Abstract
Background
Multicellular organisms are characterized by a remarkable diversity of morphologically distinct and functionally specialized cell types. Transgenic techniques for the manipulation of gene expression in specific cellular populations are highly useful for elucidating the development and function of these cellular populations. Given notable similarities in developmental gene expression between pancreatic β-cells and serotonergic neurons, we examined the pattern of Cre-mediated recombination in the nervous system of a widely used mouse line, Pdx1-cre (formal designation, Tg(Ipf1-cre)89.1Dam), in which the expression of Cre recombinase is driven by regulatory elements upstream of the pdx1 (pancreatic-duodenal homeobox 1) gene.
Methods
Single (hemizygous) transgenic mice of the pdx1-creCre/0 genotype were bred to single (hemizygous) transgenic reporter mice (Z/EG and rosa26R lines). Recombination pattern was examined in offspring using whole-mount and sectioned histological preparations at e9.5, e10.5, e11.5, e16.5 and adult developmental stages.
Results
In addition to the previously reported pancreatic recombination, recombination in the developing nervous system and inner ear formation was observed. In the central nervous system, we observed a highly specific pattern of recombination in neuronal progenitors in the ventral brainstem and diencephalon. In the rostral brainstem (r1-r2), recombination occurred in newborn serotonergic neurons. In the caudal brainstem, recombination occurred in non-serotonergic cells. In the adult, this resulted in reporter expression in the vast majority of forebrain-projecting serotonergic neurons (located in the dorsal and median raphe nuclei) but in none of the spinal cord-projecting serotonergic neurons of the caudal raphe nuclei. In the adult caudal brainstem, reporter expression was widespread in the inferior olive nucleus. In the adult hypothalamus, recombination was observed in the arcuate nucleus and dorsomedial hypothalamus. Recombination was not observed in any other region of the central nervous system. Neuronal expression of endogenous pdx1 was not observed.
Conclusions
The Pdx1-cre mouse line, and the regulatory elements contained in the corresponding transgene, could be a valuable tool for targeted genetic manipulation of developing forebrain-projecting serotonergic neurons and several other unique neuronal sub-populations. These results suggest that investigators employing this mouse line for studies of pancreatic function should consider the possible contributions of central nervous system effects towards resulting phenotypes.
Background
The development of methods for the experimental manipulation of gene expression in vivo has revolutionized the study of biology. Transgenes which drive expression of recombinases within specific cell types and/or at specific developmental time points are valuable tools for understanding the development and physiology of organ systems in vivo [1]. One such system, the mammalian brain, is a remarkably complex and heterogeneous structure comprised of many highly specialized and often rare cell types. Serotonergic neurons, which comprise a tiny fraction of all neurons in the mammalian brain, play an important and unique role in many physiological functions, including the regulation of affect in humans [2]. These neurons are themselves anatomically and functionally diverse, although the molecular, developmental and physiological basis for this diversity is not completely understood [2,3]. Recently, the advent of transgenic methods to express recombinases in all or subsets of serotonergic neurons has provided new insights into the diverse origins and functions of these neurons [4-6].
Serotonergic neurons and pancreatic insulin-producing β-cells exhibit a remarkably similar and specific cascade of transcription factor expression during development, involving the expression of nkx2.2, lmx1b, and nkx6.1 [7,8]. In the pancreas, pdx1, a homeodomain transcription factor, plays a critical role in specifying the fate of the early pancreatic primordium and, later in development, is required for successful β-cell development [9]. We hypothesized that regulatory elements which control pdx1 expression might be active in the developing brain and might be applied to genetically target serotonergic neurons and/or other neuronal cell types. We therefore examined the developmental pattern of Cre-mediated recombination in the nervous system using a widely used mouse line, Pdx1-cre (formal designation, Tg(Ipf1-cre)89.1Dam) [10-12]. This mouse line has been employed in at least 30 published studies, as it exhibits robust recombination in the developing endocrine pancreas [13,10-41]. Using two Cre reporter lines, Z/EG (Tg(CAG-Bgeo/GFP)21Lbe) and rosa26R (Gt(ROSA)26Sortm1Sor) [42,43,12], we found that that Pdx1-cre also exhibits developmental recombination in the inner ear; in rostral serotonergic neurons; in the hypothalamus; and in non-serotonergic neurons of the caudal hindbrain.
Materials and methods
Mice
Strain information is summarized in Table 1. Pdx1-cre and Z/EG mouse lines were maintained as independent colonies of hemizygous transgenic mice; the rosa26R mouse line was maintained as homozygous mutant mice. Strain background was mixed for all lines. Pdx1-cre mice were kindly provided by D. Melton; Z/EG and rosa26R mice were obtained from Jackson Labs. To generate experimental animals, transgenic hemizygous mice from the Pdx1-cre line (genotype pdx1-creCre/0) were bred with hemizygous transgenic mice from the Z/EG line (ZegGFP/0) or homozygous transgenic mice from the rosa26R line (rosa26LacZ/LacZ). Offspring genotypes were obtained in accord with expected Mendelian ratios. Offspring of the following genotypes were used for analysis: pdx1-creCre/0; ZegGFP/0 (experimental), pdx1-cre0/0; ZegGFP/0 (control); pdx1-creCre/0; rosa26LacZ/+ (experimental) and pdx1-cre0/0; rosa26LacZ/+ (control). Mice were housed on a 12-hr light-dark cycle in a controlled climate and were fed ad lib with Purina LabDiet 5053 mouse chow. All studies involving mice were approved by the UCSF Institutional Animal Care and Use Committee.
Table 1.
Transgenic mouse lines and relevant genotypes employed
| Formal designation | Common designation | Transgene design | Trans-gene insertion | Purpose | WT allele | Trans-genic allele | Geno-types analyzed | Initial reference | MGI ID |
|---|---|---|---|---|---|---|---|---|---|
| Tg(Ipf1-cre)89.1Dam | Pdx1-cre | 5.5-kb portion of the mouse pdx1 promoter fused to cre cassette | Random insertion (pro-nuclear injection | Expression of Cre is driven by regulatory elements which regulate pdx1 expression | 0 | Cre | Cre/0 (hemi-zygote); 0/0 (wild-type control) | Develop-ment 2002, 129(10):2447-2457 | 2684317 |
| Tg(CAG-Bgeo/GFP) 21Lbe |
Z/EG | Ubiquitous recombinant promoter, followed by LoxP-flanked LacZ cassette, followed by GFP cassette | Random insertion with screening for high expression in ES cells (ES cell electro-poration) | When cre is expressed in a cell, LacZ cassette is excised, leading to GFP expression in that cell and all daughter cells | 0 | GFP | GFP/0 (hemi-zygote) | Genesis 2000, 28(3-4):147-155 | 3046177 |
| Gt(ROSA)26 Sortm1Sor |
Rosa26R | LoxP-flanked stop cassette followed by LacZ cassette | Targeted to the ubiquitously expressing Gt(ROSA)26Sor locus | When cre is expressed in a cell, stop cassette is excised, leading to LacZ expression in that cell and all daughter cells | + | LacZ | LacZ/+ (hetero-zygote) | Nature Genetics 1999, 21(1):70-71 | 1861932 |
Genotyping
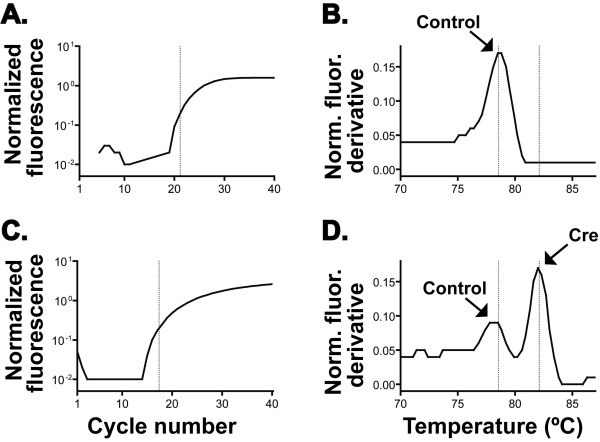
Ear punches or embryonic tails were digested in strip tubes with 0.05 U proteinase K (03115887001, Roche) in 50 μL of DirectPCR Lysis Reagent (402-E, Viagen Biotech) diluted with 50 μL water. 5 μL PCRs were performed using SYBR GreenER PCR mix (Invitrogen), primers (concentration depends on the assay, generally 200 nM) and 0.15 μL of heat-inactivated genomic DNA solution. Thermal cycling was performed on an ABI 7300 instrument with SYBR detection as follows: 95°C for 10 min; 95°C for 15 s followed by 60°C for 1 min (40 cycles); 95°C for 15 s; 60°C for 15 s; followed by a melting curve step with a 2% ramp rate from 60°C to 95°C. Allele-specific PCR products were identified using melting curve analysis as described in Results and Figure 1. Primer sequences and concentrations for assays are provided in Table 2.
Figure 1.
Transgenic mouse genotyping using multiplex allele-specific PCR and melting curve analysis. PCR was performed in an optical cycler (ABI 7300) using 1-10 ng genomic DNA from mice of the indicated genotypes and a reagent mix containing SYBR GreenER. Amplification plots (A, C) and melting curves (B, D) are shown. Primers were designed to amplify 2 specific products: a genomic control product, generated from any genomic mouse template; and a transgene-specific product, generated only from genomic templates containing a cre transgene. Normalized fluorescence (y-axis, A &C) is the baseline-subtracted ratio of SYBR signal to ROX (passive reference dye) signal during amplification cycling (A, C). Normalized fluorescence derivative (y-axis, B & D) is the 2nd derivative of normalized fluorescence during the melting curve step. Dotted lines (A, C) indicate cycle threshold. A & B. Genomic DNA from a wild-type mouse; note robust amplification of the genomic control product with a single melting peak (allowing the distinction of a negative result from a failed PCR). C & D. Genomic DNA from a pdx1-creCre/0 mouse; note robust amplification with 2 distinct melting peaks corresponding to the control and cre-specific products. Arrows indicate presence of the genomic control product (Control) and the transgene-specific product (Cre).
Table 2.
Primer sequences and reaction conditions for gel-free genotyping assays
| Assay name | Target locus | Primer 1 | Conc. (nM) | Primer 2 | Conc. (nM) | Primer 3 | Conc. (nM) | Primer 4 | Conc. (nM) | Expected results |
|---|---|---|---|---|---|---|---|---|---|---|
| Z/EG | Any transgene generated using the pCAGG construct (e.g., Z/EG line) | TCGA TGCA GGAT AACTT CGT |
400 | GGT ACC GTC GACT GCA GAAT |
400 | AGC AGC AGG CAG GGC TTT |
50 | GTCT GGA CAC GGG AGC ACTT |
50 | Primers 3 & 4 generate a single peak in all samples (control product, gdf); primers 1 & 2 generate an additional, lower Tm peak (transgene-specific product) in ZegGFP/0 or ZegGFP/GFP samples. |
| Cre | Any transgene containing cre (e.g., Pdx1-cre line) | ACATT TGGG CCAG CTAAA CAT |
200 | CGG CATC AAC GTTT TCTT TT |
200 | GGC GAG AGC AGA GTGT GGA T |
200 | AAGT CGG CAG GCA CAG GAG |
200 | Primers 3 & 4 generate a single peak in all samples (control product, k17); primers 1 & 2 generate an additional, higher Tm peak (transgene-specific product) in any sample with a cre transgene. |
| PdxCre | cre fused to 5' regulatory region of pdx1 gene (Pdx1-cre line) | TAAG GCCT GGCT TGTA GCTC |
200 | ACC GGT AATG CAG GCA AAT |
200 | AGC AGC AGG CAG GGC TTT |
30 | GTCT GGA CAC GGG AGC ACTT |
30 | Primers 3 & 4 generate a single peak in all samples (control product); primers 1 & 2 generate an additional, lower Tm peak (transgene-specific product) in pdx1-creCre/0 or pdx1-creCre/Cre samples. |
| Rosa26 | Any rosa26 allele targeted using a standard targeting allele (e.g., Rosa26R line) | GCGC GCGC GCGT GATC TGCA ACTC CAGT CTTTC |
200 | GCG CGC GCG CGC GCG CGC GCC ACAC CAG GTTA GCC TTTA AGC |
200 | GAC AGG ATAA GTAT GAC ATCA TCAA GG |
200 | Primers 1 & 2 generate a single peak in samples containing the wild-type rosa26 allele; primers 2 & 3 generate a lower Tm peak (transgene-specific product) in samples containing a targeted allele; both peaks are observed in heterozygote samples. |
Dissection and histology
4 mice or embryos were analyzed for each genotype and developmental stage. Timed matings were carried out with embryonic day 0.5 considered to be midday of the day of discovery of a vaginal plug. For whole-mount preparations, e10.5 embryos were dissected and briefly fixed in 4% para-formaldehyde in phosphate-buffered saline (PBS), then incubated overnight at 37°C in β-galactosidase (LacZ) staining media (10 mM Tris-HCl pH 7.4, 5 mM K4FeCN6, 5 mM K3FeCN6, 2 mM MgCl2 and 0.8 mg/ml X-gal (Invitrogen)). For embryonic samples, embryos were dissected at the appropriate stage and immediately embedded and frozen without fixation in Optimal Cutting Temperature media (Tissue-Tek). Embedded embryos were sectioned (transverse, 20 μm) on a cryostat. For adult samples, mice were deeply anesthetized and then perfused with phosphate-buffered saline (PBS) followed by 4% para-formaldehyde in PBS; brains were removed, cryoprotected in 30% sucrose in PBS and sectioned (50 μm, saggital) on a freezing microtome. Immunostaining was performed on-slide for embryonic samples and free-floating for adult tissues. Sections were incubated in blocking solution (4% goat serum, 2% BSA and 0.1% Triton-X-100 in PBS) for 1 h; incubated with primary antibody diluted in blocking solution for 18 h at 4°C; washed in PBS; incubated for 2 h with appropriate secondary anti-IgG antibodies conjugated to Alexa 488 or Alexa 594 dyes; and washed and mounted in Vectashield media (Vector Laboratories). Tyramide signal amplification (TSA) reagents (Invitrogen) were used as per manufacturer's instructions. For β-galactosidase staining of sections, sections were incubated overnight in staining media as described above. Slides were imaged using a confocal, upright epifluorescence or brightfield dissection microscope. Red and green fluorescence spectra were captured separately and appropriate control experiments were performed to confirm specificity and lack of cross-reactivity in labeling.
Antibodies
The following primary antibodies and dilutions were used: chicken α-GFP, 1:1000 (Aves Labs); rabbit α-serotonin, 1:6000 (Immunostar); mouse α-TPH (tryptophan hydroxylase), 1:200 (Sigma); mouse α-Nkx2.2 (Developmental Studies Hybridoma Bank, clone 74.5A5); mouse α-Mash1, 1:100 (BD Transduction); rabbit α-Pdx1 (generated in lab??), 1:1000; and mouse α-Isl1, 1:50 (Developmental Studies Hybridoma Bank, clone 40.206).
Results and discussion
As the efficiency of Cre-mediated recombination is not necessarily identical across different target loci, we intercrossed Pdx1-cre transgenic mice with mice from two distinct Cre reporter lines. The transgenes and genotypes analyzed are summarized in Table 1 and in the Methods. In brief, the reporter lines function as follows: the Z/EG line carries a single-copy transgene containing a strong and ubiquitous recombinant promoter, followed by a β-galactosidase and transcriptional stop cassette flanked by loxP sites, followed by a GFP cassette [42]. In Cre-negative cells, β-galactosidase only is expressed; in Cre-expressing lineages, the β-galactosidase cassette is excised, permitting expression of GFP. The Rosa26R mouse line harbors a transgene inserted by targeted mutagenesis into the ubiquitously expressed rosa26 locus; this transgene consists of a loxP-flanked stop cassette followed by a β-galactosidase cassette [43]. In Cre-negative cells, no reporter β-galactosidase transgene is expressed; in Cre-expressing lineages, the stop cassette is excised, permitting expression of β-galactosidase.
Standard polymerase chain reaction (PCR) genotyping protocols for the mouse lines used have been described previously [10,42,43]. We adapted previously published methods for single nucleotide polymorphism detection [44-50] to develop a gel-free genotyping method based on multiplex PCR and discrimination of allele-specific products using SYBR-Green-detected melting curve analysis. Small-product multiplex PCR reactions are performed using an optical cycler with the inclusion of SYBR Green dye, which fluoresces in the presence of double-stranded DNA. At the end of the amplification reaction, PCR products corresponding to specific alleles are detected by progressively heating the reaction and plotting the derivative of SYBR fluorescence; annealing and melting of a specific product generates a peak at a specific melting temperature. The PCR reaction well is never opened and no gels are required. Melting curve peaks (position and shape) can be manipulated using simple, inexpensive primer modification [45], such that this approach can readily address most PCR based, multiplex genotyping applications. This method confers significant advantages over existing methods, including higher throughput, uniform and robust PCR conditions, low cost and reduction of post-PCR contamination. Representative data, from an assay used to genotype mice of the Pdx1-cre line, is provided in Figure 1. Detailed instructions for assay design and implementation are available upon request (also see [45,44-50]). Primer sequences and reaction conditions are provided in Table 2.
Under our experimental conditions, reporter expression of GFP (Z/EG line) or LacZ (rosa26R line) provided a specific marker of Cre-mediated recombination. No reporter expression was evident in pdx1-cre0/0; ZegGFP/0 and pdx1-cre0/0; rosa26LacZ/+ littermate control tissues (Figures 2C; 3A; 4A; C; E; G). GFP and LacZ expression patterns described below were observed in pdx1-creCre/0; ZegGFP/0 and pdx1-creCre/0; rosa26LacZ/+ mice or embryos. For each genotype and time point, comparable patterns of GFP or LacZ expression were observed in all analyzed animals (n = 4).
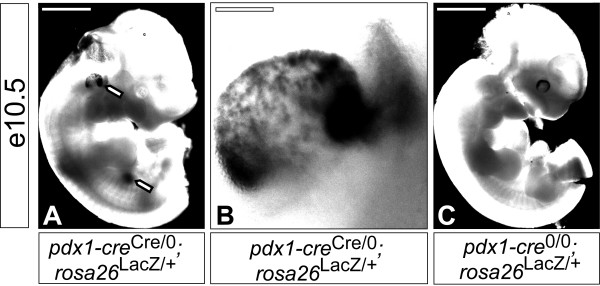
Figure 2.
Cre-mediated recombination in the pancreatic primordium and inner ear in the e10.5 embryo. Whole-mount images of a pdx1-creCre/0; rosa26LacZ/+ embryo (A & B) and a pdx1-cre0/0; rosa26LacZ/+ embryo (C) processed for β-galactosidase activity. β-galactosidase activity was observed in the pancreatic primordium (bottom arrow, left panel) and inner ear formation (top arrow, A; region in higher magnification in B). β-galactosidase activity was not evident in pdx1-cre0/0; rosa26LacZ/+ control embryos (C). Scale bars, 1 mm (A and C) and 150 μm (B).
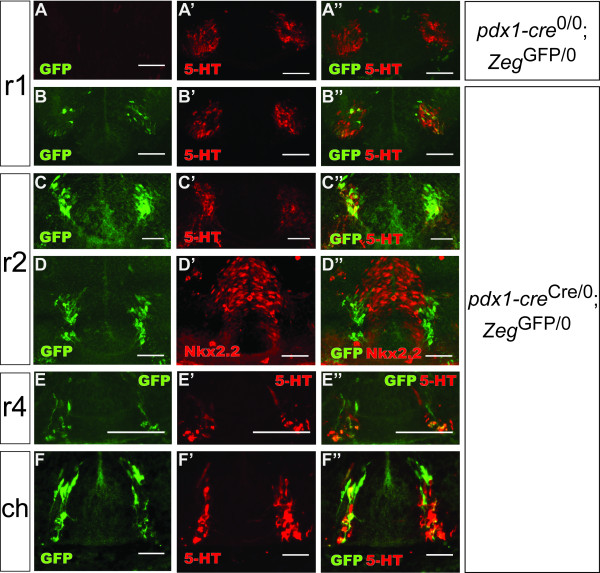
Figure 3.
Cre-mediated recombination coincides with serotonergic neurogenesis in the e11.5 embryo. Epifluorescence images of the e11.5 developing hindbrain of embryos, transversely sectioned, immunostained for GFP (green) and 5-HT or Nkx2.2 (red). In pdx1-creCre/0; ZegGFP/0 embryos, GFP was always expressed in or adjacent to newborn serotonergic neurons. Both serotonergic and non-serotonergic neurons expressed GFP in the rostral hindbrain (B, C, D) (rhombomere 1, r1; rhombomere 2, r2) and caudal hindbrain (ch) (F). The degree of co-expression of GFP and the serotonergic phenotype was greatest in the rostral hindbrain, with little overlap in the caudal hindbrain and sparse GFP expression in rhombomere 4 (r4) (E). GFP was not expressed in the Nkx2.2+ progenitor zone (D). GFP expression was not evident in sections from a pdx1-cre0/0; ZegGFP/0 embryo (A). Scale bars, 100 μm.
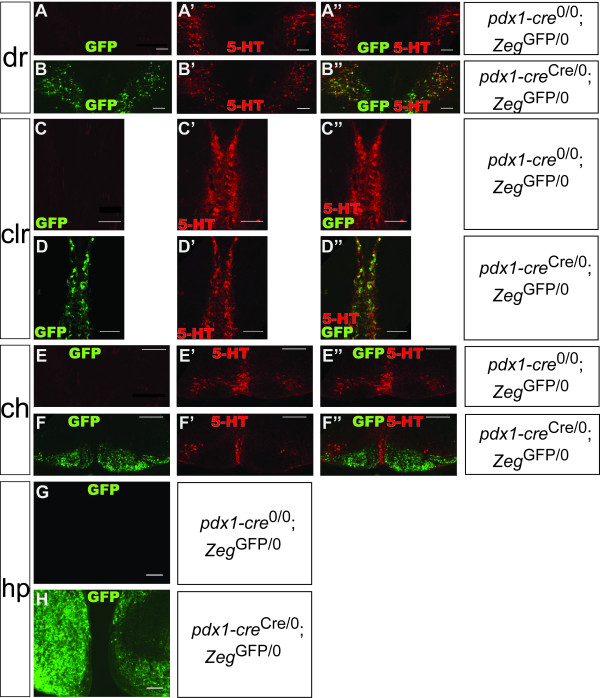
Figure 4.
Cre-mediated recombination in the hindbrain and diencephalon in the e16.5 embryo. Epifluorescence images from pdx1-creCre/0; ZegGFP/0 (B, D, F, H) and pdx1-cre0/0; ZegGFP/0 (A, C, E, G) e16.5 embryos, transversely sectioned, immunostained for GFP (green) and 5-HT (red). GFP was expressed in the dorsal raphe nucleus (dr) (B), caudal linear raphe (clr) (D), caudal hindbrain (ch) (F) and hypothalamus (hp) (H). In the rostral hindbrain, GFP expression occurred in the serotonergic dorsal raphe and caudal linear nuclei (B, D). In the caudal hindbrain, GFP expression was observed in the non-serotonergic inferior olive nucleus, adjacent to serotonergic raphe nuclei (F). GFP expression was not evident in sections from pdx1-cre0/0; ZegGFP/0 control embryos (A, C, E, G). Scale bars, 100 μm.
Recombination was first detected at e10.5 in the pancreas (Figure 2A), as reported [10], and in the inner ear formation (patchy expression in the developing anterior and posterior semicircular canal region with enriched expression in two anterior and posterior medial domains) (Figures 2A &2B). The earliest recombination in the central nervous system was observed at e11.5, in the hindbrain (Figure 3) and in the diencephalon (Figure 5).
Figure 5.
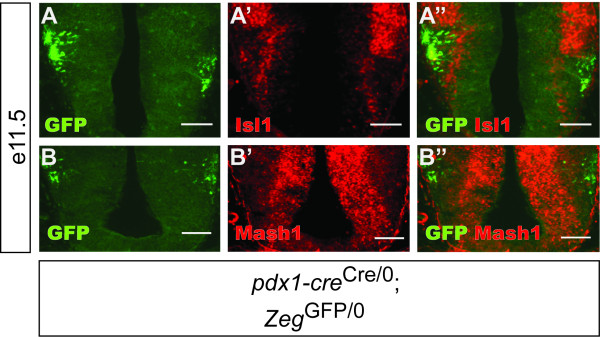
Cre-mediated recombination in the ventral diencephalon in the e11.5 embryo. Epifluorescence images of the e11.5 diencephalon of pdx1-creCre/0; ZegGFP/0 embryos, transversely sectioned, immunostained for GFP (green) and Isl1 (A) or Mash1 (B) (red). GFP was not expressed at the ventral surface near the floor plate adjacent to the Isl1+ Mash1+ neurogenic zone. Scale bars, 100 μm.
In the e11.5 hindbrain, GFP expression was observed to spatiotemporally coincide with serotonergic neurogenesis. In rhombomeres 1 (r1) and 2 (r2), GFP was observed exclusively within a ventral zone where serotonin neurons are first observed in the developing brain[8] (Figures 3B, C, D). In r1 and r2, most GFP+ cells were newborn serotonergic neurons, as identified by serotonin (5-HT) immunoreactivity, although a small minority of GFP-labeled cells appeared to be 5-HT- and some 5-HT+ cells were GFP-, suggesting that recombination in this cell lineage was mosaic at this time point (Figures 3B, C, E). GFP expression was observed adjacent to but not in the Nkx2.2+ progenitor zone (Figure 3D), suggesting that Cre is first expressed as serotonergic cells differentiate and migrate out of this progenitor zone. In rhombomere 4 (r4), serotonergic neurons are not generated[8], although 5-HT+ fibers can be detected (distinguished from cell bodies by morphology, anatomical location and lack of DAPI staining); in r4, GFP expression was observed in most 5-HT+ fibers and rare 5-HT- cell bodies (Figure 3E). In the caudal hindbrain, serotonin and reporter immunoreactivity was always observed in the same sections, but rarely within the same cells; cellular GFP immunoreactivity was observed immediately dorsal to most 5-HT+ neurons (Figure 3F). In general, we observed a rostral-caudal gradient of overlap between GFP expression and the serotonergic phenotype in the hindbrain.
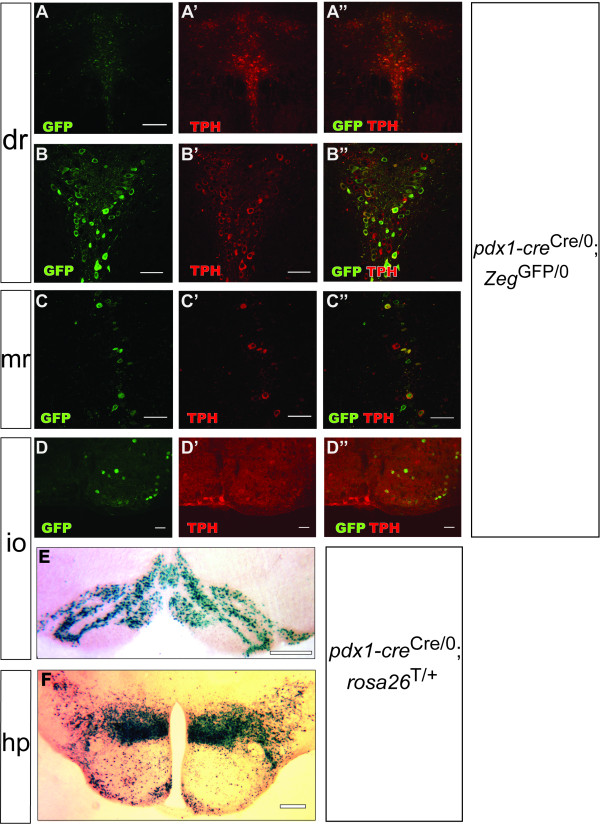
The recombination pattern observed in the e11.5 hindbrain predicted the pattern we observed at later stages of development. At e16.5, reporter expression in the rostral hindbrain was restricted to the rostral raphe nuclei, particularly the dorsal raphe nucleus (Figure 4B) and caudal linear raphe nucleus (Figure 4D). In the caudal hindbrain, reporter expression was observed in the non-serotonergic inferior olive nucleus but not in adjacent serotonergic neurons (Figure 4F). In adult tissues, confocal microscopy was employed for analysis of hindbrain sections in order to more rigorously analyze the co-expression of GFP and the serotonergic phenotype. In the dorsal raphe nucleus, which is generated in r1, the most rostral portion of the developing hindbrain [4], the large majority of serotonergic neurons (identified by immunoreactivity for tryptophan hydroxylase, or TPH) expressed GFP, and all GFP-positive cells were serotonergic (Figures 6B, B', B''). In the median raphe nucleus, which is generated in r1, r2 and r3 [4], there was partial overlap between GFP and 5-HT expression (Figures 6C, C', C''). In the caudal hindbrain, 5-HT and GFP expression were completely non-overlapping, although occurring in the same sections; recombination was restricted to the inferior olive nucleus (Figures 6D, D', D'', E). Interestingly, the inferior olive can be similarly labeled using a Cre line in which the cre was introduced into the locus for ptf1a, a transcription factor which interacts with pdx1 during early pancreatic development [51,52]. Given the anatomical localization of GFP+ 5-HT+ cells in the adult, it is likely that a vast majority of forebrain-projecting serotonergic neurons, and virtually no spinal-cord-projecting serotonergic neurons, exhibit Cre-mediated recombination in the Pdx1-cre line[2]. These data provide further evidence that caudal and rostral serotonergic neurons, though generated through highly similar developmental processes [8], exhibit distinct patterns of gene expression regulation [3]. This Pdx1-cre mouse line may be a useful resource for investigators interested in manipulating gene expression in serotonergic neurons projecting to the forebrain but not to the brainstem and spinal cord.
Figure 6.
Cre-mediated recombination in forebrain-projecting serotonergic neurons, inferior olive neurons and hypothalamic neurons in the adult brain. A-D: Individual optical sections obtained using confocal imaging of saggital sections from adult pdx1-creCre/0; ZegGFP/0 mice. A: Wide-field image of the serotonergic dorsal raphe nucleus (dr) demonstrating extensive and anatomically restricted expression of TPH and GFP in this structure. B: Higher-magnification image of the dorsal raphe nucleus: a large majority of TPH+ neurons express GFP and that all GFP+ cells in this region are serotonergic neurons. C: In the median raphe nucleus (mr), there was partial overlap between GFP and TPH expression. D: In the caudal hindbrain, GFP expression was observed in the inferior olive nucleus (io), adjacent to but not overlapping with serotonergic raphe nuclei. E-F: Brightfield images of saggital sections obtained from adult pdx1-creCre/0; rosa26LacZ/+ mice, processed for LacZ activity. E: The inferior olive nucleus was labeled with LacZ. F: Multiple nuclei of the hypothalamus, notably the dorsomedial, lateral and arcuate nuclei, were labeled with LacZ. Scale bars: 80 μm (A); 60 μm (B, C, D); 200 μm (E); 150 μm (F).
In the e11.5 diencephalon, recombination occurred, as in the hindbrain, in a restricted ventral zone of the neural tube, adjacent to neurogenic zones (here identified by Isl1 and Mash1 expression) (Figure 5). This developmental pattern resulted in GFP and LacZ expression in specific, anatomically defined nuclei of the hypothalamus, as could be observed at e16.5 (Figure 4H) and especially in adult sections. In the adult hypothalamus, reporter expression occurred in the arcuate nucleus, dorsomedial nucleus and lateral hypothalamus (Figure 6F). These regions of the hypothalamus are all critically involved in the in vivo regulation of metabolic functions such as glucose homeostasis [53]. The dorsomedial hypothalamic nucleus is relatively poorly characterized at the molecular level, and to our knowledge no transgenic mouse line has been reported which exhibits specific transgene expression in this sub-region of the hypothalamus. The possible existence of specific pdx1 regulatory sequences directing expression in the dorsomedial hypothalamic nucleus could be used to generate such transgenic mouse lines for the study of this important hypothalamic cell population. Interestingly, many hypothalamic neurons share specialized physiological attributes with β-cells, such as glucose sensing[54], and Cre transgenes generated using the insulin and ptf1a promoters produce recombination in the hypothalamus [51,55].
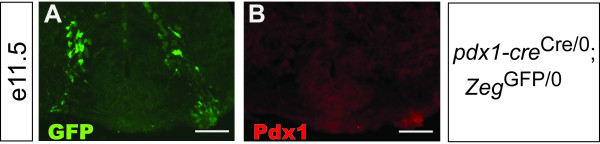
Widespread expression of endogenous pdx1 in the rat brain has been reported [56,57]. Our results suggested a more restricted pattern of endogenous pdx1 expression might occur in the mouse central nervous system. We therefore attempted to detect expression in the mouse brain of pdx1 at various developmental stages using immunofluorescence. We were unable to detect endogenous pdx1 expression at any time point, including the earliest time point at which Cre-mediated recombination was observed: tyramide signal amplification of Pdx1 immunoreactivity was attempted in pdx1-creCre/0; ZegGFP/0 e11.5 r1 tissue. No detectable signal was observed (Figure 7B), despite robust expression of GFP (Figure 7A) and reliable detection of Pdx1 in the adult pancreas under these conditions.
Figure 7.
Lack of detectable expression of endogenous pdx1 in the mouse hindbrain. Section from the r1 region of a pdx1-creCre/0; ZegGFP/0 embryo, immunostained for GFP (A) and Pdx1 (B) using TSA amplification. No detectable expression of Pdx1 was observed, despite robust expression of GFP. Scale bars, 100 μm.
While this work was under review, two publications reported neuronal transgene expression in a variety of mouse lines used for the study of pancreatic development and function [58,59]. These results are consistent with and complementary to our results. Using the Pdx1-cre mouse line employed in our study, Wicksteed et al and Song et al report a similar pattern of Cre-mediated recombination in the developing and adult hypothalamus and brainstem. Furthermore, Wicksteed et al observed a similar pattern of recombination in an independently generated mouse line in which Cre expression is driven by a similar construct incorporating regulatory sequences upstream of the pdx1 gene, suggesting that the expression patterns we observe cannot be due solely to insertion site effects. They also report a lack of galactosidase activity in mice bearing a heterozygous LacZ insertion at the pdx1 locus and a lack of amplification of endogenous pdx1 transcript from the brain (both consistent with our observations, herein and unpublished). Taken together, their results indicate that pdx1 regulatory elements can drive highly specific neuronal expression, but that endogenous pdx1 is not expressed in the mouse brain, likely due to the activity of a repressor element not contained in the Pdx1-cre transgene constructs currently employed. The reported widespread expression of endogenous pdx1 reported in the rat brain may reflect a difference between gene regulation between rat and mouse or methodological differences between the rat and mouse studies [56,57].
Conclusions
We report here, using the widely used Pdx1-cre line, a highly specific pattern of Cre-mediated recombination in the central nervous system and inner ear. This Cre line, and the regulatory sequences that direct Cre expression, may be a valuable resource for investigators seeking to manipulate gene expression in specific subsets of neurons, such as forebrain-projecting serotonergic neurons and neurons of the dorsomedial hypothalamus. To our knowledge, no other Cre mouse lines have been described to exhibit a pattern of hypothalamic recombination comparable to that we observed in the Pdx1-Cre line. However, the fact that recombination was observed in several hypothalamic nuclei which may have very different physiological functions (such as the arcuate and dorsomedial nuclei), as well as in the pancreas, may require further characterization of pdx1 regulatory elements in order to maximize the utility of pdx1 regulatory elements for this purpose.
At present, several transgenic methods have been developed which generate specific Cre-mediated recombination in serotonergic neurons [4,5,60]. Two of these methods involve the use of bacterial artificial chromosomes containing regulatory elements or genes expressed in serotonergic neurons, and unlike the Pdx1-cre line, result in recombination in the vast majority of serotonergic neurons [60,5]. However, like the Pdx1-cre line, these Cre transgenes may also result in recombination in other cell types, such as thalamocortical neurons [60] and pancreatic β-cells (Ohta, German et al, in submission). A technically sophisticated method has been described which allows for highly specific targeted recombination in subsets of serotonergic neurons; however, this method may be impractical for some investigators due to the complexity of genetic manipulations required [4].
It is important to note that the brain regions which exhibit recombination in the Pdx1-cre mouse line are well known to have important roles in in vivo metabolic function, including glucose homeostasis [61,53]. Investigators employing this line to express Cre in the developing pancreas should consider the possibility that concomitant Cre expression in the central nervous system may play a significant role in resulting in vivo phenotypes. Conversely, investigators who plan to use this mouse line to manipulate neuronal gene expression should consider the possible effects on pancreatic gene expression and function. These considerations are particularly salient when analyzing behavior and physiology in adult mice.
In conclusion, these data are consistent with the idea that common patterns of gene expression in pancreatic β-cells, serotonergic neurons and hypothalamic neurons contribute to their highly specialized and, in many cases, similar physiology.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
GH, MB, AL, MSG and LT conceived of experiments; GH and AL performed experiments; GH and AL analyzed data; GH wrote the manuscript; all authors edited the manuscript.
Contributor Information
Gerard Honig, Email: ghonig@gmail.com.
Angela Liou, Email: angela.liou@gmail.com.
Miles Berger, Email: berger.miles@gmail.com.
Michael S German, Email: mgerman@diabetes.ucsf.edu.
Laurence H Tecott, Email: larry.tecott@ucsf.edu.
Acknowledgements
Elaine J. Carlson, Elaine Storm, John L. Rubenstein, Katherine Shim and Janet Lau provided advice and reagents. L. Bogdanova assisted with mouse genotyping. This project was supported by the Sandler Foundation and the Howard Hughes Medical Institute (GH).
References
- Branda CS, Dymecki SM. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev Cell. 2004;6(1):7–28. doi: 10.1016/S1534-5807(03)00399-X. [DOI] [PubMed] [Google Scholar]
- Jacobs BL, Azmitia EC. Structure and function of the brain serotonin system. Physiol Rev. 1992;72(1):165–229. doi: 10.1152/physrev.1992.72.1.165. [DOI] [PubMed] [Google Scholar]
- Wylie CJ, Hendricks TJ, Zhang B, Wang L, Lu P, Leahy P, Fox S, Maeno H, Deneris ES. Distinct transcriptomes define rostral and caudal serotonin neurons. J Neurosci. 2010;30(2):670–684. doi: 10.1523/JNEUROSCI.4656-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen P, Farago AF, Awatramani RB, Scott MM, Deneris ES, Dymecki SM. Redefining the serotonergic system by genetic lineage. Nat Neurosci. 2008;11(4):417–419. doi: 10.1038/nn2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MM, Wylie CJ, Lerch JK, Murphy R, Lobur K, Herlitze S, Jiang W, Conlon RA, Strowbridge BW, Deneris ES. A genetic approach to access serotonin neurons for in vivo and in vitro studies. Proc Natl Acad Sci USA. 2005;102(45):16472–16477. doi: 10.1073/pnas.0504510102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JC, Cook MN, Carey MR, Shen C, Regehr WG, Dymecki SM. Linking genetically defined neurons to behavior through a broadly applicable silencing allele. Neuron. 2009;63(3):305–315. doi: 10.1016/j.neuron.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson ME, Scheel D, German MS. Gene expression cascades in pancreatic development. Mechanisms of Development. 2003;120(1):65–80. doi: 10.1016/S0925-4773(02)00333-7. [DOI] [PubMed] [Google Scholar]
- Pattyn A, Vallstedt A, Dias JM, Samad OA, Krumlauf R, Rijli FM, Brunet JF, Ericson J. Coordinated temporal and spatial control of motor neuron and serotonergic neuron generation from a common pool of CNS progenitors. Genes Dev. 2003;17(6):729–737. doi: 10.1101/gad.255803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AM, Gonez LJ, Naselli G, Macdonald RJ, Harrison LC. Conditional expression demonstrates the role of the homeodomain transcription factor Pdx1 in maintenance and regeneration of beta-cells in the adult pancreas. Diabetes. 2005;54(9):2586–2595. doi: 10.2337/diabetes.54.9.2586. [DOI] [PubMed] [Google Scholar]
- Gu G, Dubauskaite J, Melton DA. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129(10):2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Database. http://www.informatics.jax.org/
- Bult CJ, Eppig JT, Kadin JA, Richardson JE, Blake JA. The Mouse Genome Database (MGD): mouse biology and model systems. Nucleic Acids Res. 2008. pp. D724–728. [DOI] [PMC free article] [PubMed]
- Aguirre AJ, Bardeesy N, Sinha M, Lopez L, Tuveson DA, Horner J, Redston MS, DePinho RA. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17(24):3112–3126. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardeesy N, Aguirre AJ, Chu GC, Cheng KH, Lopez LV, Hezel AF, Feng B, Brennan C, Weissleder R, Mahmood U, Hanahan D, Redston MS, Chin L, Depinho RA. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci USA. 2006;103(15):5947–5952. doi: 10.1073/pnas.0601273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardeesy N, Cheng KH, Berger JH, Chu GC, Pahler J, Olson P, Hezel AF, Horner J, Lauwers GY, Hanahan D, DePinho RA. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006;20(22):3130–3146. doi: 10.1101/gad.1478706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano DA, Sekine S, Hebrok M. Primary cilia deletion in pancreatic epithelial cells results in cyst formation and pancreatitis. Gastroenterology. 2006;131(6):1856–1869. doi: 10.1053/j.gastro.2006.10.050. [DOI] [PubMed] [Google Scholar]
- Collombat P, Hecksher-Sorensen J, Krull J, Berger J, Riedel D, Herrera PL, Serup P, Mansouri A. Embryonic endocrine pancreas and mature beta cells acquire alpha and PP cell phenotypes upon Arx misexpression. J Clin Invest. 2007;117(4):961–970. doi: 10.1172/JCI29115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikura J, Hosoda K, Iwakura H, Tomita T, Noguchi M, Masuzaki H, Tanigaki K, Yabe D, Honjo T, Nakao K. Notch/Rbp-j signaling prevents premature endocrine and ductal cell differentiation in the pancreas. Cell Metab. 2006;3(1):59–65. doi: 10.1016/j.cmet.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Fujikura J, Hosoda K, Kawaguchi Y, Noguchi M, Iwakura H, Odori S, Mori E, Tomita T, Hirata M, Ebihara K, Masuzaki H, Fukuda A, Furuyama K, Tanigaki K, Yabe D, Nakao K. Rbp-j regulates expansion of pancreatic epithelial cells and their differentiation into exocrine cells during mouse development. Dev Dyn. 2007;236(10):2779–2791. doi: 10.1002/dvdy.21310. [DOI] [PubMed] [Google Scholar]
- Gupta D, Jetton TL, Mortensen RM, Duan SZ, Peshavaria M, Leahy JL. In Vivo and in Vitro Studies of a Functional Peroxisome Proliferator-activated Receptor {gamma} Response Element in the Mouse pdx-1 Promoter. J Biol Chem. 2008;283(47):32462–32470. doi: 10.1074/jbc.M801813200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiser PW, Cano DA, Landsman L, Kim GE, Kench JG, Klimstra DS, Taketo MM, Biankin AV, Hebrok M. Stabilization of beta-catenin induces pancreas tumor formation. Gastroenterology. 2008;135(4):1288–1300. doi: 10.1053/j.gastro.2008.06.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hezel AF, Gurumurthy S, Granot Z, Swisa A, Chu GC, Bailey G, Dor Y, Bardeesy N, Depinho RA. Pancreatic LKB1 deletion leads to acinar polarity defects and cystic neoplasms. Mol Cell Biol. 2008;28(7):2414–2425. doi: 10.1128/MCB.01621-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashchenko CY, Duan SZ, Usher MG, Mortensen RM. PPAR-gamma knockout in pancreatic epithelial cells abolishes the inhibitory effect of rosiglitazone on caerulein-induced acute pancreatitis. Am J Physiol Gastrointest Liver Physiol. 2007;293(1):G319–326. doi: 10.1152/ajpgi.00056.2007. [DOI] [PubMed] [Google Scholar]
- Kojima K, Vickers SM, Adsay NV, Jhala NC, Kim HG, Schoeb TR, Grizzle WE, Klug CA. Inactivation of Smad4 accelerates Kras(G12D)-mediated pancreatic neoplasia. Cancer Res. 2007;67(17):8121–8130. doi: 10.1158/0008-5472.CAN-06-4167. [DOI] [PubMed] [Google Scholar]
- Lammert E, Gu G, McLaughlin M, Brown D, Brekken R, Murtaugh LC, Gerber HP, Ferrara N, Melton DA. Role of VEGF-A in vascularization of pancreatic islets. Curr Biol. 2003;13(12):1070–1074. doi: 10.1016/S0960-9822(03)00378-6. [DOI] [PubMed] [Google Scholar]
- Lee JY, Hennighausen L. The transcription factor Stat3 is dispensable for pancreatic beta-cell development and function. Biochem Biophys Res Commun. 2005;334(3):764–768. doi: 10.1016/j.bbrc.2005.06.162. [DOI] [PubMed] [Google Scholar]
- Elghazi Lynda, W AJ, G AP, H B, G DH, B-M E. Generation of a reporter mouse line expressing Akt and EGFP upon Cre-mediated recombination. genesis. 2008;46(5):256–264. doi: 10.1002/dvg.20388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn FC, Skewes-Cox P, Kosaka Y, McManus MT, Harfe BD, German MS. MicroRNA Expression Is Required for Pancreatic Islet Cell Genesis in the Mouse. Diabetes. 2007;56(12):2938–2945. doi: 10.2337/db07-0175. [DOI] [PubMed] [Google Scholar]
- Miralles F, Hebrard S, Lamotte L, Durel B, Gilgenkrantz H, Li Z, Daegelen D, Tuil D, Joshi RL. Conditional inactivation of the murine serum response factor in the pancreas leads to severe pancreatitis. Lab Invest. 2006;86(10):1020–1036. doi: 10.1038/labinvest.3700457. [DOI] [PubMed] [Google Scholar]
- Murtaugh LC, Law AC, Dor Y, Melton DA. {beta}-Catenin is essential for pancreatic acinar but not islet development. Development. 2005;132(21):4663–4674. doi: 10.1242/dev.02063. [DOI] [PubMed] [Google Scholar]
- Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proc Natl Acad Sci USA. 2003;100(25):14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrep N, Wang J, Miyatsuka T, German MS. Signals from the neural crest regulate beta-cell mass in the pancreas. Development. 2008;135(12):2151–2160. doi: 10.1242/dev.015859. [DOI] [PubMed] [Google Scholar]
- Pasca di Magliano M, Sekine S, Ermilov A, Ferris J, Dlugosz AA, Hebrok M. Hedgehog/Ras interactions regulate early stages of pancreatic cancer. Genes Dev. 2006;20(22):3161–3173. doi: 10.1101/gad.1470806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour PA, Freude KK, Tran MN, Mayes EE, Jensen J, Kist R, Scherer G, Sander M. SOX9 is required for maintenance of the pancreatic progenitor cell pool. Proc Natl Acad Sci USA. 2007;104(6):1865–1870. doi: 10.1073/pnas.0609217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siveke JT, Einwachter H, Sipos B, Lubeseder-Martellato C, Kloppel G, Schmid RM. Concomitant pancreatic activation of Kras(G12D) and Tgfa results in cystic papillary neoplasms reminiscent of human IPMN. Cancer Cell. 2007;12(3):266–279. doi: 10.1016/j.ccr.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Stanger BZ, Datar R, Murtaugh LC, Melton DA. Direct regulation of intestinal fate by Notch. Proc Natl Acad Sci USA. 2005;102(35):12443–12448. doi: 10.1073/pnas.0505690102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanger BZ, Stiles B, Lauwers GY, Bardeesy N, Mendoza M, Wang Y, Greenwood A, Cheng K-h, McLaughlin M, Brown D, DePinho RA, Wu H, Melton DA, Dor Y. Pten constrains centroacinar cell expansion and malignant transformation in the pancreas. Cancer Cell. 2005;8(3):185–195. doi: 10.1016/j.ccr.2005.07.015. [DOI] [PubMed] [Google Scholar]
- Ueki K, Okada T, Hu J, Liew CW, Assmann A, Dahlgren GM, Peters JL, Shackman JG, Zhang M, Artner I, Satin LS, Stein R, Holzenberger M, Kennedy RT, Kahn CR, Kulkarni RN. Total insulin and IGF-I resistance in pancreatic beta cells causes overt diabetes. Nat Genet. 2006;38(5):583–588. doi: 10.1038/ng1787. [DOI] [PubMed] [Google Scholar]
- Wang S, Zhang J, Zhao A, Hipkens S, Magnuson MA, Gu G. Loss of Myt1 function partially compromises endocrine islet cell differentiation and pancreatic physiological function in the mouse. Mech Dev. 2007;124(11-12):898–910. doi: 10.1016/j.mod.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Ackermann AM, Gusarova GA, Lowe D, Feng X, Kopsombut UG, Costa RH, Gannon M. The FoxM1 transcription factor is required to maintain pancreatic beta-cell mass. Mol Endocrinol. 2006;20(8):1853–1866. doi: 10.1210/me.2006-0056. [DOI] [PubMed] [Google Scholar]
- Zhang W, Feng D, Li Y, Iida K, McGrath B, Cavener DR. PERK EIF2AK3 control of pancreatic beta cell differentiation and proliferation is required for postnatal glucose homeostasis. Cell Metab. 2006;4(6):491–497. doi: 10.1016/j.cmet.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28(3-4):147–155. doi: 10.1002/1526-968X(200011/12)28:3/4<147::AID-GENE90>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21(1):70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Akey JM, Sosnoski D, Parra E, Dios S, Hiester K, Su B, Bonilla C, Jin L, Shriver MD. Melting curve analysis of SNPs (McSNP): a gel-free and inexpensive approach for SNP genotyping. Biotechniques. 2001;30(2):358–362. doi: 10.2144/01302tt05. [DOI] [PubMed] [Google Scholar]
- Wang J, Chuang K, Ahluwalia M, Patel S, Umblas N, Mirel D, Higuchi R, Germer S. High-throughput SNP genotyping by single-tube PCR with Tm-shift primers. Biotechniques. 2005;39(6):885–893. doi: 10.2144/000112028. [DOI] [PubMed] [Google Scholar]
- Doan L, Monuki ES. Rapid Genotyping of Mouse Tissue Using Sigma's Extract-N-Amp Tissue PCR Kit. J Vis Exp. 2008. [DOI] [PMC free article] [PubMed]
- Germer S, Higuchi R. Single-Tube Genotyping without Oligonucleotide Probes. Genome Res. 1999;9(1):72–78. [PMC free article] [PubMed] [Google Scholar]
- Krizhanovsky V, Golenser E, Ben-Arie N. Genotype identification of Math1/LacZ knockout mice based on real-time PCR with SYBR Green I dye. Journal of Neuroscience Methods. 2004;136(2):187–192. doi: 10.1016/j.jneumeth.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Papp AC, Pinsonneault JK, Cooke G, Sadee W. Single nucleotide polymorphism genotyping using allele-specific PCR and fluorescence melting curves. Biotechniques. 2003;34(5):1068–1072. doi: 10.2144/03345dd03. [DOI] [PubMed] [Google Scholar]
- Sakurai T, Kamiyoshi A, Watanabe S, Sato M, Shindo T. Rapid zygosity determination in mice by SYBR Green real-time genomic PCR of a crude DNA solution. Transgenic Research. 2008;17(1):149–155. doi: 10.1007/s11248-007-9134-7. [DOI] [PubMed] [Google Scholar]
- Yamada M, Terao M, Terashima T, Fujiyama T, Kawaguchi Y, Nabeshima Y, Hoshino M. Origin of climbing fiber neurons and their developmental dependence on Ptf1a. J Neurosci. 2007;27(41):10924–10934. doi: 10.1523/JNEUROSCI.1423-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino M, Nakamura S, Mori K, Kawauchi T, Terao M, Nishimura YV, Fukuda A, Fuse T, Matsuo N, Sone M, Watanabe M, Bito H, Terashima T, Wright CV, Kawaguchi Y, Nakao K, Nabeshima Y. Ptf1a, a bHLH transcriptional gene, defines GABAergic neuronal fates in cerebellum. Neuron. 2005;47(2):201–213. doi: 10.1016/j.neuron.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Saper CB. Staying awake for dinner: hypothalamic integration of sleep, feeding, and circadian rhythms. Prog Brain Res. 2006;153:243–252. doi: 10.1016/S0079-6123(06)53014-6. full_text. [DOI] [PubMed] [Google Scholar]
- Levin BE, Routh VH, Kang L, Sanders NM, Dunn-Meynell AA. Neuronal glucosensing: what do we know after 50 years? Diabetes. 2004;53(10):2521–2528. doi: 10.2337/diabetes.53.10.2521. [DOI] [PubMed] [Google Scholar]
- Xu AW, Kaelin CB, Takeda K, Akira S, Schwartz MW, Barsh GS. PI3K integrates the action of insulin and leptin on hypothalamic neurons. J Clin Invest. 2005;115(4):951–958. doi: 10.1172/JCI24301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz PT, Perez-Villamil B, Rivera A, Moratalla R, Vallejo M. Pancreatic homeodomain transcription factor IDX1/IPF1 expressed in developing brain regulates somatostatin gene transcription in embryonic neural cells. J Biol Chem. 2000;275(25):19106–19114. doi: 10.1074/jbc.M000655200. [DOI] [PubMed] [Google Scholar]
- Perez-Villamil B, Schwartz PT, Vallejo M. The pancreatic homeodomain transcription factor IDX1/IPF1 is expressed in neural cells during brain development. Endocrinology. 1999;140(8):3857–3860. doi: 10.1210/en.140.8.3857. [DOI] [PubMed] [Google Scholar]
- Wicksteed B, Brissova M, Yan W, Opland DM, Plank JL, Reinert RB, Dickson LM, Tamarina NA, Philipson LH, Shostak A, Bernal-Mizrachi E, Elghazi L, Roe MW, Labosky PA, Myers MM, Jr, Gannon M, Powers AC, Dempsey PJ. Conditional gene targeting in mouse pancreatic {beta}-cells: Analysis of ectopic Cre transgene expression in the brain. Diabetes. 2010. in press . [DOI] [PMC free article] [PubMed]
- Song J, Xu Y, Hu X, Choi B, Tong Q. Brain expression of Cre recombinase driven by pancreas-specific promoters. Genesis. 2010. in press . [DOI] [PMC free article] [PubMed]
- Gong S, Doughty M, Harbaugh CR, Cummins A, Hatten ME, Heintz N, Gerfen CR. Targeting Cre Recombinase to Specific Neuron Populations with Bacterial Artificial Chromosome Constructs. J Neurosci. 2007;27(37):9817–9823. doi: 10.1523/JNEUROSCI.2707-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Jones JE, Kohno D, Williams KW, Lee CE, Choi MJ, Anderson JG, Heisler LK, Zigman JM, Lowell BB, Elmquist JK. 5-HT2CRs expressed by pro-opiomelanocortin neurons regulate energy homeostasis. Neuron. 2008;60(4):582–589. doi: 10.1016/j.neuron.2008.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]