Summary
Mammalian non-coding micro RNAs (miRNAs) are a class of gene regulators that have been linked to immune system function. Here, we have investigated the role of miR-155 during an autoimmune inflammatory disease. Consistent with a positive role for miR-155 in mediating inflammatory responses, Mir155−/− mice were highly resistant to experimental autoimmune encephalomyelitis (EAE). miR-155 functions in the hematopoietic compartment to promote the development of inflammatory T cells including the T helper 17 (Th17) cell and Th1 cell subsets. Furthermore, the major contribution of miR-155 to EAE was CD4+ T cell intrinsic, whereas miR-155 was also required for optimum dendritic cell production of cytokines that promoted Th17 cell formation. Our study shows that one aspect of miR-155 function is the promotion of T cell-dependent tissue inflammation, suggesting that miR-155 might be a promising therapeutic target for the treatment of autoimmune disorders.
Introduction
The mammalian inflammatory response has evolved to control infection by microbial pathogens before the onset of sepsis and death, while also playing important roles in tissue repair (Medzhitov, 2008). Despite its utility, when the inflammatory response is activated inappropriately, it may be directed against specific self-tissue antigens and cause serious disease. The outcome can be debilitating to important organ systems and is the underlying cause of widespread human autoimmune disorders.
Recent work has revealed that IL-17 producing inflammatory CD4+ T cells, or T helper 17 (Th17) cells, are critical mediators of chronic, autoimmune inflammation (Bettelli et al., 2006; Ivanov et al., 2006). Th17 cell development is driven by cytokines produced primarily by cells of the innate immune system, including transforming growth factor-β (TGF-β) interleukin-6 (IL-6), IL-23 and IL-1 (Bettelli et al., 2006; Langrish et al., 2005; Veldhoen et al., 2006). The impact of Th17 cells was first made evident in mice, where over expression of IL-17 led to increased granulopoiesis in vivo (Schwarzenberger et al., 1998). Subsequent studies demonstrated that inhibition of IL-17 in mice can ameliorate several autoimmune disorders including experimental autoimmune encephalomyelitis (EAE), collagen induced arthritis (CIA), and inflammatory bowel disease (IBD) (Ivanov et al., 2006; Komiyama et al., 2006; Murphy et al., 2003).
Micro-RNAs (miRNAs) are a novel class of non-coding RNAs that modulate gene expression at the posttranscriptional level, and are involved in regulating several aspects of inflammation (O'Connell et al., 2010; Xiao and Rajewsky, 2009). Specific miRNAs, such as miR-146a, miR-155 and miR-132, were initially shown to be upregulated during the macrophage inflammatory response (O'Connell et al., 2007; Taganov et al., 2006). The functional impact of certain miRNAs on inflammation has been demonstrated in vivo. Mice deficient in miR-223, a miRNA that is enriched in myeloid cells, display elevated granulocyte numbers and increased immunity against fungi (Johnnidis et al., 2008). Other studies have found important roles for miRN As in lymphocytes. For instance, enforced expression of the miR-17-92 cluster in T cells or specific deletion of Dicer in T regulatory (Treg) cells both trigger lethal autoimmune conditions (Chong et al., 2008; Liston et al., 2008; Xiao et al., 2008; Zhou et al., 2008).
miR-155 was among the first miRNAs linked to inflammation by virtue of its potent upregulation in multiple immune cell lineages by Toll like receptor (TLR) ligands, inflammatory cytokines, and specific antigens (Haasch et al., 2002; O'Connell et al., 2007; Taganov et al., 2006; Thai et al., 2007). A wide variety of immunologically relevant targets of miR-155 have been reported, implying distinct roles in mammalian immunity. Among these roles, miR-155 has been shown to be important for immunoglobulin (Ig) class switching to IgG in B cells via targeted repression of activation induced cytidine deaminase (AID) and the transcription factor PU.1 (Dorsett et al., 2008; Rodriguez et al., 2007; Thai et al., 2007; Vigorito et al., 2007). The fitness of T regulatory (Treg) cells is influenced by direct repression of suppressor of cytokine signaling 1 (SOCS1) by miR-155 (Lu et al., 2009). In myeloid cells, over expression of miR-155 drives a myeloproliferative disorder through a mechanism involving reduced src homology 2 domain-containing inositol-5-phosphatase 1 (SHIP1) expression, suggesting that miR-155 is acting as a positive regulator of inflammation (O'Connell et al., 2009; O'Connell et al., 2008). Despite these reported functions of miR-155 in both innate and adaptive immune cells, to date there has been little genetic evidence that endogenously expressed miR-155 actually impacts inflammatory responses in vivo. In the present study, we investigated the role that miR-155 might play during antigen specific inflammatory responses against self-tissues.
Results
Mir155−/− mice are resistant to EAE induced by myelin oligodendrocyte glycoprotein35–55 (MOG35–55)
To identify a possible role for miR-155 in mediating tissue specific autoimmune inflammation, a mouse model of EAE was used. Both wild-type (WT) and Mir155−/− mice were immunized with 100 µg of the myelin oligodendrocyte glycoprotein (MOG) peptide35–55 emulsified in complete Freund’s adjuvant (CFA) followed by administration of pertussis toxin. As anticipated, WT mice first displayed neurologic symptoms approximately 9 days post-immunization, with peak disease severity on day 14 (average clinical score of 2.1), and 100% disease incidence (Figures 1A and 1B). In contrast, Mir155−/− mice exhibited a later onset of symptoms on day 11, with a low peak disease severity on day 15 (average clinical score of 0.3). Unlike the WT controls, the disease incidence in Mir155−/− mice was only 60% (Figures 1A and 1B). On day 25, mice were sacrificed and underwent further evaluation including tissue histological analysis. Hematoxylin and eosin (H&E) brain cross-sections were scored for disease severity (Figure 1C and 1D). As expected, WT mice suffered from heavy perivascular congestion, parenchymal infiltration and focal meningeal lymphocytosis. However, brain tissue from Mir155−/− mice showed minimal histologic evidence of inflammation consistent with the mild clinical manifestation of EAE (Figures 1C and 1D). Furthermore, upon analyzing the draining lymph nodes (LNs) and spleens from both groups of mice, we found decreased overall cellularity in LNs from Mir155−/− mice, and compositionally fewer CD11b+ myeloid cells in Mir155−/− spleens (Figures 1E and 1F). Both of these observations are consistent with a reduced inflammatory condition in Mir155−/− mice.
Figure 1. Mir155−/− mice are resistant to EAE induced by MOG35–55.
A. EAE was induced in Mir155+/+ and Mir155−/− mice by immunizing both groups with 100 µg of the MOG35–55 peptide followed by administration of pertussis toxin. Their disease severity was scored regularly based upon clinical symptoms (n=10). Data represent three independent experiments. B. Disease incidence was assessed for each group (n=10). C. Representative H&E stained brain sections from Mir155+/+ or Mir155−/− mice harvested on day 25 post-immunization. D. Average histology score for each group (n=4). E. Number of live LN cells (left) and their lineage composition was assessed by flow cytometry (right) using LNs from Mir155+/+ and Mir155−/− mice 25 days after immunization (n=4). F. Number of splenocytes (left) and their lineage composition as determined by flow cytometry (right) using spleens from both groups 25 days after immunization (n=4). G. WT mice were lethally irradiated and reconstituted with Mir155+/+ or Mir155−/− BM. 4 months later, expression of miR-155 in LPS activated splenic B cells was assessed. H. MOG35–55-induced EAE was induced in mice with WT or Mir155−/− hematopoietic cells and disease was scored over a time course (n=5–7). I. 12 days following induction of EAE in WT mice with MOG35–55, splenocytes were harvested and cultured in 20 µg/ml MOG35–55 and 20ng/ml IL-12 p70 for 48 hours. Cells were then washed and 25×106 cells were injected intravenously into Mir155+/+ and Mir155−/− mice followed by administration of pertussis toxin. Mice were monitored regularly and disease severity was scored (n=5). Data represent two independent experiments. Error bars represent +/−SEM and * denotes statistical significance with a p value of <0.05 according to a student’s two-tailed t-test. See also Figure S1
Lethally irradiated WT C57BL/6 mice were next reconstituted with either Mir155+/+ or Mir155−/− bone marrow (BM) cells. After 4 months, proper engraftment and localization of the miR-155 deficiency to the hematopoietic compartment was confirmed by assaying miR-155 amounts in activated splenic B cells (Figure 1G). Following induction of EAE, mice with Mir155+/+ hematopoietic cells exhibited a faster and more severe disease phenotype than mice containing Mir155−/− hematopoietic cells (Figure 1H). In a separate experiment, 25 × 106 WT encephalitogenic splenocytes from day 12 EAE WT mice were transferred into WT or Mir155−/− hosts, which were monitored for the presence of clinical symptoms. Both groups began to show symptoms by day 8 post-adoptive transfer, and had comparable disease scores throughout the 22 day time course (Figure 1I). Furthermore, both cohorts had a disease incidence of 100% (Figure S1). Taken together, these data demonstrate that miR-155 functions in the hematopoietic compartment to promote EAE.
Mir155−/− mice exhibit defective inflammatory T cell development during EAE
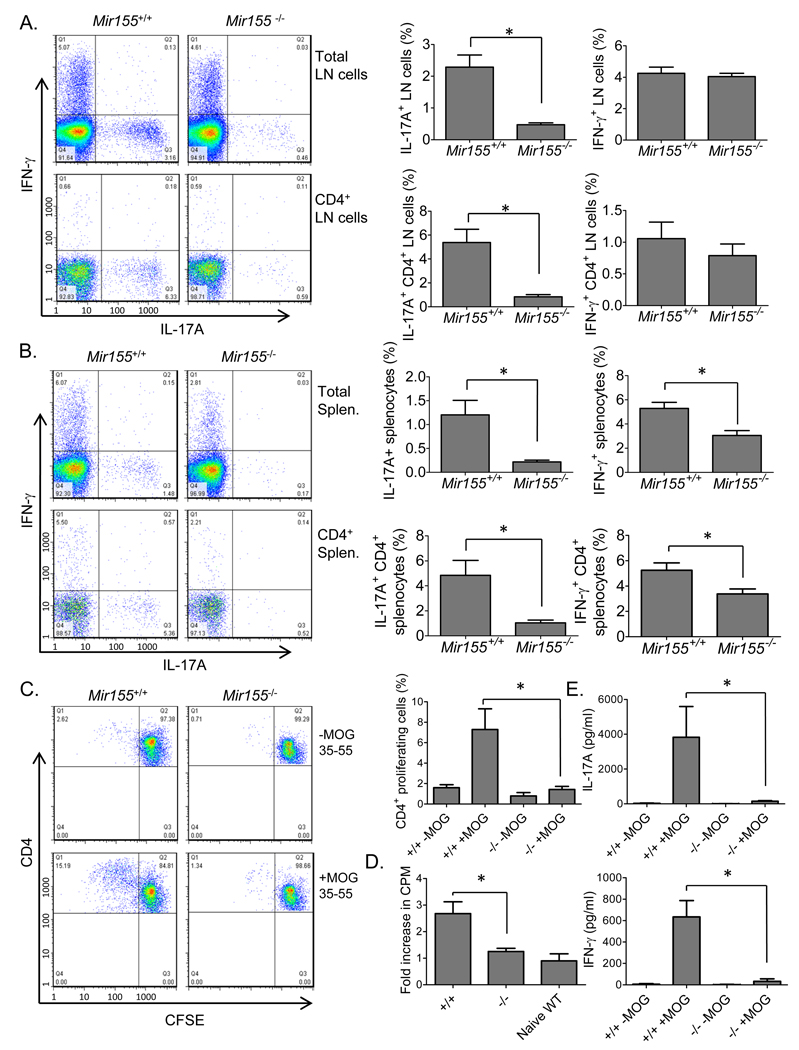
Th17 and Th1 cells are hematopoietic cells that develop during tissue specific inflammatory responses and play a pivotal role in enhancing inflammation (Littman and Rudensky, 2010). Therefore, we examined lymph nodes (LNs) and splenocytes from WT and Mir155−/− mice for the presence of IL-17 (Th17) or interferon-γ (IFN-γ) (Th1) producing CD4+ T cells during EAE. On day 25 post-immunization with MOG35–55, Mir155−/− mice had substantially diminished amounts of Th17 cells in both their LNs and spleens compared to WT mice (Figures 2A and 2B). Moderately reduced amounts of IFN-γ producing Th1 CD4+ cells were also found in the spleens but not LNs of MOG35–55 immunized mice in the absence of miR-155 (Figures 2A and 2B)The total numbers of these inflammatory T cell populations in the spleen and LNs was also similarly reduced in Mir155−/− versus Mir155+/+ mice 25 days after immunization with MOG35–55 (Figure S2).
Figure 2. Mir155−/− mice exhibit defective inflammatory T cell development during EAE.
Mir155+/+ and Mir155−/− mice were harvested 25 days following immunization with MOG35–55. A. Intracellular staining was conducted to identify total lymph node cells (top) and CD4+ lymphocytes (bottom) producing IL-17A and/or IFN-γ (n=4). B. Splenocytes were analyzed as in (A) (n=4). C. Mir155+/+ or miR155−/− splenocytes harvested from mice 25 days after EAE induction were labeled with CFSE. CFSE loss by CD4+ proliferating cells from both groups was assayed by flow cytometry following restimulation with MOG35–55 (20 µg/ml) for 72 hours (n=4). D. 3[H] thymidine incorporation was also assays using replicate cultures (n=4). 2 naïve WT mice were also included as controls. E. Production of IL-17A and IFN-γ by cells from (C.) was determined by ELISA (n=4). Data represent two independent experiments. Error bars represent +/−SEM and * denotes statistical significance with a p value of <0.05 according to a student’s two-tailed t-test. +/+ = Mir155+/+; −/− = Mir155−/−. See also Figure S2.
The in vitro recall response to the MOG35–55 peptide by WT and Mir155−/− CD4+ T cells from the spleens of EAE mice was also assessed. Carboxyfluorescein succinimidyl ester (CFSE)-labeled splenocytes from day 25 EAE mice were restimulated in vitro with 20 µg/ml of MOG35–55 or cultured in medium alone for 72 hours followed by flow cytometric analysis to determine the extent of CD4+ proliferation as determined by dilution of CFSE. We found that WT CD4+ T cells underwent cell divisions following exposure to MOG35–55, while Mir155−/− CD4+ T cells had a substantially reduced proliferative response to the same peptide (Figure 2C). In parallel, 3[H] thymidine incorporation assays using total splenocytes produced sim ilar differences (Figure 2D). Tissue culture supernatants from these experiments were assayed for IL-17A and IFN-γ production by ELISA in response to MOG35–55 stimulation. MOG35–55 stimulated Mir155−/− splenocytes showed minimal production of both of these cytokines compared to WT splenocytes further demonstrating a defective CD4+ T cell driven recall response to antigen (Figure 2E).
We next investigated whether a defect in inflammatory T cell development could also be detected during the initial onset, or induction phase, of EAE. Mice were harvested on day 13 post immunization with MOG35–55 and the LNs from Mir155−/− mice had reduced numbers of live cells compared to WT controls, while the brains (CNS) and spleens from the two groups had similar total cell numbers (Figure S3). Inflammatory T cell development in the brains, LNs and spleens was next assessed. Mir155−/− mice had substantial reductions in both the absolute numbers of Th17 cells and the percentage of Th17 cells among total CD4+ T cells in their brains compared to Mir155+/+ control mice (Figures 3A and 3B). IFN-γ producing Th1 cells were present at lower absolute numbers in the brains of Mir155−/− mice, while the proportion of Th1 T cells among total CD4+ T cells was equivalent between Mir155−/− and Mir155+/+ brains (Figure 3B). BIC (the non-coding RNA that gives rise to miR-155) expression was detected by quantitative PCR (qPCR) in Mir155+/+ but not Mir155−/− splenocytes, and deficiencies in both IL-17A and IL-23 p19 mRNAs were also observed (Figure 3C). Intracellular staining revealed diminished numbers of Th17 and Th1 CD4+ T cells in Mir155−/− spleens (Figure 3D). The recall response to MOG35–55 was also tested using splenocytes from day 13 EAE mice. Mir155−/− splenocytes exhibited diminished proliferation during this assay (Figure 3E). Defective production of IL-17A, IFN-γ, IL-6 and granulocyte-monocyte-colony stimulating factor (GM-CSF) by MOG35–55 restimulated Mir155−/− encephalitogenic splenocytes was evident (Figure 3F). Similar deficiencies in Th17 and Th17 cells were also observed in the LNs from Mir155−/− mice at this same timepoint (Figure 3G and 3H). These data indicate that the development of inflammatory T cells in Mir155−/− mice is defective during the early, induction phase of EAE.
Figure 3. miR-155 is required for inflammatory T cell development during the induction phase of EAE.
Mir155+/+ and Mir155−/− mice were harvested 13 days following immunization with MOG35–55. A. Mononuclear cells were purified from Mir155+/+ and Mir155−/− brains and intracellular staining was conducted to identify CD4+ lymphocytes producing IL-17A and/or IFN-γ (n=5). B. Total numbers of Th17 and Th1 cells in the brain, in addition to the percentage of Th17 and Th1 cells among total CD4+ T cells is shown on the right (n=5). C. WT and Mir155−/− splenocytes were analyzed for expression of BIC, IL-17A and IL-23 p19 mRNA by qPCR (n=5) and D. intracellular staining was used to determine the number of Th17 and Th1 cells (n=5). E. Splenocytes were restimulated with MOG35–55 and E. proliferation was assayed by 3[H] thymidine incorporation (n=5) and F. the production of IL-17A, IFN-γ, IL-6 and GM-CSF measured by ELISA (n=5). G. Expression of BIC and IL-17A mRNA in the LNs was assayed by qPCR (n=5) and H. the number of Th17 and Th1 cells was also quantified by flow cytometry (n=5). Error bars represent +/−SEM and * denotes statistical significance with a p value of <0.05 according to a student’s two-tailed t-test. +/+ = Mir155+/+; −/− = Mir155−/−. See also Figure S3.
A recent report found reduced numbers of T regulatory cells in Mir155−/− mice under steady state conditions (Kohlhaas et al., 2009; Lu et al., 2009). Consistent with these findings, we also observed lower Treg cell amounts in both the LNs and spleens of Mir155−/− mice compared to WT controls during EAE (Figure S2). However, Mir155−/− Treg cells are not functionally defective compared to WT Tregs on a per cell basis (Kohlhaas et al., 2009; Lu et al., 2009). Thus, the reduced EAE inflammation in Mir155−/− mice seems unlikely to be related to the Treg cell population in these animals.
Also in agreement with earlier studies (Rodriguez et al., 2007; Thai et al., 2007; Vigorito et al., 2007), we found reduced titers of anti-MOG35–55 IgG antibodies in Mir155−/− mice during EAE (Figure S2). Since it has been reported that B cells, and therefore antibodies, are dispensable specifically for MOG35–55 driven EAE (Hjelmstrom et al., 1998), it is likely that the observed antibody deficit does not account for the reduced disease severity seen in Mir155−/− mice.
Mir155−/− mice have reduced foot pad inflammation during delayed type hypersensitivity (DTH)
Another Th17 dependent model of inflammation was next used to assess whether Mir155−/− mice have a general deficit in mediating inflammatory responses to specific antigens (Ghilardi et al., 2004). WT and Mir155−/− mice were immunized with Keyhole Limpet Hemocyanin (KLH) in complete Freund’s adjuvant (CFA), and 8 days later challenged in one footpad with KLH and in the contralateral footpad with saline. After 48 hours footpad thickness was measured to assess the DTH response. As expected, WT mice had a substantial increase in footpad inflammation following KLH administration compared to saline alone (Figure 4A). In contrast, Mir155−/− mice exhibited reduced amounts of swelling in response to KLH as compared to WT mice (Figure 4A). Splenocytes and LNs were also harvested on day 10, and Mir155−/− LNs had significantly fewer total cells than WT controls, consistent with a blunted inflammatory response (Figure 4B). Cells from both organs were restimulated with KLH for 3 days. While LN cell and splenocyte proliferative differences were not observed in response to stimulation with KLH (Figure 4C), substantial reductions in IL-17A, IFN-γ and IL-6 production were seen in Mir155−/− versus WT splenocytes and LN cells during recall responses (Figure 4D). These data reveal a general role for miR-155 in mediating antigen and tissue specific inflammation and point to a consistent defect in inflammatory T cell production.
Figure 4. Mir155−/− mice have reduced foot pad inflammation during DTH.
Mir155+/+ and Mir155−/− mice were immunized with 100 µg of KLH in CFA and 8 days later injected with 50 µg of KLH in one footpad and PBS in the other. A. Increases in footpad inflammation were measured for both groups (n=5). B. Total numbers of splenocytes and LN cells was assessed (n=5). C. Proliferation of splenocytes and LN cells following in vitro restimulation with KLH was determined by assaying 3[H] thymidine incorporation (n=5). D. Production of IL-17A, IFN-γ and IL-6 from the cells in (C.) was determined by ELISA (n=5). Error bars represent +/−SEM and * denotes statistical significance with a p value of <0.05 according to a student’s t-test. +/+ = Mir155+/+; −/−= Mir155−/−.
A T cell intrinsic role for miR-155 in the development of inflammatory T cells during EAE
Inflammatory T cells must be able to properly receive and coordinate the signals provided by specific inflammatory cytokines that mediate their development. Thus, we tested whether miR-155 expression by CD4+ T cells is involved in their ability to be skewed towards the Th17 lineage in vitro. CD4+ splenic T cells were isolated from Mir155+/+ and Mir155−/− mice and cultured in the presence of CD3 and CD28 antibodies with and without the addition of the Th17 skewing factors IL-6 and TGF-γ. Following four days of culture, we found that Mir155−/− CD4+ T cells were defective in their ability to produce Th17 cells compared to WT controls as assayed by intracellular staining of IL-17A (Figures 5A and 5B). The same cell populations and culture conditions produced similar amounts of IFN-γ+ Th1 cells despite the miR-155 deficiency (Figure 5A and 5B). As in previous reports (Haasch et al., 2002), we also observed upregulation of miR-155 in activated CD4+ T cells (Figure 5C), and detected expression of BIC and miR-155 in CD4+ T cells grown in conditions that promote Th17 development (Figure 5D). Reduced expression of IL-17A mRNA was measured in Mir155−/− compared to WT CD4+ T cells under Th17 skewing conditions (Figure 5D). These results reveal a T cell intrinsic role for miR-155 in promoting the in vitro development of Th17 cells.
Figure 5. miR-155 expression by CD4+ T cells is necessary for proper Th17 cell development in vitro.
A. CD4+ T cells were isolated from Mir155+/+ or Mir155−/− spleens and cultured in the presence of plate bound CD3 and soluble CD28 antibodies, with (Th17 cell) and without (Th0 cell) IL-6 (50 ng/ml) and TGF-β (2 ng/ml). After 96 hours, expression of IL-17A and IFN-γ was assayed by intracellular staining following by flow cytometry. B. Results from a representative experiment are represented graphically (n=2). Data represent three independent experiments. C. Expression of Mir155 was measured by qPCR before and after activation with CD3 and CD28 antibodies (n=3). D. Expression of BIC, Mir155, and IL-17A in WT and Mir155−/− CD4+ T cells was assayed by qPCR following 96 hours of culture with CD3 and CD28 antibodies alone, or in Th17 cell skewing conditions (n=2). Data represent two independent experiments. Error bars represent +/−SEM.
To test whether miR-155 plays a T cell intrinsic role in driving inflammatory T cell development during EAE in mice, we adoptively transferred 6×106 purified naïve Mir155+/+ or Mir155−/− CD4+ T cells into Rag1−/− recipients and induced EAE 24 hours later. Mice receiving Mir155+/+ CD4+ T cells had a substantially more severe and accelerated disease course compared to mice receiving Mir155−/− CD4+ T cells (Figure 6A). Reduced percentages of Th17, and to a lesser extent Th1, CD4+ T cells was observed in the spleens and LNs from mice that received Mir155−/− CD4+ T cells compared to those engrafted with Mir155+/+ CD4+ T cells (Figures 6B and 6C).
Figure 6. Expression of miR-155 by CD4+ T cells is required for proper development of inflammatory T cells during EAE.
A. 5×106 WT or Mir155−/− CD4+ T cells from naïve mice were injected i.v. into Rag1−/− recipients, EAE was induced with MOG35–55 24 hours later, and disease was scored over a time course (n=5–6). Data represent two independent experiments. B. Mice were harvested and engraftment of CD3+CD4+ T cells was assayed by flow cytometry using splenocytes (top). Expression of IL-17A and IFN-γ by CD4+ cells in the spleens and LNs was assayed by intracellular staining followed by flow cytometry. A representative plot from the LNs is shown (bottom). C. The averages of 5–6 mice per group are shown graphically. D. Mir155+/+ and Mir155−/− mice were injected with 1×107 WT CD45.1+CD4+ naïve T cells, and EAE was induced 24 hours later (n=5). Disease symptoms were scored over a time course. Data represent two independent experiments. E. Mice were harvested and CD4+ T cells in the brains were analyzed by flow cytometry to detect cells expressing CD45.1, IL-17A and IFN-γ. F. The average of 5 mice per group from (E.). Error bars represent +/−SEM and * denotes statistical significance with a p value of <0.05 according to a student’s two-tailed t-test. +/+ = Mir155+/+; −/− = Mir155−/−. See also Figure S4.
To examine whether miR-155 expression specifically in CD4+ T cells could restore disease severity in Mir155−/− mice, we adoptively transferred 1×107 naïve CD45.1+CD4+ T cells into both WT and Mir155−/− recipients and induced EAE 24 hours later. Although Mir155+/+ mice began to show clinical symptoms a few days before Mir155−/− mice, both groups exhibited similar disease scores for most of the time course (Figure 6D). The brain was harvested on day 23 post-immunization, and the development of Th17and Th1 cells was determined by intracellular staining for IL17A and IFN-γ, respectively (Figures 6E, 6F and S4). While both mouse groups had roughly equivalent percentages of both Th17 and Th1 cells among the total CD4+ T cells in the brain, these cellular subsets were comprised predominately of the adoptively transferred CD45.1+CD4+ WT T cells in the CNS of Mir155−/− mice. This bias occurred despite roughly similar amounts of both WT and Mir155−/− CD4+ T cells being present in the brains of Mir155−/− EAE mice (Figures 6E and 6F). Conversely, these same inflammatory T cell populations were comprised largely of endogenous origin (CD45.1−CD4+ Mir155+/+ T cells) in the CNS of Mir155+/+ mice (Figures 6E and 6F). These data demonstrate that miR-155 expression by CD4+ T cells is critical for the proper development of inflammatory T cells subsets in the CNS and that this accounts for a majority of miR-155’s contribution to EAE.
miR-155 expression in lipopolysaccharide (LPS)-activated, GM-CSF-derived myeloid dendritic cells is necessary for proper production of Th17 relevant inflammatory cytokines
Due to the lag in EAE phenotype development when WT CD4+ T cells were administered to Mir155−/− mice, it is possible that miR-155 also functions in non-T cell immune cell types to promote inflammatory T cell development. For encephalitogenic Th17 cells to develop they must receive signals from relevant inflammatory cytokines, such as IL-6 and IL-23, which are produced by GM-CSF-derived DCs (Gutcher and Becher, 2007). Therefore, we examined the impact of miR-155 on the gene expression profile of LPS-activated DCs, which express high levels of miR-155 (Figures 7A and 7B). Total RNA was collected from purified WT or Mir155−/− DCs after 20 hours of LPS treatment and subjected to a microarray analysis. Several targets of miR-155 were expressed at higher amounts in Mir155−/− vs. WT control DCs (Figure 7C). Among these, SHIP1 and SOCS1 have been shown to be directly targeted by miR-155 (Androulidaki et al., 2009; Lu et al., 2009; O'Connell et al., 2009), and to function by negatively regulating cytokine production by DCs (An et al., 2005; Shen et al., 2004). Their elevated expression in Mir155−/− DCs was confirmed using quantitative PCR (qPCR) and by Western blotting (Figure 7D). Consistent with the elevated expression of these negative regulators, decreased expression of several inflammatory cytokine genes including IL-6, IL-23 p19 and IL-12 and IL-23 p40 was observed in Mir155−/− DCs (Figure 5C). These results were confirmed by qPCR and ELISA, which also detected a subtle decrease in tumor necrosis factor-α (TNF-α) production (Figures 7E and 7F). To further corroborate these findings, miR-155 was overexpressed in GM-CSF-derived DCs using a retroviral vector described previously (O'Connell et al., 2009), and higher amounts of IL-6, IL-23 p19, IL-12 and IL-23 p40, and TNF-α mRNA expression were observed following LPS treatment (Figure 7G). We also tested whether Mir155−/− DCs were defective in their ability to induce CD4+ T cell proliferation following presentation of cognate antigens. Both Mir155+/+ and Mir155−/− DCs induced equivalent proliferation of 2D2 or OT2 CD4+ T cells which recognize MOG35–55 or an ovalbumin peptide, respectively (Figure H). Taken together, these experiments demonstrate that miR-155 promotes DC expression of specific cytokines required for inflammatory T cell development.
Figure 7. miR-155 expression by LPS-activated, GM-CSF-derived myeloid dendritic cells is necessary for proper production of Th17 cell relevant inflammatory cytokines.
A. CD11c+ DCs were derived using GM-CSF at 20 ng/ml. B. Expression of BIC (top) and mature miR-155 (bottom) before and after 20 hours of LPS stimulation (100 ng/ml) was assayed using qPCR. C. Total RNA was next used for a microarray analysis to determine mRNA expression differences between Mir155+/+ and Mir155−/− LPS treated DCs. Several selected targets of miR-155 were expressed in higher amounts in Mir155−/− DCs, while a subset of selected proinflammatory cytokines were expressed at lower amounts. Red = higher expression and Green = lower expression in the Mir155−/− vs. Mir155+/+ DCs. D. Expression of SHIP1 and SOCS1 mRNAs was assessed by qPCR and by Western blotting (n=3). E. qPCR was also used to assay expression of IL-23 p19, IL-6, IL-12p40 and TNF-α mRNA amounts (n=3). Data represent two independent experiments. F. Concentrations of the cytokines from (E) in the culture supernatants were determined by ELISAs (n=3). Data represent two independent experiments. G. GM-CSF-derived DCs overexpressing miR-155, a miR-155 “seed” mutant or a control vector were stimulated with LPS for 20 hours and expression of IL-23 p19, IL-6, IL- 12p40 and TNF-α mRNA was assayed by qPCR. Data represent two independent experiments. H. Proliferation of 2D2 or OT2 CD4+ T cells in response to their respective antigens presented by WT or Mir155−/− DCs was assessed by assaying 3[H] thymidine incorporation (n=3). Data represent two independent experiments. Error bars represent +/−SEM and * denotes statistical significance with a p value of <0.05 according to a student’s two-tailed t-test.
Discussion
We have shown here that miR-155 plays an important role in driving chronic inflammation that is inappropriately directed at tissue specific antigens, a destructive process that is at the heart of human autoimmune diseases. At the cellular level, Mir155−/− mice exhibit defective inflammatory T cell development during the induction phase of autoimmunity. This appears to be largely due to miR-155 function in CD4+ T cells, and may also involve insufficient production of inflammatory cytokines by DCs. While diminished inflammatory T cell development is beneficial in the context of autoimmunity, it is detrimental to mammals following infection by certain pathogens. Thus, our observations also suggest a protective role for miR-155 in response to infection.
MiR-155 expression is dramatically increased in CD4+ T cells upon their activation suggesting functional importance for this miRNA in activated T cells (Haasch et al., 2002). Our results expand upon previous work, which demonstrated that miR-155 can impact Th1 cell and Th2 cell lineage skewing in vitro (Rodriguez et al., 2007; Thai et al., 2007), by finding a novel role for miR-155 in Th17 cell biology both in vitro and in vivo. Recently, inhibition of miR-326 using a miRNA “sponge” was shown to reduce EAE symptoms by preventing Th17 cell differentiation also through a T cell intrinsic mechanism (Du et al., 2009). Thus, multiple miRNAs appear to directly regulate inflammatory T cell development, as suggested by early studies analyzing Dicer deficient T cells (Muljo et al., 2005).
Many direct targets of miR-155 in CD4+ T cells have been identified, some of which impact Th cell lineage decisions (Rodriguez et al., 2007). For instance, c-Maf is targeted by miR-155 and functions as a promoter of Th2 cell development (Rodriguez et al., 2007), while SOCS1 is repressed by miR-155 in both FoxP3+CD4+ Treg cells and FoxP3− CD4+ T cells and impacts Treg cell fitness (Lu et al., 2009). In general, a complex picture of how miR-155 directs T cell developmental pathways is emerging and it appears to involve many targets and pathways. This may also be true for miR-155’s positive role in Th17 cell development. On one hand, miR-155 might function to block the inhibitory impact of cytokines such as IL-4 and IFN-γ on the Th17 cell differentiation pathway. miR-155 has been shown to limit production of IL-4 by CD4+ T cells through repression of c-Maf (Rodriguez et al., 2007), while IFN-γR mRNA is directly targeted by miR-155 in CD4+ T cells (Banerjee et al., 2010). However, we found that WT CD4+ T cells restore EAE disease severity following their adoptive transfer into Mir155−/− mice. This argues against a major role for elevated production of a secreted inhibitory molecule like IL-4 being responsible for the reduced EAE observed in Mir155−/− mice because it would also inhibit the transferred WT T cells through a paracrine mechanism.
Alternatively, it is possible that miR-155 can positively regulate signaling pathways that promote Th17 cell formation. This could be achieved via repression of proteins that function to negatively regulate signaling pathways that are activated by TGF-β, IL-6 and IL-23. While it is unclear whether miR-155 directly regulates these pathways in CD4+ T cells, recent studies found that miR-155 targets Sma and Mad related protein 5 (Smad5) in B cell lymphoma cells (Rai et al., 2010), and activates signal transducer and activator of transcription 3 (Stat3) in breast cancer cells (Jiang et al., 2010), factors involved in TGF-β and IL-6 signaling, respectively. Moving forward, the identity and relative importance of each target of miR-155 during the development of different T cell lineages, including Th17 cells, should continue to be investigated, and may reveal a differential importance for unique targets of miR-155 depending on the T cell subset.
Interestingly, a previous study found that Mir155−/− FoxP3−CD4+ T cells were at a competitive disadvantage in the presence of WT FoxP3−CD4+ T cells under steady state conditions (Lu et al., 2009). During our EAE phenotype “rescue” experiments Mir155−/− CD4+ T cells also showed reduced fitness compared to the adoptively transferred WT CD4+ T cells. Thus, the role of miR-155 in conferring CD4+ T cell fitness extends to inflammatory settings where miR-155 is also required for proper inflammatory T cell differentiation. These reductions in fitness and differentiation by effector T cells may explain why Mir155−/− mice do not succumb to spontaneous systemic autoimmunity or suffer from heightened EAE symptoms as a consequence of their reduced Treg cell numbers (Lu et al., 2009).
Similar to Mir155−/− mice, GM-CSF-deficient mice are also resistant to EAE (McQualter et al., 2001) and GM-CSF derived DCs have recently been linked to Th17 cell development during autoimmunity by functioning as an important source of the Th17 cytokines IL-6 and IL-23 (Sonderegger et al., 2008). Our present work demonstrates that miR-155 is upregulated in GM-CSF-derived DCs and functions to enhance production of these cytokines. This may contribute to the defects in Th17 cell development and tissue specific inflammation observed in the Mir155−/− mice. Additionally, because Th17 cells are a primary source of GM-CSF during EAE (Ponomarev et al., 2007), they are thought to reinforce this chronic inflammatory condition by generating additional DCs. Thus, the defective production of Th17 cells in Mir155−/− mice during EAE may also explain the reduced amounts of GM-CSF observed during our splenocyte recall response experiments. Together, our data suggest a role for miR-155 in regulating both the production and function of GM-CSF-derived DCs in the promotion of autoimmune inflammation.
Elevated expression of miR-155 has been observed in brain lesions from multiple sclerosis (MS) patients (Junker et al., 2009) and in synovial samples from patients with rheumatoid arthritis (RA) (Stanczyk et al., 2008). Human trials have found that inhibition of the IL-23-IL-17 inflammatory axis using blocking antibodies can reduce the severity of psoriasis and rheumatoid arthritis (Steinman, 2010). Based upon this expression profile and our current study, miR-155 may be an effective therapeutic target in the treatment of a range of autoimmune conditions where Th17 cells have been shown to drive disease. However, delivery of miRNA inhibitors to specific cell types in vivo remains a difficult challenge.
In addition to inflammatory T cells, many autoimmune conditions, including human MS, RA, and Systemic Lupus Erythematosus (SLE), also involve the actions of auto-antibodies that have been shown to exacerbate diseases. Beyond its role in mediating inflammatory T cell development as identified in our current study, previous reports have clearly shown that miR-155 is important for production of antigen-specific IgGs (Rodriguez et al., 2007; Thai et al., 2007; Vigorito et al., 2007). Therefore, modulation of miR-155 may be able to treat conditions that depend both upon humoral and cell-mediated immune mechanisms, making it a versatile therapeutic target.
Experimental Procedures
Mice
All experiments were approved by the Caltech Institutional Animal Care and Use Committee (IACUC). Mir155+/+, Mir155−/−, Rag1−/−, CD45.1+, OT2 and 2D2 mice are all on a C57BL/6 genetic background. For bone marrow reconstitutions, mice were conditioned with 1000 Rads using a Cs137 source prior to injection of donor BM.
Mouse models of EAE and DTH
For induction of EAE, mice were injected subcutaneously (s.c.) into the base of the tail with a volume of 200 µl containing 100 µg/ml MOG35–55 peptide (GenScript) emulsified in complete Freund’s adjuvant (CFA). Mice were also injected intraperitoneally (i.p). with 200 ng of pertussis toxin on days 0 and 2, and clinical symptoms scored regularly according to the following criteria: 0 – No symptoms, 0.5 - Partially limp tail, 1 - Completely limp tail, 1.5 – Impaired righting reflex, 2 – Hind limb paresis, 2.5 – Hind-limb paralysis, 3 – Forelimb weakness, 4 – Complete paralysis, 5 – Death. For induction of DTH responses, Keyhole limpet hemocyanin (KLH) was purchased from Calbiochem. Mice were immunized s.c. at the base of the tail with 100 µg KLH in 200 µl CFA. To assess DTH, all mice involved in the studies were given 50 µg KLH in 50 µl PBS intradermally into the left foot pad and 50 µl phosphate buffered saline (PBS) alone in the right foot pad 8 days after the immunization. 2 days later, foot pad swelling was measured with a micrometer.
Cell culture and reagents
DCs were derived from WT or Mir155−/− RBC-depleted bone marrow using rGM-CSF (ebioscience) at a concentration of 20 ng/ml in complete RPMI (supplemented with 10% FBS, 100 units/ml penicillin, 100 units/ml streptomycin, 50 µM beta-mercaptoethanol). Cells were cultured at 5% CO2 and 37°C in a humidified incubator. DCs were stimulated with E. coli LPS (Sigma) at a concentration of 100 ng/ml. For Th17 cell skewing, CD4+ splenocytes were cultured in complete RPMI, plate bound CD3 antibodies, and soluble CD28 antibodies (2 µg/ml), IL-6 (50 ng/ml) and TGF-β (2 ng/ml) (Biolegend) for 96 hours. Splenocytes or LN cells were also cultured in complete RPMI during restimulation with relevant antigens. The MOG35–55 peptide was synthesized by Genscript. KLH was obtained from Calbiochem. For CFSE experiments, 25 × 106 splenocytes were labeled in 5 µM CFSE for 10 minutes at 37°C, washed and cultured. Cellular proliferation was also assayed by pulsing cells with 3[H] thymidine (1 µCi/well) for the final 18 hrs. For co-culture assays, WT or Mir155+/+ DCs were pulsed with the MOG35–55 peptide or Ovalbumin and used to activated purified 2D2 or OT2 CD4+ T cells, respectively, at a 1:10 ratio. For some experiments, splenocytes were obtained from day 12 EAE WT mice and cultured with 20 µg/ml MOG35–55 and 20 ng/ml IL-12 for 2 days before cells were washed and injected intravenously.
Intracellular staining and flow cytometry
To detect intracellular expression of IL-17A, IFN-γ or FoxP3 in CD4+ splenocytes, LNs, or brains cells (purified using Percoll) were first treated with 750 ng/ml ionomycin and 50 ng/ml phorbol myristate acetate (PMA) (Calbiochem) in the presence of 0.5 µl of GolgiPlug (BD Biosciences) for 4–5 hours at 37°C. Cells were subsequently surface stained using CD4+ antibodies and then permeabilized and fixed in 100 µL of eBioscience Perm-Fix solution overnight at 4°C. Cells were washed once in perm wash buffer (eBioscience) and then stained with 0.3 µg of fluorophore-conjugated anti-IL-17A, IFN-γ or FoxP3 (eBioscience) for 20 minutes at 4°C. Fluorophore-conjugated monoclonal antibodies specific to CD11b (Mac1), CD3ε or B220 (eBioscience) were used to stain red blood cell (RBC)-lysed splenocytes or LN cells. Antibodies recognizing CD11c (eBioscience) were also used to stain in vitro derived DCs. After washing, stained cells were assayed using a BD FACSCalibur flow cytometer and results further processed using FlowJo software.
Microarray and qPCR
Total RNA was isolated from magnetic-activated cell separation (MACS) sorted, LPS activated CD11c+ myeloid DCs derived from WT or Mir155−/− BM using Trizol (Invitrogen) per manufacturer’s instructions. Global mRNA expression amounts were next assayed using the Affymetrix total mouse genome array V 2.0 as described previously (O'Connell et al., 2008), and the data was analyzed further using Rosetta Resolver software (GEO accession number GSE23641). Sybrgreen-based quantitative realtime PCR (qPCR) was conducted using the 7300 Realtime PCR system (Applied Biosystems, Foster City, CA) to assay BIC, SHIP1, SOCS1, IL-17A, IL-6, IL-23 p19, IL-12 and IL-23 p40, TNF-α and L32 mRNA amounts using gene specific primers (sequences available upon request). Mature miR-155 and sno202 RNA amounts were assayed using specific Taqman probes from Applied Biosystems. For all experiments, mRNA was normalized to L32 and miRNA to sno202.
ELISAs
To detect protein expression of GM-CSF, IL-6, IL-17A, IFN-γ, IL-23 p19 and p40, IL-12 and 23 p40 and TNF-α ELISAs were performed using cytokine specific kits from eBioscience and carried out according to the manufacturer’s instructions. Serum IgG antibodies against MOG35–55 were assayed by plating serial dilutions of mouse serum on plates coated with MOG35–55 and specific antibodies detected using biotinylated anti-mouse IgG antibodies and Streptavidin horseradish peroxidase (HRP) (Southern Biotech).
Western blotting
Cellular extract was size fractionated using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting was performed as described. Specific antibodies were used to detect SHIP1, SOCS1 and β-actin.
Retrovirus production and infections
Murine stem cell virus (MSCV)-based retroviruses expressing murine miR-155 were prepared as described previously (O'Connell et al., 2009), and used to spin infect freshly isolated WT bone marrow. Immediately following, cells were cultured in GM-CSF containing medium until day 7 before LPS stimulation.
Histological examination of central nervous system tissues
Brains and spinal cords from EAE mice were dissected and fixed in formaldehyde for 48 hours. Tissue sections were next prepared, stained with H&E and visualized with a Nikon Eclipse 50i microscope, and photographed using a Spot® Digital Camera and software. Sections were scored by a pathologist blinded to the genotype of the tissue or the clinical severity of disease according to the following criteria: 0 - no sign of infiltrate, 1 - perivascular congestion (light), 2 - perivascular congestion (heavy), 3 - perivascular congestion (heavy) and parenchymal infiltrate, 4 - focal meningeal lymphocytosis, 5 - extensive sclerosis.
Statistical analysis
Statistical significance was determined by performing a two-tailed t-test. P values <0.05 were considered significant.
Supplementary Material
Acknowledgements
R.M.O. was funded in part by the Irvington Institute Fellowship Program of the Cancer Research Institute, and by award number K99HL102228 from the National Heart, Lung and Blood Institute. D.A.K. was supported by the National Institutes of Health – Building Interdisciplinary Research Careers in Women's Health (BIRCWH) center at UCLA (K12 HD001400). A.A.C. was funded by the Graduate Research Fellowship Program of the National Science Foundation. D.S.R. was funded by award number 1K08CA133521 from the National Cancer Institute. J.L.R. is a Merck Fellow of the Jane Coffin Child’s Memorial Fund. This work was also supported by NIH grant 1R01AI079243-01. We would like to thank Alan Bradley for providing us with Mir155−/− animals.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- An H, Xu H, Zhang M, Zhou J, Feng T, Qian C, Qi R, Cao X. Src homology 2 domain-containing inositol-5-phosphatase 1 (SHIP1) negatively regulates TLR4-mediated LPS response primarily through a phosphatase activity- and PI-3K-independent mechanism. Blood. 2005;105:4685–4692. doi: 10.1182/blood-2005-01-0191. [DOI] [PubMed] [Google Scholar]
- Androulidaki A, Iliopoulos D, Arranz A, Doxaki C, Schworer S, Zacharioudaki V, Margioris AN, Tsichlis PN, Tsatsanis C. The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity. 2009;31:220–231. doi: 10.1016/j.immuni.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A, Schambach F, DeJong CS, Hammond SM, Reiner SL. Micro-RNA-155 inhibits IFN-gamma signaling in CD4+ T cells. Eur J Immunol. 2010;40:225–231. doi: 10.1002/eji.200939381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, Kuchroo VK. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- Chong MM, Rasmussen JP, Rudensky AY, Littman DR. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J Exp Med. 2008;205:2005–2017. doi: 10.1084/jem.20081219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett Y, McBride KM, Jankovic M, Gazumyan A, Thai TH, Robbiani DF, Di Virgilio M, San-Martin BR, Heidkamp G, Schwickert TA, et al. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–638. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S, Li Z, Wu Z, Pei G. MicroRNA miR-326 regulates T(H)-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol. 2009 doi: 10.1038/ni.1798. [DOI] [PubMed] [Google Scholar]
- Ghilardi N, Kljavin N, Chen Q, Lucas S, Gurney AL, De Sauvage FJ. Compromised humoral and delayed-type hypersensitivity responses in IL-23-deficient mice. J Immunol. 2004;172:2827–2833. doi: 10.4049/jimmunol.172.5.2827. [DOI] [PubMed] [Google Scholar]
- Gutcher I, Becher B. APC-derived cytokines and T cell polarization in autoimmune inflammation. J Clin Invest. 2007;117:1119–1127. doi: 10.1172/JCI31720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasch D, Chen YW, Reilly RM, Chiou XG, Koterski S, Smith ML, Kroeger P, McWeeny K, Halbert DN, Mollison KW, et al. T cell activation induces a noncoding RNA transcript sensitive to inhibition by immunosuppressant drugs and encoded by the proto-oncogene, BIC. Cell Immunol. 2002;217:78–86. doi: 10.1016/s0008-8749(02)00506-3. [DOI] [PubMed] [Google Scholar]
- Hjelmstrom P, Juedes AE, Fjell J, Ruddle NH. B-cell-deficient mice develop experimental allergic encephalomyelitis with demyelination after myelin oligodendrocyte glycoprotein sensitization. J Immunol. 1998;161:4480–4483. [PubMed] [Google Scholar]
- Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Jiang S, Zhang HW, Lu MH, He XH, Li Y, Gu H, Liu MF, Wang ED. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 2010;70:3119–3127. doi: 10.1158/0008-5472.CAN-09-4250. [DOI] [PubMed] [Google Scholar]
- Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- Junker A, Krumbholz M, Eisele S, Mohan H, Augstein F, Bittner R, Lassmann H, Wekerle H, Hohlfeld R, Meinl E. MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain. 2009;132:3342–3352. doi: 10.1093/brain/awp300. [DOI] [PubMed] [Google Scholar]
- Kohlhaas S, Garden OA, Scudamore C, Turner M, Okkenhaug K, Vigorito E. Cutting edge: the Foxp3 target miR-155 contributes to the development of regulatory T cells. J Immunol. 2009;182:2578–2582. doi: 10.4049/jimmunol.0803162. [DOI] [PubMed] [Google Scholar]
- Komiyama Y, Nakae S, Matsuki T, Nambu A, Ishigame H, Kakuta S, Sudo K, Iwakura Y. IL-17 plays an important role in the development of experimental autoimmune encephalomyelitis. J Immunol. 2006;177:566–573. doi: 10.4049/jimmunol.177.1.566. [DOI] [PubMed] [Google Scholar]
- Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, McClanahan T, Kastelein RA, Cua DJ. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med. 2005;201:233–240. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston A, Lu LF, O'Carroll D, Tarakhovsky A, Rudensky AY. Dicer-dependent microRNA pathway safeguards regulatory T cell function. J Exp Med. 2008;205:1993–2004. doi: 10.1084/jem.20081062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littman DR, Rudensky AY. Th17 and regulatory T cells in mediating and restraining inflammation. Cell. 2010;140:845–858. doi: 10.1016/j.cell.2010.02.021. [DOI] [PubMed] [Google Scholar]
- Lu LF, Thai TH, Calado DP, Chaudhry A, Kubo M, Tanaka K, Loeb GB, Lee H, Yoshimura A, Rajewsky K, Rudensky AY. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30:80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQualter JL, Darwiche R, Ewing C, Onuki M, Kay TW, Hamilton JA, Reid HH, Bernard CC. Granulocyte macrophage colony-stimulating factor: a new putative therapeutic target in multiple sclerosis. J Exp Med. 2001;194:873–882. doi: 10.1084/jem.194.7.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R. Origin and physiological roles of inflammation. Nature. 2008;454:428–435. doi: 10.1038/nature07201. [DOI] [PubMed] [Google Scholar]
- Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CA, Langrish CL, Chen Y, Blumenschein W, McClanahan T, Kastelein RA, Sedgwick JD, Cua DJ. Divergent pro- and antiinflammatory roles for IL-23 and IL-12 in joint autoimmune inflammation. J Exp Med. 2003;198:1951–1957. doi: 10.1084/jem.20030896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci U S A. 2009;106:7113–7118. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- O'Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, Paquette RL, Baltimore D. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci U S A. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev ED, Shriver LP, Maresz K, Pedras-Vasconcelos J, Verthelyi D, Dittel BN. GM-CSF production by autoreactive T cells is required for the activation of microglial cells and the onset of experimental autoimmune encephalomyelitis. J Immunol. 2007;178:39–48. doi: 10.4049/jimmunol.178.1.39. [DOI] [PubMed] [Google Scholar]
- Rai D, Kim SW, McKeller MR, Dahia PL, Aguiar RC. Targeting of SMAD5 links microRNA-155 to the TGF-beta pathway and lymphomagenesis. Proc Natl Acad Sci U S A. 2010;107:3111–3116. doi: 10.1073/pnas.0910667107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzenberger P, La Russa V, Miller A, Ye P, Huang W, Zieske A, Nelson S, Bagby GJ, Stoltz D, Mynatt RL, et al. IL-17 stimulates granulopoiesis in mice: use of an alternate, novel gene therapy-derived method for in vivo evaluation of cytokines. J Immunol. 1998;161:6383–6389. [PubMed] [Google Scholar]
- Shen L, Evel-Kabler K, Strube R, Chen SY. Silencing of SOCS1 enhances antigen presentation by dendritic cells and antigen-specific anti-tumor immunity. Nat Biotechnol. 2004;22:1546–1553. doi: 10.1038/nbt1035. [DOI] [PubMed] [Google Scholar]
- Sonderegger I, Iezzi G, Maier R, Schmitz N, Kurrer M, Kopf M. GM-CSF mediates autoimmunity by enhancing IL-6-dependent Th17 cell development and survival. J Exp Med. 2008;205:2281–2294. doi: 10.1084/jem.20071119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanczyk J, Pedrioli DM, Brentano F, Sanchez-Pernaute O, Kolling C, Gay RE, Detmar M, Gay S, Kyburz D. Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 2008;58:1001–1009. doi: 10.1002/art.23386. [DOI] [PubMed] [Google Scholar]
- Steinman L. Mixed results with modulation of TH-17 cells in human autoimmune diseases. Nat Immunol. 2010;11:41–44. doi: 10.1038/ni.1803. [DOI] [PubMed] [Google Scholar]
- Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–189. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, Das PP, Miska EA, Rodriguez A, Bradley A, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Jeker LT, Fife BT, Zhu S, Anderson MS, McManus MT, Bluestone JA. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J Exp Med. 2008;205:1983–1991. doi: 10.1084/jem.20080707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.