Abstract
Measurements of population dynamics are ubiquitous in experiments with microorganisms. Studies with microbes elucidating adaptation, selection, and competition rely on measurements of changing populations in time. Despite this importance, quantitative methods for measuring population dynamics microscopically, with high time resolution, across many replicates remain limited. Here we present a new noninvasive method to precisely measure microbial spatiotemporal population dynamics based on digital in-line holographic (DIH) microscopy. Our inexpensive, replicate DIH microscopes imaged hundreds of swimming algae in three dimensions within a volume of several microliters on a time scale of minutes over periods of weeks.
INTRODUCTION
Microbial populations are tractable experimental systems for studying adaptation, evolution, and ecology. Microbial systems benefit from short generation times, small sizes, large populations, and the ability to survive in well-defined environments. Populations of microorganisms are amenable to the study of processes across time scales, from phenotypic changes within a single generation to mutation and selection across many generations in a large population. Studies on microbial populations have elucidated fundamental biological processes including rapid adaptation to environmental conditions1 and evolutionary divergence in population density and genotype across replicates.2
Existing methods for measuring population dynamics, such as plating or flow cytometry, are invasive and labor intensive. As a result, these methods can perturb dynamics and have limited temporal resolution. Noninvasive bulk optical methods such as optical density or fluorescence achieve high temporal resolution but cannot resolve single cells and have limited dynamic range. They can also be confounded by changes in cell morphology, fluorescence intensity, or aggregation, especially over long time scales.
Ideally, measurements of microbial population dynamics should resolve single cells, be minimally invasive, achieve high temporal resolution and large dynamic range, and be performed on replicate populations over long periods of time. With the exception of recent microfluidic advances for following small populations of bacteria,3, 4 no general method has been presented to satisfy these constraints.
Holography is a technique for reconstructing three-dimensional images from a two-dimensional interference pattern, the hologram.5 Digital holography records the hologram on a digital image sensor and reconstructs the image numerically. Here we employ lensless digital in-line holographic (DIH) microscopy to measure microbial population dynamics. DIH microscopy permits the detection of single cells throughout a microliter sized imaging volume rapidly and noninvasively. DIH microscopy is simple, cost effective, and reliable, permitting the construction of replicate microscopes.
Our implementation of digital holographic microscopy is lensless, requiring no objective for magnification, and inline so the scattered and reference wave share a common optical path. Lensless holography uses a diverging spherical wave for illumination while restricting the sample to regions close to the source so that the digital sensor adequately samples interference fringes without magnification.6, 7, 8 Here we produce this spherical wave using a gradient index (GRIN) lens. Unlike a pinhole, the GRIN lens requires no precise alignment and is mechanically robust. Further, inline holographic microscopy employs a single optical path so light sources of comparatively short coherence length, such as laser diodes, can be used.7 Laser diodes obviate the need for mechanical shutters that reduce reliability. For samples of sufficiently low optical density, such as microbial populations in liquid culture, inline holography is applicable.
Digital in-line holographic microscopy has been used to measure trajectories of chemotaxing algae and to study swimming behavior between predator and prey.6, 7 DIH microscopy has not been used to measure microbial population dynamics despite clear advantages over existing methods. A significant technical obstacle to measuring population dynamics by DIH microscopy is optical variations across the relatively large imaged region. Here we address these variations by empirically measuring attenuation across the imaged volume and subsequently normalizing reconstructed intensities to allow global image segmentation. This approach is independent of cell density and is thus better suited to measuring population dynamics over a large dynamic range of cell density compared to local variance based methods.9
We constructed ten DIH microscopes, each incorporating independent control of hologram acquisition, temperature, and white light intensity for studying microbial photoresponse. Our method is well suited for a variety of model microorganisms including Escherichia coli, Chlamydomonas reinhardtii, and Tetrahymena thermophila. We describe the apparatus, acquisition and analysis procedures, optical properties of the imaging, and calibrations of the density measurement, and present data from experiments with the ∼5 μm unicellular alga Chlamydomonas reinhardtii.
APPARATUS
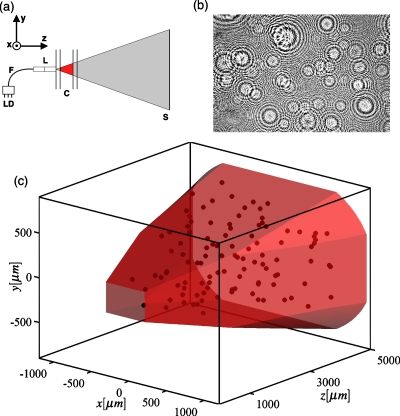
The optical system is depicted in Fig. 1a. The optical axis of the system is in the +z direction [Fig. 1a]. A 635 nm 1 mW laser diode (Blue Sky Research, Malpitas, CA) is coupled to a single mode fiber. The fiber is terminated in a precision glass ferrule (Aria Technologies, Livermore, CA). The ferrule is coupled to a custom lens assembly consisting of a glass spacer and a custom pitch 0.5 numerical aperture (NA) GRIN lens (GRINTech GmbH, Jena, Germany), which focuses the beam from the fiber to a point on the distal face of the lens, from which it diverges with a 1∕e2 NA of 0.27. The beam illuminates a sample held in an antireflective coated quartz cuvette with interior dimensions 5×10×40 mm3 of 1.25 mm thick quartz (Starna, Atascadero, CA). The lens contacts one 10×40 mm2 wall of the cuvette, with optical gel matching the refractive index of the quartz in the interface (Cargille Laboratories, Cedar Grove, NJ). The focus of the GRIN lens is nominally 2 mm below the center of the 10 mm dimension of the cuvette and centered in the x direction.
Figure 1.
(a) Schematic of the DIH microscope. A 500 μs pulse of 635 nm light triggered from the laser diode (LD) passes through a single mode optical fiber (F) which is coupled to a GRIN lens (L). An approximately spherical wave originating at the lens focus passes through the cuvette (C) and is incident on the sensor (S). (b) An example of a normalized hologram. (c) Three dimensional view of the imaging volume with centroid locations of 110 algae from the hologram in (b). The intersection of two regions sets the shape of the imaging volume (shaded): the region where the signal-to-background is sufficient after maximum intensity normalization and the projection of the sensor to the focal point of the GRIN lens.
A complementary metal-oxide-semiconductor (CMOS) sensor (3474×2314 square pixels, 6.4 μm pitch, Canon Digital Rebel XT) placed 46.5 mm from the beam focus recorded the hologram. The sensor was extracted from the body of the camera and the infrared filter was removed. The sensor was mounted in the microscope housing and attached to the camera body via ribbon cables shielded by copper tape common to the camera ground. Images were triggered and acquired by the computer via open-source software (GPHOTO, www.gPhoto.org) and RAW formatted images were decoded to 16 bit integers (DCRAW, www.cybercom.net/~dcoffin/dcraw/). The acquisition interval is limited to 4 s by the data transfer speed between the camera and computer.
Two linearized thermistors (Omega Engineering, Stamford, CT) are located in the aluminum cuvette holder of each microscope. The voltage from the thermistor is amplified then sampled by the computer through an analog-to-digital converter (ADC) (LabJack, Lakewood, CO) with a rms noise of 0.003 °C in a 1 Hz bandwidth. Thermistors in all microscopes are calibrated to a reference, reducing systematic error to 0.003 °C. The thermistor voltage is input to a proportional-integral-differential (PID) feedback circuit that drives a Peltier element (Custom Thermoelectric, Bishopville, MD) to provide temperature control of the aluminum housing. The standard deviation of the temperature across microscopes is 0.02 °C. Between 19 and 26 °C the time constant of the PID temperature feedback is 90 s.
Two 1 W, warm white, light-emitting diodes (LEDs) (Luxeon) illuminate the sample from above and below. Two diffusive screens are placed between each LED and the sample.10 A digital-to-analog converter (LabJack) provides independent computer control over the two LEDs. The LED illumination is measured by four photodiodes around the cuvette, which are amplified then recorded by an ADC (LabJack). The response of each photodiode amplifier pair is calibrated to a reference to within 5%. The illumination level can be modulated at up to 1 Hz. Temperature stability is independent of changes in illumination.
Microscope housings are machined from aluminum (www.emachineshop.com), housed in a light-tight cardboard box and mounted on silicone elastomers for vibration isolation. All microscopes are maintained in a temperature-stabilized room.
DATA ACQUISITION AND IMAGE ANALYSIS
The reference beam intensity must be measured for normalization of acquired holograms.10 This is accomplished by projecting the median intensity of several holograms acquired at low cell density (<4×104∕mL for C. reinhardtii), typically taken at the beginning of an experiment. During an experiment, the photodiode and thermistor voltages are measured once per second and written to the computer’s hard drive. The LEDs are turned off for less than 1 s for each acquisition, which consists of a 200 ms exposure by the camera sensor and a 500 μs pulse of the laser. The hologram is downloaded to the computer’s hard drive where it is reconstructed and processed. Reconstruction is performed in ∼320 planes parallel to the sensor with a spacing of 17 μm. The reconstructed amplitude is converted to intensity.
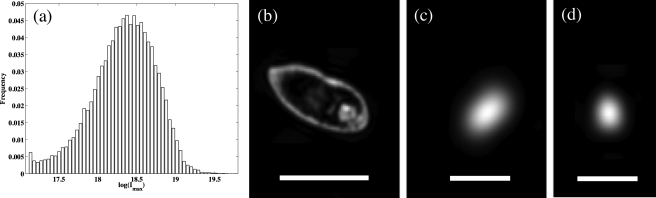
In the implementation of DIH microscopy employed here, a phase approximation is used that permits the evaluation of the diffraction integral by fast Fourier transform. This approximation imparts aberration on the reconstruction that is corrected by considering the effects of the phase approximation analytically.10 However, there are additional factors, some of which are difficult to measure, such as the refractive index of a C. reinhardtii cell, that impart a spatial dependence in the intensity of scatterers within the imaging volume. To address this, we estimated the spatial dependence of reconstructed algal intensities in a control experiment and derived an empirical normalization. For the control experiment, bright objects were segmented at thresholds from a decreasing geometric series of intensities. After each segmentation, the thresholded signals were subtracted from the hologram for reconstruction and segmentation at the next threshold. The maximum intensity of scatterers exhibited a factor of 6 decrease from small to large lateral distance from the optical axis and a factor of 3 decrease from the cuvette wall near the lens to the opposite wall. To quantify these variations, we fit the spatial distribution of log(Imax) (where Imax is the maximum intensity) to a function of the three spatial coordinates. In subsequent experiments, a multiplier calculated from this fit is used to normalize reconstructed intensities. The distribution of log(Imax) normalized by this method shows a good separation of signal due to C. reinhardtii from background, as depicted in Fig. 2a. This allows identification of algae with a global threshold on maximum intensity.
Figure 2.
(a) Histogram of normalized maximum intensities for C. reinhardtii. [(b)–(d)] Reconstructed intensities for Tetrahymena thermophila, Chlamydomonas reihardtii, and Escherichia coli (left to right). The scale bars are 40, 5, and 5 μm. Contrasts are adjusted to show detail.
After normalization and global segmentation, the centroid locations of algae are extracted and the size in voxels, and mean and max intensity are recorded. The reconstructions and analysis are performed on a four core 2.4 GHz CPU (Intel Core 2 Quad 6600), requiring about 400 s per hologram at algal densities below 105∕mL.11
PERFORMANCE
To characterize the resolution of our imaging system, we measured the full width at half maximum of the intensity profile of 1.1 μm diameter polystyrene beads and found 1.7 μm in x, 2.3 μm in y, and 41.5 μm in z (parallel to the optical axis, Fig. 1). Imaging of a 200 μm pitch grid on the distal interior face of a cuvette found no spurious magnification or distortions (<2 μm).
Example images of Tetrahymena thermophila, Chlamydomonas reinhardtii, and Escherichia coli are shown in Figs. 2b, 2c, 2d. T. thermophila is a ∼50 μm motile ciliate that is commonly used as a model organism. E. coli is a ∼1×2μm2 rod-shaped bacterium. Signal from T. thermophila and C. reinhardtii is well above the background, while the signal-to-background ratio for E. coli is 50-fold lower than for algae and approaches the sensitivity limit of the microscope. The sensitivity is limited by laser intensity fluctuations and reflections from the sensor. Within our implementation, the sensitivity can be improved by acquiring two holograms within a short time interval and reconstructing and processing the difference or by filtering the spurious signal computationally. Due to the high signal-to-background ratio for C. reinhardtii, this was not necessary for the measurements presented here. However, to reliably image E. coli, suppressing background signal from reflections and laser fluctuations by differencing images or filtering is necessary.
Each reconstruction provides a three-dimensional image of a ∼7 μL volume within the 2 mL cuvette [Fig. 1c]. Geometric differences between microscopes and cuvettes result in variability in the measured volume between microscopes and therefore systematic error in measurements of replicate populations. To characterize this error, we compared densities of suspensions of 5.2 μm diameter polystyrene beads measured by flow cytometry and DIH microscopy. We measured three bead suspensions of known density in all ten microscopes and found variability in the effective volume of 4% (6.76±0.13 μL). To precisely characterize a single microscope, we measured four bead densities in triplicate obtaining an effective volume of 6.65±0.14 μL, comparable to the 6.86 μL volume calculated from reconstructed images.10 The discrepancy likely arises from small systematic errors in the density measurement by flow cytometry.
With increasing density, out-of-focus light between adjacent algae degraded the signal-to-background ratio. Setting a maximum classification error of 5% therefore limited the measurement to densities below 105 cells∕mL, corresponding to ∼675 cells in the imaging volume. We measured algal densities as high as 5×105∕mL by restricting the considered volume to ∼1 μL along the optical axis where the signal-to-background ratio is larger.10
Each microscope incorporated independent control over temperature and illumination, the physical parameters relevant for the population dynamics of algae. For each microscope, the temperature at the feedback thermistor was stable to ±0.004 °C over 30 days. For systems with constant illumination the LED intensity was stable to ±0.5% over the 30-day experiment discussed below. Real time acquisition and analysis was reliable for at least a month.
POPULATION DYNAMICS OF CHLAMYDOMONAS REINHARDTII
To demonstrate the capabilities of our instrument, we measured the population dynamics of replicate populations of C. reinhardtii. This organism has been used widely as a model for microbial motility, metabolism, and photosynthesis.12, 13 First, we measured population dynamics in nine replicate growing populations of algae over a period of 60 h. A second experiment measured six replicates under conditions of nitrogen deprivation for 30 days. To our knowledge, these experiments constituted the first measurements of microbial population dynamics by digital holography.
Frozen samples of C. reinhardtii (UTEX 2244 mt+, University of Texas Culture Collection) were thawed, washed, and resuspended in 20 mL of sterile Sager-Granick medium (Chlamydomonas center, www.chlamy.org) in 125 mL Erlenmeyer flasks. Axenic cultures were grown under continuous illumination at approximately 8000 lx at room temperature with shaking at 200 rpm.
Experiments were started from cultures which had reached a density of 106 cells∕mL as measured by hemocy-tometry. Cultures were diluted to 5000 cells∕mL and quartz cuvettes were filled with 2 mL of this dilution, sealed with Teflon stoppers, and placed in an incubator providing constant illumination and temperature. Cuvettes were then loaded serially into the microscopes over a period of 90 min. The temperature of the aluminum housing was constant at 25.00±0.02 °C. During this experiment the illumination from the LEDs was zero for the first 12 h and 1100±60 lx combined, symmetrically, from above and below for the remainder of the experiment. For this measurement, holograms were acquired every 12 min.
For the second, long-term measurement, algae were grown as described above and washed once and resuspended in medium with the ammonium concentration reduced 450-fold from 11.2 mM to 25 μM. Ammonium is the sole nitrogen source in the medium. The mean illumination intensity for all systems was 800±40 lx, again symmetrically from above and below. About this mean, fluctuations in illumination were 0%, 10%, and 30% of the mean, each in duplicate. Fluctuations were uniform and white to 1 Hz and were sustained for the duration of the experiment. The temperature of the aluminum housing was constant at 25.00±0.02 °C. Due to the slow population dynamics in this experiment, measurements were made every 26 min.
For both experiments, populations were tested for contamination at the end of the experiment by plating, in replicate, in four conditions. No evidence of contamination was observed in either experiment. At the end of the long-term experiment, viability was confirmed by flow cytometry. C. reinhardtii rapidly loses chlorophyll autofluorescence upon lysis;14 in all populations, strong chlorophyll fluorescence was observed. Viability was further confirmed using the SYTOX® Green [Invitrogen (Carlsbad, CA) 10 nM, 5 min incubation] viability stain.
Population dynamics
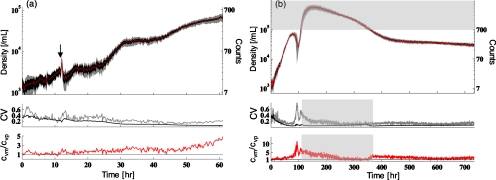
Population dynamics from the first experiment are shown in Fig. 3a. A doubling in the density during an initial dark period of 12 h was followed by a transient response to the increase in illumination and a reproducible increase in the density by a factor of 30 over 48 h (doubling time of 9.6 h). The measured densities are remarkably reproducible across all nine systems [Fig. 3a]. We compared the measured coefficient of variation cvm=σs∕⟨d⟩s, where ⟨d⟩s and σs are the mean density and the standard deviation across systems, to the mean variation within systems assuming counts are Poisson distributed at each point with a mean at the observed density, cvp.10 The ratio cvm∕cvp is typically larger than 1 [Fig. 3a, lower panel] and characterizes system-to-system variability beyond what we expect from counting noise alone. The divergence results from system-to-system variations in the trend with respect to the mean dynamics rather than increasing fluctuations in the measurement. Fluctuations in density within each system remain Poisson distributed throughout the experiment.10
Figure 3.
(a) Dynamics of replicate populations of C. reinhardtii, nine replicates in rich medium. Upper panel: mean dynamics (line) shown with the standard deviation across replicates (gray) and the mean fluctuation within systems assuming Poisson statistics (black). The illumination was turned on at 12 h as indicated by the arrow. Middle panel: the coefficient of variation (CV) across replicates (cvm, gray) and the mean CV assuming Poisson statistics (cvp, black). Lower panel: the ratio cvm∕cvp. The straight black line denotes cvm∕cvp=1, where variation between systems is equal to the variation we expect from counting statistics alone. (b) Dynamics of six replicate nitrogen-deprived populations with fluctuating illumination. Panels are the same as in (a). Mean illumination is constant. Fluctuations in illumination vary across replicates from no fluctuation to 30% of the mean. Upper panel: points within the shaded region are above the calibrated maximum algal density of 105∕mL. Here, densities as high as 5×105 were determined from counts within in a ∼1 μL volume (Ref. 10). Middle and lower panels: shaded regions indicate where cvm,cvp and cvm∕cvp were estimated from counts above the maximum calibrated density.
In the second, long-term experiment, we followed six replicate populations of algae under nitrogen deprivation over a period of 30 days [Fig. 2b]. Here the mean illumination was constant but the amplitude of fluctuations was varied across systems as discussed above. Despite variable fluctuations in illumination, we observed highly reproducible dynamics [Fig. 3b]. All six systems exhibited complex dynamics: initial fast increase in density by a factor 50 (doubling time of 13 h) before a simultaneous drop by a factor 5, followed by a second sharp increase, and for the rest of the experiment by a gradual decrease, which for the last 300 h became very slow. Remarkably, between 400 and 700 h, we observed a mean cvm∕cvp ratio of 1.45 [Fig. 3b, lower panel], indicating that the variability across systems exceeded counting noise by only 45%. Turning off the illumination disrupted these stable dynamics.10
The biology of this reproducibility in C. reinhardtii population dynamics, despite fluctuations in illumination, remains to be elucidated. However, the data demonstrate the long-term stability of our apparatus.
Spatial distributions
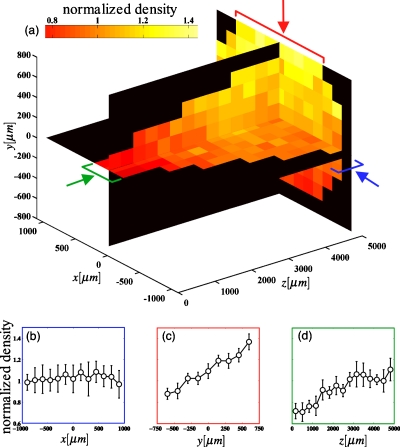
In addition to temporal dynamics, DIH microscopy measures the spatial structure of microbial populations within the imaging volume [Fig. 4a]. For the first algae experiment [Fig. 3a], we observed spatial gradients in the y and z directions that were reproducible and stable across replicate microscopes. No gradient was detected in the x direction [Fig. 4b]. The gradient in y, of 50% over 1.5 mm, [Fig. 4c] was likely due to gravitaxis, while the gradient in z, approximately a factor of 2 smaller, possibly arose from thermal gradients that we have not attempted to eliminate. The biological or physical mechanisms resulting in spatial structure in these populations are beyond the scope of this work. However, we note that reducing density gradients by stirring or eliminating thermal gradients could be implemented without loss of generality. Spatial distributions during the 30-day acquisition were similar to those observed in Fig. 4, indicating that a reproducible physical mechanism is likely responsible for the observed spatial gradients.
Figure 4.
Spatial density profiles for C. reinhardtii. For each microscope, the imaged volume was divided into 150×150×332 μm3 bins in x, y, and z, respectively. We then sum the counts within each bin across the entire time series shown in Fig. 2a, resulting in an average of ∼50 counts∕bin in each microscope. For each bin we compute the normalized density, the ratio of the number of counts to the number of counts we expect assuming a uniform spatial distribution (a) presents the average normalized density across systems for three planes within the imaging volume. (b)–(d) present profiles in x, y, and z, respectively, as indicated by the arrows in (a). Bars indicate bins that are included in the average. Gravity points in the −y direction. Error bars are standard error.
CONCLUSIONS
The technique presented here combines long-term stability and small systematic error (imaged volume: ±2%, illumination intensity: ±5%, temperature: ±0.02 °C) with a noninvasive, rapid measurement that resolves single cells. This permits the quantitative study of biological sources of variability between replicate populations over long periods of time without neglecting short timescale dynamics. Further, this variability can be quantified with respect to perturbations in light or temperature or changes in initial conditions such as genetic or chemical changes.
The primary restrictions of the measurement are the low optical density of the sample, incompatibility with fluorescence, and the computation time necessary for analysis. The device is readily adapted to higher cell densities by shortening the optical path length or using longer wavelengths that scatter less. The maximum cell density that can be imaged scales better than 1∕L, where L is the optical path length. Reducing the path length to 0.5 mm would permit the detection of algal densities of at least 5×106∕mL, which is close to the maximum algal density in standard growth medium. Finally, species or strain discrimination cannot be done with fluorescence but must rely on morphological features. The computation could be sped up by a factor of 10 or more using a graphics-processing unit or a computer cluster.
This work establishes DIH microscopy as a noninvasive, low-cost method for the quantitative study of population dynamics in replicate populations of microorganisms. The method has a broad range of applications, including long-term monitoring of microbial growth, adaptation of populations to environmental changes, quantitative comparison of genetic perturbations to growth, and spatial organization. In contrast to existing methodologies, our measurement will noninvasively elucidate the effects of these processes on the timescale of minutes over periods of days or weeks.
ACKNOWLEDGMENTS
We would like to acknowledge members of our laboratory for comments on the manuscript and fruitful discussions, Dr. Susan K. Dutcher for help with C. reinhardtii, and Herbert Stuermer of GRINTech GmbH for help designing custom GRIN lenses. S.K. acknowledges funding through a Helen Hay Whitney Postdoctoral fellowship.
References
- Lenski R., Rose M., Simpson S., and Tadler S., Am. Nat. 138, 1315 (1991). 10.1086/285289 [DOI] [Google Scholar]
- Finkel S. and Kolter R., Proc. Natl. Acad. Sci. U.S.A. 96, 4023 (1999). 10.1073/pnas.96.7.4023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagaddé F. K., Song H., Ozaki J., Collins C. H., Barnet M., Arnold F. H., Quake S. R., and You L., Mol. Syst. Biol. 4, 187 (2008). 10.1038/msb.2008.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagadde F., You L., Hansen C., Arnold F., and Quake S., Science 309, 137 (2005). 10.1126/science.1109173 [DOI] [PubMed] [Google Scholar]
- Gabor D., Nature (London) 161, 777 (1948). 10.1038/161777a0 [DOI] [PubMed] [Google Scholar]
- Sheng J., Malkiel E., Katz J., Adolf J., and Belas R., Proc. Natl. Acad. Sci. U.S.A. 104, 17512 (2007). 10.1073/pnas.0704658104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Jericho M., Meinertzhagen I. A., and Kreuzer H. J., Proc. Natl. Acad. Sci. U.S.A. 98, 11301 (2001). 10.1073/pnas.191361398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson M., Sebesta M., Bengtsson B., Pettersson S. G., Egelberg P., and Lenart T., Opt. Lasers Eng. 41, 553 (2004). 10.1016/S0143-8166(02)00214-2 [DOI] [Google Scholar]
- Sheng J., Malkiel E., and Katz J., Appl. Opt. 45, 3893 (2006). 10.1364/AO.45.003893 [DOI] [PubMed] [Google Scholar]
- See supplementary material at http://dx.doi.org/10.1063/1.3473937E-RSINAK-81-031008for a detailed description of the reconstruction, image analysis, calibration, and C. reinhardtii time series.
- Frigo M. and Johnson S., Proc. IEEE 93, 216 (2005). 10.1109/JPROC.2004.840301 [DOI] [Google Scholar]
- Kassen R. and Bell G., Heredity 80, 732 (1998). 10.1046/j.1365-2540.1998.00329.x [DOI] [Google Scholar]
- Collins S. and Bell G., Nature (London) 431, 566 (2004). 10.1038/nature02945 [DOI] [PubMed] [Google Scholar]
- Harris E. H., The Chlamydomonas Sourcebook (Elsevier, New York, 2009), Vols. 1–3. [Google Scholar]