Abstract
The endogenous circadian clock ensures daily rhythms in diverse behavioral and physiological processes, including locomotor activity and sleep/wake cycles, but also food intake patterns. Circadian rhythms are generated by an internal clock system which synchronizes these daily variations to the day/night alternance. In addition, circadian oscillations may be reset by the time of food availability in peripheral metabolic organs. Circadian rhythms are seen in many metabolic pathways (glucose and lipid metabolism, ..) and endocrine secretions (insulin, ..). As a consequence, misalignment of the internal timing system vs environmental zeitgebers (light for instance), as experienced during jetlag or shift work, may result in disruption of physiological cycles of fuel utilization or energy storage. A large body of evidence from both human and animal studies now points to a relationship between circadian disorders and altered metabolic response, suggesting that circadian and metabolic regulatory networks are tightly connected. After a review of the current understanding of the molecular circadian core clock, we will discuss the hypothesis that clock genes themselves link the core molecular clock and metabolic regulatory networks. We propose that the nuclear receptor and core clock component Rev-erbα behaves as a gatekeeper to timely coordinate the circadian metabolic response.
Keywords: Animals; Biological Clocks; physiology; CLOCK Proteins; physiology; Circadian Rhythm; physiology; Humans; Metabolic Networks and Pathways; physiology; Metabolic Syndrome X; physiopathology; Nuclear Receptor Subfamily 1, Group D, Member 1; physiology
Keywords: circadian rhythm, metabolic disorders, Rev-erb-α, biological clock, metabolic syndrome
1. Introduction: circadian rhythms are daily fluctuations in behavioral and physiological processes
Circadian rhythms are daily fluctuations with a period of approximately one day (‘circa diem’) observed in many physiological processes and behaviour. They are driven by an endogenous clock and are defined as cycles persisting when organisms are isolated from environmental cues (‘free-running’), the subjected circadian ‘night’ and ‘day’ being in this case predicted by the endogenous oscillator. Obvious circadian rhythms are the sleep and wake alternance, as well as daily variations in body temperature, blood pressure and heart rate. In humans, sleep onset is accompanied by a fall in body temperature, while blood pressure and heart rate start to increase in the late dark phase to prepare for the awakening. Along the same line, catabolic functions are turned on during the active awaken phase, whereas anabolic processes usually take place during the rest phase. These rhythms allow the organism to anticipate, adapt and optimize its metabolic, hormonal and locomotor activity to predictable environmental daily changes imposed by the rising and setting of the sun (27).
In mammals, a central ‘master’ clock residing in the suprachiasmatic nuclei (SCN) of the hypothalamus integrates information from light and synchronizes our physiology to the day/night alternance (Figure 1). In turn, it coordinates many ‘slave oscillators’ in the rest of the body through still poorly defined neuroendocrine signals. These peripheral oscillators convey output signals from the master clock and integrate additional circadian cues such as food availability to drive local circadian rhythms in intermediary metabolites (such as NAD+/NADH) or in key, often rate-limiting, enzymes involved in local physiology, such as in metabolic pathways. Indeed, many metabolic functions are subjected to circadian variations, ranging from lipid and carbohydrate metabolism, hormone (insulin, leptin, cortisol, growth hormone) secretion, and, less intuitive though primordial, gene transcription (58). This ensures that nutrient transporters and metabolic enzymes are produced at appropriate times (i.e. stimulation of catabolic pathways during the wake period to match fuel demands) and allows temporal separation of divergent processes such as glycolysis and gluconeogenesis. Thus, there is a strong interaction between circadian control and metabolism, a concept which is further supported by the observation that circadian disruptions induced either by phase shift (as observed in shiftworkers) or due to genetic alteration in the clock machinery (sequence polymorphisms present in certain clock genes or genetically-induced mutations) are associated to increased incidence of metabolic disorders in humans and murine models.
Figure 1.

The ‘master’ clock residing in the suprachiasmatic nucleus (SCN) of the hypothalamus drives rhythmic behaviour (sleep/wake, locomotor activity, circadian feeding pattern) and synchronizes many peripheral clocks, which are thought to be important for local rhythmic activity, for instance of hepatic metabolism, cardiovascular function and the endocrine response. Food is also an important time cue for peripheral clocks. Transcriptome analysis in peripheral organs found 3–20% rhythmic genes (58), including genes involved in lipid and glucose metabolism and most of the transcription factors of the nuclear receptor family (86), suggesting a direct interaction between circadian behaviour and metabolic transcriptional networks.
2. Molecular basis of circadian control
The central clock and peripheral oscillators share a common molecular architecture, conserved between species, and consist of transcriptional/(post)translational events generating and sustaining circadian rhythms. In mammals, CLOCK (Circadian locomotor output cycles kaput), Bmal1 (Brain and Muscle ARNT like protein 1), and the CLOCK paralog NPAS2 (78), activate transcription of various target genes including the per (Period) and cry (Cryptochrome) genes (Figure 2). In turn, the proteins Per and Cry repress CLOCK/Bmal1-mediated gene transactivation, thereby shutting down their own transcription. This allows a new cycle to start. An additional feedback loop, thought to improve the robustness of the former one, involves the nuclear receptors Rev-erbα and RORα. CLOCK/Bmal1 activates Rev-erbα transcription resulting in daily fluctuations of Rev-erbα, which, in turn, represses Bmal1 (60). RORα, another nuclear receptor and component of the clock machinery, competes with Rev-erbα for the binding to the Bmal1 promoter RORE/RevRE site, and activates its transcription (1; 28; 69). Therefore, the two nuclear receptors Rev-erbα and RORα act as a regulatory loop and occupancy of Bmal1 promoter by either Rev-erbα or RORα is crucial for a proper timing of the core clock machinery. More recently, the nuclear receptor co-factor and metabolic regulator Peroxisome Proliferator-Activated Receptor γ (PPARγ) co-activator (PGC)1α, whose expression cycles in liver and muscle, has been suggested to potentiate RORα activation of Rev-erbα and Bmal1 transcription and as such to be involved in circadian rhythm regulation (46).
Figure 2.
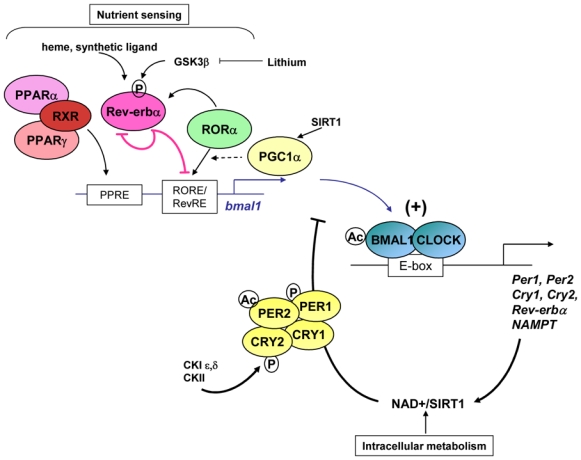
Generation and maintenance of circadian rhythms require a complex interplay of transcriptional/translational feedback regulatory loops. The CLOCK/Bmal1 heterodimer activates transcription of the Per and Cry genes via E boxes in their promoter. Per and Cry form a repressive complex which inhibits CLOCK/Bmal1 transcriptional activity, leading to their own repression. An interlocked regulatory loop is formed by Rev-erbα and RORα which compete for the binding to a RORE/RevRE and subsequent regulation (repression vs activation, respectively) of Bmal1. Rhythmic expression of Rev-erbα drives cyclic occupancy of this RORE. Post-translational modification (phosphorylation, ubiquitination, acetylation, and degradation by the proteasome) plays important role for the proper timing of the clock. Intracellular metabolism (through NAD+/SIRT1) and nutrients (through binding to nuclear receptors) impinge on the clock machinery. CK, Casein Kinase.
Several reports have challenged the concept of transcriptionally-mediated cellular rhythmicity. Using an inhibitor of transcription, Schibler and colleagues (17) show that inhibition of RNA polymerase II-dependent transcription only diminished the amplitude but did not abolish gene expression circadian oscillations in cultured fibroblasts, highlighting the importance of nontranscriptional events in the regulation of the clock machinery. Along the same line, Bmal1 oscillations are not required for the circadian clock function (45) and phosphorylation, independently of any transcriptional event, is a crucial step for circadian rhythm generation and amplitude in cyanobacteria (56).
Beside the core set of clock genes described above, post-translational modifications such as phosphorylation, sumoylation and acetylation of the clock components are important for an appropriate timing to nearly 24 hours. Protein phosphorylation by the casein kinase 1ε (CK1ε) (20) and CKII (44; 79; 80) influences Per and Cry as well as Bmal1 protein stability and activity, and thus the circadian period (24). For instance, missense mutation of CK1ε results in a circadian period shortened to 20h in the tau hamsters (47). Inhibition of glycogen synthase kinase (GSK)3 β, which stabilizes the Rev-erbα protein, with lithium leads to rapid proteasomal degradation of Rev-erbα and activation of Bmal1 (88). GSK3 β-mediated stabilization of Rev-erbα appears a crucial event for circadian rhythm initiation, maintenance and synchronisation after serum shock (88). Additionally, SUMOylation has been shown to affect Bmal1 activity, protein ubiquitination and degradation, thereby being essential for proper circadian rhythmicity (12; 40). Along the same line, acetylation of histones and the chromatin compaction state participate in the regulation of the clock machinery (21). CLOCK itself was shown to function as an acetyltransferase on both histones (18) and Bmal1 (29). Finally, Lazar and colleagues have demonstrated that Rev-erbα regulates Bmal1 activity after recruiting the nuclear co-repressor (NCoR)/HDAC3 complex to the RevRE/RORE present in its promoter (87).
3. Peripheral clocks and metabolism
The discovery that clock genes are expressed in vivo in organs such as the liver, adipose tissue, aortas, etc.. has been instrumental in the identification of clock machineries in peripheral organs (3; 58; 68; 90). In the liver, 3–20% of the transcripts are cyclic, the majority of which are involved in key metabolic events (58; 75). Although the observation that clock genes cycle in periphery does not necessarily prove the existence of peripheral clock systems, it can be hypothesized that, at least in some tissues, peripheral pacemakers are able on their own to drive and orchestrate local circadian variations in metabolic transcription clusters. Indeed, disruption of the hepatic molecular clock via overexpression of the clock machinery component Rev-erbα specifically in the liver - leaving the central pacemaker intact - results in nearly complete (90%) dampening of circadian variations of the hepatic transcriptome (34). However, 10% of the transcriptome, ie. 31 transcripts including Per2, still cycle indicating that rhythmic expression of these transcripts can be driven by both systemic cues and local oscillators. Interestingly, Lamia et al. have described the metabolic phenotype of total vs liver-specific Bmal1-deficient mice (35). Although some transcripts still cycle in the liver of hepato-specific Bmal1 −/− mice, indicating again that systemic cues are important in driving local oscillations, liver-specific, but not whole-body, Bmal1-deficient mice suffer from hypoglycemia and display altered liver circadian expression of genes important in the glucose metabolic pathway, supporting the concept that the lack of a functional liver clock, in presence of an intact central oscillator, leads to impaired glucose homeostasis.
Food availability is a dominant ‘zeitgeber’ for peripheral clocks and changes in the time of food availability re-synchronize peripheral clocks independently of the master SCN clock (15; 75) (Figure 1). Interestingly, food-derived lipid-soluble nutrients and hormones can activate transcription factors of the nuclear receptor superfamily, which comprises numerous regulators of metabolic pathways such as Rev-erbα and RORα, and also the PPARs, or the glucocorticoid receptor among many others. A gene expression analysis has revealed that 28 out of 49 of them display tissue-specific circadian expression in liver, white and brown adipose tissue, and skeletal muscle (86), highlighting the possibility that nuclear receptors may trigger circadian expression of their target genes (see below).
4. Circadian misalignment and metabolic disorders
From the above data, it appears that a proper alignment of the circadian clock with environmental ‘zeitgebers’ is required for normal, ‘healthy’ metabolic regulation. Martino and colleagues have recently reported that circadian desynchrony in casein kinase-1 ε mutant hamsters (due to misalignment between the endogenous clock period which is shortened by 4 hours, and environmental day/night cues) induced cardiomyopathy and renal disease, and that adjusting the light/dark cycle to the shortened endogenous circadian period of these animals normalizes their behavioural and locomotor patterns and reverses this abnormal phenotype (50). In humans a progressive desynchrony protocol was applied to 10 adults by subjecting them to 28-h days for 10 days, with a 14h/14h sleep/wake alternance (70). Circadian misalignment led to decreased leptin throughout the entire cycle, increased glucose despite increased insulin, suggesting decreased insulin sensitivity, and increased blood pressure. Even more striking was the observation that forced desynchrony resulted in exacerbated postprandial glucose excursion, with a maximal disturbance during maximal misalignment (ie 180° phase shift obtained after 3 extended days).
Along the same line, numerous association studies have demonstrated that chronic desynchronization experienced by shift workers increases the risk to develop obesity, type 2 diabetes, hyperlipidemia, high blood pressure and cardiovascular disease (16; 32; 76; 77). In addition, shift-workers display an altered post-prandial response during night-time work period characterized by exaggerated increase in glucose, insulin and triglyceride levels following a test meal (48).
Altogether these observations reflect the now recognized relationship between the circadian clock and metabolism, and emphasize that disruption of circadian rhythms leads to metabolic complications and higher risk to obesity and type 2 diabetes, with its constellation of adverse cardiovascular events.
5. Molecular evidences that the molecular clock system impinges on metabolic pathways, and reciprocally
5.1. Core-clock genes connect to metabolism
Clock genes cycle and thus might regulate circadian rhythms in many tissues, including peripheral metabolic tissues (such as liver, adipose tissue, skeletal muscle and pancreas), and in central nuclei known to be involved in nutrient sensing and food-entrainment (such as the mediobasal nucleus of the hypothalamus). The concept that clock genes themselves participate in metabolic regulation stemmed from a study from Turek and colleagues published in 2005 which revealed that clockΔ19 mutant mice are hyperphagic, become obese and develop hyperlipidemia and hyperglycemia upon high fat feeding (81) (Table 1). Interestingly, clock mutant mice on a Jcl-ICR background are resistant to diet-induced obesity likely due to impaired dietary lipid absorption (57). In addition, Bmal1−/− mice display altered circadian plasma glucose oscillations, as well as glucose intolerance (67). Bmal1 expression is induced during the differentiation of 3T3L1 preadipocytes into mature adipocytes, and seems to participate in the control of adipogenesis (72).
Table 1.
Metabolic consequences of genetically-induced or spontaneous mutations in different clock genes in murine models
| Genotype | Metabolic phenotype | Reference | |
|---|---|---|---|
| clockΔ19 mutant | C57Bl/6 genetic background | Fed a regular chow diet: Increased BW gain and energy intake; Altered circadian pattern in food intake and energy expenditure; Hypercholesterolemia, hypertriglyceridemia, hyperglycemia and hypoinsulinemia; Increased leptin levels during the light phase; Altered circadian pattern of hypothalamic expression of neuropeptides involved in food intake regulation (ghrelin, orexin, CART). Fed a high fat diet: Further increase in BW, energy intake, TG, cholesterol, and glucose levels; adipocyte hypertrophy and hepatic steatosis |
(81) |
| Decreased gluconeogenesis (when fed a regular chow diet) | (67) | ||
| Jcl-ICR genetic background | Moderate weight gain on a high fat diet due to impaired dietary lipid absorption | (57) | |
| Bmal1 −/− | Whole-body deficiency | Decreased BW (when fed a regular chow diet) | (51) |
| Blunted gluconeogenesis, altered circadian variations of blood glucose (decreased at CT4, increased at CT16, vs wt) when fed a regular chow diet | (67) | ||
| Increased fat mass, glucose intolerance, decreased insulin levels when fed a regular chow diet. | (35) | ||
| Liver-specific deficiency | Normal feeding behaviour, normal fat mass (as compared to wt); fasting hypoglycemia accompanied by altered circadian pattern of expression of glut -2, LPK, PEPCK and G6PT.(regular chow diet) | (35) | |
| Total KO + muscle rescue | Normalization of BW (regular chow diet) | (51) | |
| Rev-erbα −/− | Increased plasma TG levels, abnormal bile acid metabolism, altered circadian expression pattern of bile acid metabolism-related genes in liver (when fed a regular rodent chow diet) | (19; 65) | |
| Staggerer (sg/sg) | Increased liver apolipoprotein C-III expression and TG levels, decreased HDL-cho (when fed a regular chow diet) and aggravated atherosclerosis development. | (49; 64) | |
| Reduced fat depots when fed a regular chow diet, and resistance to high fat diet-induced obesity likely due to compromised feeding capacity. | (38) | ||
BW, body weight; TG, triglyceride; cho, cholesterol; HDL-cho, high density lipoprotein-cholesterol; glc, glucose; CART, cocaine and amphetamine-regulated transcript; CT, circadian time (CT 0 = light on/CT 12, light off); wt, wild-type; glut -2, hepatic glucose transporter; PEPCK, phosphoenolpyruvate carboxy kinase; G6P-t, glucose-6-phosphatase-translocase, LPK, L-pyruvate kinase.
In 2008, Lamia and colleagues reported the metabolic phenotype of mice with a liver-specific Bmal1 deletion (35). Interestingly, these mice have normal locomotor activity and feeding behaviour, as compared with total Bmal1 deficiency which results in loss of circadian locomotor and feeding pattern. Liverspecific Bmal1 KO mice have altered circadian glucose homeostasis and altered circadian expression of genes involved in glucose homeostasis, especially Glut-2, neoglucogenic genes (glucose-6-phosphatase-translocase and phosphoenolpyruvate carboxy kinase) and L-pyruvate kinase, resulting in hypoglycemia during the fasting period (35). These data suggest that the hepatic clock plays a role in maintaining adequate circadian glucose availability. Alternatively, and because whole-body Bmal1 deficiency results in normal fasting glycemia but glucose intolerance, desynchronization between hepatic and other endogenous clocks may have resulted in an inappropriate or desynchronized hepatic glucose response.
Emerging evidences demonstrate that mutations or polymorphisms in clock genes are associated to features of the metabolic syndrome in humans as well. Clock genes are expressed in human tissues and cells (6; 26) and are linked to the metabolic syndrome. Several Clock polymorphisms have been identified which are associated with body weight and increased susceptibility to obesity (71; 73). Bmal1 has also been associated to type 2 diabetes and hypertension in humans (85).
5.2. SIRT1/NAD+
Asher et al. (5) and Nakahata et al. (54) have shown that the NAD+-dependent histone deacetylase sirtuin (SIRT)1, whose activity and/or expression level cycles, counter-regulates CLOCK and drives cyclic expression of Bmal1, Per2 and Cry1. Nakahata et al. (55) and Ramsey et al., (63) went on by demonstrating that, in turn, CLOCK/Bmal1 regulates the expression of the nicotinamide phosphoribosyltransferase (NAMPT), the rate-limiting enzyme of the NAD+ synthetic pathway also known as visfatin. Interestingly, SIRT1 plays a crucial metabolic role and SIRT1 protein levels are increased upon starvation in the liver. SIRT1 interacts with PGC1α to enhance the gluconeogenic pathway (66). In addition, SIRT1 is important in adipocyte physiology and, upon food withdrawal, it antagonizes the action of PPARγ, one of the master regulators of adipogenesis, on genes mediating fat storage such as the aP2 promoter, thereby promoting fat mobilization in starved mice (59). SIRT1 also interferes with the adipogenic process, and triggers lipolysis in differentiated adipose cells (59). To catalyze the removal of an acetyl group SIRT1 uses NAD+ which directly reflects the metabolic status of the cell. Caloric restriction, by increasing the content of NAD+, stimulates SIRT1 activity (8). Moreover, SIRT1 activity and NAD+ metabolism are modulated by AMPK, a ‘fuel gauge’ that senses the intracellular AMP/ATP ratio (11). This implies that not only the clock modulates metabolism, but also the other way around, that the metabolic status of the cell influences the clock machinery. Indeed, it has been demonstrated that murine models of genetically- (obese ob/ob and diabetic db/db mice) or high fat feeding-induced obesity and diabetes have disrupted circadian expression of clock and clockregulated genes and altered locomotor activity, feeding pattern and sleep regulation (31; 33; 36; 37).
5.3. Nuclear receptors
Beside Rev-erbα, which will be discussed below, several nuclear receptors (such as PPARα and PPARγ) and the nuclear receptor co-regulator PGC1α show strong circadian pattern in numerous tissues suggesting that they may link nutrient sensing and circadian control of metabolism. PGC1α has been shown to enhance Bmal1 and Rev-erbα transcription through RORα transcriptional activity potentiation, and liver-specific PGC1α-deficiency leads to arrhythmicity (46). Interestingly, PGC1α has been shown to play a pivotal role in the response to fasting, a condition under which its expression increases in the liver. In this organ, PGC1α triggers the fasting-induced activation of the gluconeogenic pathway and fatty acid oxidation, and mice deficient for PGC1α display hypoglycaemia and hepatic steatosis (43; 61). The fasting/feeding transition is one of the most predictable daily changes. It implies coordinated metabolic adjustments in several metabolic tissues in order to maintain fuel supply and energy homeostasis at all times. Because PGC1α interacts with different nuclear receptors and transcription factors, it may be part of the coordinated response necessary to face these metabolic challenges.
Feeding time is a dominant zeitgeber of the peripheral clock (15), and food access restriction inverses the phase of circadian expression in mouse liver. Remarkably, glucocorticoids have been suggested to interfere with that process since mice harbouring a hepato-specific deletion of the glucocorticoid receptor invert their circadian liver gene expression more rapidly after the shift of food availability to day-time (39). With a different protocol, other data have been reported indicating restricted feeding but not corticosterone injection may entrain the liver clock in rat (74).
Nutrients derived from food, in particular fatty acids, may activate PPARα, a key regulator of fatty acid metabolism whose expression oscillates diurnally in liver (42). Interestingly, PPARα binds to the Bmal1 promoter and regulates its expression (10). The CLOCK/Bmal1 heterodimer reciprocally regulates PPARα (30). Finally, PPARα increases Rev-erbα expression in human liver cells (25). Thus nuclear receptors may entrain circadian variations in their target genes and thus participate to the circadian control of metabolic pathways.
6. Rev-erbα: a clock gene and nuclear receptor transducing clock signals into metabolic regulations
6.1. Rev-erbα: a new target in circadian disorders?
Mice deficient for Rev-erbα display a 0.5h shorter period and more heterogeneously distributed period length. They are also more sensitive to light pulse-induced phase shift compared to wild-type controls (60). Heme has been identified as a natural Rev-erbα ligand which potentiates the recruitment of the nuclear co-repressor NCoR, thereby enhancing Rev-erbα-mediated transcriptional repression of its target genes (62; 89). The Drosophila homolog of Rev-erbα, called E75, had already been reported to bind heme, which regulates E75 stability and transcriptional activity. Interestingly, intracellular heme levels oscillate and may therefore affect the transcriptional repressive activity of Reverbα. A synthetic ligand for Rev-erbα has been recently developed, which increases the recruitment of NCoR and Rev-erbα transcriptional activity (52). Interestingly, this compound induces circadian phase shift in vitro. This indicates that one can modulate Rev-erbα activity and that this modulation impacts on circadian rhythms, identifying Rev-erbα as an interesting pharmacological target for phase-resetting drugs. One might also speculate that other ‘orphans’ such as the ROR isotypes, might be adopted in the future. This would offer the possibility to affect the ROR/Rev-erb regulatory loop pharmacologically, supposedly in a tissue specific manner. It is also noteworthy that the synthetic Rev-erbα ligand has been shown to induce phase advance as well as phase delay in vitro, depending on the abundance of Rev-erbα. This observation indicates that treatment response to Rev-erbα ligands will ultimately depend on the time of administration. Thus, the discovery that Rev-erbα acts as a liganded nuclear receptor opens a new area of research of more selective Rev-erbα modulators.
6.2. Rev-erbα links circadian clock to energy homeostasis
Rev-erbα is also an important metabolic regulator expressed in a circadian manner in tissues such as liver, adipose tissue, muscle and pancreas. Indeed, Rev-erbα modulates lipid, glucose and bile acid metabolism, adipogenesis and the inflammatory reaction. Intuitively then, it is likely that Rev-erbα may relay circadian signals into metabolic and inflammatory regulatory responses and vice versa (Figure 3).
Figure 3.
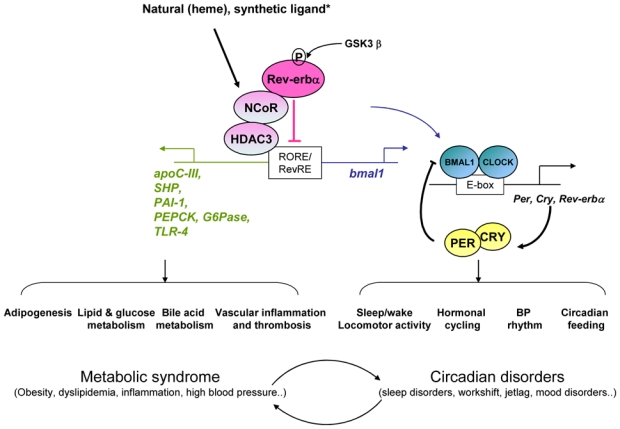
Rev-erbα binds to RORE/RevRE sites located in the promoter of its target genes. Rev-erbα is critical for the proper timing of the core clock machinery. Rev-erbα also regulates metabolic functions including lipid and bile acid metabolism, adipogenesis, gluconeogenesis, the inflammatory response and thrombosis. Rev-erbα ligands modify its transcriptional activity and suggest that Rev-erbα may constitute a promising target for the treatment of circadian alterations of metabolic pathways. * synthetic ligand: see Meng et al. BP, Blood pressure.
6.2.1. Rev-erbα participates in lipid and glucose metabolism
Rev-erbα down-regulates the expression of liver apolipoprotein C-III, a constituent of very low density lipoproteins, and Rev-erbα-deficient mice exhibit a dyslipidemia characterized by increased triglyceride levels (19; 65). Rev-erbα has also been suggested to repress the expression of elovl3, a gene encoding a very long chain fatty acid elongase (4). These data indicate that Rev-erbα modulates both systemic and hepatic lipid metabolism. It also influences de novo glucose synthesis. Indeed, Lazar and colleagues have demonstrated that heme binding to Rev-erbα, resulting in enhanced transcriptional activity, leads to increased repression of the gluconeogenic Pepck gene in vitro in human hepatoma HepG2 cells (89). It remains unclear whether Rev-erbα affects circadian variations in glucose levels. Nonetheless, these data indicate that modulation of Rev-erbα activity is likely to affect lipid and glucose metabolism in a circadian fashion.
6.2.2. Rev-erbα connects the adipose tissue clock and adipogenesis
Altered circadian patterns due to work shift or sleep disorders are associated with increased BMI. It is tempting to speculate that chronic alteration in circadian variations of various adipogenic regulators such as PPARγ, or other genes involved in lipid uptake might result in abnormal circadian pattern of adipocyte physiology with subsequent changes in differentiation, lipid storage and adipokine secretion (9). Rev-erbα expression cycles in adipose tissue (90), and is induced during the adipogenic process and by PPARγ activation by rosiglitazone in rat adipose tissue (13; 22). Ectopic over-expression of Rev-erbα in 3T3L1 pre-adipocytes promotes their differentiation into mature adipocytes and enhances lipid storage (22; 83). In a recent paper, Lazar and colleagues demonstrated that impaired recruitment of the NCoR/HDAC3 complex to the Rev-erbα response element present in the Bmal1 promoter results in aberrant circadian pattern of the canonical clock genes leading, in turn, to altered diurnal expression of genes involved in metabolism (2). Strikingly, this was associated in vivo to a lean phenotype (NCoR mutant mice having less adipose tissue) and ameliorated metabolic phenotype. This does not exclude that Rev-erbα may exert both circadian-independent functions in the adipose tissue, eg on adipogenesis, and a circadian role through the control of the rhythmic alternation between fat storage and disposal.
6.2.3. Rev-erbα participates in the circadian control of bile acid metabolism
Bile acids are biologically active molecules which serve for lipid solubilization in the intestinal lumen and modulate lipid and glucose metabolism (41). The cholesterol 7α-hydroxylase (Cyp7A1) gene, which encodes the first and ratecontrolling enzyme of the major bile acid biosynthetic pathway, exhibits a strong diurnal pattern of expression (14). We have recently shown that Rev-erbα crosstalks with the bile acid receptor FXR for the regulation of the Small Heterodimer Partner (Shp) and modulates Cyp7A1 expression through altered circadian expression of the two CYP7A1 regulators Shp and E4BP4 (also known as Nfil3) (19). The albumin site D-binding protein (DBP) is a clock output protein which, together with the hepatocyte leukaemia factor (HLF) and the thyrotroph embryonic factor (TEF), belongs to the PARbZip family of transcription factors. These well conserved mammalian proteins display strong circadian variations in numerous tissues, and play important metabolic roles. In particular, they are important for xenobiotic detoxification. Since DBP and E4BP4 bind to and regulate common sets of target genes (82), one might speculate that Rev-erbα may play a more broaden role in regulating clock output signals as well as specific catabolic pathways.
6.3. Rev-erbα and cardio-vascular diseases
The cardiovascular system displays circadian variations in numerous outputs such as blood pressure, heart rate, thrombolysis cascade, etc.. and myocardial infarction and stroke are known to peak early in the morning. Rev-erbα has been shown to modulate metabolism of triglyceride-rich lipoproteins which are associated to increased risk of cardio-vascular disease, thereby possibly affecting the development of atherosclerosis (65). On the other hand, atherosclerosis is also characterized by an inflammatory status of the vascular wall and the presence of macrophages within the lesion. Rev-erbα is present in vascular wall cells including macrophages (7; 23; 53). In human macrophages, Rev-erbα represses the induction of toll like receptor (TLR)-4, the receptor of lipopolysaccharide (LPS), thereby diminishing the production of cytokines in response to LPS. These data demonstrate an anti-inflammatory role of Reverbα (23) and suggest a positive impact of Rev-erbα on atherosclerotic lesions. Along the same line, Rev-erbα has been identified as a potent negative regulator of plasminogen activator inhibitor (PAI)-1, an important inhibitor of the fibrinolysis cascade that may promote the development of atherothrombosis (84). In humans, PAI-1 oscillates diurnally with a zenith in the early morning which coincides with acute thrombotic and cardiovascular events such as myocardial infarction. GSK3β enhances PAI-1 repression via phosphorylation and stabilization of the Rev-erbα protein, and a phosphorylated form of Rev-erbα dampens PAI-1 oscillations. This suggests that Rev-erbα may affect the circadian expression levels and rhythmicity of PAI-1, and affect the fibrinolysis cascade in a circadian manner.
7. Future perspectives
Although the biological functions of Rev-erbα are still not completely understood, it plays crucial role in the core clock regulation on the one hand, and, on the other hand, in lipid and adipogenic control, the inflammatory response, and has been suggested to influence the gluconeogenic pathway. It is therefore likely that Rev-erbα mediates metabolic circadian rhythmicity. Yet, except for the shorter circadian period displayed by the Rev-erbα-deficient mice, we still have little knowledge on how Rev-erbα connects circadian disorders to metabolic pathologies. A better characterization of the biological functions of Rev-erbα in metabolism in vivo will contribute to an improved understanding of the integration between circadian and metabolic pathways.
Recently, several studies have identified novel natural (heme) and synthetic ligands for Rev-erbα and revealed the possibility to increase Rev-erbα activity through enhanced recruitment of its co-repressor NCoR. In addition, Rev-erbα modulation was proven to be efficient in phase-advance or delay circadian rhythms, at least in vitro. Therefore, Rev-erbα ligands may be useful in resetting the clock after jetlag or work shift, and re-entrain and put in resonance our body clocks. One can also hypothesize that the time of administration of a drug affecting Rev-erbα activity will be an important factor to consider for its therapeutic efficacy. Given the broaden role of Rev-erbα in lipid metabolism, adipogenesis and vascular wall patho-physiology, Rev-erbα represents a promising target for the treatment of metabolic abnormalities resulting from chronic aberrant circadian rhythms. The next step will be to determine whether these ligands may affect its capacity to regulate target genes in vivo and normalize circadian disorder-related metabolic abnormalities.
Acknowledgments
The authors acknowledge funding supports from INSERM, Contrat Plan Etat Région (CPER), the Région Nord Pas-de-Calais/FEDER, the Sixth EU Research Framework Program Diabesity (contract LSHM-CT-2003-503041), COST-action BM0602
Reference List
- 1.Akashi M, Takumi T. The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nat Struct Mol Biol. 2005;12:441–448. doi: 10.1038/nsmb925. [DOI] [PubMed] [Google Scholar]
- 2.Alenghat T, Meyers K, Mullican SE, Leitner K, Adeniji-Adele A, Avila J, Bucan M, Ahima RS, Kaestner KH, Lazar MA. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature. 2008;456:997–1000. doi: 10.1038/nature07541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ando H, Yanagihara H, Hayashi Y, Obi Y, Tsuruoka S, Takamura T, Kaneko S, Fujimura A. Rhythmic messenger ribonucleic acid expression of clock genes and adipocytokines in mouse visceral adipose tissue. Endocrinology. 2005;146:5631–5636. doi: 10.1210/en.2005-0771. [DOI] [PubMed] [Google Scholar]
- 4.Anzulovich A, Mir A, Brewer M, Ferreyra G, Vinson C, Baler R. Elovl3: a model gene to dissect homeostatic links between the circadian clock and nutritional status. J Lipid Res. 2006;47:2690–2700. doi: 10.1194/jlr.M600230-JLR200. [DOI] [PubMed] [Google Scholar]
- 5.Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through Per2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 6.Balmforth AJ, Grant PJ, Scott EM, Wheatcroft SB, Kearney MT, Staels B, Marx N. Inter-subject differences in constitutive expression levels of the clock gene in man. Diab Vasc Dis Res. 2007;4:39–43. doi: 10.3132/dvdr.2007.004. [DOI] [PubMed] [Google Scholar]
- 7.Barish GD, Downes M, Alaynick WA, Yu RT, Ocampo CB, Bookout AL, Mangelsdorf DJ, Evans RM. A Nuclear Receptor Atlas: macrophage activation. Mol Endocrinol. 2005;19:2466–2477. doi: 10.1210/me.2004-0529. [DOI] [PubMed] [Google Scholar]
- 8.Bordone L, Guarente L. Calorie restriction, SIRT1 and metabolism: understanding longevity. Nat Rev Mol Cell Biol. 2005;6:298–305. doi: 10.1038/nrm1616. [DOI] [PubMed] [Google Scholar]
- 9.Bray MS, Young ME. Circadian rhythms in the development of obesity: potential role for the circadian clock within the adipocyte. Obes Rev. 2007;8:169–181. doi: 10.1111/j.1467-789X.2006.00277.x. [DOI] [PubMed] [Google Scholar]
- 10.Canaple L, Rambaud J, Dkhissi-Benyahya O, Rayet B, Tan NS, Michalik L, Delaunay F, Wahli W, Laudet V. Reciprocal regulation of brain and muscle Arnt-like protein 1 and peroxisome proliferator-activated receptor alpha defines a novel positive feedback loop in the rodent liver circadian clock. Mol Endocrinol. 2006;20:1715–1727. doi: 10.1210/me.2006-0052. [DOI] [PubMed] [Google Scholar]
- 11.Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. AMPK regulates energy expenditure by modulating NAD(+) metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cardone L, Hirayama J, Giordano F, Tamaru T, Palvimo JJ, Sassone- Corsi P. Circadian clock control by SUMOylation of Bmal1. Science. 2005;309:1390–1394. doi: 10.1126/science.1110689. [DOI] [PubMed] [Google Scholar]
- 13.Chawla A, Lazar MA. Induction of Rev-ErbA alpha, an orphan receptor encoded on the opposite strand of the alpha-thyroid hormone receptor gene, during adipocyte differentiation. J Biol Chem. 1993;268:16265–16269. [PubMed] [Google Scholar]
- 14.Chiang JY, Miller WF, Lin GM. Regulation of cholesterol 7 alphahydroxylase in the liver. Purification of cholesterol 7 alpha-hydroxylase and the immunochemical evidence for the induction of cholesterol 7 alpha-hydroxylase by cholestyramine and circadian rhythm. J Biol Chem. 1990;265:3889–3897. [PubMed] [Google Scholar]
- 15.Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di Lorenzo L, De Pergola G, Zocchetti C, L’Abbate N, Basso A, Pannacciulli N, Cignarelli M, Giorgino R, Soleo L. Effect of shift work on body mass index: results of a study performed in 319 glucose-tolerant men working in a Southern Italian industry. International Journal of Obesity. 2003;27:1353–1358. doi: 10.1038/sj.ijo.0802419. [DOI] [PubMed] [Google Scholar]
- 17.Dibner C, Sage D, Unser M, Bauer C, d’Eysmond T, Naef F, Schibler U. Circadian gene expression is resilient to large fluctuations in overall transcription rates. EMBO J. 2009;28:123–134. doi: 10.1038/emboj.2008.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 19.Duez H, van der Veen JN, Duhem C, Pourcet B, Touvier T, Fontaine C, Derudas B, Bauge E, Havinga R, Bloks VW, Wolters H, van der Sluijs FH, Vennstrom B, Kuipers F, Staels B. Regulation of bile acid synthesis by the nuclear receptor Rev-erbalpha. Gastroenterology. 2008;135:689–698. doi: 10.1053/j.gastro.2008.05.035. [DOI] [PubMed] [Google Scholar]
- 20.Eide EJ, Vielhaber EL, Hinz WA, Virshup DM. The circadian regulatory proteins Bmal1 and cryptochromes are substrates of casein kinase I epsilon. Journal of Biological Chemistry. 2002;277:17248–17254. doi: 10.1074/jbc.M111466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Etchegaray JP, Lee C, Wade PA, Reppert SM. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421:177–182. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- 22.Fontaine C, Dubois G, Duguay Y, Helledie T, Vu-Dac N, Gervois P, Soncin F, Mandrup S, Fruchart JC, Fruchart-Najib J, Staels B. The orphan nuclear receptor Rev-Erbalpha is a peroxisome proliferator-activated receptor (PPAR) gamma target gene and promotes PPARgamma-induced adipocyte differentiation. J Biol Chem. 2003;278:37672–37680. doi: 10.1074/jbc.M304664200. [DOI] [PubMed] [Google Scholar]
- 23.Fontaine C, Rigamonti E, Pourcet B, Duez H, Duhem C, Fruchart JC, Chinetti-Gbaguidi G, Staels B. The nuclear receptor Rev-erbalpha is a liver X receptor (LXR) target gene driving a negative feedback loop on select LXRinduced pathways in human macrophages. Mol Endocrinol. 2008;22:1797–1811. doi: 10.1210/me.2007-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- 25.Gervois P, Chopin-Delannoy S, Fadel A, Dubois G, Kosykh V, Fruchart JC, Najib J, Laudet V, Staels B. Fibrates increase human Rev-erbalpha expression in liver via a novel peroxisome proliferator-activated receptor response element. Mol Endocrinol. 1999;13:400–409. doi: 10.1210/mend.13.3.0248. [DOI] [PubMed] [Google Scholar]
- 26.Gomez-Abellan P, Hernandez-Morante JJ, Lujan JA, Madrid JA, Garaulet M. Clock genes are implicated in the human metabolic syndrome. Int J Obes (Lond) 2008;32:121–128. doi: 10.1038/sj.ijo.0803689. [DOI] [PubMed] [Google Scholar]
- 27.Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728– 742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guillaumond F, Dardente H, Giguere V, Cermakian N. Differential control of Bmal1 circadian transcription by Rev-erb and ROR nuclear receptors. J Biol Rhythms. 2005;20:391–403. doi: 10.1177/0748730405277232. [DOI] [PubMed] [Google Scholar]
- 29.Hirayama J, Sahar S, Grimaldi B, Tamaru T, Takamatsu K, Nakahata Y, Sassone-Corsi P. CLOCK-mediated acetylation of Bmal1 controls circadian function. Nature. 2007;450:1086–1090. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- 30.Inoue I, Shinoda Y, Ikeda M, Hayashi K, Kanazawa K, Nomura M, Matsunaga T, Xu H, Kawai S, Awata T, Komoda T, Katayama S. CLOCK/Bmal1 is involved in lipid metabolism via transactivation of the peroxisome proliferator-activated receptor (PPAR) response element. J Atheroscler Thromb. 2005;12:169–174. doi: 10.5551/jat.12.169. [DOI] [PubMed] [Google Scholar]
- 31.Jenkins JB, Omori T, Guan Z, Vgontzas AN, Bixler EO, Fang J. Sleep is increased in mice with obesity induced by high-fat food. Physiol Behav. 2006;87:255–262. doi: 10.1016/j.physbeh.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 32.Karlsson B, Knutsson A, Lindahl B. Is there an association between shift work and having a metabolic syndrome? Results from a population based study of 27,485 people. Occup Environ Med. 2001;58:747–752. doi: 10.1136/oem.58.11.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, Turek FW, Bass J. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 34.Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. Systemdriven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci U S A. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laposky AD, Bradley MA, Williams DL, Bass J, Turek FW. Sleep-wake regulation is altered in leptin-resistant (db/db) genetically obese and diabetic mice. Am J Physiol Regul Integr Comp Physiol. 2008;295:R2059–R2066. doi: 10.1152/ajpregu.00026.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laposky AD, Shelton J, Bass J, Dugovic C, Perrino N, Turek FW. Altered sleep regulation in leptin-deficient mice. Am J Physiol Regul Integr Comp Physiol. 2006;290:R894–R903. doi: 10.1152/ajpregu.00304.2005. [DOI] [PubMed] [Google Scholar]
- 38.Lau P, Fitzsimmons RL, Raichur S, Wang SC, Lechtken A, Muscat GE. The orphan nuclear receptor, RORalpha, regulates gene expression that controls lipid metabolism: staggerer (SG/SG) mice are resistant to diet-induced obesity. J Biol Chem. 2008;283:18411–18421. doi: 10.1074/jbc.M710526200. [DOI] [PubMed] [Google Scholar]
- 39.Le Minh N, Damiola F, Tronche F, Schutz G, Schibler U. Glucocorticoid hormones inhibit food-induced phase-shifting of peripheral circadian oscillators. EMBO J. 2001;20:7128–7136. doi: 10.1093/emboj/20.24.7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee J, Lee Y, Lee MJ, Park E, Kang SH, Chung CH, Lee KH, Kim K. Dual modification of Bmal1 by SUMO2/3 and ubiquitin promotes circadian activation of the CLOCK/Bmal1 complex. Mol Cell Biol. 2008;28:6056–6065. doi: 10.1128/MCB.00583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lefebvre P, Cariou B, Lien F, Kuipers F, Staels B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol Rev. 2009;89:147–191. doi: 10.1152/physrev.00010.2008. [DOI] [PubMed] [Google Scholar]
- 42.Lemberger T, Saladin R, Vazquez M, Assimacopoulos F, Staels B, Desvergne B, Wahli W, Auwerx J. Expression of the peroxisome proliferator-activated receptor alpha gene is stimulated by stress and follows a diurnal rhythm. J Biol Chem. 1996;271:1764–1769. doi: 10.1074/jbc.271.3.1764. [DOI] [PubMed] [Google Scholar]
- 43.Lin J, Wu PH, Tarr PT, Lindenberg KS, St Pierre J, Zhang CY, Mootha VK, Jager S, Vianna CR, Reznick RM, Cui L, Manieri M, Donovan MX, Wu Z, Cooper MP, Fan MC, Rohas LM, Zavacki AM, Cinti S, Shulman GI, Lowell BB, Krainc D, Spiegelman BM. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 44.Lin JM, Kilman VL, Keegan K, Paddock B, Emery-Le M, Rosbash M, Allada R. A role for casein kinase 2 alpha in the Drosophila circadian clock. Nature. 2002;420:816–820. doi: 10.1038/nature01235. [DOI] [PubMed] [Google Scholar]
- 45.Liu AC, Tran HG, Zhang EE, Priest AA, Welsh DK, Kay SA. Redundant function of Rev-erbalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 2008;4:e1000023. doi: 10.1371/journal.pgen.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu C, Li SM, Liu TH, Borjigin J, Lin JD. Transcriptional coactivator PGC-1a integrates the mammalian clock and energy metabolism. Nature. 2007;447:477–4U4. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- 47.Lowrey PL, Shimomura K, Antoch MP, Yamazaki S, Zemenides PD, Ralph MR, Menaker M, Takahashi JS. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science. 2000;288:483–492. doi: 10.1126/science.288.5465.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lund J, Arendt J, Hampton SM, English J, Morgan LM. Postprandial hormone and metabolic responses amongst shift workers in Antarctica. J Endocrinol. 2001;171:557–564. doi: 10.1677/joe.0.1710557. [DOI] [PubMed] [Google Scholar]
- 49.Mamontova A, Seguret-Mace S, Esposito B, Chaniale C, Bouly M, Delhaye- Bouchaud N, Luc G, Staels B, Duverger N, Mariani J, Tedgui A. Severe atherosclerosis and hypoalphalipoproteinemia in the staggerer mouse, a mutant of the nuclear receptor RORalpha. Circulation. 1998;98:2738–2743. doi: 10.1161/01.cir.98.24.2738. [DOI] [PubMed] [Google Scholar]
- 50.Martino TA, Oudit GY, Herzenberg AM, Tata N, Koletar MM, Kabir GM, Belsham DD, Backx PH, Ralph MR, Sole MJ. Circadian rhythm disorganization produces profound cardiovascular and renal disease in hamsters. Am J Physiol Regul Integr Comp Physiol. 2008;294:R1675–R1683. doi: 10.1152/ajpregu.00829.2007. [DOI] [PubMed] [Google Scholar]
- 51.McDearmon EL, Patel KN, Ko CH, Walisser JA, Schook AC, Chong JL, Wilsbacher LD, Song EJ, Hong HK, Bradfield CA, Takahashi JS. Dissecting the functions of the mammalian clock protein Bmal1 by tissuespecific rescue in mice. Science. 2006;314:1304–1308. doi: 10.1126/science.1132430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meng QJ, McMaster A, Beesley S, Lu WQ, Gibbs J, Parks D, Collins J, Farrow S, Donn R, Ray D, Loudon A. Ligand modulation of Reverbalpha function resets the peripheral circadian clock in a phasic manner. J Cell Sci. 2008;121:3629–3635. doi: 10.1242/jcs.035048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Migita H, Morser J, Kawai K. Rev-erbalpha upregulates NF-kappaBresponsive genes in vascular smooth muscle cells. FEBS Lett. 2004;561:69–74. doi: 10.1016/S0014-5793(04)00118-8. [DOI] [PubMed] [Google Scholar]
- 54.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian Control of the NAD+ Salvage Pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 57.Oishi K, Atsumi G, Sugiyama S, Kodomari I, Kasamatsu M, Machida K, Ishida N. Disrupted fat absorption attenuates obesity induced by a high-fat diet in Clock mutant mice. FEBS Lett. 2006;580:127–130. doi: 10.1016/j.febslet.2005.11.063. [DOI] [PubMed] [Google Scholar]
- 58.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 59.Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado DO, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor Rev-erbalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 61.Puigserver P. Tissue-specific regulation of metabolic pathways through the transcriptional coactivator PGC1-alpha. Int J Obes (Lond) 2005;29 (Suppl 1):S5–S9. doi: 10.1038/sj.ijo.0802905. [DOI] [PubMed] [Google Scholar]
- 62.Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, Burris LL, Khorasanizadeh S, Burris TP, Rastinejad F. Identification of heme as the ligand for the orphan nuclear receptors Rev-erbalpha and Reverbbeta. Nat Struct Mol Biol. 2007;14:1207–1213. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, Takahashi JS, Imai SI, Bass J. Circadian Clock Feedback Cycle Through NAMPT-Mediated NAD+ Biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Raspe E, Duez H, Gervois P, Fievet C, Fruchart JC, Besnard S, Mariani J, Tedgui A, Staels B. Transcriptional regulation of apolipoprotein C-III gene expression by the orphan nuclear receptor RORalpha. J Biol Chem. 2000;276:2865–2871. doi: 10.1074/jbc.M004982200. [DOI] [PubMed] [Google Scholar]
- 65.Raspe E, Duez H, Mansen A, Fontaine C, Fievet C, Fruchart JC, Vennstrom B, Staels B. Identification of Rev-erbalpha as a physiological repressor of apoC-III gene transcription. J Lipid Res. 2002;43:2172–2179. doi: 10.1194/jlr.m200386-jlr200. [DOI] [PubMed] [Google Scholar]
- 66.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 67.Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, FitzGerald GA. Bmal1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rudic RD, McNamara P, Reilly D, Grosser T, Curtis AM, Price TS, Panda S, Hogenesch JB, FitzGerald GA. Bioinformatic analysis of circadian gene oscillation in mouse aorta. Circulation. 2005;112:2716–2724. doi: 10.1161/CIRCULATIONAHA.105.568626. [DOI] [PubMed] [Google Scholar]
- 69.Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 70.Scheer FA, Hilton MF, Mantzoros CS, Shea SA. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci U S A. 2009;106:4453–4458. doi: 10.1073/pnas.0808180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Scott EM, Carter AM, Grant PJ. Association between polymorphisms in the Clock gene, obesity and the metabolic syndrome in man. Int J Obes (Lond) 2008;32:658–662. doi: 10.1038/sj.ijo.0803778. [DOI] [PubMed] [Google Scholar]
- 72.Shimba S, Ishii N, Ohta Y, Ohno T, Watabe Y, Hayashi M, Wada T, Aoyagi T, Tezuka M. Brain and muscle Arnt-like protein-1 (Bmal1), a component of the molecular clock, regulates adipogenesis. Proc Natl Acad Sci U S A. 2005;102:12071–12076. doi: 10.1073/pnas.0502383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sookoian S, Gemma C, Gianotti TF, Burgueno A, Castano G, Pirola CJ. Genetic variants of Clock transcription factor are associated with individual susceptibility to obesity. Am J Clin Nutr. 2008;87:1606–1615. doi: 10.1093/ajcn/87.6.1606. [DOI] [PubMed] [Google Scholar]
- 74.Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- 75.Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 76.Suwazono Y, Dochi M, Sakata K, Okubo Y, Oishi M, Tanaka K, Kobayashi E, Kido T, Nogawa K. A longitudinal study on the effect of shift work on weight gain in male Japanese workers. Obesity (Silver Spring) 2008;16:1887–1893. doi: 10.1038/oby.2008.298. [DOI] [PubMed] [Google Scholar]
- 77.Suwazono Y, Dochi M, Sakata K, Okubo Y, Oishi M, Tanaka K, Kobayashi E, Nogawa K. Shift work is a risk factor for increased blood pressure in Japanese men: a 14-year historical cohort study. Hypertension. 2008;52:581–586. doi: 10.1161/HYPERTENSIONAHA.108.114553. [DOI] [PubMed] [Google Scholar]
- 78.Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tamaru T, Hirayama J, Isojima Y, Nagai K, Norioka S, Takamatsu K, Sassone-Corsi P. CK2alpha phosphorylates Bmal1 to regulate the mammalian clock. Nat Struct Mol Biol. 2009;16:446–448. doi: 10.1038/nsmb.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tsuchiya Y, Akashi M, Matsuda M, Goto K, Miyata Y, Node K, Nishida E. Involvement of the protein kinase CK2 in the regulation of mammalian circadian rhythms. Sci Signal. 2009;2:ra26. doi: 10.1126/scisignal.2000305. [DOI] [PubMed] [Google Scholar]
- 81.Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, Eckel RH, Takahashi JS, Bass J. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ueda HR, Hayashi S, Chen WB, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nature Genetics. 2005;37:187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 83.Wang J, Lazar MA. Bifunctional role of Rev-erbalpha in adipocyte differentiation. Mol Cell Biol. 2008;28:2213–2220. doi: 10.1128/MCB.01608-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang J, Yin L, Lazar MA. The orphan nuclear receptor Rev-erb alpha regulates circadian expression of plasminogen activator inhibitor type 1. J Biol Chem. 2006;281:33842–33848. doi: 10.1074/jbc.M607873200. [DOI] [PubMed] [Google Scholar]
- 85.Woon PY, Kaisaki PJ, Braganca J, Bihoreau MT, Levy JC, Farrall M, Gauguier D. Aryl hydrocarbon receptor nuclear translocator-like (Bmal1) is associated with susceptibility to hypertension and type 2 diabetes. Proc Natl Acad Sci U S A. 2007;104:14412–14417. doi: 10.1073/pnas.0703247104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 87.Yin L, Lazar MA. The orphan nuclear receptor Rev-erbalpha recruits the NCoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Mol Endocrinol. 2005;19:1452–1459. doi: 10.1210/me.2005-0057. [DOI] [PubMed] [Google Scholar]
- 88.Yin L, Wang J, Klein PS, Lazar MA. Nuclear receptor Rev-erbalpha is a critical lithium-sensitive component of the circadian clock. Science. 2006;311:1002– 1005. doi: 10.1126/science.1121613. [DOI] [PubMed] [Google Scholar]
- 89.Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, Lazar MA. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- 90.Zvonic S, Ptitsyn AA, Conrad SA, Scott LK, Floyd ZE, Kilroy G, Wu X, Goh BC, Mynatt RL, Gimble JM. Characterization of peripheral circadian clocks in adipose tissues. Diabetes. 2006;55:962–970. doi: 10.2337/diabetes.55.04.06.db05-0873. [DOI] [PubMed] [Google Scholar]


