Abstract
γ-Crystallin genes are specifically expressed in the eye lens. Their promoters constitute excellent models to analyse tissue-specific gene expression. We investigated murine Cryge/f promoters of different length in lens epithelial cell lines. The most active fragment extends from position –219 to +37. Computer analysis predicts homeodomain and paired-domain binding sites for all rodent Crygd/e/f core promoters. As examples, we analysed the effects of Prox1 and Six3, which are considered important transcription factors involved in lens development. Because of endogenous Prox1 expression in N/N1003A cells, a weak stimulation of Cryge/f promoter activity was found for PROX1. In contrast, PROX1 stimulated the Crygf promoter 10-fold in CD5A cells without endogenous PROX1. In both cell lines Six3 repressed the Crygf promoter to 10% of its basal activity. Our cell transfection experiments indicated that Cryg expression increases as Six3 expression decreases. Prox1 and Six3 act antagonistically on regulation of the Crygd/e/f promoters. Functional assays using randomly mutated γF-crystallin promoter fragments define a Six3-responsive element between –101 and –123 and a Prox1-responsive element between –151 and –174. Since Prox1 and Six3 are present at the beginning of lens development, expression of Crygd/e/f is predicted to remain low at this time. It increases as Six3 expression decreases during ongoing lens development.
INTRODUCTION
The γ-crystallins are recognised as structural proteins, expressed specifically in the eye lens of mammals and other vertebrates (with the exception of birds). The γ-crystallins are encoded by a cluster of six genes, Cryga–Crygf (for reviews see 1,2). In the mouse, the Cryg genes are expressed from embryonic day (E) 13.5 onwards in primary fibre cells and later on in secondary fibre cells, but not in epithelial cells (3,4). Mutations in the Cryg genes have been reported for mouse and man and it is commonly accepted that these mutations are causative for a variety of lens opacities (5–8).
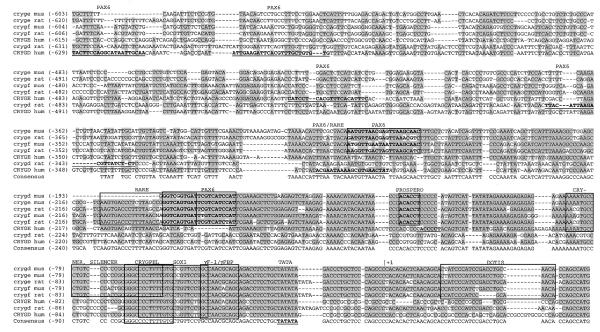
Because of the unique expression in the lens, the regulation of Cryg gene expression has been studied in various laboratories. The highly conserved proximal promoter region in the rat Cryge/f and mouse Crygd/e/f genes is characterised by binding sites for transcription factors and by a variety of sequence elements with remarkable features but without known functions (Fig. 1).
Figure 1.
The Crygd/e/f promoters of mammals. The mouse, rat and human Crygd/e/f promoters were compared by AlignX. Identical stretches are underlined in grey. The mouse and rat Crygd/e/f promoters are highly conserved between position –230 (of the mouse Crygf sequence) and the translational start site. It is commonly accepted that these sequences are necessary for lens-specific expression. The following sequence elements can be observed in all of them (positions refer to the mouse Crygf sequence): RARE, retinoic acid response element, –208/–183 (9,10); CRYNER, γ-crystallin nested repeats, –87/–59 (11); SILENCER, –76/–58 (12,13); CRYGPEL, common γ-crystallin promoter element, –67/–54 (14); SOX1, Sox1-binding site, –63/–44 (15); γF-1/γFBP, γF-crystallin binding protein, 46/–36 (16–18); TATA box, –23/–18; DOTIS, downstream of transcription initiation site, +15/+35 (19,20). Novel putative binding sites for Pax6 and Prospero predicted by MatInspector Professional are boxed in bold. The GenBank/EMBL accession nos of the aligned sequences are: mouse Crygf, M11039 (21); rat Crygf, M19357 (22); mouse Crygd, M16512 (23); mouse Cryge, X57855 (40); rat Cryge, M19359 (22); human CRYGF, K03009 (24); human CRYGD, K03005 (24); rat Crygd, M19359 (22), human CRYGE, K03007 (24); human CRYGD and ψCRYGE, AC018961 (25).
Additionally, there are differences in the response of cell culture systems and transgenic mice using the same promoter region. For example, the fragment –67/+45 of the proximal Crygf promoter is sufficient for lens-specific expression in transgenic mice (18). However, in N/N1003A lens cells the smallest promoter eliciting activity is the –226/+45 fragment (26).
On the other hand, a broad variety of transcription factors have been demonstrated to be involved in proper regulation of lens development and differentiation (27). However, only a few of them have been investigated with respect to their function in the regulation of γ-crystallin encoding genes.
Pax6 is referred to as a ‘master control gene’ of eye development regulating several crystallin genes (28–30). Prox1 is expressed in the mouse lens placode at E9.5, in the lens vesicle (E10.5), in the anterior, proliferating epithelium and in differentiating fibre cells (from E12 onward). In Prox1 knock-out mutants lens fibre cell elongation is affected and Crygd expression is decreased, whereas Cryge/f expression remains constant (31).
Six3 is important for early eye development, as demonstrated recently by its ectopic expression in the ear placode of medaka fish. This ectopic expression leads to formation of a morphologically intact lens in the ear placode, including expression of crystallins (32). Moreover, Six3 was found to be expressed at E6.5 in the head fold, a region later forming the mouse eye anlagen; however, in the lens it was not detected after E18 (33). Therefore, we investigated the role of those three transcription factors in the regulation of Cryg gene expression in some detail. We could demonstrate that Prox1 stimulates the Cryge/f promoter ∼10-fold, but Six3 represses it to near background level. Pax6 is obviously without effect on Cryge/f expression. Based upon these data, we suggest an antagonistic model of Prox1 and Six3 action at the Cryge/f promoter.
MATERIALS AND METHODS
Cell lines
Human lens epithelial cell line CD5A was established from epithelial cells of donor lenses. The cells were immortalised using adenovirus 12–SV40 hybrid virus, kindly provided by J. S. Rhim (Bethesda, MD) (34). Virus was produced in CV1 cells and stored at –80°C as culture supernatant. Virus was added directly to the lens epithelium upon arrival in the laboratory; the cells were monitored for growth over a 2–3 week period. Once the cells had migrated and covered >50% of the surface of a 24-well plate, they were trypsinised into a single well of a 6-well plate. These were allowed to grow to confluence and then passaged into a 25 cm2 flask. Cells were increased in number by passaging into larger flasks until ready for freezing down (typically passage 5) from a 175 cm2 flask.
Mouse lens epithelial cell line αTN4 and mouse fibroblast-like cell line NIH 3T3 were cultured in DMEM under standard conditions; for the N/N1003A cell line EMEM with 10% rabbit serum was used as described previously (35,36).
PCR and western blotting
RNA was prepared from organs and cell lines using the RNAeasy system (Qiagen, Hilden, Germany). An aliquot of 5 µg total RNA was transcribed to first strand cDNA with a Ready To Go™ T-primed First Strand Kit (Pharmacia Biotech, Freiburg, Germany).
PCR conditions using a Robocycler (Stratagene, Amsterdam, The Netherlands) for 15–40 cycles of denaturation at 95°C, with annealing and extension at 72°C, each for 45 s, have been previously described (37). PCR products were resolved on 3–5% agarose gels.
A 70 bp fragment of the 3′-untranslated region (UTR) of murine Six3 cDNA was amplified from the first strand cDNA and Six3-pcDNA3.1 using the primer pair 5′-AGAACAAACCGAAATCAGGATAC-3′ and 5′-CACACTCCCACCCCAGCCAA-3′; the annealing temperature was 51°C. Primer binding sites are conserved in mouse and human. PCR products were sequenced directly. Furthermore, aliquots of RNA of the cells and organs were subjected to similar PCR reactions. No 70 bp fragment was amplified. Therefore, genomic contamination of the RNA preparations can be excluded.
A 108 bp fragment encompassing parts of exons IV and V of the human PROX1 gene (38) was amplified from first strand cDNA and Prox1-pcDNA3 using the primer pair 5′-AATGACTTTGAGGTTCCAGAGAGATTCCTG-3′ and 5′-CAAAGATGTTGATCCTTCCTGGAAGAAG-3′, a conserved sequence in mouse and human; annealing was at 52°C.
For western blot analysis 10 µg of each cell preparation was electrophoresed through a SDS–7% polyacrylamide gel and electroblotted onto a PVDF membrane. The membrane was blocked overnight in 5% powdered milk in phosphate-buffered saline and incubated with a 1:1000 dilution of an antibody raised against the homeodomain and C-terminal domain (amino acids 546–736) of human PROX1 (39). Peroxidase-conjugated goat anti-rabbit IgG (Dianova, Hamburg, Germany) was used at 1:10 000 in TBST (10 mM Tris, pH 8.0, 150 mM NaCl, 0.05% Tween-20). Peroxidase was detected by incubation for 15 min in a 15 ml volume comprising 10 ml H2O, 5 ml 50 mM Na3PO4, pH 7.5, 750 mM NaCl, 500 mM imidazole, 0.25% Tween-20 and 5 mg diaminobenzidine, 100 µl CoCl2 (40 µg/ml) and 10 µl 30% H2O2.
Reporter plasmids and expression systems
5′-Deletions of the Cryge promoter (40) were cloned into the reporter gene vector pBLCAT6 (41) or its derivative pEK0CAT (42). The resulting constructs represented fragments (–629/+37), (–514/+37), (–325/+37), (–219/+37), (–163/+37), (–124/+37), (–77/+37), (–25/+37) and (–629/+8) of the Cryge promoter. The mouse (–226/+45) Crygf promoter plasmid pγ226LucII (15) as well as the promoter-less construct pPLLucII (43) were kindly provided by Y. Kamachi (Nagoya, Japan). Control reporter vector pRL-SV40 was purchased from Promega (Heidelberg, Germany).
Pax6 cDNA subcloned in the pBluescriptKS(+) vector was kindly provided by R. Balling (Neuherberg, Germany). After digestion with BamHI and DraI it was subcloned into the eukaryotic expression vector pSG5 (Stratagene, Heidelberg, Germany); the resulting plasmid is referred to as pSG-Pax6.
For expression of Six3, the EcoRI fragment of plasmid pC5Six3 containing the full-length Six3 cDNA (kindly provided by P. Gruss, Göttingen, Germany) was subcloned into the pcDNA3.1 expression vector. The complete Prox1 coding sequence was cloned into the pcDNA3 expression vector (Invitrogen).
The generated plasmids, Prox1-pcDNA3 and Six3-pcDNA3.1, were expressed in vitro using the reticulocyte lysate of the TNT® Quick Coupled Transcription/Translation System (Promega) containing [35S]methionine. An aliquot of 1 µl of lysate was electrophoresed through a 7% SDS–polyacrylamide gel, the gel was dried under vacuum and radioactivity was detected overnight on a Fuji Imaging Plate using a PhosphorImager SI (Amersham Pharmacia Biotech).
Eukaryotic cell lines N/N1003A and CD5A were also used to express Pax6, PROX1 and Six3.
Random mutagenesis of the γF-crystallin core promoter
To detect regions of functional interest, mutations were randomly introduced into the γF-crystallin promoter between positions –214 and +48 using the PCR random mutagenesis protocol according to Wan et al. (44). The forward PCR primer (5′-CAC CTG GAT CCT CTA CAG TCG AGG CCC AAG CTA CAT C-3′) contains a BamHI site for cloning, whereas the reverse PCR primer (5′-GAG GCC AAG CTT CGC TGG TGT TGG CAG GTC AGA TGG-3′) has a HindIII restriction site. The PCR buffers were exactly as described (44); the MnSO4 concentrations used were 0.1, 0.2 and 0.8 mM for clones 1–15, 16–36 and 37–40 respectively. The PCR products were cloned into the pPLLucII vector (43) using the BamHI and HindIII restriction sites and transfected into DH5α bacteria. Forty clones (out of 96) had an insert and have been characterised by sequencing in both directions using an ABI-3100 sequencer (PE Biosystems, Weiterstadt, Germany); the forward primer was 5′-AAG CTT CGC TGG TGT TGG-3′ and the reverse primer 5′-GGA TCC TCT AGA GTC GAG GC-3′; DNA was prepared using a plasmid NucleoSpin column (Macherey Nagel, Düren, Germany).
Transfection and reporter gene assay
For chloramphenicol acetyltransferase (CAT) reporter gene assays, 7 × 105 cells per 35 mm diameter dish were seeded. Twenty-four hours later cells were transfected with 3 or 4 µg plasmid DNA plus 0.2 µg pCMVβ (Clontech, Heidelberg, Germany) using LipofectAMINE (Gibco, Eggenstein, Germany) or DOSPER (Roche, Mannheim, Germany). Seventy-two hours after transfection cells were harvested and 100 µg cell extract was assayed for CAT using a CAT-ELISA kit (Roche). Additionally, 10 µg were assayed for β-galactosidase activity for internal standardisation.
For luciferase (Luc) reporter gene assays 1.5 × 105 cells were cultivated in 12-well plates for 24 h and transfected by the calcium phosphate precipitation method. Each dish received 2 µg pγ226LucII reporter plasmid and 0.02 µg pRL-SV40 control plasmid and various amounts of the Six3-pcDNA3.1 or Prox1-pcDNA3 expression plasmid, respectively, or the parental plasmid pcDNA3.1.
For analysis of the randomly mutated core promoter fragments, calcium phosphate transfection into the CD5A cell line was performed using 2 µg mutated promoter fused to the Luc reporter gene, 0.4 µg effector (either Prox1-pcDNA3 or Six3-pcDNA3.1) and 0.02 µg pRL-SV40 for transfection control.
Cells were harvested 48 h after transfection and cellular extracts were prepared by multiple freeze/thaw cycles. The extracts were assayed in triplicate with the Dual-Luciferase Reporter Assay System (Promega) and the standard deviations calculated.
DNA interaction
For affinity binding and precipitation, fragments (–226/+46, –214/–165, –164/+115, –114/–65, –64/–15, –14/+36, –184/–145, –134/–95, –84/–45 and –34/+6) of the Crygf promoter were Biotin-16-ddUTP (Roche) end-labelled and immobilised on uniform, paramagnetic polystyrene beads via Streptavidin covalently attached to the bead surface (Deutsche Dynal, Hamburg, Germany). An aliquot of 0.5 µg immobilised DNA sequence was incubated with 1 µl of reticulocyte lysate containing [35S]methionine-labelled Six3 in 20 µl of binding buffer (50 mM Tris–HCl, pH 7.9, 0.1 mM EDTA, 0.01% NP13 detergent, 1 mM DTT, 10% glycerol, 70 mM NaCl) containing 250 ng poly(dI·dC) and 3 µg bovine serum albumin. In preliminary experiments Six3 did not bind to the (–114/–65) Crygf promoter fragment. Therefore, we added a 50-fold molar excess of non-immobilised (–114/–65) fragment as non-specific competitor. Binding was for 20 min at 30°C. Pellets were washed four times in binding buffer containing an additional 200 mM KCl and electrophoresed on SDS–polyacrylamide gels according to standard procedures.
In control experiments binding and washing were carried out as described above using the immobilised (–226/+45) Crygf promoter fragment. We eluted with a 50-fold molar excess of non-immobilised Crygf fragments, to displace Six3 from the (–226/+45) Crygf promoter. In particular, Six3 specifically binds to the (–226/+45) Crygf promoter fragment and also to those Crygf fragments that contain a specific Six3-binding site. Because of the excess of specific competitor Crygf fragments, Six3 is displaced from the immobilised (–226/+45) Crygf promoter and is eluted in the supernatant. In contrast, an excess of non-specific Crygf fragments do not compete with the (–226/+45) Crygf promoter. Six3 remains bound on the immobilised (–226/+45) Crygf promoter and stays in the pellet. Supernatant and pellet were electrophoresed on SDS–polyacrylamide gels according to standard procedures.
RESULTS
Comparison of several Crygd/e/f promoters: prediction of novel transcription factor-binding sites
The Crygd/e/f promoters of mouse, rat and man were analysed for transcription factor-binding sites using MatInspector Professional (45). The alignments and the common binding sites are summarised in Figure 1. Since the promoter of the human ψCRYGF gene lacks typical promoter features (24), it was not included in this comparison.
The analysed Crygd/e/f promoters show high homology, especially in the proximal region, implicating various common promoter elements. All of them contain a Sox1- and a γFBP-binding site. Since human CRYGD and ψCRYGE and rat Crygd do not contain the Silencer and Cryner elements, as well as the RARE element, it is concluded that these particular promoters could be different to the others. These rodent promoters for Crygd/e/f were defined as a separate subfamily.
Some 35 additional transcription factor-binding sites were identified using MatInspector Professional (http://genomatix.gsf.de). All were conserved in positions for proximal promoters of the rodent Crygd/e/f subfamily. Of these, seven were binding sites for proteins containing a homeodomain or a paired domain and are located in the region –211 to –105. These included Pax6- and Prox1-binding sites. Pax6 and PROX1 are important for lens development and fibre cell differentiation and so provided the focus for the present studies.
The region –198 to –186 is predicted as a binding site for the paired domain of Pax6; it partly overlaps with the retinoic acid response element (RARE). A similar module is also present in the human CRYGD promoter, but at a more upstream position. In fact, all distal Crygd/e/f promoters have predicted Pax6-binding sites, but at different positions.
The region –125 to – 131 contains a predicted binding site for the Drosophila protein Prospero (46). Prospero is closely related to the vertebrate homologue Prox1 (47), which is required for lens fibre cell elongation (31), suggesting that PROX1 could be important for Cryd/e/f core promoter activity.
It is the combination of transcription factors that is important for lens development and other initial analyses of the core promoter have identified a large number of putative sites for transcription factors. To positively identify other transcription factors involved in regulating the Crygd/e/f core promoter, we chose to study the effect of Six3 because of the recent demonstration of its importance in eye development (33). Our experimental approach was to monitor Crygd/e/f promoter activity after overexpression of Pax6, Prox1 and Six3 in lens epithelial cell lines.
Characterisation of the rodent Crygd/e/f promoter
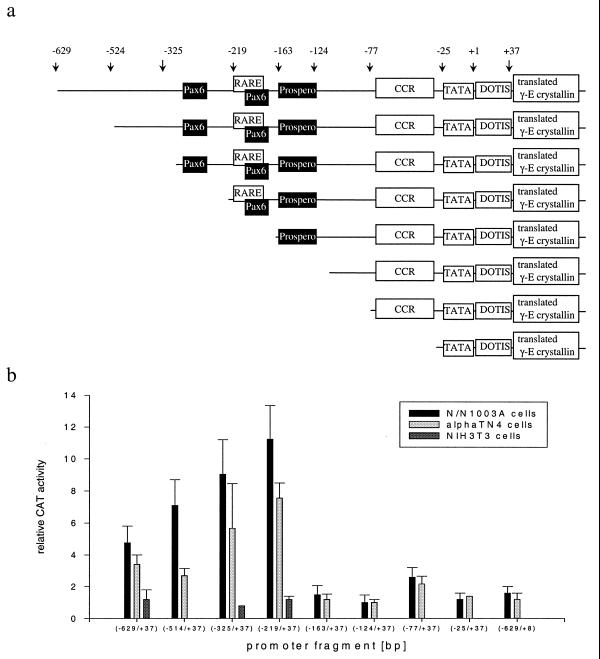
As examples for the Crygd/e/f promoters we used several deletion constructs of either the mouse Cryge or Crygf promoter (Fig. 2a) driving the CAT or Luc gene as reporter. These were transfected into lens epithelial cell lines N/N1003A, αTN4 and CD5A; non-lens derived NIH 3T3 cells were used as a control. In N/N1003A and αTN4 cells the entire (–629/+37) Cryge promoter shows 4–5 times higher activity than the promoter-less reporter vector. Deletion of the fragment –629 to –219 results in an additional 2.5-fold increase in promoter activity; fragment (–219/+37) is the most active and further deletions produced fragments not significantly different in activity from the promoter-less reporter vector. The only exception is the (–77/+37) fragment, which has 2.6-fold the background activity. Therefore, the (–219/+37) Cryge element is defined as the core promoter of the Crygd/e/f subfamily (Fig. 2b).
Figure 2.
Deletions at the Cryge promoter. (a) Deletion constructs. Schematic overview of the deletion constructs of the Cryge promoter cloned into the reporter gene CAT or Luc. Putative binding sites for Pax6 and Prospero are given in black. CCR is the γ-crystallin common region (21), including the Cryner, Silencer, Crygpel and γF-1 elements as well as the Sox1- and γFBP-binding sites, as described in Figure 1. (b) Effects of deletion on the Cryge promoter. Transfection experiments were performed with N/N1003A or αTN4 lens epithelial cells or fibroblast-like NIH 3T3 cells. In all transfections a CAT construct was co-transfected with pCMVβ vector for internal standarisation of efficiency of gene transfer. Cell extracts were measured for CAT expression by CAT ELISA and for β-galactosidase activity. All data were normalised to the promoter-less plasmid, which was set as 1.
A better response for the basal promoter was obtained in the rabbit cell line N/N1003A as compared to the murine cell line αTN4. This was selected for our detailed studies. In line with the features of the Cryge promoter, a 10-fold elevation of basal activity was observed for the (–226/+45) Crygf promoter in rabbit N/N1003A cells and also in human CD5A cells. Activation of the promoter fragment was lens cell specific, as no activity was detected in the fibroblast cell line NIH 3T3.
Influence of Pax6 on the Cryge promoter
We investigated the ability of Pax6 to stimulate the entire Cryge promoter by co-transfection of increasing amounts into the rabbit N/N1003A cell line. No statistically significant difference was observed between cells co-transfected or not with the Pax6 expression plasmid (data not shown). Since Pax6 was shown (42) to be expressed in all cell lines derived from the lens (N/N1003A, αTN4 and NKR) or of neuronal origin (PC12, U87 and U373), exogenous addition of Pax6 might not influence the endogenous effect.
Function of Prox1 at the Crygf promoter
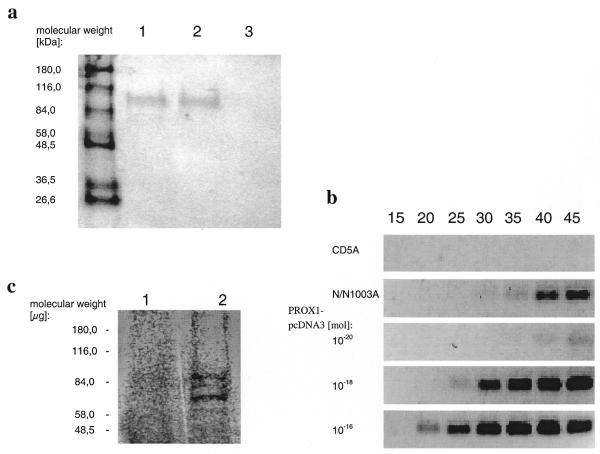
Endogenous expression of Prox1 in the cell lines used was analysed by western blotting. A band of the expected size (90 kDa) was detected in N/N1003A, but not in CD5A, cells (Fig. 3a). We confirmed these results by RT–PCR; Prox1 is expressed in N/N1003A cells, but not in CD5A cells (Fig. 3b). To obtain Prox1 protein for further investigations, Prox1 cDNA was cloned in the expression vector pcDNA3. The PROX1-pcDNA3 plasmid was transcribed and translated in reticulocyte lysate (Fig. 3c). PROX1 protein was detected in transfected CD5A cells (Fig. 3a). Therefore, we used the CD5A cell line (which does not express endogenous Prox1) to test the effect of Prox1 on the Crygf promoter.
Figure 3.
Expression of PROX1. (a) Western blot analysis of PROX1 in N/N1003A and CD5A cells. Lysates of N/N1003A cells (lane 1), CD5A cells (lane 3) and PROX1-pcDNA3-transfected CD5A cells (lane 2) were immunostained with antibody raised against the C-terminal part (amino acids 546–736) of human Prox1. The correct 90 kDa band was recognised in N/N1003A cells and in transiently PROX1-pcDNA3-transfected CD5A cells. Untransfected CD5A cells show no endogenous PROX1 expression. (b) Endogenous expression of PROX1 in lens epithelial cells. A 108 bp fragment encoding parts of exon IV and exon V of PROX1 could be detected in N/N1003A cells, but not in the CD5A cell line. For comparison, increasing amounts of PROX1-pcDNA3 were used as template. (c) In vitro expression of PROX1. PROX1-pcDNA3 was transcribed and translated with reticulocyte lysate containing [35S]methionine. A 90 kDa band for PROX1 is apparent (lane 2). Shorter polypeptides are expressed when the second or third AUG was used as the initiation codon. As a control in vitro expression was performed with plasmid pcDNA3 without the cDNA sequence coding for PROX1 (lane 1).
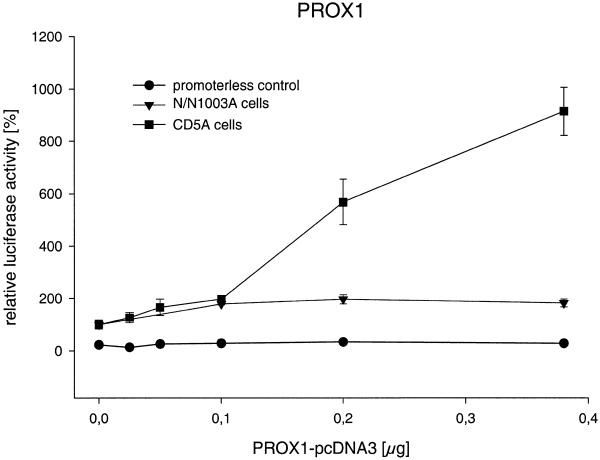
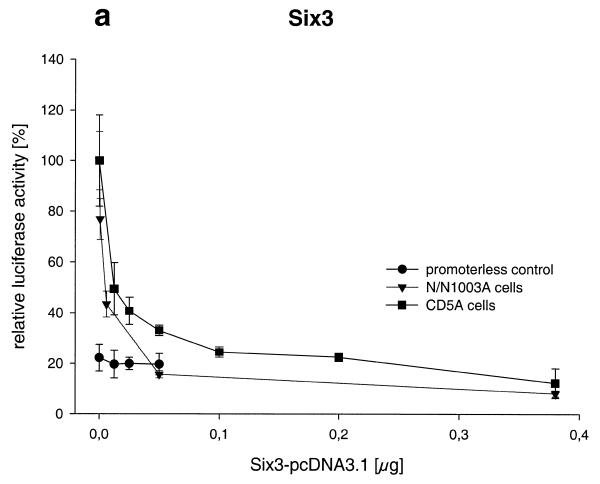
In transient co-transfection experiments we investigated the ability of Prox1 to regulate the Crygf promoter. Therefore, increasing amounts of Prox1-pcDNA3 were co-transfected with pγ226LucII. Prox1 activates the Crygf promoter 10-fold with a sigmoid dose–response curve; to avoid an overloading effect of the transfection system, no more than 0.4 µg Prox1 DNA was used (Fig. 4). In contrast to CD5A cells, only a 2-fold activation of the promoter without a clear dose-dependent relationship was observed in N/N1003A cells. This difference might be caused by endogenous expression of Prox1 in N/N1003A cells. Prox1 did not affect pPLLucII, indicating promoter-specific effects.
Figure 4.
PROX1 activates the Crygf promoter. Increasing amounts of PROX1 were co-transfected into CD5A and N/N1003A cells. Prox1 stimulates the Crygf promoter 2-fold in N/N1003A and 10-fold in CD5A cells as compared to its basal activity. To transfect a constant amount of DNA, the increasing amount of PROX1-pcDNA3 was compensated for by decreasing of the amount of empty vector pcDNA3. At the origin of the graph only pcDNA3 was used. In control experiments using a reporter plasmid without the Crygf promoter and with PROX1 overexpression in CD5A cells no alteration in relative luciferase activity was observed.
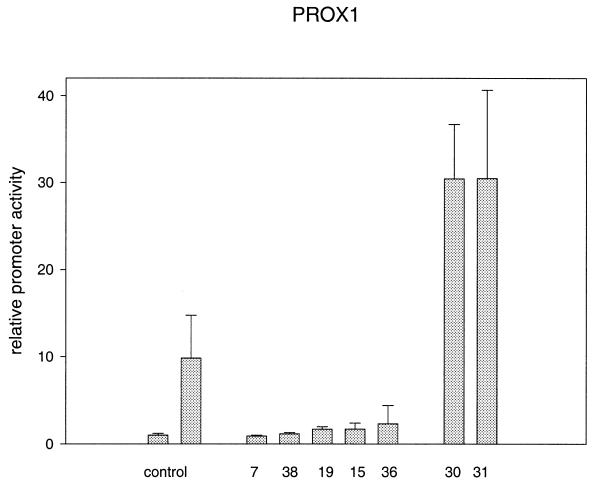
To test whether the predicted Prospero-binding site is responsible for stimulation of the Crygf promoter, we used a set of 36 mutated promoter fragments. The mutations were randomly distributed within the entire proximal core promoter fragment (–214/+48). As demonstrated in Figure 5, clones 7, 15, 19, 36 and 38 lost their ability to stimulate the Crygf promoter. The observed reporter gene activity corresponds to the control level without Prox1 stimulation. Four of these mutations were localised within an interval between –151 and –174. Clone 15 affects the RARE element, which was demonstrated previously to be important for Crygf expression (9,10). Some other mutations found within this region obviously do not influence Prox1-dependent promoter activity (clones 13, 14, 17 and 29).
Figure 5.
Altered Prox1 activation of mutated γF-crystallin promoters. The left control shows γF-crystallin promoter activity, which was co-transfected with the pcDNA3.1 expression vector not containing PROX1. In the right control, 29 of 36 point mutated γF-crystallin promoters are activated by PROX1, on average to 10-fold of their basal activities. On transfecting five of 36 mutated plasmids (7, 38, 29, 15 and 36) Prox1 produced no or reduced activation of the promoters as compared to their basal activity. In transfection experiments of two (30 and 31) of 36 mutated plasmids a 30-fold activation was observed. All mutations are represented in Figure 10.
Clones 30 and 31 additionally enhance the stimulatory activity of Prox1 3-fold (i.e. 30-fold over the control). Mutation 31 is very close to the inhibitory mutation 19, but another mutation two bases downstream is without effect on Prox1 stimulation (clone 4). Therefore, all five of these mutations (7, 19, 31, 36 and 38) define a Prox1-responsive element (PRORE) between positions –151 and –174. The other mutation enhancing the Prox1 stimulatory effect (clone 30) is localised at position –68 within the silencer element (12,13); it is suggested that destruction of the silencer enhances the possibility of Prox1 stimulating the promoter.
Function of Six3 at the Crygf promoter
We confirmed Six3 expression in the mouse eye at E12.5 as reported previously (33) by RT–PCR. Endogenous transcription of Six3 was also detected in human CD5A lens epithelial cell lines. The detection limit of PCR for Six3 is approximately 1000 molecules of Six3-pcDNA3.1 per reaction (Fig. 6). Six3 cDNA in rabbit N/N1003A cells could not be amplified, because the corresponding sequences of the rabbit are not yet known and conserved sequences from mouse and man could not be used successfully.
Figure 6.
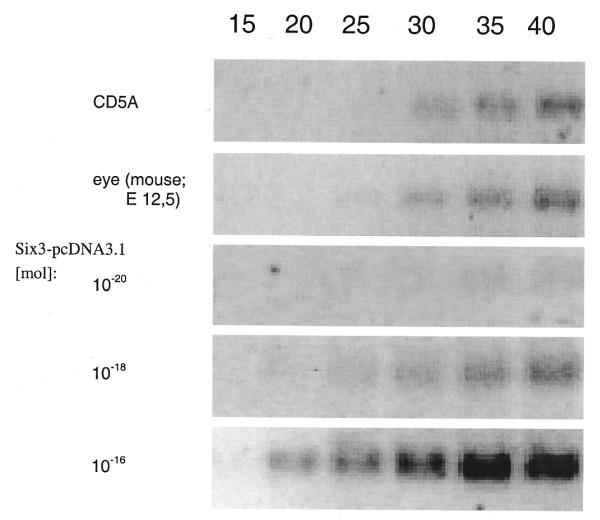
Endogenous expression of Six3 in the mouse eye and human CD5A cell line. A 70 bp 3′-UTR was amplified from the CD5A cell line and the mouse eye at E12.5. For comparison, increasing amounts of Six3-pcDNA3.1 were used as template.
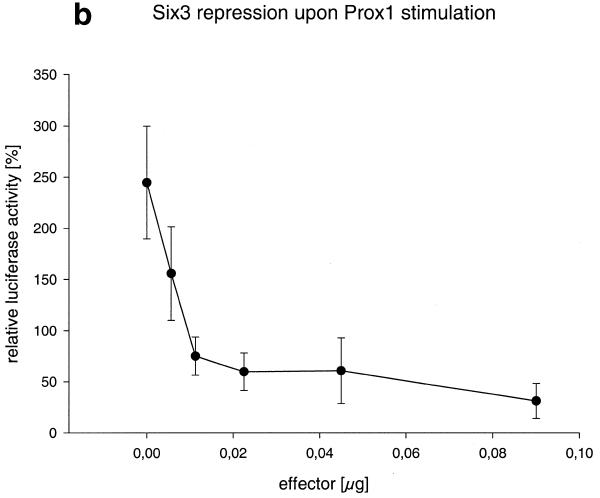
As described for Prox1, we investigated the influence of Six3 on Crygf expression. Increasing amounts of Six3-pcDNA3.1 were co-transfected with pγ226LucII into human CD5A lens epithelial or rabbit N/N1003A cells. As little as 6 ng of the Six3 expression plasmid repressed the activity of the Crygf promoter by 40%, compared to the promoter activity without Six3-pcDNA3.1. Larger amounts of Six3 expression vector led to total repression of the Crygf promoter (Fig. 7a). As a control, Six3 shows no effect on the promoter-less pPLLucII reporter plasmid. A similar repression of Crygf promoter activity was also observed after its stimulation by Prox1 (Fig. 7b).
Figure 7.
Six3 represses the Crygf promoter. (a) Increasing amounts of Six3 were co-transfected into CD5A and N/N1003A cells. Six3 represses the γF-cyrstallin promoter to <20% of its basal activity. To transfect a constant amount of DNA the increasing amount of Six3-pcDNA3.-1 was compensated for by decreasing the amount of the empty vector pcDNA3.-1. At the origin of the graph only pcDNA3.-1 was used. In control experiments using a reporter plasmid without the Crygf promoter and with Six3 overexpression in CD5A cells no alteration in relative luciferase activity was observed. (b) Constant amounts of the pγ226LucII (2 µg) and PROX1-pcDNA3 (300 ng) plasmids were co-transfected into N/N1003A cells together with different amounts of Six3-pcDNA3.1 (0–90 ng). 100% is the basal activity of the untreated Crygf promoter. Six3 is able to obliterate Prox1 activation.
To obtain Six3 protein in sufficient amounts for further investigations, Six3 cDNA was cloned into the expression vector pcDNA3.1. The corresponding Six3-pcDNA3.1 plasmid was transcribed and translated in reticulocyte lysate. Using SDS–PAGE, the expected 37 kDa size for full-length Six3 is observed (Fig. 8a).
Figure 8.
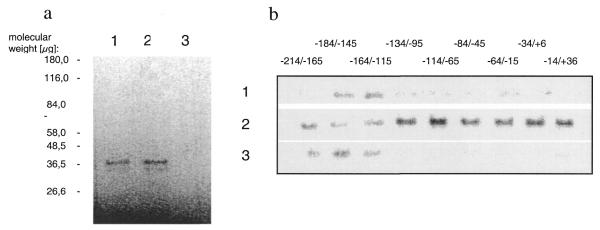
Six3 interaction with Crygf promoter DNA. (a) In vitro expression of Six3. Six3-pcDNA3.1 was transcribed and translated with reticulocyte lysate containing [35S]methionine. The correct 37 kDa band for Six3 is apparent (lanes 1 and 2). As a control in vitro expression was performed with the pcDNA3.1 plasmid without the cDNA sequence coding for Six3 (lane 4). (b) Affinity binding of Six3 to Crygf promoter sequences. The immobilised Crygf promoter sequences (–184–145) and (–164/–115) specifically precipitate Six3 (lane 1). As a control Six3 was bound to the immobilised (–226/+46) Crygf fragment. The free DNA sequences (–214/–165), (–184/–145) and (–164/–115) displace Six3 from the immobilised fraction (lane 2) into the supernatant (lane 3). Therefore, Six3 interacts with the Crygf promoter between nucleotides –214 and –115.
We investigated the ability of Six3 to interact with the Crygf core promoter by DNA precipitation. In vitro expressed Six3 was precipitated by immobilised DNA fragments (–184/–145) and (–164/–115) of the Crygf promoter (Fig. 8b, lane 1). To confirm the specificity of the interaction, oligonucleotides derived from the core promoter were used to compete off Six3 bound to the (–226/+45) Crygf promoter. Only oligonucleotides representing the sequences (–214/–165), (–184/–145) and (–164/–115) of the Crygf promoter were able to displace Six3 (Fig. 8b, lanes 2 and 3). Obviously, Six3 can be displaced from the Crygf promoter by the sequence –184/–145, but it does not bind to it. This might be caused by steric inhibition due to biotin–Streptavidin-mediated binding of the oligonucleotide to the matrix. All fragments of the Crygf promoter downstream of base pair –115 did not show any specific interaction with Six3.
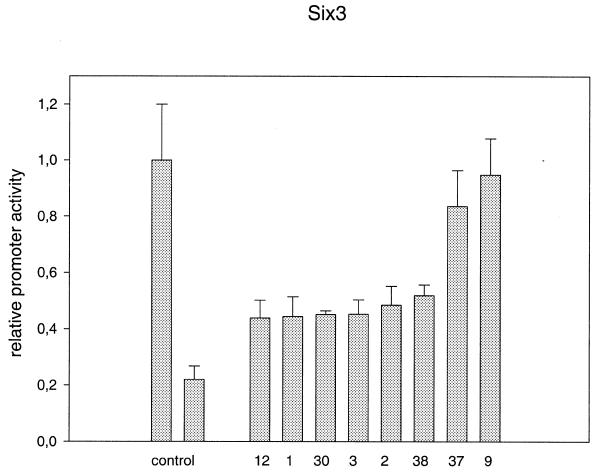
To decide whether in vitro binding to the overlapping promoter fragments has functional relevance, the same 36 clones as used for definition of the PRORE were used for transient transfections with Six3. Loss of the inhibitory action of Six3 was used to demonstrate an important function of the corresponding bases. As outlined in Figure 9, two clones (9 and 37) led to a complete loss of Six3 repressor activity. Both clones have mutations in the interval between –121 and –117. Moreover, six other clones (1–3, 12, 30 and 38) show only about half of the Six3 repressor activity of the wild-type promoter. The corresponding positions were all between –101 and –123 except for clone 2, which obviously destroys the Sox1-binding site. Since all other clones do not demonstrate an effect on Six3 repressor activity, the Six3-responsive element (SIRE) can be defined as between positions –101 and –123.
Figure 9.
Reduced Six3 repression in mutated γF-crystallin promoters. The left control shows γF-crystallin promoter activity when co-transfected with the pcDNA3.1 expression vector not containing Six3. In the right control, 28 of 36 point mutated γF-crystallin promoters are repressed, on average to 20% of their basal activities, due to co-expression of Six3-pcDNA3.1. Transfecting eight of 36 mutated plasmids (12, 1, 30, 3, 2, 38, 37 and 9) Six3 produced diminished repression of the promoters to between 40 and 100% of their basal activities. All mutations are represented in Figure 10.
DISCUSSION
Similar regulation of rodent Crygd/e/f genes
The Crygd/e/f genes of rat, mouse and man are highly conserved (10,40). A corresponding level of conservation is also found for their proximal promoters (Fig. 1), indicating a similar regulation of this gene subfamily. In line with their homology, the Cryge and Crygf promoters show similar activities in corresponding deletion constructs using rabbit N/N1003A cells as hosts. Successive deletion of the distal part of the Cryge promoter from base pair –629 to –219 leads to an increase in promoter activity, as observed for the Crygf promoter (48). Promoter fragments missing the sequences upstream of position –219 have lost most of their activity because of the deleted RARE element (9). Mediated by this element, co-transfection with recombinant RARα and RARβ receptors enhanced activity of the Crygf promoter 25-fold (9). Moreover, Cryg genes are activated by Sox1 and Maf via corresponding promoter elements in the proximal Crygf promoter (15,49). The L-Maf-binding element (MARE; 47) is nearly identical to the previously reported γF-1 element (16); c-Maf null mutant mice do not express Cryg genes (50). However, the chicken γFBP protein (binding to the γF-1-binding site) inhibits promoter activity in reporter gene assays (17). The mouse orthologue of γFBP, Hic1, is expressed in a variety of embryonic tissues, but not in the eye (51).
Pax6 is considered one of the most important genes in lens development because of its induction of ectopic eye formation in Drosophila (52) and because of the series of Small eye mutations, which do not develop eyes at all in the homozygous condition (53). In contrast to former observations (55), we detected a potential Pax6-binding site in the Crygd/e/f promoters using the computer program MatInspector Professional (45) for promoter analysis. However, from our co-transfection experiments no indication of either an activating or an inhibitory influence of Pax6 can be deduced. Even if N/N1003A cells express endogenous Pax6, a small stimulatory effect should be expected, as seen for PROX1. Moreover, in the same cell line Cvekl and colleagues (28) demonstrated that the αA-crystallin promoter could be stimulated by overexpression of Pax6. Therefore, we conclude that the predicted binding site is not active in the core Cryge/f promoter.
Prox1 is an activator of the Crygd/e/f genes
Using the MatInspector program (45), we found a Prospero-binding site in all Crygd/e/f promoters. The homologous gene in mammals, Prox1, seems to be very important for lens development and differentiation, because it is expressed in the mouse lens from the placode stage onward. Moreover, in Prox1–/– mice lens fiber cell elongation is affected (31). Our cell culture data using the CD5A lens epithelium cell line strongly support the idea that Prox1 is an important activator of Cryg expression. However, our random mutagenesis screen for promoter activity demonstrates that the predicted Prospero-binding site (–125/–131) is not responsible for the function of Prox1 at the Crygf promoter. A mutation within this region does not change the stimulatory activity of Prox1, whereas mutations in a more upstream region between positions –151 and –174 lead to either a complete loss of the stimulatory activity or to a significant increase. Therefore, we have defined this region as the Prox1-responsive element (PRORE).
All mutations affecting stimulation of the Crygf promoter by PROX1 are conserved in the Crygd/e/f promoters of the mouse and the Cryge/f promoters of the rat. The only exception is clone 31, which has a G instead of an A in the Crygd promoter of the rat. This position, being responsible for higher stimulation by PROX1 in clone 31 (A→T), might also have an effect on Crygd expression, which could not be observed in Prox1–/– mice. These Prox1–/– mice lost only Crygd expression; Cryge/f expression was observed. Even if no quantitative expression data in Prox1–/– mice are yet available (31), additional activators are obviously important for Cryge and Crygf expression.
Six3 represses Crygd/e/f gene expression
The data presented here demonstrate a function of Six3 interaction with the Crygf promoter resulting in a significant decrease as compared to its basal activity. Furthermore, Six3 is able to obliterate the activator PROX1. Six3 repression of the Crygf promoter explains the inversely related expression pattern of Six3 and γ-crystallins during lens development (33). The start of Cryg expression correlates with decreased expression of Six3, which is not expressed in lens fibres later than E14.5 (Fig. 10).
Figure 10.
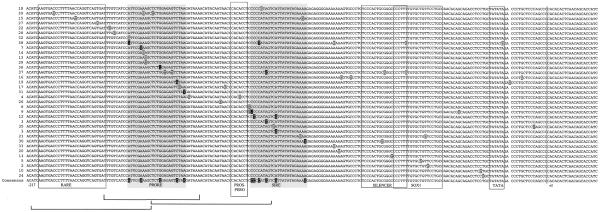
Alignment of mutated γF-crystallin promoters. For orientation, the transcription initiation site, the TATA box, the RARE, Silencer and Sox1 elements and the Prospero site are boxed. Point mutations are highlighted in grey. Point mutations affecting PROX1 activation (Fig. 5) or Six3 repression (Fig. 9) are marked in black. The resulting PRORE and SIRE are boxed in grey. Brackets show sequences which were shown to bind Six3 in vitro (Fig. 8b).
An interesting question might be whether Six3 can be understood as the silencer interacting with the corresponding element defined by Peek et al. (12,13). However, our data do not support this hypothesis, since the silencer element is located between positions –76 and –58. Our DNA binding studies defined a Six3-binding region between –214 and –115. Using a random mutagenesis screen for the Crygf promoter, we could define a SIRE in the region between –101 and –123. In this region mutations in the Crygf promoter change the repressive effect of Six3 significantly; mutations outside obviously do not influence Six3 function on the Crygf promoter. All positions of the mutated clones are conserved in the Crygd/e/f promoters of the mouse and the Cryge/f promoters of the rat. The expanded region of in vitro DNA binding and the shorter region defined by the cell culture experiments might be explained by the assumption that these positions might not be accessible under in vivo conditions.
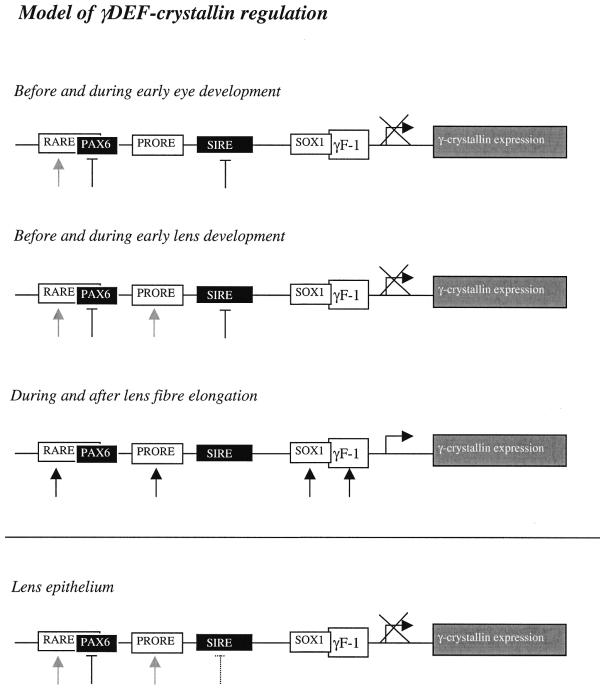
Our current model of Cryg gene activation is summarised in Figure 11. Cryg genes are not expressed during early eye development and lens formation due to inhibition by Six3. As soon as Six3 disappears from the lens fibres, Cryg genes are switched on, activated by Prox1 (as well as by c-Maf and Sox1). The concerted actions of Prox1, c-Maf and Sox1 are enhanced when repression by Six3 is removed. In contrast, in lens epithelium Prox1 is expressed, however, no Cryg expression occurs. The reason for this is absence of the activators L/c-Maf and Sox1. Outside the eye lens c-Maf, Sox1 and Prox1 are not co-expressed, therefore no Cryg expression can be observed in other tissues.
Figure 11.
Model of Crygd/e/f regulation during lens development. The retinoic acid receptors (interacting with the RARE element), Prox1 and Sox1 are activators, while Pax6 and Six3 are inhibitors of the Crygd/e/f promoters. The retinoic acid receptors, Pax6 (E8.0) and Six3 (E6.5) are expressed before eye development. Prox1 (E9.5, interacting with the PRORE element) is apparent even before lens development. Crygd/e/f are not expressed during early lens development due to the repressor Six3 (interacting with the SIRE). During lens fibre cell differentiation an additional activator, Sox1, is expressed. The repressor Six3 disappears. Therefore, Cryg expression begins in the fibre cells. In lens epithelium cells γ-crystallins cannot be observed during lens development because of the presence of Six3 (until E18.0).
Acknowledgments
ACKNOWLEDGEMENTS
We gratefully acknowledge the receipt of clones and cell lines from R. Balling (Neuherberg, Germany), P. Gruss (Göttingen, Germany), Y. Kamachi (Nagoya, Japan), J. Reddan (Oakland, CA), J.S. Rhim (Bethesda, MD) and P. Russell (Bethesda, MD). Oligonucleotides were synthesised by Utz Linzner (GSF-AG BIODV). This work was supported, at least in part, by an ARC project grant (1061) from the British Council to R.A.Q. and a grant from the German Academic Exchange Service (DAAD) to J.G. (313-ARC-X).
References
- 1.Graw J. (1997) The crystallins: genes, proteins and diseases. Biol. Chem., 378, 1331–1348. [PubMed] [Google Scholar]
- 2.Wistow G. and Piatigorsky,J. (1987) Recruitment of enzymes as lens structural proteins. Science, 236, 1554–1556. [DOI] [PubMed] [Google Scholar]
- 3.Van Leen R.W., Breuer,M.L., Lubsen,N.H. and Schoenmakers,J.G. (1987) Developmental expression of crystallin genes: in situ hybridization reveals a differential localization of specific mRNAs. Dev. Biol., 123, 338–345. [DOI] [PubMed] [Google Scholar]
- 4.Santhiya S.T., Abd-alla,S.M., Loster,J. and Graw,J. (1995) Reduced levels of gamma-crystallin transcripts during embryonic development of murine Cat2nop mutant lenses. Graefes Arch. Clin. Exp. Ophthalmol., 233, 795–800. [DOI] [PubMed] [Google Scholar]
- 5.Cartier M., Breitman,M.L. and Tsui,L.C. (1992) A frameshift mutation in the gamma E-crystallin gene of the Elo mouse. Nature Genet., 2, 42–45. [DOI] [PubMed] [Google Scholar]
- 6.Klopp N., Favor,J., Loster,J., Lutz,R.B., Neuhauser-Klaus,A., Prescott,A., Pretsch,W., Quinlan,R.A., Sandilands,A., Vrensen,G.F. and Graw,J. (1998) Three murine cataract mutants (Cat2) are defective in different gamma-crystallin genes. Genomics, 52, 152–158. [DOI] [PubMed] [Google Scholar]
- 7.Heon E., Priston,M., Schorderet,D.F., Billingsley,G.D., Girard,P.O., Lubsen,N. and Munier,F.L. (1999) The gamma-crystallins and human cataracts: a puzzle made clearer. Am. J. Hum. Genet., 65, 1261–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith R.S., Hawes,N.L., Chang,B., Roderick,T.H., Akeson,E.C., Heckenlively,J.R., Gong,X., Wang,X. and Davisson,M.T. (2000) Lop12, a mutation in mouse Crygd causing lens opacity similar to human Coppock cataract. Genomics, 63, 314–320. [DOI] [PubMed] [Google Scholar]
- 9.Tini M., Otulakowski,G., Breitman,M.L., Tsui,L.C. and Giguere,V. (1993) An everted repeat mediates retinoic acid induction of the gamma F-crystallin gene: evidence of a direct role for retinoids in lens development. Genes Dev., 7, 295–307. [DOI] [PubMed] [Google Scholar]
- 10.Tini M., Fraser,R.A. and Giguere,V. (1995) Functional interactions between retinoic acid receptor-related orphan nuclear receptor (ROR alpha) and the retinoic acid receptors in the regulation of the gamma F-crystallin promoter. J. Biol. Chem., 270, 20156–20161. [DOI] [PubMed] [Google Scholar]
- 11.Stoger T., Augusteyn,R.C. and Graw,J. (1997) The Cryner element in the murine gamma-crystallin promoters interacts with lens proteins. Ophthalmic Res., 29, 161–171. [DOI] [PubMed] [Google Scholar]
- 12.Peek R., van der Logt.P., Lubsen,N.H. and Schoenmakers,J.G. (1990) Tissue- and species-specific promoter elements of rat gamma-crystallin genes. Nucleic Acids Res., 18, 1189–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peek R., Kraft,H.J., Klok,E.J., Lubsen,N.H. and Schoenmakers,J.G. (1992) Activation and repression sequences determine the lens-specific expression of the rat gamma D-crystallin gene. Nucleic Acids Res., 20, 4865–4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graw J., Liebstein,A., Pietrowski,D., Schmitt-John,T. and Werner,T. (1993) Genomic sequences of murine gamma B- and gamma C-crystallin-encoding genes: promoter analysis and complete evolutionary pattern of mouse, rat and human gamma-crystallins. Gene, 136, 145–156. [DOI] [PubMed] [Google Scholar]
- 15.Kamachi Y., Sockanathan,S., Liu,Q., Breitman,M., Lovell-Badge,R. and Kondoh,H. (1995) Involvement of SOX proteins in lens-specific activation of crystallin genes. EMBO J., 14, 3510–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Q.R., Tini,M., Tsui,L.C. and Breitman,M.L. (1991) Interaction of a lens cell transcription factor with the proximal domain of the mouse gamma F-crystallin promoter. Mol. Cell. Biol., 11, 1531–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Q., Shalaby,F., Puri,M.C., Tang,S. and Breitman,M.L. (1994) Novel zinc finger proteins that interact with the mouse gamma F-crystallin promoter and are expressed in the sclerotome during early somitogenesis. Dev. Biol., 165, 165–177. [DOI] [PubMed] [Google Scholar]
- 18.Goring D.R., Bryce,D.M., Tsui,L.C., Breitman,M.L. and Liu,Q. (1993) Developmental regulation and cell type-specific expression of the murine gamma F-crystallin gene is mediated through a lens-specific element containing the gamma F-1 binding site. Dev. Dyn., 196, 143–152. [DOI] [PubMed] [Google Scholar]
- 19.Pietrowski D., Durante,M.J., Liebstein,A., Schmitt-John,T., Werner,T. and Graw,J. (1994) Alpha-crystallins are involved in specific interactions with the murine gamma D/E/F-crystallin-encoding gene. Gene, 144, 171–178. [DOI] [PubMed] [Google Scholar]
- 20.Pietrowski D. and Graw,J. (1997) Autokinase activity of alpha-crystallin inhibits its specific interaction with the DOTIS element in the murine gamma D/E/F-crystallin promoter in vitro. Biol. Chem., 378, 1183–1186. [PubMed] [Google Scholar]
- 21.Lok S., Breitman,M.L., Chepelinsky,A.B., Piatigorsky,J., Gold,R.J. and Tsui,L.C. (1985) Lens-specific promoter activity of a mouse gamma-crystallin gene. Mol. Cell. Biol., 5, 2221–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.den Dunnen J.T., van Neck,J.W., Cremers,F.P., Lubsen,N.H. and Schoenmakers,J.G. (1989) Nucleotide sequence of the rat gamma-crystallin gene region and comparison with an orthologous human region. Gene, 78, 201–213. [DOI] [PubMed] [Google Scholar]
- 23.Murer-Orlando M., Paterson,R.C., Lok,S., Tsui,L.C. and Breitman,M.L. (1987) Differential regulation of gamma-crystallin genes during mouse lens development. Dev. Biol., 119, 260–267. [DOI] [PubMed] [Google Scholar]
- 24.Meakin S.O., Breitman,M.L. and Tsui,L.C. (1985) Structural and evolutionary relationships among five members of the human gamma-crystallin gene family. Mol. Cell. Biol., 5, 1408–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Birren B., Linton,L., Nusbaum,C. and Lander,E. (1999) EMBL accession no AC018961.
- 26.Lok S., Stevens,W., Breitman,M.L. and Tsui,L.C. (1989) Multiple regulatory elements of the murine gamma 2-crystallin promoter. Nucleic Acids Res., 17, 3563–3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jean D., Ewan,K. and Gruss,P. (1998) Molecular regulators involved in vertebrate eye development. Mech. Dev., 76, 3–18. [DOI] [PubMed] [Google Scholar]
- 28.Cvekl A., Sax,C.M., Li,X., McDermott,J.B. and Piatigorsky,J. (1995) Pax-6 and lens-specific transcription of the chicken delta 1-crystallin gene. Proc. Natl Acad. Sci. USA, 92, 4681–4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gopal-Srivastava R., Cvekl,A. and Piatigorsky,J. (1996) Pax-6 and alphaB-crystallin/small heat shock protein gene regulation in the murine lens. Interaction with the lens-specific regions, LSR1 and LSR2. J. Biol. Chem., 271, 23029–23036. [DOI] [PubMed] [Google Scholar]
- 30.Richardson J., Cvekl,A. and Wistow,G. (1995) Pax-6 is essential for lens-specific expression of zeta-crystallin. Proc. Natl Acad. Sci. USA, 92, 4676–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wigle J.T., Chowdhury,K., Gruss,P. and Oliver,G. (1999) Prox1 function is crucial for mouse lens-fibre elongation. Nature Genet., 21, 318–322. [DOI] [PubMed] [Google Scholar]
- 32.Oliver G., Loosli,F., Koster,R., Wittbrodt,J. and Gruss,P. (1996) Ectopic lens induction in fish in response to the murine homeobox gene Six3. Mech. Dev., 60, 233–239. [DOI] [PubMed] [Google Scholar]
- 33.Oliver G., Mailhos,A., Wehr,R., Copeland,N.G., Jenkins,N.A. and Gruss,P. (1995) Six3, a murine homologue of the sine oculis gene, demarcates the most anterior border of the developing neural plate and is expressed during eye development. Development, 121, 4045–4055. [DOI] [PubMed] [Google Scholar]
- 34.Rhim J.S., Trimmer,R., Arnstein,P. and Huebner,R.J. (1981) Neoplastic transformation of chimpanzee cells induced by adenovirus type 12–simian virus 40 hybrid virus. Proc. Natl Acad. Sci. USA, 78, 313–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krausz E., Augusteyn,R.C., Quinlan,R.A., Reddan,J.R., Russell,P., Sax,C.M. and Graw,J. (1996) Expression of Crystallins, Pax6, Filensin, CP49, MIP, and MP20 in lens-derived cell lines. Invest. Ophthalmol. Vis. Sci., 37, 2120–2128. [PubMed] [Google Scholar]
- 36.Reddan J.R., Chepelinsky,A.B., Dziedzic,D.C., Piatigorsky,J. and Goldenberg,E.M. (1986) Retention of lens specificity in long-term cultures of diploid rabbit lens epithelial cells. Differentiation, 33, 168–174. [DOI] [PubMed] [Google Scholar]
- 37.Grimm C., Chatterjee,B., Favor,J., Immervoll,T., Loster,J., Klopp,N., Sandulache,R. and Graw,J. (1998) Aphakia (ak), a mouse mutation affecting early eye development: fine mapping, consideration of candidate genes and altered Pax6 and Six3 gene expression pattern. Dev. Genet., 23, 299–316. [DOI] [PubMed] [Google Scholar]
- 38.Zinovieva R.D., Duncan,M.K., Johnson,T.R., Torres,R., Polymeropoulos,M.H. and Tomarev,S.I. (1996) Structure and chromosomal localization of the human homeobox gene Prox 1. Genomics, 35, 517–522. [DOI] [PubMed] [Google Scholar]
- 39.Tomarev S.I., Zinovieva,R.D., Chang,B. and Hawes,N.L. (1998) Characterization of the mouse Prox1 gene. Biochem. Biophys. Res. Commun., 248, 684–689. [DOI] [PubMed] [Google Scholar]
- 40.Graw J., Coban,L., Liebstein,A. and Werner,T. (1991) Murine gamma E-crystallin is distinct from murine gamma 2-crystallin. Gene, 104, 265–270. [DOI] [PubMed] [Google Scholar]
- 41.Boshart M., Kluppel,M., Schmidt,A., Schutz,G. and Luckow,B. (1992) Reporter constructs with low background activity utilizing the cat gene. Gene, 110, 129–130. [DOI] [PubMed] [Google Scholar]
- 42.Krausz E. and Graw,J. (1996) A new cat reporter gene vector designed for rapid and efficient cloning of PCR products. Gene, 177, 99–102. [DOI] [PubMed] [Google Scholar]
- 43.Kamachi Y. and Kondoh,H. (1993) Overlapping positive and negative regulatory elements determine lens-specific activity of the delta 1-crystallin enhancer. Mol. Cell. Biol., 13, 5206–5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wan L., Twitchett,M.B., Eltis,L.D., Mauk,A.G. and Smith,M. (1998) In vitro evolution of horse heart myoglobin to increase peroxidase activity. Proc. Natl Acad. Sci. USA, 95, 12825–12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quandt K., Frech,K., Karas,H., Wingender,E. and Werner,T. (1995) MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res., 23, 4878–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hassan B., Li,L., Bremer,K.A., Chang,W., Pinsonneault,J. and Vaessin,H. (1997) Prospero is a panneural transcription factor that modulates homeodomain protein activity. Proc. Natl Acad. Sci. USA, 94, 10991–10996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tomarev S.I., Sundin,O., Banerjee-Basu,S., Duncan,M.K., Yang,J.M. and Piatigorsky,J. (1996) Chicken homeobox gene Prox 1 related to Drosophila prospero is expressed in the developing lens and retina. Dev. Dyn., 206, 354–367. [DOI] [PubMed] [Google Scholar]
- 48.Meakin S.O., Reddan,J.R., Tsui,L.C. and Breitman,M.L. (1989) A rabbit lens epithelial cell line supports expression of an exogenous crystallin gene characteristic of lens fiber cell differentiation. Exp. Eye Res., 48, 131–137. [DOI] [PubMed] [Google Scholar]
- 49.Ogino H. and Yasuda,K. (1998) Induction of lens differentiation by activation of a bZIP transcription factor, L-Maf. Science, 280, 115–118. [DOI] [PubMed] [Google Scholar]
- 50.Kawauchi S., Takahashi,S., Nakajima,O., Ogino,H., Morita,M., Nishizawa,M., Yasuda,K. and Yamamoto,M. (1999) Regulation of lens fiber cell differentiation by transcription factor c-Maf. J. Biol. Chem., 274, 19254–19260. [DOI] [PubMed] [Google Scholar]
- 51.Grimm C., Sporle,R., Schmid,T.E., Adler,I.D., Adamski,J., Schughart,K. and Graw,J. (1999) Isolation and embryonic expression of the novel mouse gene Hic1, the homologue of HIC1, a candidate gene for the Miller-Dieker syndrome. Hum. Mol. Genet., 8, 697–710. [DOI] [PubMed] [Google Scholar]
- 52.Halder G., Callaerts,P. and Gehring,W.J. (1995) Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science, 267, 1788–1792. [DOI] [PubMed] [Google Scholar]
- 53.Hill R.E., Favor,J., Hogan,B.L., Ton,C.C., Saunders,G.F., Hanson,I.M., Prosser,J., Jordan,T., Hastie,N.D. and van Heyningen,V. (1991) Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature, 354, 522–525. [DOI] [PubMed] [Google Scholar]