Abstract
Chromatin fibers encountered in various species and tissues are characterized by different nucleosome repeat lengths (NRL) of the linker DNA connecting the nucleosomes. While single cellular organisms and rapidly growing cells with high protein production have short NRL ranging from 160 to 189 base pairs (bp), mature cells usually have longer NRL ranging between 190 and 220 bp. Recently, various experimental studies have examined the effect of NRL on the internal organization of chromatin fiber. Here we investigate by mesoscale modeling of oligonucleosomes the folding patterns for different NRL, with and without linker histone, under typical monovalent salt conditions using both one-start solenoid and two-start zigzag starting configurations. We find that short to medium NRL chromatin fibers (173 to 209 bp) with linker histone condense into irregular zigzag structures, and that solenoid-like features are viable only for longer NRL (226 bp). We suggest that medium NRL are more advantageous for packing and various levels of chromatin compaction throughout the cell cycle than their shortest and longest brethren; the former (short NRL) fold into narrow fibers, while the latter (long NRL) arrays do not easily lead to high packing ratios due to possible linker DNA bending. Moreover, we show that the linker histone has a small effect on the condensation of short-NRL arrays but an important condensation effect on medium-NRL arrays which have linker lengths similar to the linker histone lengths. Finally, we suggest that the medium-NRL species, with densely packed fiber arrangements, may be advantageous for epigenetic control because their histone tail modifications can have a greater effect compared to other fibers due to their more extensive nucleosome interaction network.
Keywords: chromatin, mesoscale modeling, linker histone, fiber condensation
1. Introduction
The genome of every living organism contains the complete information and guidelines required for the organism’s growth and development. Intriguingly, the DNA storage and manipulation mechanisms have to satisfy two antagonistic requirements: a high compaction ratio, and facile access to the genome. Understanding how the internal organization of DNA achieves both factors is crucial for our understanding of the most basic cellular processes.
DNA storage in eukaryotic cells is achieved through the chromatin fiber. The basic chromatin building block is the nucleosome: a histone core composed of 4 pairs of protein dimers (histone proteins H2A, H2B, H3, and H4) around which 147 base pairs (bp) of DNA are wound 1.75 turns [1]. Every histone dimer has two protruding tails (H2A has four) which are highly positively charged and thus readily available for interactions with the DNA polyelectrolyte.
The length of the DNA wrapped around nucleosome core (147 bp) plus the length of the DNA linker (or ‘linker’ for short therein) connecting each nucleosome to the next (nucleosome repeat length – NRL) varies within and between organisms (see Table 1) [2, 3]. While some simple organisms have short DNA linkers ranging from 18 to 45 base pairs, the typical DNA linker length for mature transcriptionaly inactive eukaryotic cells, between 50 and 60 bp, leads to NRL values between 197 and 207 bp. Table 1 shows that rapidly growing cells with high protein production are associated with a relatively short NRL ranging from ∼ 160 to 189 bp; these include unicellular organisms, embryonic stem (ES), and tumor cells. Mature cells tend to have longer NRL ranging between 190 and 220 bp. An exception to this trend is NRL in rat neurons which is long before birth, 200 bp, and drops to 170 bp and less later on [4]. Longer NRL chromatin (220 bp) appears in starfish [3] which resides in higher salt environments.
Table 1.
Nucleosome repeat length (NRL) within and between organisms.
| Species/tissue | NRL |
|---|---|
| Aspergillus nidulans [111] | 154 |
| Rat neuron [112] | 162 |
| Saccharomyces cerevisiae [113] | 165 |
| Neurospora crassa [114] | 170 |
| H1c, H1d, H1e null ES [115] | 174 |
| Amoebae [116] | 176 |
| Chinese hamster ovary (CHO) cells [2] | 178 |
| Plasmodia [116] | 181 |
| HeLa cells [2] | 188 |
| Hepatoma cells [2] | 188 |
| Teratoma cells [2] | 188 |
| P815 cells (mouse mastocytes) [2] | 188 |
| Myoblast cells [2] | 189 |
| CV1 cells (African green monkey) [2] | 189 |
| Wild-type mouse ES cells [115] | 189 |
| H10, H1c, H1e null mouse thymus [115] | 189 |
| BHK (Syrian hamster kidney) [2] | 190 |
| Rat kidney primary culture [2] | 191 |
| H10, H1c, H1e null mouse liver [115] | 191 |
| Rat bone marrow [2] | 192 |
| Rat fetal liver (14 days) [2] | 193 |
| Wild-type mouse liver [115] | 195 |
| Wild-type mouse thymus [115] | 196 |
| Rat liver [2] | 196 |
| Rat kidney [2] | 196 |
| Syrian hamster liver [2] | 196 |
| Syrian hamster kidney [2] | 196 |
| Chick oviduct [2] | 196 |
| Rat glia [112] | 201 |
| Chicken erythrocyte [117] | 212 |
| Echinoderm sperm [3] | ∼ 220 |
Monovalent (K+, Na+) and divalent (Mg2+) ions, as well as linker histone (LH) proteins H1/H5, are essential for chromatin fiber compaction [3]. Chromatin depleted of H1 is decondensed, with a decreased sedimentation velocity [5]. At low ionic strengths, this leads to a more open and randomly organized, beads-on-a-string form of the chromatin fiber [6–8]. Chambon and collaborators [9] showed that the lack of H1 in the presence of highly concentrated monovalent ions (0.6 M) is associated with chromatin with very short NRL. Recent experimental data also show a strong linear relationship between the number of LHs H1 per nucleosome and NRL [3]. H1 in living cells binds dynamically to both euchromatin and heterochromatin, in a ‘stop-and-go’ mode [10], and switches its carrier nucleosome every several minutes [11]. Low H1 stoichiometry and short NRL also characterize newly replicated eukaryotic HeLa cells (NRL = 165 bp) [12]. H1 concentrations and NRL values in those cells rapidly evolve to the values present in mature chromatin [13]. The same behavior was observed in Ehrlich ascites tumor cells (EAT) [14], suggesting a relationship between these factors and certain tumors.
The detailed structure of the chromatin fiber has been a puzzle for more than 3 decades [15, 16]. Early on, the first proposed structure for the 30 nm chromatin fiber was a one-start helix (solenoid) where every nucleosome is in contact with its immediate neighbors, i ± 1 [6, 17]. The DNA linkers in this model are bent in the fiber interior. Such linker bending offers a relatively constant fiber width for different NRL and can easily produce a 30 nm fiber with a packing ratio of 6 to 8 nucleosomes per 11 nm of fiber length. However, in this solenoid model, the role of LH is not clarified because the wide angle between the bent linkers of entering and exiting nucleosomes generally excludes close interactions with the LH, though interactions between LH and non-parental DNA linkers are possible.
The second major type of proposed model for the 30 nm chromatin fiber is a two-start helix (zigzag structure) in which straight DNA linkers crisscross the fiber axis and thus promote i ± 2 interactions between nucleosomes [18–22]. The straight linkers make the width of zigzag fiber model more strongly dependent on the NRL. In addition, the LH in this model has a clearly defined role, to attract the DNA linkers exiting/entering the parent nucleosomes and form rigid stems [7].
Since 1980, many studies have supported aspects of both models. The early results by Langmore and collaborators [19] based on electron micrographs supported the two-start cross-linker model. Their measurements indicated a strong linear relationship between the linker length and the fiber width for both Necturus, where the DNA linker length is 48 bp and fiber width is 31 nm, and Thyone, with DNA linker length of 87 bp and fiber width 39 nm, in a buffer with monovalent and divalent ions. Later, the same group produced similar results using cryo-EM imaging [20]. They also showed that highly compacted chromatin fiber has solid centers [23], which supported the idea that DNA linkers cross the fiber axis.
Electron tomography [21] showed that chromatin fibers under moderate salt concentration (0.15 M NaCl) have asymmetric zigzag structures determined by the properties of the nucleosome-linker unit. Irregularity in fiber structure is supported by many modeling studies [24–26].
The correlated breaks in DNA produced by ionizing radiation offer an indirect way to view arrangements of nucleosomes [27]. The end-labeled fragments induced by correlated breaks and separated by gel electrophoresis exhibited characteristic peaks at 78 bp (one helical turn around histone core) and between 175 and 450 bp; these values reflect the positions of nearest neighbor nucleosomes and suggest a zigzag organization for chromatin.
The influence of LH on chromatin structure in higher eukaryotes was investigated by Woodcock and collaborators [7]. They showed that LH leads to formation of a zigzag-promoting stem motif by mediating the close contact of the exiting and entering linker DNA. More recently, EM imaging combined with sedimentation coefficient measurements by Routh et al. [28] demonstrated that short-NRL (167 bp) arrays form narrow fibers (21 nm diameter) in the presence of LH. For medium-NRL arrays (197 bp), highly compact 30-nm fibers result.
Later, X-ray crystallography made an important contribution to the high-resolution nucleosome structures [1, 29], by producing a low resolution image of a cross-linked tetranucleosome [22, 30], which supported a two-start zigzag. However, that structure was based on a fiber with short linkers (20 bp) and without LH.
The structure of chromatin has also been probed by disulfide bridging [31]. In such experiments, H2A/H2B and H4 histones are targets for cysteine replacement because their tail bases are crucial for compaction via interaction with H2A/H2B tails of neighboring nucleosomes [1]. Following endonuclease cleavage, the initial 10 to 12 nucleosome fibers were reduced to 5 to 6 nucleosome constructs which constitute the individual starts of the two-start configurations. This result was interpreted as evidence for zigzag configurations because one-start solenoids would preserve the initial connectivity (i ± 1 contacts) and retain the 12-nucleosome repeat pattern.
Initially, the one-start helical structure also had its proponents [6, 32– 35], and recent results by Rhodes and collaborators have renewed this view [28, 36–39]. These results suggest that for longer NRL with Mg2+ and LH chromatin folds into an interdigitated one-start helix. The studies show that the chromatin fiber has a fairly constant diameter of 33–35 nm for moderate-length linkers (NRL between 177 and 207 bp) and 44 nm for long linkers (NRL between 217 to 237 bp). The moderate to long NRL fibers they analyzed have a high packing ratio, indicative of solenoid-like topology: 11 nucleosomes per 11 nm for moderate-length linkers and 15 to 17 nucleosomes for long-linker arrays [28, 37]. However, for short NRL (167 bp), their experiments [28, 39] indicate that chromatin adopts a two-start helical arrangement with less compact (6.1 nucleosomes per 11 nm) and thinner (21 nm of diameter) fibers.
Recent computational studies using a coarse-grained model described by several tunable parameters such as the linker DNA opening angle and twisting angle between successive nucleosomes [25] found periodic patterns in fiber dimensions for NRL from 202 to 222 bp, a strong effect of NRL on the viable chromatin conformations (two-start and three-start were found), and increased structural irregularity for NRL > 214 bp. While such patterns agree with X-ray scattering studies [19], they differ from the above cited work [37]. Modeling based on EM measurements of reconstituted fibers, however, show a range of possible conformations as NRL changes [26]; the authors emphasize “the multiplicity of fiber structures!” tuned by the NRL. Moreover, Monte Carlo simulations of coarse-grained models of chromatin with NRL ranging from 155 to 211 bp have revealed densely compacted fibers with possible one, two, and three-start structures [40].
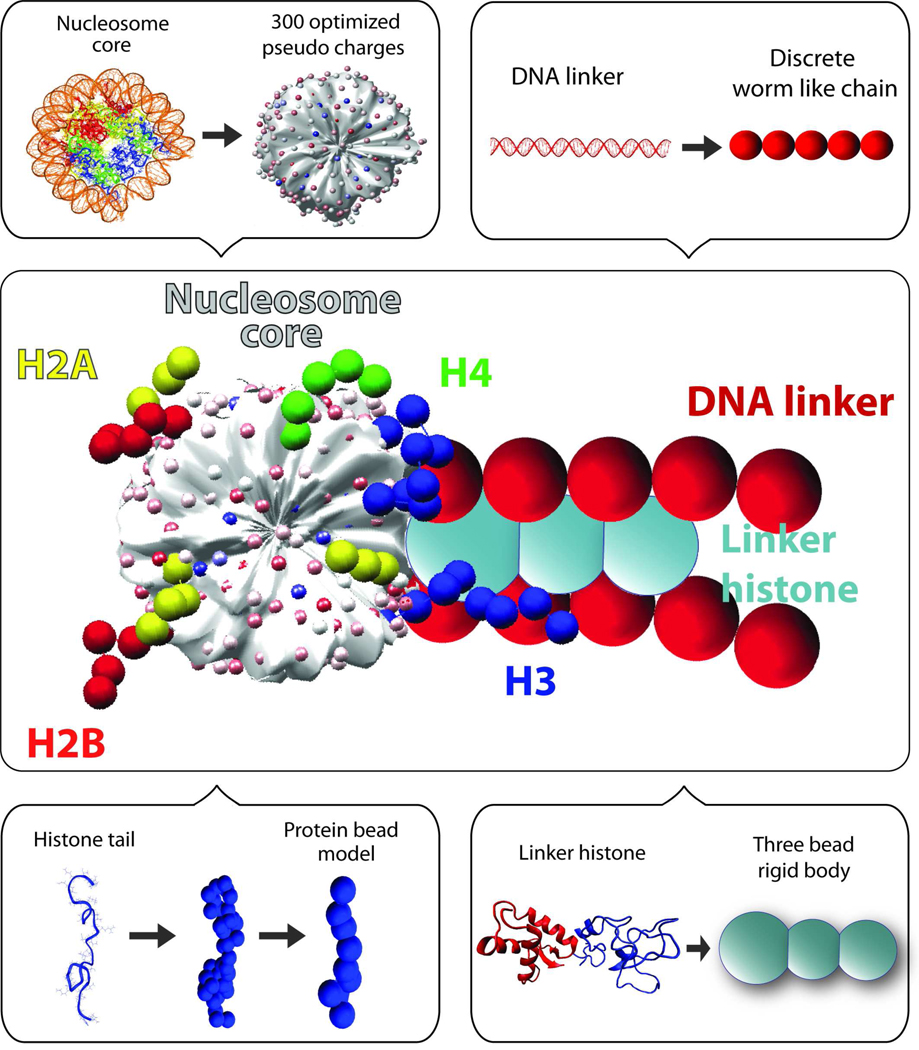
Over the past few years, we have developed a mesoscale model for studying chromatin structure [41–45] (see Figure 1). Our mesoscale model essentially captures basic physics of chromatin such as its electrostatics, DNA and nucleosome mechanics, structural irregularity and histone tail flexibility, and averages out other effects: protein/DNA sequence effects, hydrogen-bonding, atomistic fluctuations, and solvation effects [45]. The model details, including simulation methods, validation studies, and prior applications, were recently presented in Arya and Schlick [45], where the role of histone tails in compacting fiber structure was analyzed. In our recent study in collaboration with experimentalists [46], we examined the effect of linker histones and divalent ions (the latter by a first-order approximation) on chromatin structure. The crosslinking experimental procedure and modeling both provided evidence for an organized, compact zigzag model at monovalent salt with LH and an ordered zigzag accented with some bent linker DNA at divalent salt conditions. The heterogeneous nature of chromatin emerged as an important feature that helps condense chromatin as well as possibly transition the 30-nm fiber into higher-order condensed forms [46]. Many other modeling studies of nucleosomes and oligonucleosomes have been reported, e.g., [25, 26, 47–54]. Each model is suitable for a different level of questions and resolution, and all have advanced our understanding of chromatin organization and experimental structures.
Figure 1.
Mesoscale model of the basic chromatin building block. The nucleosome core surface with wrapped DNA without histone tails is modeled as an irregularly shaped rigid body with 300 optimized pseudo surface charges (smallest white, pink, magenta, and blue spheres). The linker DNA (large red spheres) is treated using the discrete worm like chain model. The histone tails are coarse grained as bead models (red, yellow, green, and blue medium spheres). The LH is modeled as 3 charged beads rigidly connected to the nucleosome (turquoise spheres).
Here we present results of our mesoscale chromatin model for chromatin structure as a function of linker DNA length (NRL = 173, 182, 191, 200, 209, 218, or 226 bp) and LH. For each condition, we start simulations from solenoid and zigzag structures and compare structural features of the converged fibers. We use mainly 24-core arrays as typically studied in a laboratory [55], at monovalent ion concentrations of 0.15 M. Additionally, we show representative results for different monovalent ion concentrations, with/without Mg2+, for three NRL, as well as representative snapshots of 48-core arrays. The effect of magnesium ions on a fixed linker DNA length was presented separately [46], and an initial study for two NRL values with divalent ions was presented elsewhere [56].
Our present results show that nucleosome arrays with short NRL tend to fold into conformations with intense two-start interactions regardless of LH presence. Specifically, arrays with very short DNA linkers (173 bp), with and without LH, and arrays with short DNA linkers (182 bp) without LH, form narrow ladder-like structures in which cores i interact mainly with their i ± 1 and i ± 2 neighbors. Arrays with short DNA linkers (182 bp) and LH form slightly thicker fibers with intense i ± 2 and i ± 3 contacts.
The presence of LH (roughly the length of 30 bp [49]) has the strongest structural effect on arrays with medium NRL (i.e., for NRL 191–209 bp). In these arrays, the linker DNA length is not much greater than twice the LH length, and this promotes formation of the rigid stem. Independent of the starting (solenoid or zigzag) conformation, arrays with medium NRL and LH fold into compact two-start configurations characterized by strong i ± 2 and moderate i ± 5 interactions, which reflect their tightly packed two-start structure. This result suggests that by promoting the formation of the DNA stem for medium-sized DNA linkers, LH straightens and stiffens the DNA linkers and in turn destabilizes solenoid-like features.
Our investigations also indicate that, in the absence of LH, medium (191 to 209 bp) to long (218 to 226 bp) NRL encourage chromatin structural heterogeneity. Chromatin fibers with these DNA linkers without LH fold into loose structures with either solenoid-like or zigzag-like features. LH cannot prevent long DNA linkers from bending in their middle section and, thereby, promotes a wide-variety of nucleosome neighbors to come into close contact. Arrays with very long linkers (NRL = 226 bp) and LH, can adopt either a topology with dominant zigzag features characterized by strong i ± 2 and i ± 5 contacts or heteromorphic topology in which i ± 3 and i ± 5 neighbors interact intensely followed by i ± 2 and higher-order pairs. The heterogeneous structure of longer NRL arrays makes packing into a tight fiber architecture more difficult due to their much larger accessible configurational space.
We also address the role of histone tails for various nucleosome repeat lengths. We show that the tails may be evolutionary optimized for NRL between 191 and 209 bp, which are the lengths usually encountered in Nature.
Finally, we also show that fiber compaction increases with increasing monovalent ion concentration and inclusions of divalent ions for medium to long NRL, where the linker length is long enough to reorganize the nucleosomes to allow close spatial proximity while also avoiding steric clashes.
2. Results
2.1. Overall analyses
For each NRL we examine (Table 2), we perform thorough analysis for 24-core oligonucleosomes at monovalent salt concentration (CS) of 0.15 M. For each NRL, we use four conditions (combinations of interdigitated solenoid or zigzag starting forms with/without LH; see Supplemental Figure S1 for starting forms). For each of these four conditions, 12 trajectories of length 35 to 50 million steps are performed with combinations of 4 random initial seeds and 3 twist deviations (0, ±12°) about the mean DNA twist (see Table 2). Our MC simulations converge rapidly as indicated by convergence of global and local quantities (see Figure S2) as also analyzed previously [56]. We use the last 5 million steps for ensemble averages and statistical analyses (Section 4.4). For visualization purposes, we additionally conduct a limited number of simulations for 48-core oligonucleosomes, as shown in Figures 2 and 3, as a function of NRL. We also conduct simulations in three other conditions for selected NRL: low monovalent salt: CS = 0.01 M; high monovalent salt CS = 0.2 M; and both monovalent and divalent ions: CS = 0.15 M monovalent salt with moderate divalent ions, to obtain general trends of altered ionic environment.
Table 2.
DNA twisting angles as a function of nucleosome repeat length (NRL).
| Number of segments, nS | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
|---|---|---|---|---|---|---|---|
| Linker length, (bp) | 26.47 | 35.29 | 44.12 | 52.94 | 61.76 | 70.59 | 79.41 |
| Closest nucleosome repeat length (bp) | 173 | 182 | 191 | 200 | 209 | 218 | 226 |
| Number of turns, τns | 2.57 | 3.43 | 4.28 | 5.14 | 6 | 6.85 | 7.71 |
| Deviation from integral turns, int(τns) − τnS | −0.57 | −0.43 | −0.28 | −0.14 | 0 | −0.85 | −0.71 |
| Whole linker twist, nSϕns (= int(τns) − τns in °) | 154.82 | 205.20 | 259.20 | 309.60 | 0 | 54.00 | 104.40 |
| Whole linker twist, nSϕns (radian ∈ [−π, π]) | 2.7021 | −2.7018 | −1.7593 | 0.8778 | 0 | 0.9425 | 1.8221 |
| Twist per segment, ϕns (radian) | 0.9007 | −0.6754 | −0.3519 | −0.1463 | 0 | 0.1178 | 0.2025 |
Figure 2.
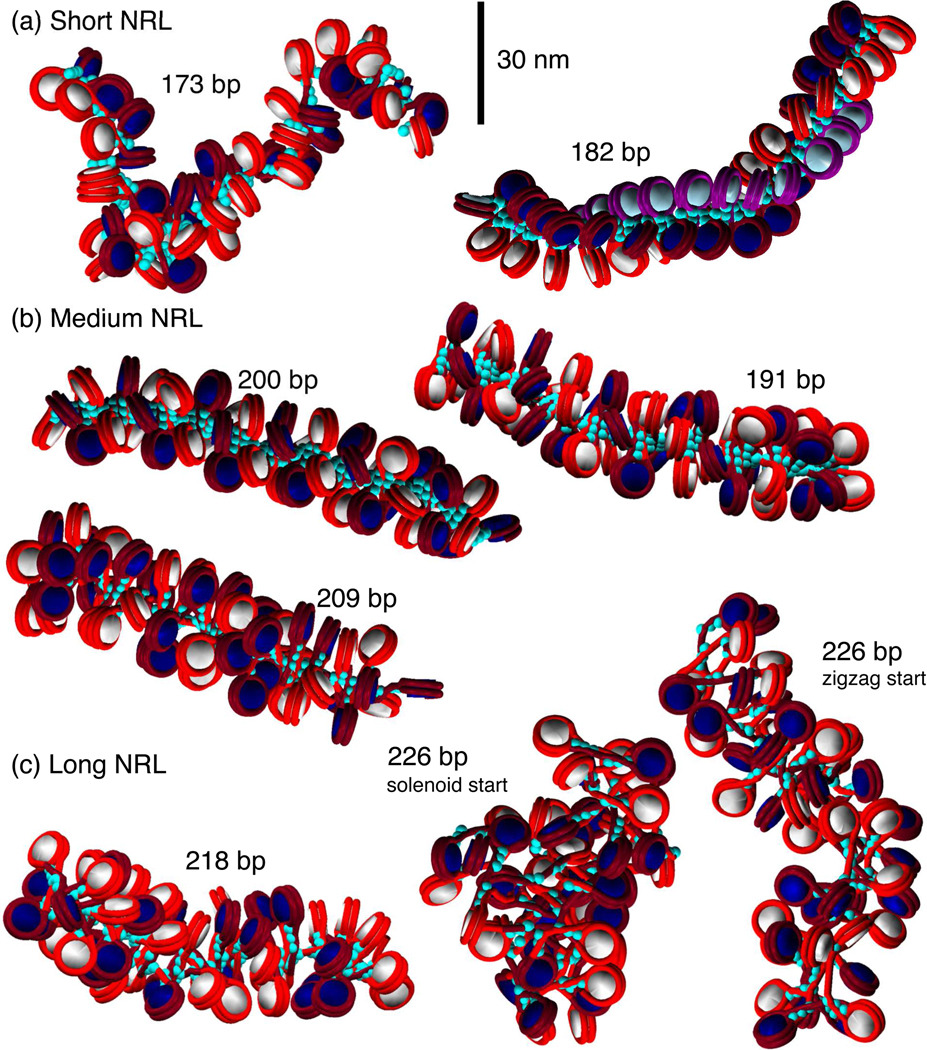
Space filling models based on MC simulations of 48-unit oligonucleosome chains of all NRL compacted at 0.15 M monovalent salt with LH (turquoise beads). Alternating nucleosomes are colored white and navy, with corresponding wrapped DNA as red and burgundy. In the 182-bp array, nucleosomes i, i + 1 and i + 2 are colored white, navy, and light blue, with corresponding DNA as red, burgundy, and purple, to highlight the three-start structure. LH is turquoise.
Figure 3.
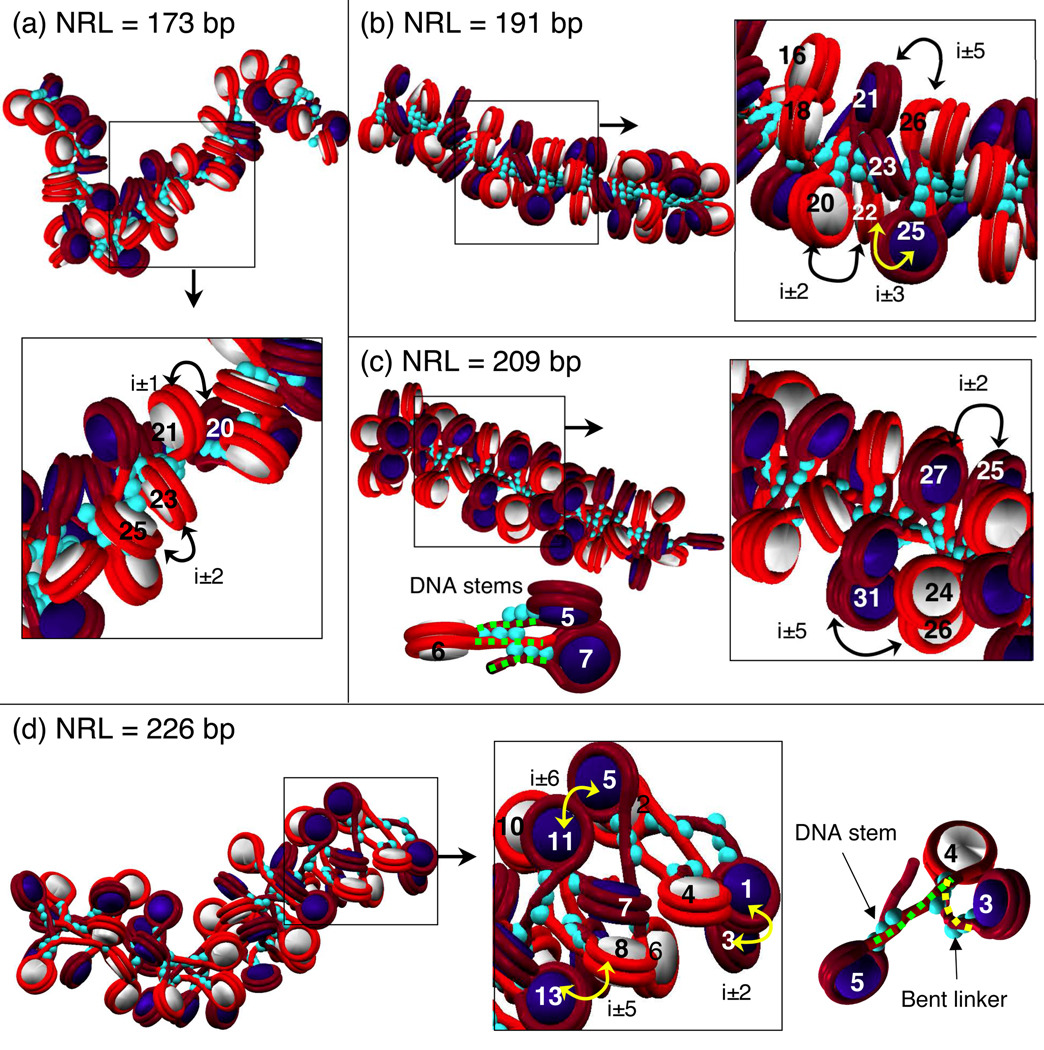
Selected models from Figure 2 analyzed for internucleosome contacts and linker DNA bending. (a) Arrays with very short NRL (173 bp) and LH fold into a narrow structure with low linear packing ratio regardless LH presence. (b) and (c) Arrays with medium long NRL (191 and 209 bp) and linker histone fold into zigzag structures with straight linkers and DNA linker stems. (d) Arrays with longest NRL (226 bp) fold into irregular structures with both DNA stems and bent linkers. Alternating nucleosomes are colored white and navy, with correspondingly wrapped DNA as red and burgundy. LH is turquoise.
Important to our overall analysis is a description of the internal organization of nucleosomes (see Figure 3 for illustration of contacts) in a chromatin fiber by a two-dimensional interaction-intensity matrix, I′(i, j) (Supplemental Figure S3). This matrix measures the intensity of histone-tail mediated interactions between nucleosomes i and j, as described in detail in Section 4.5.1. For an NC-core array, the accompanying normalized one-dimensional projection depicts the relative intensity of interactions between nucleosomes separated by k neighbors (Figure 4) [43]. Thus, the ideal two-start zigzag configuration has dominant i ± 2 and moderate i ± 5 interactions (Figure S1) [22], while the ideal 6-nucleosomes per turn solenoid model has dominant i ± 1 and i ± 6 interactions (see Figure 3 in [46]). The interdigitated solenoid model, which we use here, is characterized by i ± 5 and i ± 6 interactions [37, 57] (Figure S1).
Figure 4.
Internucleosomal interactions patterns at 0.15 M monovalent salt. Interaction intensities versus nucleosome position separation k for 24-core arrays: (a) Short NRL (173 and 182 bp) without LH, (b) Short NRL (173 and 182 bp) with LH, (c) Medium NRL (191, 200, and 209 bp) without LH, (d) Medium NRL (191, 200, and 209 bp) with LH, (e) Long NRL (218 and 226 bp) without LH, (f) Long NRL (218 and 226 bp) with LH. Results for trajectories started from idealized zigzag and interdigitated solenoid conformations are shown separately.
Other quantities that characterize fiber structure are the ensemble averages of the: dimer distance between consecutive nucleosomes, triplet distance between i ± 2 nucleosome neighbors, DNA bending angle between the vector ‘leaving’ one nucleosome and the vector ‘entering’ its consecutive neighbor (as introduced in Ref. [45]), triplet angle between the geometric centers of three consecutive nucleosomes, and dihedral angle between the geometric centers of four consecutive nucleosomes. All these quantities are detailed in Section 4.6 and Supplemental Figure S4. The dimer and triplet distances reflect the proximity of i ± 1 and i ± 2 neighbors respectively; zigzag structures are characterized by smaller triplet than dimer distances, and for solenoid forms it is the reverse. The bending angle helps describe linker DNA bending: the angle is larger for solenoid fibers with bent linkers than for zigzag arrays. Classic zigzag fibers also have small average triplet angles that allow strong i ± 2 interactions, while solenoid fibers have very wide triplet angles by construction. The average dihedral angle is 180° when the distance between nucleosomes i and i + 3 is maximal. As the fiber components reorient bringing nucleosomes i and i + 3 closer, this dihedral angle decreases. Thus, while small dihedral angles indicate close proximity of four consecutive nucleosomes, large dihedral angles signal weak interactions between nucleosomes separated by 2 or more neighbors.
Additionally, we describe the degree of compaction of the oligonucleosomes by the sedimentation coefficient (S20,w), overall nucleosomal packing ratios (number of nucleosomes per 11 nm of fiber length), fiber width, fiber volume, percentage of ‘filled’ fiber volume, and curvature of the fiber axis (see Sections 4.7 and 4.8 for details).
Figures 4, 5, and 6 show the main results for 24-core arrays: internucleosome interaction patterns and geometrical features. Figure 7 assesses chromatin compactness as function of the salt concentration, and Figure 8 describes tail mediated interactions. All these features will be detailed in the subsections below.
Figure 5.
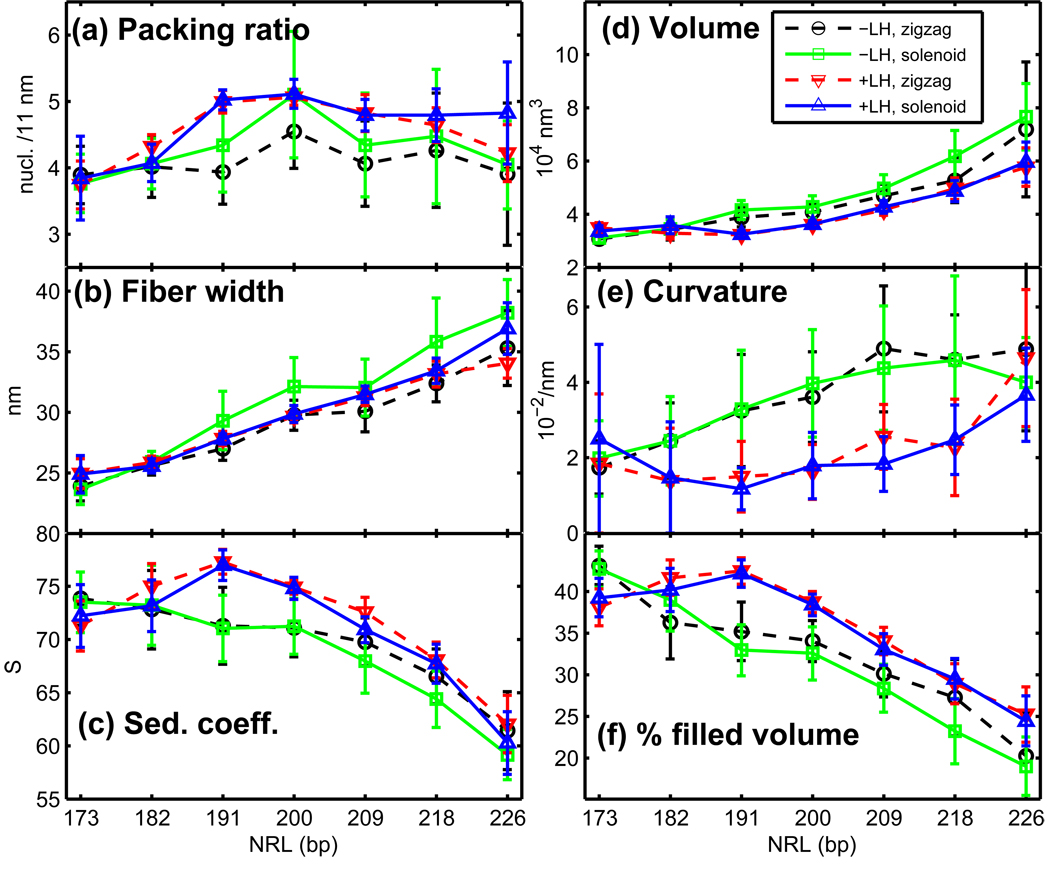
Chromatin fiber dimensions as a function of NRL. (a) Nucleosome linear packing ratio, (b) fiber width, and (c) sedimentation coefficients, (d) fiber volume, (e) fiber curvature, and (f) percentage of filled volume, all as functions of nucleosome repeat length for 24-core oligonucleosomes. Results shown for simulations started from zigzag configuration and modeled without LH, started from solenoid without LH, started from zigzag with LH, started from solenoid with LH at 0.15 M monovalent salt. Nucleosome packing ratios are measured as a number of nucleosomes per 11 nm of fiber axis length. The fiber width is calculated as an average distance of nucleosomes (+ nucleosome half radius) from the fiber axis (Supplemental Figure S6).
Figure 6.
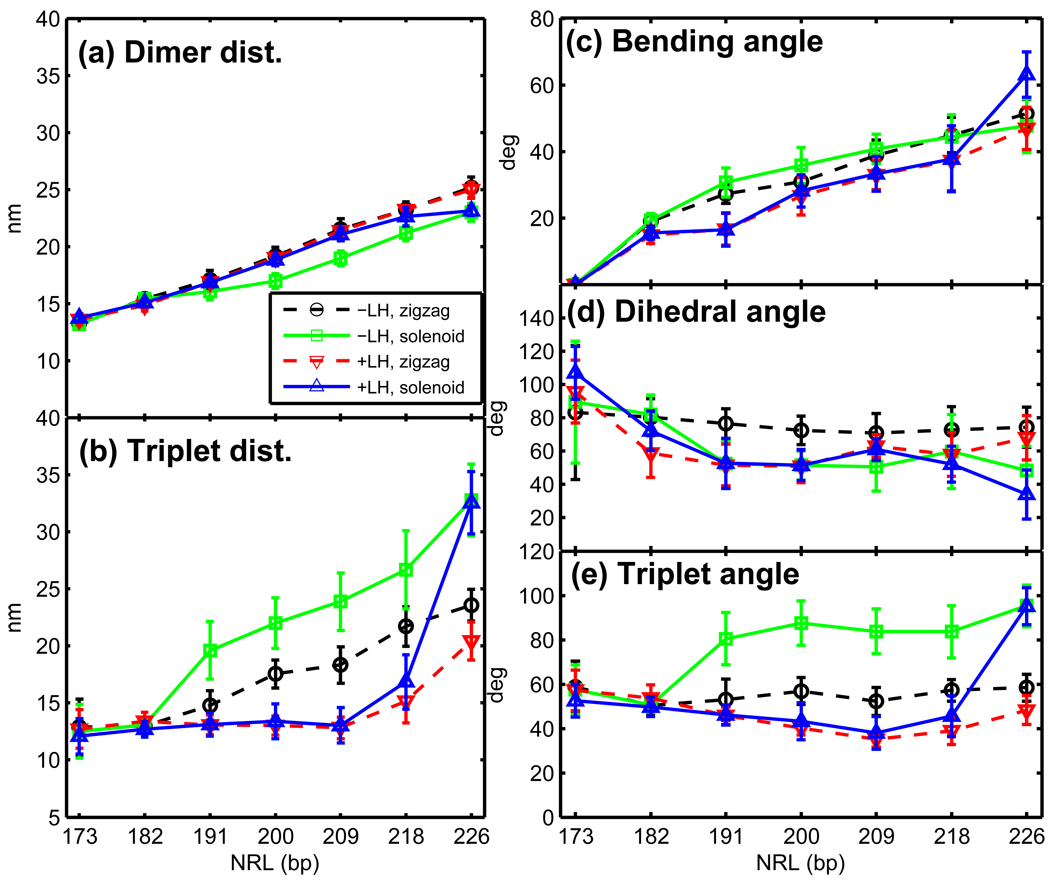
Geometric parameters for different NRL systems 24-core arrays: (a) dimer distances (between i ± 1 nucleosome neighbors), (b) triplet distances (between i ± 2 neighbors), (c) bending angles, (d) dihedral angles, and (e) triplet angles for 24-core arrays at 0.15 M monovalent salt. The bending angle is defined as the angle between vectors passing through first two and last two DNA linker beads. The triplet angle is an angle between the geometric centers of three consecutive nucleosomes. The dihedral angle is the angle between two planes defined by four consecutive nucleosomes. See Supplemental Figure S4.
Figure 7.
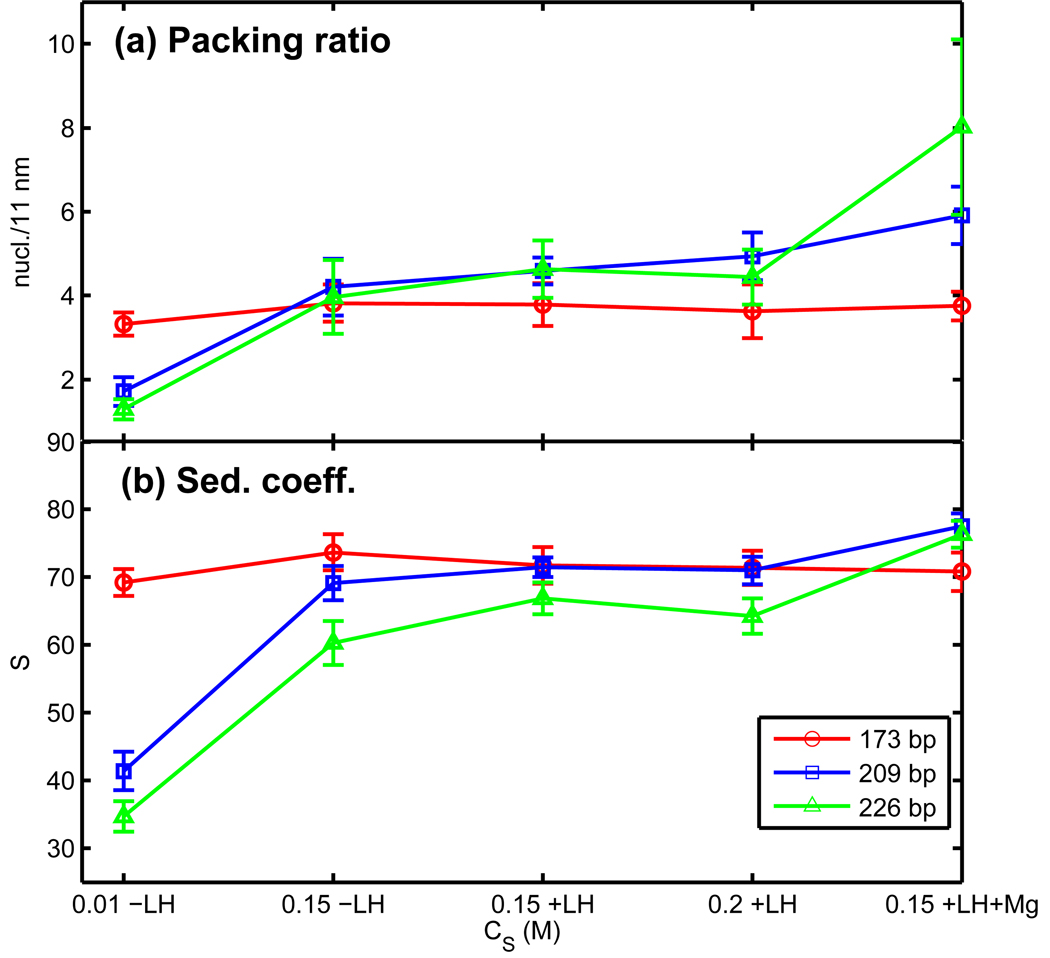
Chromatin fiber measurements as a function of the salt environment for 24 oligonucleosomes: (a) Nucleosome linear packing ratio and (b) sedimentation coefficients. Ensemble averages over trajectories started from zigzag and interdigitated solenoid configurations. The three new salt environment corresponds to monovalent CS = 0.01 M without LH (−LH), monovalent CS = 0.2 M with LH (+LH) and moderate monovalent salt at CS = 0.15 M with LH and divalent ions (+LH+Mg).
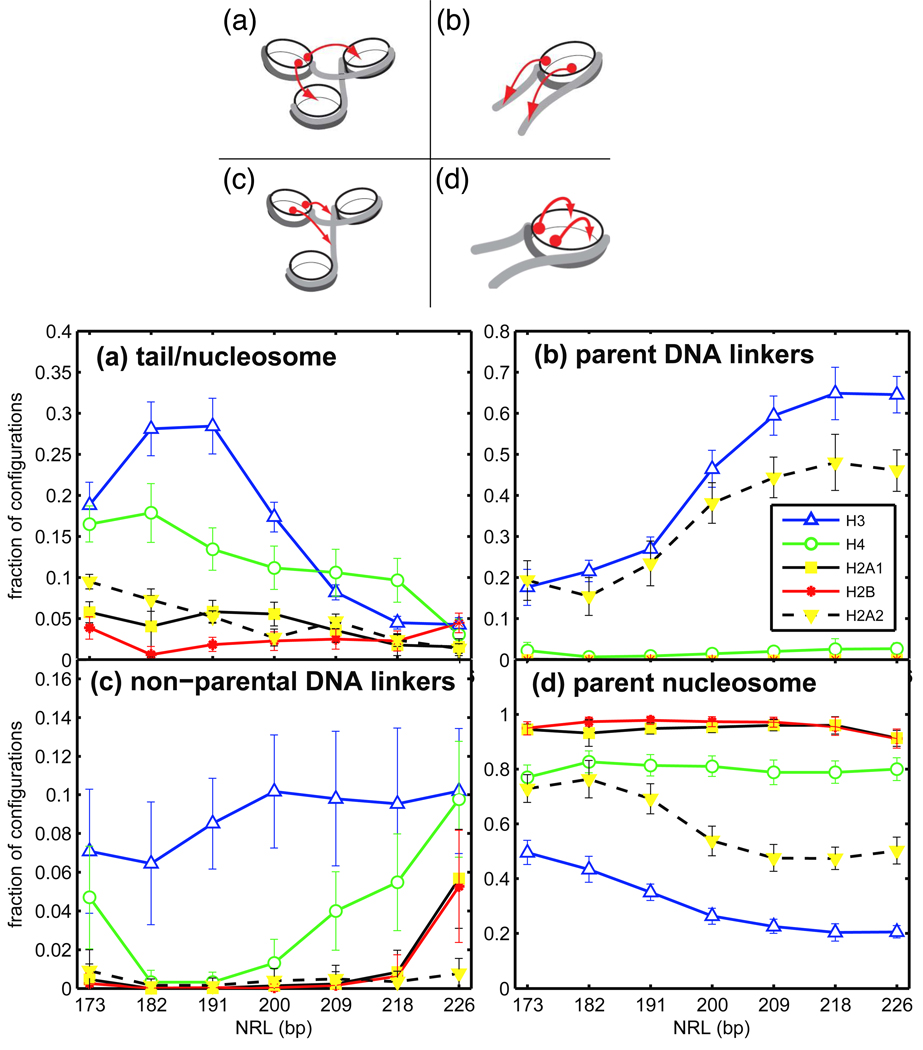
Figure 8.
Frequency analyses and cartoon images of different tail interactions in 24-core oligonucleosomes with linker histone at 0.15 M monovalent salt: (a) nucleosome/nucleosome interactions, (b) interactions with parent linker DNA, (c) interactions with non-parent DNA linkers, and (d) interactions with parent nucleosome. H2A1 and H2A2 denote N-termini and C-termini, respectively, of the H2A tails.
2.2. Overall fiber structure as a function of a DNA linker length
Overall, we recognize three separate NRL ranges: (a) short NRL: 173 bp to 182 bp, (b) medium NRL: 191 bp to 209 bp, and (c) long NRL: 218 to 226 bp. The fibers with shortest NRL fold into narrow ladder-like structures regardless of the presence of LH and show no increase in packing ratio upon the addition of LH. Arrays with medium NRL with LH strongly resemble shapes of highly compacted arrays visualized by the EM-assisted nucleosome interaction capture (EMANIC) technique [46]. These are regularly shaped with dominant two-start zigzag configurations. Arrays with medium NRL also show the highest linear packing density increase and sedimentation coefficients upon addition of LH. For arrays with medium NRL, LH leads to a moderate increase in the fiber diameter. In contrast, fibers with long NRL with LH have a more heterogeneous structure, with both bent DNA linkers and straight DNA linkers due to stems formed by the interaction of the exiting/entering DNA with LH. The DNA linkers tend to bend because the total length of two linker histones (∼ 60 bp) is shorter than the length of a single DNA linker (see Figure 3 for illustration on a 226-bp array, where linker DNA length is 79 bp).
2.2.1. Short NRL: 173 and 182 bp
Our simulations suggest that chromatin fibers with the shortest (NRL = 173 bp) DNA linkers, both with and without LH, fold into a two-start ladder-like structure characterized by strong i±1 and i±2 interactions (Figures 3a and 4a,b), a low linear packing density (∼ 3.8 nucleosomes/11 nm; Figure 5a), and a narrow width (∼ 24–25 nm; Figure 5b), regardless of the starting conformation. Further support for a ladder-like structure for these arrays comes from the large dihedral angle observed regardless of LH presence (Figure 6d).
The interaction patterns for arrays with 182-bp NRL without LH also show strong i ± 1 and i ± 2 contacts and large dihedral angles (∼ 80°), resembling the arrays with the shortest linker DNA described above (Figures 4a and 6d).
The addition of LH to the 182-bp arrays slightly straightens the linker DNAs (as indicated by a small decrease in the bending angles; Figure 6c) and reorganizes the nucleosome contacts, increasing the relative intensity of i ± 2 and i ± 3 interactions at the expense of that of i ± 1 (Figure 4b). This indicates that 182-bp arrays with LH fold into a fiber that interconverts between a two-start and a three-start structure. LH also reduces the dihedral angles to ∼ 65° consistent with the increase of i ± 3 contacts (Figure 6d). The addition of LH, however, does not increase fiber compaction significantly; the packing ratios, sedimentation coefficients, fiber width, fiber volume and % filled volume remain almost constant (Figure 5). The triplet angles also remain constant (50–60°; Figure 6e) regardless of the starting conformation.
2.2.2. Medium NRL: 191, 200 and 209 bp
The structure of medium-NRL arrays depends on the presence of LH. Without LH, these arrays exhibit structural heterogeneity. When started from zigzag configurations, they converge to arrays with irregular zigzag-like features: intense i ± 2, notable i ± 1, and higher contacts (Figure 4c), medium dihedral angles (∼ 70°; Figure 6d), and medium triplet angles (< 65°; Figure 6e). When started from solenoid configurations, they converge to structures with irregular solenoid-like characteristics: intense i ± 1, and i ± 2 contacts combined with multiple other prominent higher-order interactions (Figure 4c), large triplet angles (> 80°; Figure 6e), and smaller dihedral angles (∼ 50°; Figure 6d).
When LH is added to medium-NRL arrays, solenoid interactions are destabilized in favor of zigzag-like forms. Simulations started from both zigzags and solenoids form a compact zigzag fiber with uniform features that include straight DNA linkers (reduced bending angles) that stabilize a DNA stem (see Figure 3c for stem illustration). Independent of NRL, this zigzag fiber has dominant i±2 interactions and moderate i±5 contacts (Figures 3b,c and 4d), consistent with smaller triplet than dimer distances (Figure 6a,b). These fibers exhibit the smallest dihedral (∼ 50°; Figure 6d) and triplet (< 50°; Figure 6e) angles. The tightness of these medium-NRL with LH fibers is also supported by the relatively high packing ratios (∼ 5.0 nucleosomes/11 nm), sedimentation coefficients, and the % of filled volumes, as well as small curvature (straighter fibers) observed in Figure 5.
2.2.3. Long NRL: 218 and 226 bp
Long DNA linker arrays exhibit structural diversity. Without LH, both zigzag and solenoid-like features are viable. Namely, arrays started from zigzag conformations converge to an irregular two-start structure with dominant i ± 2 and i ± 3 interactions and intense i ± 5 contacts (Figure 4e), as well as medium dihedral and triplet angles (Figure 6); arrays started from solenoid converge to structures with zigzag and solenoid characteristics. Specifically, the 218-bp arrays started from solenoid possess equally intense i ± 2 and i ± 5 interactions, while the 226-bp arrays have stronger i ± 5 and i ± 6 interactions followed by i ± 2 contacts (Figure 4e). Both have small dihedral angles and large triplet angles (Figure 6), characteristic of solenoid conformations.
The effect of LH in favoring a zigzag structure weakens for long NRL. This is because the DNA linker is much longer than twice the size of the LH. While LH still triggers the formation of a DNA stem, increasing the intensity of i ± 2 interactions (Figure 4f), it cannot prevent the long DNA linkers from bending in their middle section (see Figure 3d for illustration), and this promotes a wider range of nucleosome interactions. The 218-bp arrays with LH started from zigzag and solenoid conformations converge to different structures with dominant i ± 2 and intense i ± 3 and i ± 5 interactions. The different relative intensities of these interactions between the structures started from solenoid and zigzag reflects the structural diversity favored by long DNA linkers.
Thus we observe that, as the NRL increases, the effectiveness of LH in forming a DNA stem and promoting two-start contacts decreases. In our longest NRL systems (226 bp) with LH, both solenoid-like and zigzag-like characteristics are viable (Figure 4f). When started from zigzag, the 226-bp with LH arrays converge to a structure with dominant i ± 2 interactions, intense i ± 5 contacts, and small dihedral and triplet angles, all of which are consistent with a compact zigzag structure. However, when started from solenoid, the 226-bp arrays with LH lead to heteromorphic structures with zigzag interaction patterns (intense i ± 3 and i ± 5, and strong i ± 2 contacts), but small dihedral angles (> 40°; Figure 6d), and large triplet angles (> 90°; Figure 6e), consistent with the solenoid form.
The long linker lengths of these fibers, however, restrict their compaction relative to medium-NRL fibers. This is reflected by their smaller packing ratios, sedimentation coefficients, and % of filled volume in Figure 5.
2.3. Linker histone role
As discussed above, the interaction patterns and linear packing density (Figure 4a,b and Figure 5) of the shortest linker arrays (NRL = 173 bp) weakly depend on the presence of the LH because the LH’s length is longer than the DNA linker and the DNA stem cannot form. As the NRL increases to 182 bp, the LH induces DNA stem formation [7] (Figure 2) because the DNA linker and LH become comparable in size. However, as for the 173-bp arrays, LH cannot increase fiber compaction; the DNA linkers in arrays with these low NRL values reorient themselves to avoid collision with neighboring cores as well as linker DNAs when LH is present. The nucleosome reorientation also prohibits the formation of one-start solenoid structures.
LH has the strongest influence on arrays with medium NRL (NRL between 191 and 209 bp). Addition of LH to arrays with this NRL ranges encourages a zigzag organization and increases compaction. These linker lengths are sufficiently long to form a DNA stem and consequently reorganize in a compact zigzag structure avoiding steric clashes (Figure 3b,c)
The effect of LH is smaller for arrays with longer NRL values (218 and 226 bp) because the LH’s length is less than half the DNA linker’s length. This allows the base pairs in the center of the linker DNA to bend in the folded fibers (Figures 3d and 6c). This bending, in turn, increases the i ± 3 and i ± 5 interactions at the cost of the i ± 2 interactions.
2.4. Role of ionic concentration
The results reported above are for 0.15 M monovalent salt concentration (CS). The compaction of chromatin depends on the ionic environment. To show that fiber compaction increases in high monovalent ionic concentrations and upon addition of divalent ions, we present representative data for 24-core arrays in three additional conditions: (i) low monovalent salt (CS = 0.01 M) without LH, (ii) CS = 0.2 M with LH, and (iii) CS = 0.15 M with Mg2+ and LH. The divalent ion treatment is a simple first-order approximation as introduced in [45] implemented by reducing the DNA persistence length to 30 nm and allowing the DNA linker beads to touch one another (see details in Methods). Figure 7 shows the increase in packing ratio and sedimentation coefficients as a function of the salt environment for NRL = 173, 209, and 226 bp. Representative snapshots for 48-core arrays are presented in Supplemental Figure S5 to illustrate the change in compaction at different salt environments. The compaction of the 173-bp arrays does not increase because the linkers are too short. The compaction in the 182-bp and 226-bp arrays increases at higher monovalent salt and most significantly upon addition of magnesium ions. Thus, we expect the values shown in Figures 4–6 to change accordingly in these altered salt environments.
2.5. Role of histone tails
Three types of tail interactions are responsible for bridging contacts: (i) nucleosome/nucleosome interactions, (ii) tail interactions with parental DNA, and (iii) tail interactions with non-parental DNA. The H3/H4 tails are known to have a key role in chromatin compaction [43, 58–66] while H2A1/H2A2/H2B tails are involved in histone core aggregation [67], transcription control, and possible mediation of inter-fiber interactions due to their position on the periphery of the nucleosome [43] (H2A1 and H2A2 are, respectively, the N-termini and C-termini of the H2A tails).
In our simulations, we dissect the role of the different tails in fiber bridging contacts as function of the NRL by measuring the fraction of configurations that specific tails are attached to a chromatin component (i.e., parent core, non-parent core, parent DNA linker, or non-parent linker). A tail is considered to be attached to a component if it is closer than the excluded volume distance for tail/particle interactions (1.8 nm) (Section 4.5.2).
2.5.1. Tail/nucleosome interactions (H3 and H4 dominant)
For compact chromatin with LH and linkers comparable to the LH size (173–200 bp), the H3 tails mediate the largest number of internucleosomal contacts, followed by H4 (Figure 8a). In these arrays, the H3 and H4 tails spend 19%–28% and 11%–18% of their time interacting with other cores, respectively. The time spent by tails mediating internucleosome interactions can be analyzed in light of the internucleosomal contact patterns (Figure 4). In the 173-bp with LH arrays, nucleosomes spend 39% of time in the vicinity of their immediate sequential neighbors (i± 1) and thus 61% with i±2 and higher contacts (Figure 4b).
As NRL increases to 200 bp, the nucleosomes spend 81%–97% of the time interacting with i ± 2 and higher neighbors (Figure 4c). Interactions with immediate neighbors are not essential for fiber bridging in moderate-NRL arrays because their DNA linkers hardly bend. Effectively, this means that the H3 tails in 173-bp arrays spend only 12% of the time in contact with i ± 2 and higher neighbors, while for the 182, 191, and 200-bp with LH fibers they spend 14%, 26%, and 16% of their time mediating fiber-bridging interactions, respectively.
The interactions of H3 tails with non-parental cores diminish with the increase of the DNA linker length (Figure 8a). The H4 tails, on the other hand, spend > 10% of their time mediating internucleosome interactions in arrays with NRL up to 218 bp and take over as the most important for internucleosome interactions at 209 and 218 bp. Such internucleosome contacts contribute to fiber bridging.
The H3 and H4 contact probabilities in arrays with longest NRL fall to less than 5% due to looser conformations. Interactions of other tails (H2A and H2B) are negligible.
2.5.2. Interactions with parent DNA linker (H3 and H2A2 dominant)
The tail interactions with parent linker DNAs are mostly mediated by the H3 and H2A2 tails (Figure 8b). Their proximity to the linker DNA, length, and highly positive charge allow them to screen the electrostatic repulsion among the negatively charged DNA linkers. Besides the dominant H3 and H2A2 tails, the shorter H4 tails interact moderately with parental DNA because they are close to the entry/exit positions of the parental DNA linkers. All these three types of tails, in cooperation with LH, create a positively charged region which neutralizes the negatively charged DNA linkers. The screening of the electrostatic repulsion between linkers enables formation of a DNA stem [7].
The efficiency of the H3 tails in screening electrostatic repulsions between parental DNA linkers strongly depends on the linker length. For short NRL (173 bp), the H3 tails have intense interactions with neighboring nucleosomes by construction and consequently reduced interactions with parental DNA linkers. The intensity of the H3 tails interactions rises with the increase of the DNA linker length and reaches a maximum for NRL between 209 and 226 bp. The lengths (4.7 nm) and placement of the H2A2 tails close to the nucleosome dyad axis allow them to interact with parental DNA strongly at a wide range of linker lengths.
2.5.3. Interactions with non-parental DNA linkers (H3 dominant)
The tail interactions with non-parental DNAs in fibers with LH are mostly mediated by the H3 tails (Figure 8c) because they are sufficiently long (12.6 nm in our model). The interaction intensity reaches its peak at NRL = 200 bp. The 200-bp with LH fibers have the highest packing ratios overall and are also among the most compact as reflected by their high sedimentation coefficient.
Other tails are much shorter (4.7 to 7.8 nm) and positioned at the periphery of the nucleosome, and this restricts their interaction with non-parental DNA linkers.
2.5.4. Interactions with parent nucleosome
The short H2A1 and H2B tails are not involved significantly in fiber bridging. They spend most of the time in the vicinity of the parent nucleosome (Figure 8d). Other tails (H2A2/H3/H4) spend less time interacting with their parent nucleosome because they are involved in interactions with other nucleosomes and with DNA.
2.6. Configurational homogeneity
As discussed above, the structural organization of chromatin can be characterized through the analysis of the spatial orientation of consecutive nucleosomes (two, three, and four; see Section 4.6 and Supplemental Figure S4). In a compact zigzag configuration, the i and i ± 2 cores are close, which implies small triplet distances and angles; the straight linkers produce small bending angles; and the dihedral angles are expected to be small if the fiber is compact enough to bring nucleosomes i and i + 3 into intimate contact. In comparison, a compact solenoid fiber would have small dimer distances; larger triplet distances and angles; and, if compact enough, also small dihedral angles that allow cores separated by more than 2 neighbors to interact.
Our analysis confirms that arrays with short linkers (NRL = 173 bp) are homogeneous, regardless of LH presence or starting conformation; all quantities computed are statistically equivalent for the four conditions. As expected, these arrays show small dimer and triplet distances and medium triplet angles. Their dihedral angles are large and have wider error bars due to narrow fiber widths and short average distances between nucleosomes. The size of all error bars decreases upon the addition of LH, confirming that LH promotes structural homogeneity.
Arrays with medium NRL (191 to 209 bp) and LH prefer zigzag configurations characterized by smaller average triplet than dimer distances; small bending, triplet, and dihedral angles; and smaller error bars (Figure 6). These quantities show that LH strongly stabilizes zigzag fiber arrangements. Medium-NRL arrays without LH are heterogeneous and can adopt loose zigzag (with smaller triplet than dimer distances, larger bending angles, medium triplet angles, and large dihedral angles) or solenoid (smaller dimer than triplet distances, larger bending angles, large triplet angles, and small dihedral angles) conformations.
Arrays with long NRL (218–226 bp) and LH exhibit an increased heterogeneity when compared to their shorter brethren (Figure 6). The 218-bp arrays with LH converge to two states characterized by dominant zigzag interactions; each state has slightly different average values of the dimer and triplet distances and triplet and dihedral angles. Moreover, the higher error bars in the bending angles reflect these differences. For 226-bp arrays with LH, the large bending angles in the trajectories started from solenoid indicate that some entering/exiting DNA linkers are widely separated, with linker histones not necessarily interacting with both parental nucleosome linkers. This can be explained from the decreased ability of linker histones to produce DNA stems for long DNA linkers (Figure 3d).
Note that as shown in Supplemental Figure S2, the 24-core short-NRL ensembles converge by 20 million MC steps, while the 24-core long-NRL systems converge by 40 million MC steps, for both starting conformations. Thus, the different viable fibers discussed above reflect actual heterogeneity in the chromatin fiber architecture and the existence of multiple minima in certain conditions rather than the lack of convergence; such variations in chromatin structure are expected for a floppy polymer in solution [26, 46]. Our simulations also suggest that there is a large energetic cost for structural interconversion between solenoid and zigzag states for long NRL.
In sum, our results indicate that zigzag conformations are always viable and that solenoid-like characteristics are viable in either chromatin without LH or in systems with long DNA linkers. Even within one form, there are substantial fluctuations in internal geometric values. Our results also support the recent experimental findings that chromatin fibers are spontaneously dynamic even when compact [68].
3. Discussion
Our modeling reveals that, in the presence of LH, fibers with a wide range of NRL have strong i ± 2 interactions, consistent with the classical zigzag configuration. Very short linker arrays (NRL = 173 bp) have strong i ± 1 and i ± 2 interactions with or without LH, simply by construction. They also exhibit much wider dihedral angles than the longer NRL fibers, commensurate with their narrow widths and ladder-like structure regardless of LH presence (Figures 3, 5, and 6). These characteristics underscore the primary role of LH as a bridge between neighboring DNA linkers. Our observation is consistent with the experimentally measured stoichiometry of H1 in wild type embryonic stem cells [3]. Even smaller values are found in simple unicellular organisms like yeast [69]. Further, in-vivo experiments show that H1 does not have a crucial role in such organisms, i.e., its substantial reduction causes minor phenotypical changes [70, 71].
We also find that LH has a notable structural effect in fibers with slightly longer NRL (182 bp). Here, LH forms the DNA stem, decreasing the exposure of the DNA, and enhancing the intensity of i ± 2 and i ± 3 contacts. LH, however, does not increase the packing ratio or sedimentation coefficient at the moderate monovalent ion concentration considered (0.15 M).
For fibers with medium NRL (191 to 209 bp), LH tends to favor zigzag structures. Higher packing ratios can be achieved only with fibers of medium NRL in the presence of LH. These fibers also show the highest increase in sedimentation coefficients and linear packing ratios, and the smallest dihedral angles, suggesting that LH is evolutionary optimized for NRL between 191 and 209 bp, most often encountered in Nature [2].
Our packing ratios are smaller than the experimentally reported values for several reasons. First, our model does not account for ion correlation effects beyond simple screening as discussed previously [45], and most of our results correspond to moderate monovalent salt. We have shown that for medium to long NRL the packing ratios are very sensitive to the ionic environment: higher concentrations of monovalent salt as well as added divalent ions increase compaction significantly (Figure 7). Second, our LH model is a simple geometric one and does not consider explicit electrostatic interactions with non-parental DNA linkers or the nucleosome, nor the binding-unbinding of LH. While this description accurately reproduces the most important structural role of LH — formation of the DNA stem — further model refinements could help describe these other effects. It should also be noted that experimental measurements for packing ratios are conducted manually and differ from the procedure we use (Supplemental Figure S6). Namely, manual procedures consider the chromatin fiber as a flat two-dimensional object, while our procedure considers the chromatin as a three-dimensional entity.
The longer linker lengths (NRL = 218 bp and higher) are less often encountered in Nature. Our modeling suggests that those arrays have complex internal structure with multiple stable conformations and small relative packing ratios.
Though short DNA linkers are not desirable as they do not offer high packing density, they have one advantage over the longer linkers. Nucleosome arrays with short linkers expose their DNA to transcription and replication machinery; their higher standard deviations of dihedral angles indicates that they are less stable and more prone to opening, as experimentally shown [68]. The ladder-like structures require just one or two displacements to expose DNA to the environment.
Facile access to the DNA may be important in simpler organisms, such as yeast, slime molds and ciliates, which have a very short life span and very high reproduction rates [2, 72, 73]. These organisms require access to all protein sequences in one cell. Interestingly, chromatin can fold into a compact structure without LH when linkers are short [28].
Simpler organisms also have smaller concentrations of LH per nucleosome than found in higher organisms [3]. Moreover, covalent histone tail modifications which increase the attraction between nucleosomes have much stronger influence on chromatin arrays with medium linkers, characterized by i ± 2 and i ± 5 interactions, compared to very short (NRL = 173 bp or less) or long linkers (NRL = 218 bp or longer). Our interaction-intensity averages indicate that, in short linker arrays with LH, a tail modification will likely affect only its nearest neighbors. In contrast, a tail modification in a highly compacted chromatin fiber with medium long linkers could affect multiple nonadjacent nucleosomes. This scenario might explain the versatility of the epigenetic control in higher organism.
4. Computational Methods
4.1. Model overview
Our multiscale mesoscopic model was recently detailed in a study of the role of histone tails [45] along with summary of prior validation studies [24, 42, 43]. In the sections below, we present the main features of the model, including modeling of chromatin’s structural elements, treatment of ionic screening, details of energy terms, and validation of model. We also summarize the Monte Carlo conformational sampling algorithm, chromatin simulation program, and data analysis tools.
The mesoscopic model incorporates all key structural elements of chromatin represented at various levels of accuracy (see Figure 1) using different modeling strategies: the nucleosome core with wrapped DNA excluding protruding tails is represented as an electrostatic object by Debye-Hückel charges [41, 74, 75]; the DNA linker is represented by beads in the worm-like chain model [47, 76]; and the histone tails and LH are coarse grained as beads [45]. The geometry of the basic chromatin unit is derived from available structural data [29]. Despite inherent limitations of the coarse grained approach, the combined model matches the experimentally measured sedimentation and diffusion coefficients, linear mass values, and other experimental measurements of static and dynamic properties [42, 43, 45, 46]. Here we systematically probe chromatin fiber configurations at linker lengths relevant to biology (26 to 79 bp) with 24-nucleosome arrays. We have also performed simulations for 48-core arrays for illustrative purposes.
4.1.1. Nucleosome core model
Our oligonucleosome chain contains NC nucleosome cores. Each nucleosome represents the four histone dimers without protruding tails and the 147 DNA base pairs tightly wound 1.75 times around them, as an electrostatically charged object (Figure 1). Specifically, 300 charges are evenly distributed over the nonuniform surface based on the nucleosome crystal structure (PDB code 1KX5) [29]. The irregular discrete charge optimization (DiSCO) algorithm [41] is used to define the values of those charges through a Debye-Hückel approximation of the electric field by an optimization procedure that minimizes the error between the Debye-Hückel approximation and the electric field of the full atom representation of the nucleosome core (more than 13,000 atoms) at distances > 5 Å. The optimization is achieved through the truncated-Newton TNPACK optimization routine [77–79], integrated within the DiSCO package, as described by Beard and Schlick [75] and Zhang et al. [41]. The electric field is computed using the nonlinear Poisson-Boltzmann equation solver QNIFFT 1.2 [80–82]. The atomic radii input for QNIFFT is taken from the default extended atomic radii based loosely on M. Connolly’s Molecular Surface program [83], and the charges are taken from the AMBER 1995 force field [84]. Representative charges and positions of the 300 pseudocharges within the nucleosome are given in the Supplemental Table S1 for the monovalent ion concentration CS = 0.15 M. Data for other salt concentrations are available from the authors upon request. The excluded volume of the whole nucleosome is treated through the effective excluded volumes of each charge by a Lennard-Jones potential (Supplemental Table S2).
4.1.2. DNA linker model and the oligonucleosome chain
Each nucleosome core, other than the first core, is attached to (is ‘parent of’) two DNA linkers (the ‘exiting’ and ‘entering’ linker DNA). The double stranded DNA linker connecting two adjacent nucleosome cores is modeled as an elastic worm-like chain of nb discrete spherical beads [47, 76]. Each inter-bead segment has an equilibrium length (l0) of 3 nm and each bead carries a salt concentration-dependent negative charge assigned through the Stigter’s procedure developed on the basis of the charged rod approximation [85]. The resulting DNA bead charges at monovalent salt concentrations of 0.01, 0.15 and 0.2 M are −7.54e, −24.09e, and −29.77e, respectively.
The sequence of Nc nucleosomes and nb DNA beads forms the oligonucleosome chain, starting from i = 1 for the first nucleosome to i = N (N = Nc(nb+1)) for the last linker DNA bead, as illustrated in Supplemental Figure S7a. Consistent with the crystal structure, the points of attachment of the exiting and entering linker DNA to the nucleosome define an angle θ0 about the center of the nucleosome core and are separated by a distance 2ω0 normal to the plane of the nucleosome core (see Supplemental Table S2 and Supplemental Figure S7b,c). To prevent a possible overlap between DNA beads an other components of the chromatin array, each bead is assigned an excluded volume through the Lennard-Jones potential (Supplemental Table S2). This approach significantly reduces the number of degrees of freedom (from around 800 atoms to approximately 1 bead per DNA twist). The dynamics of DNA chains are governed by the internal force field comprising of stretching, bending and twisting energy terms as described in [45] (see energy function in Section 4.2.2 below).
Within the oligonucleosome chain, each linker DNA bead and nucleosome is allowed to twist about the DNA axis. This is implemented by assigning local coordinate systems to all DNA linker beads and nucleosome cores. As detailed in Supplemental Figure S7, the coordinate system of each chain component i is specified by three orthonormal unit vectors {ai,bi,ci}, where ci = ai×bi. For each nucleosome core i, three additional coordinate systems are defined to describe the DNA bending and twisting at their points of attachment to the nucleosome: represents the direction from the attachment point of the exiting linker DNA to the center of the i + 1 DNA bead; represents the local tangent on the nucleosome core at the point of attachment of the exiting linker DNA; and represents the tangent corresponding to the entering linker DNA.
To transform the coordinate system of one linker DNA to that of the next (or to that of the entering point of attachment to the core) along the oligonucleosome chain (i.e., {ai,bi,ci} → {ai+1,bi+1,ci+1}), we define the Euler angles αi, βi, and γi as follows
| (1) |
| (2) |
and
| (3) |
The Euler angles , and are defined equivalently and transform the coordinate system of the nucleosome core to that of the exiting linker DNA (i.e., ). Further details on the Euler angles and a geometric description of the oligonucleosome chain are provided in [42, 45, 74] and the supplementary material of [74].
The 3 nm equilibrium length of each DNA inter-bead segment in our chromatin model determines the values of the NRL that we can model. For a given number of inter-bead segments, nS = nb + 1, the linker length measured in base pairs is simply computed as , where a = 0.34 nm/bp is the rise per base pair. Thus, for 3 to 9 bead segments, the linker lengths are 26.47, 35.29, 44.12, 52.94, 61.76, 70.59, and 79.41 bp (Table 2). Since the NRL is defined as the linker length plus the 147 bp of DNA wound around the nucleosome core, the DNA linker lengths we can model are closest to integer NRL of 173, 182, 191, 200, 209, 218, and 226 bp. The shortest theoretical DNA linker length of 2 segments (one DNA bead) was not considered because it is too short for the worm-like chain model. NRL longer than 226 bp were also not considered because they rarely occur in Nature.
To implement the correct non-integral twist for each DNA segment, we first estimate the actual number of turns, τnS, that each DNA linker should make according to its length by dividing the linker length over the number of base pairs per turn for DNA in chromatin (lr), i.e., . Here, we use lr = 10.3 bp/turn for DNA in chromatin, based on experimental observations [86, 87]. Note that a range of 10.2–10.5 bp/turn has been reported for DNA of chromatin, which is different than the twist for nucleosome-free DNA. The resulting τns values in Table 2 are non-integral for all the NRL studied, except for NRL = 209 bp, where the linker length corresponds to 6 full helical turns. When the length of the linker DNA corresponds to an integral number of turns, the average mean twist of that DNA section is exactly zero. However, a non-integral number of turns shifts the average twist of the DNA linker involved.
Thus, to model the different DNA linker lengths, we incorporate the appropriate equilibrium twist per DNA linker segment to accommodate non-integral numbers of DNA turns. In practice, we accomplish this by including a penalty term in the total torsional energy of the bead segments. This torsional energy is
| (4) |
where, s is the torsional rigidity of DNA, N is the number of beads in the oligonucleosome chain, ϕns is the twist deviation penalty term per segment, and αi and γi, two of the Euler angles defined above. The sum αi + γi ∈ [−π, π] gives the linker DNA twist at each bead location. Thus, subtracting ϕns from this sum of angles shifts the average linker DNA twist per segment from zero to the required value.
The values of the twist penalty term per segment are obtained as follows: first, the difference between the required number of turns for the DNA linker and an integral number of turns is calculated (e.g., int(τns) − τns); second, the obtained fractional number of turns is converted into radians ∈ [−π,π]; finally, the resulting twist deviation of the DNA linker is divided by nS to obtain the twist deviation per segment. Given that the sign of ϕns only affects the direction of the relative rotation between consecutive DNA beads, both +ϕns and −ϕns produce the same behavior in our simulations. In other words, the fractional number of turns can be computed as a difference from the higher or lower integer turn value. For example, our shortest DNA linker of 173 bp NRL is modeled by 2 beads or 3 inter-bead segments, which corresponds to an actual linker length of and τ3 = (26.47 bp)/(10.3 bp/turn) = 2.57 helical turns around the DNA axis. The difference from 2 or 3 turns (−0.57 or +0.43) yield the same twist deviation of 2.7 radians for a whole linker DNA and a penalty term per DNA segment of ϕns = 2.7/nS = 0.9 radians. For consistency, in Table 2, we define the difference as the lower integer minus the actual number of turns.
4.1.3. Flexible histone tail model
There are 10 histone tails per nucleosome core: tails belonging to N-termini of H2A (denoted H2A1), H2B, H3, and H4 histones, plus C-termini tails of H2A histones (denoted as H2A2). The histone tails are modeled as chains of spherical beads with each bead representing 5 adjacent amino acids [42, 43, 88]. Each of the two H2A1, H2A2, H2B, H3, and H4 histone tails is represented using 4, 3, 5, 8, and 5 beads, respectively, for a total of 50 tail beads per nucleosome to model the 250 or so histone tail residues that comprise each nucleosome. The lengths of the H2A1, H2A2, H2B, H3, and H4 tails are 6.2, 4.7, 7.8, 12.6, and 7.8 nm, respectively.
Each histone tail is rigidly fixed to its idealized position in the nucleosome crystal structure by a stiff spring between the core and the first tail bead (Figure 1). For tail beads not attached to the core, the stretching and bending harmonic potentials between beads and bond angles between three consecutive beads are tuned to reproduce configurational properties of the atomistic histone tails obtained via Brownian dynamics simulations [42, 88]; the derived force constants are given in Table S3–S4 in the Supplemental Material. The excluded volume of each tail bead is modeled through a Lennard-Jones potential with fixed parameters kev and σtt (Supplemental Table S2).
The electrostatic interactions of histone tails in the presence of salt are modeled by rescaling the charges to reproduce the atomistic potential. For salt concentrations of 0.01, 0.15, and 0.2 M, the scaling factors for the bead charges are 0.75, 1.12, and 1.2. The tails interact with all of the chromatin components, except for the few components listed below, by means of excluded volume and electrostatic interactions. The interactions between neighboring tail beads belonging to the same chain do not interact electrostatically with each other as their interactions are already accounted for through the intramolecular force field. To ensure that the tail-bead attachment remains as close as possible to the equilibrium location, histone tail beads directly attached to the nucleosome do not interact with the nucleosome pseudocharges [45].
4.1.4. Linker histone model
The rat H1d linker histone was the basis for the linker histone (LH) model [46]. Its structure was predicted through fold recognition and molecular modeling [49, 89]. H1d is made of 3 domains, N-terminal region of 33 residues, central globular domain of 76 residues, and highly charged C-terminal domain of 110 residues. In our model, we neglected the short, relatively uncharged N-terminal region and interpret only central globular and C-terminal domains. We model the C-terminal domain by two charged beads and globular domain by a single bead. The three beads are rigidly fixed for each nucleosome and placed on the dyad axis separated by a distance of 2.6 nm [49, 89, 90].
The DiSCO approximation developed for the nucleosome core modeling [74] was applied to assign charges to each linker bead as well [46]. The Debye-Hückel potential of a coarse grained model of each domain (globular and C-terminal) was fitted to the full atom electrostatic potentials obtained by solving the complete non linear Poisson-Boltzmann equation. Consequently, the globular bead carries an effective charge of +13.88e and each C-terminal bead carries a charge of +25.62e at 0.15 M salt. The LH also interacts electrostatically with the other chromatin components except for the nucleosome charges and non-parental DNA linkers. Unlike histone tails or DNA, the three linker histone beads hold fixed relative positions with respect to each other and their parent cores, making the core and LH a unified object that moves as a whole. Compatible with this ‘unified core-linker-histone object’, interactions between LHs and core charges and between non-parent DNA linker and LH are excluded.
Apart form electrostatic interactions, each LH bead interacts with all chromatin components except their parent nucleosome through a Lennard-Jones excluded volume potential.
4.2. Chromatin interaction energies
Below we summarize our treatment of chromatin electrostatics with monovalent and divalent ions, followed by energy terms for interactions among chromatin constituents.
4.2.1. Chromatin electrostatics with monovalent and divalent ions
Physiological salt conditions with monovalent and divalent cations are indispensable for compacting chromatin by screening the highly charged chromatin components (e.g., nucleosomal and linker DNA). We treat the counterions implicitly using mean field theories. Specifically, our DiSCO algorithm [41, 75] parameterizes the screening potential from the Poisson-Boltzmann equation (PBE) using a Debye-Hückel approximation with salt-dependent effective charges, obtained by minimizing the difference between the electric fields from PBE and the (linear) Debye-Hückel approximation using our efficient TNPACK (truncated Newton) optimization package [77, 78]. Thus, DiSCO is used to evaluate the effective charges on the nucleosome core, LHs and histone tails; the effective charges for DNA beads are obtained using an analytical method by Stigter [85]. For the nucleosome core, we typically use 300 effective-charges uniformly distributed across the nucleosome surface; this produces a robust approximation, with < 10% error in the DH approximation over a large range of salt concentrations [41, 75].
The DiSCO approach has been implemented for monovalent ions and assumes that the screening potential is independent of chromatin conformation. To treat divalent ions, we developed a first-order approximation following experimental studies on DNA bending [91, 92], which suggest a reduction of the DNA persistence length to promote linker bending. Specifically, we reduce the repulsion among linker DNA in linker/linker interactions by setting an inverse Debye length of 2.5 nm−1 to allow DNA to almost touch one another, and reduce the persistence length of the linker DNA sequences from 50 to 30 nm according to experimental findings [91, 92]. A refinement of this simple approach has recently been developed [93].
4.2.2. Chromatin energy function
The total potential energy is expressed as the sum of stretching, bending, and torsional components of linker DNA, stretching of histone tails, intramolecular bending of the histone tails, total electrostatic energy, and excluded volume terms [42]:
| (5) |
The first three terms denote stretching
| (6) |
bending,
| (7) |
and torsional energy of the linker DNA (Eq. (4)). Here h and g denote the stretching and bending rigidities of DNA, li the separation between the DNA beads, and IC denotes a nucleosome particle within the oligonucleosome chain (see parameters in Supplemental Table S2). As mentioned above, N is the total number of beads in the chromatin chain, βi and are bending angles, and l0 the equilibrium separation distance between beads of relaxed DNA.
The fourth term, EtS, represents the total stretching energy of the histone tails, composed of two terms: stretching of tail beads, and stretching of the histone tail bead from its assigned attachment site, as given by:
| (8) |
Here NT = 10NC is the total number of histone tails, Nbj is the number of beads in the j-th tail, kbjk is the stretching constant of the bond between the k-th and k + 1-th beads of the j-th histone tail, and lijk and ljk0 represent the distance between tail beads k and k + 1, and their equilibrium separation distance, respectively. In the second term, htc is the stretching bond constant of the spring attaching the histone tail to the nucleosome core, tij is the position vector of the first tail bead in the coordinate system of its parent nucleosome, and tij0 is it ideal position vector in the crystal configuration.
The fifth term, EtB, represents the intramolecular bending contribution to the histone tail energies:
| (9) |
where θijk and θjk0 represent the angle between three consecutive tail beads (k, k + 1, and k + 2), and their equilibrium angle, respectively, and kθjk is the corresponding bending force constant. The sixth term, EC, represents the total electrostatic interaction energy of the oligonucleosome. All these interactions are modeled using the Debye-Hückel potential that accounts for salt screening:
| (10) |
where qi and qj are the ‘effective’ charges separated by a distance rij in a medium with a dielectric constant of k and an inverse Debye length of 1/k, ε0 is the electric permittivity of vacuum, and ε is the dielectric constant (set to 80). As described above, the salt-dependent effective charges are calculated using DiSCO [41, 75] by matching the electric field from the PBE (solved using the DelPhi software) to the field parameterized using the Debye-Hückel form (see Eq. (10)).
The last term, EV, represents the total excluded volume interaction energy of the oligonucleosome. The excluded volume interactions are modeled using the Lennard-Jones potential and the total energy is given by:
| (11) |
where σij is the effective diameter of the two interacting beads and kij is an energy parameter that controls the steepness of the excluded volume potential. These parameters were all taken from relevant models of the components as described fully and tabulated in Supplemental Table S2–S4 and Ref. [45].
4.3. Monte Carlo sampling algorithm and model validation
Sampling of the chromatin configurations is performed by Monte Carlo simulations as developed previously [42–44]. We employ four different Monte Carlo (MC) moves (pivot, translation, rotation, and tail regrowth) to efficiently sample from the ensemble of oligonucleosome conformations at constant temperature. Global pivot moves are implemented by randomly choosing one of the linker beads or nucleosome cores, selecting a random axis passing through the chosen component, and then rotating the shorter part of the oligonucleosome about this axis by an angle chosen from a uniform distribution within [0,20]. Local translation and rotation moves also begin by choosing a randomly oriented axis passing through randomly picked linker bead nucleosome core. In a translation move, the chosen component is shifted along the axis by a distance sampled from a uniform distribution in the range [0,0.6 nm], whereas in a rotation move, it is rotated about the axis by an angle uniformly sampled from the range [0,36]. All three MC moves are accepted/rejected based on the standard Metropolis criterion. The tail regrowth move is implemented to enhance sampling of histone-tail conformations. This move employs the configurational bias MC method [94, 95] to randomly select a histone tail chain and regrow it on the other end using the Rosenbluth scheme [96]. To prevent histone tail beads from penetrating the nucleosome core during tail regrowth, the volume enclosed within the nucleosomal surface is discretized, and any insertion attempts that place the tail beads within this volume are rejected automatically. Typically, the pivot,translation, rotation, and tail regrowth moves are attempted with probabilities of 0.2, 0.1, 0.1, and 0.6, respectively [42, 43].
Our mesoscale chromatin simulation program has been validated for many experimentally-measured properties (see [24, 42, 43, 45]). These properties include salt-induced compaction of oligonucleosomes to reproduce experimental sedimentation coefficients [55] and nucleosome packing ratios [7, 97, 98]; diffusion and salt-dependent behavior of mononucleosomes, dinucleosomes, and trinucleosomes [99–101]; salt-dependent extension of histone tails measured via the tail-to-tail diameter of the core, and radius of gyration for mononucleosomes over a broad range of monovalent salt concentrations [102]; the irregular zigzag topology of chromatin fibers consistent with experimental models [7, 30, 101, 103] and its enhanced compaction upon LH binding [30]; linker crossing orientations in agreement with various experiments [7, 104–106]; and internucleosome interaction patterns consistent with cross-linking and EM experiments [46]. Importantly, the refined model with tails improved the agreement with experimental results compared to the rigid-tail model [45].
4.4. Data Collection
We conducted Monte Carlo sampling with variable linker lengths with 24 nucleosome arrays. Every experimental set (number of nucleosomes, DNA linker length, LH presence) includes a set of 24 simulations divided into two groups according to the starting configuration, zigzag and interdigitated solenoid. Each group covers the mean DNA twist angle (Table 2) and two DNA twist deviations, −12°, and +12° from the mean twist to mimic natural variations, by 4 independent Monte Carlo trajectories. The additional DNA twist variations account for natural variations. We conducted experiments with and without LH. Additionally, we conducted simulations with 48 cores for visualization purposes. The starting configurations for 48-core oligonucleosomes were generated from the compacted 24-core oligonucleosomes. The bulk of our simulations was performed under identical experimental conditions: temperature 293.15 K and 0.15 M monovalent salt concentration (Cs). To analyze salt effects, for selected NRL, three additional experimental sets were essayed (via 24 trajectories each): low monovalent salt (Cs = 0.01 M) without LH, high monovalent salt (Cs = 0.2 M) with LH, and moderate monovalent salt (CS = 0.15 M) with divalent ions and LH. Each simulation trajectory was 35 to 50 million MC steps long. The last 5 million steps were used for statistical analysis. Simulations were run on a 2.33 GHz Intel-Xeon machine. Typically, a 10-million step simulation of 24-core oligonucleosomes takes 4–6 CPU days. Convergence was monitored by global and local geometric and energetic terms (Supplemental Figure S2).
4.5. Calculation of Interaction Patterns
4.5.1. Internucleosome interactions
The internucleosome interactions matrices I′(i,j) describe the fraction of MC iterations that cores i and j are in contact with one another. Each matrix element is defined as:
| (12) |
where M is the MC configurational frame, and the mean is calculated over converged MC frames used for statistical analysis where
| (13) |
At a given MC step M, we consider nucleosomes i and j to be in contact if the shortest distance between the tail-beads directly attached to i and the tail-beads or core-charges of core j is smaller than the tail-tail (σtt) or tail-core (σtt) excluded volume distance, respectively [42]. In our computations we use this cut-off value of 1.8 nm. Figure S3 of the supplementary material shows a typical two-dimensional map (I′(i,j)) of the frequency of histone tail mediated interactions for a zigzag fiber.
These matrices can be projected into normalized one-dimensional maps
| (14) |
that depict the relative intensity of interactions between cores separated by k neighbors. These maps reveal the pattern of internucleosome interactions (dominant, moderate, weak) in a chromatin fiber, providing key insights into structural organization.
4.5.2. Tail interactions
To calculate the interactions of tails with different nucleosome components we follow a similar procedure to that described above. Namely, we measure the fraction of the time that tails of a specific kind t (t = H2A1, H2A2, H2B, H3, and H4) in a chromatin chain are ‘in contact’ with a specific component c of the chromatin chain (c = its parent nucleosome, a non-parental nucleosome, parent DNA linkers, or non-parental DNA linkers) by constructing two-dimensional matrices with the following elements
| (15) |
with the average taken over the converged MC configurations used for statistical analysis with
| (16) |
For a given frame M, we consider a specific t-kind tail of core i to be either free or in contact with only one of the N chromatin component of the oligonucleosome chain. The t-tail is in contact with a component of type c if the shortest distance between its beads and the beads or core charges of c is smaller than the shortest distance to any other type of component and also smaller than the relevant tail-component excluded volume distance (Supplemental Table S2). The resulting normalized patterns of interactions provide crucial information into the frequency by which different tails mediate chromatin interactions.
4.6. Bending, Triplet, and Dihedral Angles
The local bending angle between consecutive nucleosomes is defined as in [45] as the angle formed between the vector exiting one nucleosome and the vector entering the next nucleosome. The former connects the centers of the first two linker DNA beads and the latter those of the last two linker DNA beads (Supplemental Figure S4).
The local triplet angle for three consecutive nucleosomes is the angle defined by nucleosome centers {i,i + 1,i + 2}.
The local dihedral angle is defined for four consecutive nucleosome centers {i, i + 1, i + 2, i + 3}. See Supplemental Figure S4.
For a given MC frame, we calculate the bending, triplet, and dihedral angles of a fiber by taking the average of the local angles over all the nucleosome pairs, triplets, or quadruplets, respectively. We then repeat this procedure for each simulation frame and average the values to obtain mean bending, triplet, and dihedral angles.
4.7. Calculation of Sedimentation Coefficients
We applied the method developed by Bloomfield et al. [107, 108] to calculate the sedimentation coefficient of a given oligonucleosome array conformation, from the inter-core distances [55, 109]. The sedimentation coefficient S20,w is approximated from SNC, where
| (17) |
Here, SNC represents S20,w for a rigid structure consisting of NC nucleosomes of radius R1, Rij is the distance between the centers of two nucleosomes, and S1 is S20,w for a mononucleosome. This approach assumes spherical nucleosomes, a reasonable approximation. We use R1 = 5.5 nm and S1 = 11.1 Svedberg (1 S = 10−13 sec) as done previously [109]. Similar results can be obtained by a more complex procedure implemented in the program HYDRO [110] which calculates S20,w using the radii of both the nucleosome core particle (5.0 nm) and the DNA bead (1.5 nm).
4.8. Calculation of Fiber Packing Ratio, Curvature, and Volume
To calculate the fiber packing ratio (number of nucleosomes per 11 nm of fiber length) for each simulation frame, we first compute the length of the fiber axis passing through a chromatin fiber core (Supplemental Figure S6). At each simulation frame, we define the fiber axis as a 3-dimensional parametric curve , where (j = 1, 2, and 3) are three functions that return the center positions of the i-th nucleosome (ri1, ri2, and ri3) in the x, y, or z direction respectively. We approximate these functions with polynomials of the form
| (18) |
by fitting the data sets [rij] by a least-squares procedure. We have chosen second-order polynomials to approximate the fiber axis because higher-order polynomials tend to produce highly nonlinear fiber axis curves with small packing ratios. We determine the coefficients of the polynomial Pj(i) by minimizing the sum of the squares of the residuals lj
| (19) |
which account for the differences between a proposed polynomial fit and the observed nucleosome positions. After determining the polynomial coefficients, we use Eq. (18) to produce NC points per spatial dimension and compute the fiber length Lfiber as follows:
| (20) |
where the distances are between every two consecutive nucleosome centers. The packing ratio (# of cores per 11 nm) is then calculated as the number of cores multiplied by 11 nm/Lfiber. From the fiber axis, we define the local fiber radius for a given nucleosome core to be the perpendicular distance between a nucleosome core center and its closest linear fiber axis segment plus the nucleosome radius (Rcore = 5.5 nm). We then average over all local fiber radii in a given fiber to obtain the fiber radius at each simulation frame. Finally, we repeat this procedure for each simulation frame and average the value to obtain a mean fiber radius. The fiber width, Dfiber, is twice that value. Additionally, from the parametric definition of the fiber axis, we identify the mean curvature of the chromatin fiber at each simulation frame as:
| (21) |
where ṙax(i) ≈ (2p1,1i+p2,1, 2p1,2i+p2,2, 2p1,3i+p2,3), and r̈ax(i) ≈ 2(p1,1,p1,2,p1,3).
In calculating the fiber volume, Vfiber, for simplicity we use the fiber length and width described above and assume a cylindrical geometry.
We also approximate the percentage of filled volume or the volume occupied by the NC nucleosomes and linker DNAs divided by the total fiber volume. The volume of each nucleosome is approximated by that of a cylinder with height lcore = 5.5 nm and radius R1. The volume of each linker DNA has been approximated as that of a cylinder with diameter l0 and height equal to the segment length l0 multiplied by the number of inter-bead segments nS(e.g., for NRL = 209 bp, nS = 7 segments or 21 nm of height).
Supplementary Material
Acknowledgements
This work was supported by NSF grant MCB-0316771 and NIH grant R01 GM55164 to T. Schlick. Acknowledgment is also made to the donors of the American Chemical Society (Award PRF39225-AC4) Petroleum Research Fund and Philip Morris USA, and to Philip Morris International. Computing support from the NYU HPC USQ and Cardiac clusters is gratefully acknowledged.
Abbreviations used
- NRL
Nucleosome Repeat Length
- bp
base pairs
- LH
Linker Histone
- MC
Monte Carlo
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Compton JL, Bellard M, Chambon P. Biochemical evidence of variability in the DNA repeat length in the chromatin of higher eukaryotes. Proc. Natl. Acad. Sci. USA. 1976;73:4382–4386. doi: 10.1073/pnas.73.12.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woodcock CL, Skoultchi AI, Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 2006;14:17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- 4.Jaeger AW, Kuenzle CC. The chromatin repeat length of brain cortex and cerebellar neurons changes concomitant with terminal differentiation. EMBO J. 1982;1:811–816. doi: 10.1002/j.1460-2075.1982.tb01252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bates DL, Jonathan P, Butler G, Pearson EC, Thomas JO. Stability of the higher-order structure of chicken-erythrocyte chromatin in solution. Eur. J. Biochem. 1981;119:469–476. doi: 10.1111/j.1432-1033.1981.tb05631.x. [DOI] [PubMed] [Google Scholar]
- 6.Thoma F, Koller T, Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J. Cell Biol. 1979:403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bednar J, Horowitz RA, Grigoryev SA, Carruthers LM, Hansen JC, Koster AJ, Woodcock CL. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc. Natl. Acad. Sci. USA. 1998;95:14173–14178. doi: 10.1073/pnas.95.24.14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carruthers LM, Bednar J, Woodcock CL, Hansen JC. Linker histones stabilize the intrinsic salt-dependent folding of nucleosomal arrays: Mechanistic ramifications for higher-order chromatin folding. Biochem. 1998;37:14776–14787. doi: 10.1021/bi981684e. [DOI] [PubMed] [Google Scholar]
- 9.Spadafora C, Oudet P, Chambon P. Rearrangement of chromatin structure induced by increasing ionic strength and temperature. Eur. J. Biochem. 1979;100:225–235. doi: 10.1111/j.1432-1033.1979.tb02053.x. [DOI] [PubMed] [Google Scholar]
- 10.Leibovitch BA, Elgin SR. Heterochromatin and eurochromatin – organization, packaging, and gene regulation. Wiley-VCH; 2005. Encyclopedia of Molecular Cell Biology and Molecular Medicine, Chapter 7; pp. 137–155. ISBN-10: 3527305483. [Google Scholar]
- 11.Misteli T, Gunjan A, Hock R, Bustin M, Brown DT. Dynamic binding of histone H1 to chromatin in living cells. Nature. 2000;408:877–881. doi: 10.1038/35048610. [DOI] [PubMed] [Google Scholar]
- 12.Annunziato AT, Seale RL. Maturation of nucleosomal and nonnucleosomal components of nascent chromatin: Differential requirements for concurrent protein synthesis. Biochem. 1982;21:5431–5438. doi: 10.1021/bi00265a008. [DOI] [PubMed] [Google Scholar]
- 13.Annunziato AT, Schindler RK, Thomas CA, Jr, Seale RL. Dual nature of newly replicated chromatin. Evidence for nucleosomal and non-nucleosomal DNA at the site of native replication forks. J. Biol. Chem. 1981;256:11880–11886. [PubMed] [Google Scholar]
- 14.Bavykin S, Srebreva L, Banchev T, Tsanev R, Zlatanova J, Mirzabekov A. Histone H1 deposition and histone-DNA interactions in replicating chromatin. Proc. Natl. Acad. Sci. USA. 1993;90:3918–3922. doi: 10.1073/pnas.90.9.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tremethick DJ. Higher-order structures of chromatin: The elusive 30 nm fiber. Cell. 2007;128:651–654. doi: 10.1016/j.cell.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 16.van Holde K, Zlatanova J. Chromatin fiber structure, where is the problem now? Sem. Cell Dev. Biol. 2007;18:651–658. doi: 10.1016/j.semcdb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 17.Finch JT, Klug A. Solenoidal model for superstructure in chromatin. Proc. Natl. Acad. Sci. USA. 1976;73:1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Staynov DZ. Possible nucleosome arrangements in the higher-order structure of chromatin. Int. J. Biol. Macromol. 1983;5:3–9. [Google Scholar]
- 19.Williams SP, Athey BD, Muglia LJ, Schappe RS, Gough AH, Langmore JP. Chromatin fibers are left-handed double helices with diameter and mass per unit length that depend on linker length. Biophys. J. 1986;49:233–248. doi: 10.1016/S0006-3495(86)83637-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Athey BD, Smith MF, Rankert DA, Williams SP, Lang-more JP. The diameters of frozen-hydrated chromatin fibers increase with DNA linker length: Evidence in support of variable diameter models for chromatin. J. Cell Biol. 1990;111:795–806. doi: 10.1083/jcb.111.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horowitz RA, Agard DA, Sedat JW, Woodcock CL. The three-dimensional architecture of chromatin in situ: Electron tomography reveals fibers composed of a continuously variable zig-zag nucleosomal ribbon. J. Cell Biol. 1994;125:1–10. doi: 10.1083/jcb.125.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dorigo B, Schalch T, Kulangara A, Duda S, Schroeder RR, Richmond TJ. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306:1571–1573. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- 23.Smith MF, Athey BD, Williams SP, Langmore JP. Radial density distribution of chromatin: Evidence that chromatin fibers have solid centers. J. Cell Biol. 1990;110:245–254. doi: 10.1083/jcb.110.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun J, Zhang Q, Schlick T. Electrostatic mechanism of nucleosomal array folding revealed by computer simulation. Proc. Natl. Acad. Sci. USA. 2005;102:8180–8185. doi: 10.1073/pnas.0408867102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stehr R, Kepper N, Rippe K, Wedemann G. The effect of internucleosomal interaction on folding of the chromatin fiber. Biophys. J. 2008;95:3677–3691. doi: 10.1529/biophysj.107.120543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong H, Victor J-M, Mozziconacci J. An all-atom model of the chromatin fiber containing linker histones reveals a versatile structure tuned by the nucleosomal repeat length. PLoS ONE. 2007;2:e877. doi: 10.1371/journal.pone.0000877. 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rydberg B, Holley WR, Mian IS, Chatterjee A. Chromatin conformation in living cells: Support for a zig-zag model of the 30 nm chromatin fiber. J. Mol. Biol. 1998;284:71–84. doi: 10.1006/jmbi.1998.2150. [DOI] [PubMed] [Google Scholar]
- 28.Routh A, Sandin S, Rhodes D. Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc. Natl. Acad. Sci. USA. 2008;105:8872–8877. doi: 10.1073/pnas.0802336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davey CA, Sargent DF, Luger K, Mäder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J Mol. Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 30.Schalch T, Duda S, Sargent DF, Richmond TJ. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 31.Dorigo B, Schalch T, Kulangara A, Duda S. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306:1571–1573. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- 32.McGhee JD, Nickol JM, Felsenfeld G, Rau DC. Higher order structure of chromatin: Orientation of nucleosomes within the 30 nm chromatin solenoid is independent of species and spacer length. Cell. 1983;33:831–841. doi: 10.1016/0092-8674(83)90025-9. [DOI] [PubMed] [Google Scholar]
- 33.Butler P. A defined structure of the 30 nm chromatin fibre which accommodates different nucleosomal repeat lengths. EMBO J. 1984;3:2599–2604. doi: 10.1002/j.1460-2075.1984.tb02180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Widom J, Klug A. Structure of the 300 Å chromatin filament: X-ray diffraction from oriented samples. Cell. 1985;43:207–213. doi: 10.1016/0092-8674(85)90025-x. [DOI] [PubMed] [Google Scholar]
- 35.Godde JS, Widom J. Chromatin structure of Schizosaccharomyces pombe: A nucleosome repeat length that is shorter than the chromatosomal DNA length. J Mol. Biol. 1992;226:1009–1025. doi: 10.1016/0022-2836(92)91049-u. [DOI] [PubMed] [Google Scholar]
- 36.Huynh VA, Robinson PJ, Rhodes D. A method for the in vitro reconstitution of a defined “30 nm” chromatin fibre containing stoichiometric amounts of the linker histone. J. Mol. Biol. 2005;345:957–968. doi: 10.1016/j.jmb.2004.10.075. [DOI] [PubMed] [Google Scholar]
- 37.Robinson PJJ, Fairall L, Huynh VAT, Rhodes D. EM measurements define the dimensions of the “30-nm” chromatin fiber: Evidence for a compact interdigitated structure. Proc. Natl. Acad. Sci. USA. 2006;103:6506–6511. doi: 10.1073/pnas.0601212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson PJ, Rhodes D. Structure of the “30 nm” chromatin fibre: A key role for the linker histone. Curr. Opin. Struct. Biol. 2006;16:336–343. doi: 10.1016/j.sbi.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 39.Kruithof M, Chien F-T, Routh A, Logie C, Rhodes D, van Noort J. Single-molecule force spectroscopy reveals a highly compliant helical folding for the 30-nm chromatin fiber. Nat. Struct. Mol. Bio. 2009;16:534–540. doi: 10.1038/nsmb.1590. [DOI] [PubMed] [Google Scholar]
- 40.Aumann F, Sühnel J, Langowski J, Diekmann S. Rigid assembly and Monte Carlo models of stable and unstable chromatin structures: The effect of nucleosomal spacing. Theor. Chem. Acc. 2010;125:217–231. [Google Scholar]
- 41.Zhang Q, Beard DA, Schlick T. Constructing irregular surfaces to enclose macromolecular complexes for mesoscale modeling using the Discrete Surface Charge Optimization (DiSCO) Algorithm. J. Comp. Chem. 2003;24:2063–2074. doi: 10.1002/jcc.10337. [DOI] [PubMed] [Google Scholar]
- 42.Arya G, Zhang Q, Schlick T. Flexible histone tails in a new mesoscopic oligonucleosome model. Biophys. J. 2006;91:133–150. doi: 10.1529/biophysj.106.083006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arya G, Schlick T. Role of histone tails in chromatin folding revealed by a new mesoscopic oligonucleosome model. Proc. Natl. Acad. Sci. USA. 2006;103:16236–16241. doi: 10.1073/pnas.0604817103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arya G, Schlick T. Efficient global biopolymer sampling with end-transfer configurational bias Monte Carlo. J. Chem. Phys. 2007;126:044107. doi: 10.1063/1.2428305. [DOI] [PubMed] [Google Scholar]
- 45.Arya G, Schlick T. A tale of tails: How histone tails mediate chromatin compaction in different salt and linker histone environments. J. Phys. Chem. A. 2009;113:4045–4059. doi: 10.1021/jp810375d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grigoryev SA, Arya G, Correll S, Woodcock CL, Schlick T. Evidence for heteromorphic chromatin fibers from analysis of nucleosome interactions. Proc. Natl. Acad. Sci. USA. 2009;106:13317–13322. doi: 10.1073/pnas.0903280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Allison S, Austin R, Hogan M. Bending and twisting dynamics of short linear DNAs. Analysis of the triplet anisotropy decay of a 209 base pair fragment by Brownian simulation. J. Chem. Phys. 1989;90:3843–3854. [Google Scholar]
- 48.Wedemann G, Langowski J. Computer simulation of the 30-nm chromatin fiber. Biophys. J. 2002;82:2847–2859. doi: 10.1016/S0006-3495(02)75627-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bharath MM, Chandra NR, Rao MR. Molecular modeling of the chromatosome particle. Nucleic Acids Res. 2003;31:4264–4274. doi: 10.1093/nar/gkg481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mozziconacci J, Victor J-M. Nucleosome gaping supports a functional structure for the 30 nm chromatin fiber. J. Struct. Biol. 2003;143:72–76. doi: 10.1016/s1047-8477(03)00102-3. [DOI] [PubMed] [Google Scholar]
- 51.Korolev N, Lyubartsev AP, Nordenskiöld L. Computer modeling demonstrates that electrostatic attraction of nucleosomal DNA is mediated by histone tails. Biophys. J. 2006;90:4305–4316. doi: 10.1529/biophysj.105.080226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mühlbacher F, Schiessel H, Holm C. Tail-induced attraction between nucleosome core particles. Phys. Rev. E. 2006;74:031919. doi: 10.1103/PhysRevE.74.031919. [DOI] [PubMed] [Google Scholar]
- 53.Kepper N, Foethke D, Stehr R, Wedemann G, Rippe K. Nucleosome geometry and internucleosomal interactions control the chromatin fiber conformation. Biophys. J. 2008;95:3692–3705. doi: 10.1529/biophysj.107.121079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voltz K, Trylska J, Tozzini V, Kurkal-Siebert V, Langowski J, Smith J. Coarse-grained force field for the nucleosome from self-consistent multiscaling. J. Comp. Chem. 2008;29:1429–1439. doi: 10.1002/jcc.20902. [DOI] [PubMed] [Google Scholar]
- 55.Hansen JC, Ausio J, Stanik VH, van Holde KE. Homogeneous reconstituted oligonucleosomes, evidence for salt-dependent folding in the absence of histone H1. Biochem. 1989;28:9129–9136. doi: 10.1021/bi00449a026. [DOI] [PubMed] [Google Scholar]
- 56.Schlick T, Perišić O. Mesoscale simulations of two nucleosome-repeat length oligonucleosomes. Phys. Chem. Chem. Phys. 2009;11:10729–10737. doi: 10.1039/b918629h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Daban JR. Physical constraints in the condensation of eukaryotic chromosomes. Local concentration of DNA versus linear packing ratio in higher order chromatin structures. Biochem. 2000;39:3861–3866. doi: 10.1021/bi992628w. [DOI] [PubMed] [Google Scholar]
- 58.Tse C, Hansen JC. Hybrid trypsinized nucleosomal arrays: Identification of multiple functional roles of the H2A/H2B and H3/H4 N-termini in chromatin fiber compaction. Biochem. 1997;36:11381–11388. doi: 10.1021/bi970801n. [DOI] [PubMed] [Google Scholar]
- 59.Moore SC, Ausió J. Major role of the histones H3-H4 in the folding of the chromatin fiber. Biochem. Biophys. Res. Commun. 1997;230:136–139. doi: 10.1006/bbrc.1996.5903. [DOI] [PubMed] [Google Scholar]
- 60.Hansen JC, Tse C, Wolffe AP. Structure and function of the core histone N-termini: More than meets the eye. Biochem. 1998;37:17637–17641. doi: 10.1021/bi982409v. [DOI] [PubMed] [Google Scholar]
- 61.Dorigo B, Schalch T, Bystricky K, Richmond TJ. Chromatin fiber folding: Requirement for the histone H4 N-terminal tail. J. Mol. Biol. 2003;327:85–96. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- 62.Shogren-Knaak M, Ishii H, Sun J-M, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 63.Kan P-Y, Lu X, Hansen JC, Hayes JJ. The H3 tail domain participates in multiple interactions during folding and self-association of nucleosome arrays. Mol. Cell. Biol. 2007;27:2084–2091. doi: 10.1128/MCB.02181-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kan P-Y, Hayes JJ. Detection of interactions between nucleosome arrays mediated by specific core histone tail domains. Methods. 2007;41:278–285. doi: 10.1016/j.ymeth.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 65.Wang X, Hayes JJ. Acetylation mimics within individual core histone tail domains indicate distinct roles in regulating the stability of higher-order chromatin structure. Mol. Cell. Biol. 2008;28:227–236. doi: 10.1128/MCB.01245-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kan P-Y, Caterino TL, Hayes JJ. The H4 tail domain participates in intra- and internucleosome interactions with protein and DNA during folding and oligomerization of nucleosome arrays. Mol. Cell. Biol. 2009;29:538–546. doi: 10.1128/MCB.01343-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bertin A, Durand D, Renouard M, Livolant F, Mangenot S. H2A and H2B tails are essential to properly reconstitute nucleosome core particles. Eur. Biophys. J. 2007;36:1083–1094. doi: 10.1007/s00249-007-0212-9. [DOI] [PubMed] [Google Scholar]
- 68.Poirier MG, Oh E, Tims HS, Widom J. Dynamics and function of compact nucleosome arrays. Nat. Struct. Mol. Biol. 2009;16:938–944. doi: 10.1038/nsmb.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Freidkin I, Katcoff DJ. Specific distribution of the Saccharomyces cerevisiae linker histone homolog HHO1p in the chromatin. Nucleic Acids Res. 2001;29:4043–4051. doi: 10.1093/nar/29.19.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shen X, Yu L, Weir JW, Gorovsky MA. Linker histones are not essential and affect chromatin condensation in vivo. Cell. 1995;82:47–56. doi: 10.1016/0092-8674(95)90051-9. [DOI] [PubMed] [Google Scholar]
- 71.Shen X, Gorovsky M. Linker histone H1 regulates specific gene expression but not global transcription in vivo. Cell. 1996;86:475–483. doi: 10.1016/s0092-8674(00)80120-8. [DOI] [PubMed] [Google Scholar]
- 72.Chambers SAM, Vaughn JP, Ramsay-Shaw B. Shortest nucleosomal repeat lengths during sea urchin development are found in two-cell embryos. Biochem. 1983;22:5626–5631. doi: 10.1021/bi00293a026. [DOI] [PubMed] [Google Scholar]
- 73.D’Anna JA, Tobey RA. Changes in nucleosome repeat lengths precede replication in the early replicating Metallothionein II gene region of cells synchronized in early S phase. Biochem. 1989;28:2895–2902. doi: 10.1021/bi00433a022. [DOI] [PubMed] [Google Scholar]
- 74.Beard DA, Schlick T. Computational modeling predicts the structure and dynamics of chromatin fiber. Structure. 2001;9:105–114. doi: 10.1016/s0969-2126(01)00572-x. [DOI] [PubMed] [Google Scholar]
- 75.Beard DA, Schlick T. Modeling salt-mediated electrostatics of macromolecules: The Discrete Surface Charge Optimization algorithm and its application to the nucleosome. Biopolymers. 2001;58:106–115. doi: 10.1002/1097-0282(200101)58:1<106::AID-BIP100>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 76.Heath PJ, Gebe JA, Allison SA, Schurr JM. Comparison of analytical theory with Brownian dynamics simulations for small linear and circular DNAs. Macromolecules. 1996;29:3583–3596. [Google Scholar]
- 77.Schlick T, Fogelson A. TNPACK - A truncated Newton minimization package for large-scale problems: I. Algorithm and usage. ACM Trans. Math. Softw. 1992;18:46–70. [Google Scholar]
- 78.Schlick T, Fogelson A. TNPACK - A truncated Newton minimization package for large-scale problems: II. Implementation examples. ACM Trans. Math. Softw. 1992;18:71–111. [Google Scholar]
- 79.Xie DX, Schlick T. Efficient implementation of the truncated-newton algorithm for large-scale chemistry applications. SIAM J. Optim. 1999;10:132–154. [Google Scholar]
- 80.Gilson MK, Sharp KA, Honig BH. Calculating the electrostatic potential of molecules in solution: Method and error assessment. J. Comput. Chem. 1988;9:327–335. [Google Scholar]
- 81.Sharp KA, Honig B. Electrostatic interactions in macromolecules: Theory and applications. Annu. Rev. Biophys. Biophys. Chem. 1990;19:301–332. doi: 10.1146/annurev.bb.19.060190.001505. [DOI] [PubMed] [Google Scholar]
- 82.Sharp KA, Honig B. Calculating total electrostatic energies with the nonlinear Poisson-Boltzmann equation. J. Phys. Chem. 1990;94:7684–7692. [Google Scholar]
- 83.Connolly ML. Solvent-accessible surfaces of proteins and nucleic acids. Science. 1983;221:709–713. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- 84.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 85.Stigter D. Interactions of highly charged colloidal cylinders with applications to double-stranded DNA. Biopolymers. 1977;16:1435–1448. doi: 10.1002/bip.1977.360160705. [DOI] [PubMed] [Google Scholar]
- 86.Drew HR, Travers AA. DNA bending and its relation to nucleosome positioning. J. Mol. Biol. 1985;186:773–790. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- 87.Deng J, Pan B, Sundaralingam M. Structure of d(ITITACAC) complexed with distamycin at 1.6 Å resolution. Acta Crystallogr. D. 2003;59D:2342–2344. doi: 10.1107/s0907444903020730. [DOI] [PubMed] [Google Scholar]
- 88.Zhang Q. PhD thesis. New York: New York University; 2005. Mesoscopic, microscopic, and macroscopic modeling of protein/DNA complexes. [Google Scholar]
- 89.Bharath MM, Chandra NR, Rao MR. Predictions of an HMg-box fold in the C-terminal domain of histone H1: Insight into its role in DNA condensation. Proteins. 2002;49:71–81. doi: 10.1002/prot.10204. [DOI] [PubMed] [Google Scholar]
- 90.Sheng S, Czajkowsky DM, Shao Z. Localization of linker histone in chromatosomes by cryo-atomic force microscopy. Biophys. J. 2006;91:L35–L37. doi: 10.1529/biophysj.106.090423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Baumann CG, Smith SB, Bloomfield VA, Bustamante C. Ionic effects on the elasticity of single DNA molecules. Proc. Natl. Acad. Sci. USA. 1997;94:6185–6190. doi: 10.1073/pnas.94.12.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rouzina I, Bloomfield VA. DNA bending by small, mobile multivalent cations. Biophys. J. 1998;74:3152–3164. doi: 10.1016/S0006-3495(98)78021-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gan HH, Schlick T. Chromatin ionic atmosphere analyzed by a mesoscale electrostatic approach. 2010 doi: 10.1016/j.bpj.2010.08.023. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Frenkel D, Mooij GCAM, Smit B. Novel scheme to study structural and thermal-properties of continuously deformable molecules. J. Phy. Cond. Matt. 1992;4:3053–3076. [Google Scholar]
- 95.de Pablo JJ, Laso M, Suter UW. Simulation of polyethylene above and below the melting point. J. Chem. Phys. 1992;96:2395–2403. [Google Scholar]
- 96.Rosenbluth MN, Rosenbluth AW. Monte Carlo calculation of the average extension of molecular chains. J. Chem. Phys. 1955;23:356–359. [Google Scholar]
- 97.Williams SP, Langmore JP. Small angle X-ray scattering of chromatin. Radius and mass per unit length depend on linker length. Biophys. J. 1991;59:606–618. doi: 10.1016/S0006-3495(91)82276-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gerchman SE, Ramakrishnan V. Chromatin higher-order structure studied by neutron scattering and scanning transmission electron microscopy. Proc. Natl. Acad. Sci. USA. 1987;84:7802–7806. doi: 10.1073/pnas.84.22.7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yao J, Lowary PT, Widom J. Direct detection of linker DNA bending in defined-length oligomers of chromatin. Proc. Natl. Acad. Sci. USA. 1990;87:7603–7607. doi: 10.1073/pnas.87.19.7603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yao J, Lowary P, Widom J. Linker DNA bending induced by the core histones of chromatin. Biochemistry. 1991;30:8408–8414. doi: 10.1021/bi00098a019. [DOI] [PubMed] [Google Scholar]
- 101.Bednar J, Horowitz RA, Dubochet J, Woodcock C. Chromatin conformation and salt-induced compaction: Three-dimensional structural information from cryoelectron microscopy. J. Cell Biol. 1995;131:1365–1376. doi: 10.1083/jcb.131.6.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bertin A, Leforestier A, Durand D, Livolant F. Role of histone tails in the conformation and interactions of nucleosome core particles. Biochemistry. 2004;43:4773–4780. doi: 10.1021/bi036210g. [DOI] [PubMed] [Google Scholar]
- 103.Leuba SH, Yang G, Robert C, Samori B, van Holde K, Zlatanova J, Bustamante C. Three-dimensional structure of extended chromatin fibers as revealed by tapping-mode scanning force microscopy. Proc. Natl. Acad. Sci. USA. 1994;91:11621–11625. doi: 10.1073/pnas.91.24.11621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Toth K, Brun N, Langowski J. Chromatin compaction at the mononucleosome level. Biochemistry. 2006;45:1591–1598. doi: 10.1021/bi052110u. [DOI] [PubMed] [Google Scholar]
- 105.van Holde K, Zlatanova J. What determines the folding of the chromatin fiber? Proc. Natl. Acad. Sci. USA. 1996;93:10548–10555. doi: 10.1073/pnas.93.20.10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kepert JF, Toth KF, Caudron M, Mucke N, Langowski J, Rippe K. Conformation of reconstituted mononucleosomes and effect of linker histone H1 binding studied by scanning force microscopy. Biophys. J. 2003;85:4012–4022. doi: 10.1016/S0006-3495(03)74815-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bloomfield V, Dalton WO, van Holde KE. Frictional coefficients of multisubunit structures. I. Theory. Biopolymers. 1967;5:135–148. doi: 10.1002/bip.1967.360050202. [DOI] [PubMed] [Google Scholar]
- 108.Kirkwood JG. The general theory of irreversible processes in solutions of macromolecules. J. Polymer Sci. 1954;12:1–14. [Google Scholar]
- 109.Garcia-Ramirez M, Dong F, Ausio J. Role of the histone “tails” in the folding of oligonucleosomes depleted of histone H1. J. Biol. Chem. 1992;267:19587–19595. [PubMed] [Google Scholar]
- 110.Garcia de la Torre J, Navarro S, Lopez Martinez MC, Diazand FG, Lopez Cascales JJ. HYDRO: A computer program for the prediction of hydrodynamic properties of macromolecules. Biophys. J. 1994;67:530–531. doi: 10.1016/S0006-3495(94)80512-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Morris NR. Nucleosome structure in Aspergillus nidulans. Cell. 1976;8:357–363. doi: 10.1016/0092-8674(76)90147-1. [DOI] [PubMed] [Google Scholar]
- 112.Pearson EC, Bates DL, Prospero TD, Thomas JO. Neuronal nuclei and glial nuclei from mammalian cerebral cortex. nucleosome repeat lengths, DNA contents and H1 contents. Eur. J. Biochem. 1984;144:353–360. doi: 10.1111/j.1432-1033.1984.tb08471.x. [DOI] [PubMed] [Google Scholar]
- 113.Downs JA, Kosmidou E, Morgan A, Jackson SP. Suppression of homologous recombination by the Saccharomyces cerevisiae linker histone. Mol. Cell. 2003;11:1685–1692. doi: 10.1016/s1097-2765(03)00197-7. [DOI] [PubMed] [Google Scholar]
- 114.Noll M. Differences and similarities in chromatin structure of Neurospora crassa and higher eucaryotes. Cell. 1976;8:349–355. doi: 10.1016/0092-8674(76)90146-x. [DOI] [PubMed] [Google Scholar]
- 115.Fan Y, Nikitina T, Morin-Kensicki EM, Zhao J, Magnuson TR, Woodcock CL, Skoultchi AI. H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol. Cell. Biol. 2003;23:4559–4572. doi: 10.1128/MCB.23.13.4559-4572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Stalder J, Braun R. Chromatin structure of Physarum Polycephalum plasmodia and amoebae. FEBS letters. 1978;90:223–227. doi: 10.1016/0014-5793(78)80373-1. [DOI] [PubMed] [Google Scholar]
- 117.Bates DL, Thomas JO. Histones H1 and H5: One or two molecules per nucleosome? Nucl. Acids Res. 1981;25:5883–5894. doi: 10.1093/nar/9.22.5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.