. SAM algorithms were utilized to identify significance.
(0.10 MB XLS)
Developmentally up-regulated CNS vascular genes. A data set containing probe sets from the mouse 430–2 GeneChip statistically up-regulated in CNS vessels during development as identified by probe sets with adult GFP+:postnatal>2 with expression value in adult GFP+>200. Pericyte genes were eliminated from this dataset. SAM algorithms were utilized to identify significance.
(0.43 MB XLS)
Developmentally down-regulated CNS vascular genes. A data set containing probe sets from the mouse 430–2 GeneChip statistically down-regulated in CNS vessels during development as identified by probe sets with adult GFP+:postnatal<2 with expression value in postnatal GFP+>200. Pericyte genes were eliminated from this dataset. SAM algorithms were utilized to identify significance.
(0.40 MB XLS)
BBB enriched genes. A data set containing probe sets from the mouse 430–2 GeneChip statistically enriched in CNS endothelial cells compared with those from the CNS parenchyma as identified by probe sets with BrainGFP+:LiverGFP+>2 and BrainGFP+:LungGFP+>2 with expression value in BrainGFP+>200. Additionally separate work sheets are presented for data sets containing BrainGFP+:LiverGFP+>2 with expression value in BrainGFP+>200, and BrainGFP+:LungGFP+>2 with expression value in BrainGFP+>200. Pericyte genes were eliminated from each dataset. SAM algorithms were utilized to identify significance.
(1.01 MB XLS)
Peripheral endothelial enriched genes. A data set containing probe sets from the mouse 430–2 GeneChip statistically enriched in peripheral endothelial cells compared with those from the CNS as identified by probe sets with LiverGFP+:BrainGFP+ >2 and LungGFP+:BrainGFP+ >2 with expression value in LiverGFP+>200 and LungGFP+>200. SAM algorithms were utilized to identify significance.
(0.28 MB XLS)