Abstract
DNA methylation profiles can be used to define molecular cancer subtypes that may better inform disease etiology and clinical decision-making. This investigation aimed to create DNA methylation profiles of bladder cancer based on CpG methylation from almost 800 cancer-related genes and to then examine the relationship of those profiles with exposures related to risk and clinical characteristics. DNA, derived from formalin-fixed paraffin-embedded tumor samples obtained from incident cases involved in a population-based case-control study of bladder cancer in New Hampshire, was used for methylation profiling on the Illumina GoldenGate Methylation Bead Array. Unsupervised clustering of those loci with the greatest change in methylation between tumor and non-diseased tissue was performed to defined molecular subgroups of disease, and univariate tests of association followed by multinomial logistic regression was used to examine the association between these classes, bladder cancer risk factors and clinical phenotypes. Membership in the two most methylated classes was significantly associated with invasive disease (P < 0.001 for both class 3 and 4). Male gender (P = 0.04) and age >70 years (P = 0.05) was associated with membership in one of the most methylated classes. Finally, average water arsenic levels in the highest percentile predicted membership in an intermediately methylated class of tumors (P = 0.02 for both classes). Exposures and demographic associated with increased risk of bladder cancer specifically associate with particular subgroups of tumors defined by DNA methylation profiling and these subgroups may define more aggressive disease.
Introduction
Identification of molecularly defined subgroups of tumors holds the promise of personalized treatment strategies (1). For example, examination of RNA expression in a panel of genes, in breast cancer, is now used clinically to provide more individualized, targeted and less toxic forms of therapy (2). In addition, for understanding cancer etiology, the examination of molecular profiles of tumors has demonstrated considerable utility in delineating carcinogen exposure-associated differences in individual tumors (3,4).
Bladder cancer is the ninth most incident form of cancer in the USA with over 70 000 new cases diagnosed in 2009 (5). Seventy percent of bladder cancers are non-invasive and highly treatable, although these are more probably to recur (6). Thirty percent of bladder cancers are invasive at presentation, spreading into and through the muscular layers of the bladder and causing high rates of death from metastasis (6,7). This cancer is three to four times more common in men with tobacco smoking being the main risk factor for this disease. Other risk factors include occupational exposures, arsenic ingestion, chlorination by-products and possibly hair dye use and dietary factors (8–10).
Epigenetics is an evolving research area with potential utility for apportioning etiologic fractions as well as for designing future personalized therapies. Epigenetics involves heritable stable changes to gene expression, which are potentially reversible. These changes include DNA hypermethylation leading to gene silencing as well as DNA hypomethylation, leading to oncogene activation and genomic instability (11,12). Alterations in the DNA methylation pattern of the promoter region of cancer-related genes have been associated with risk factors, clinical presentation and outcomes of bladder cancer (13). These risk factors include smoking, arsenic, age and gender, all of which have been associated with an increased prevalence of individual gene alterations or coordinated epigenetic alteration of a small panel of genes in bladder tumors (14–18). Expanding on this concept, previous work in breast, colorectal and head and neck cancer has shown that profiles of the gene promoter methylation may define type of disease and have been associated with the etiologic and clinicopathological features of those diseases (19–21).
In this study, we sought to utilize the DNA methylation profiles of bladder cancers based on the CpG methylation of almost 1500 CpG loci associated with >800 cancer-related genes to identify molecular subgroups of the disease. We then examined the association of those subgroups with risk factors of bladder cancer in order to gain an improved understanding of the etiology of this disease. This approach may help better target prevention efforts and aid in identifying novel subtypes of bladder cancer of therapeutic interest.
Materials and methods
Subjects
A description of the study design appears in earlier reports (22,23). Briefly, bladder cancer cases were drawn from subjects enrolled in two stages of a non-consecutive population-based case–control study of bladder cancer in New Hampshire, conducted from 1994 to 1998 and from 2001 to 2004. Cases of incident bladder cancer were identified from the state cancer registry and a standardized histopathologic review was conducted by a single study pathologist (A.R.S.) to verify the diagnosis and histopathology of the cases. Formalin-fixed paraffin-embedded tumor tissue was obtained from a subset of the cases in the overall study. In addition, non-diseased bladder epithelium (n = 5) was obtained from individuals without cancer through the National Disease Research Interchange. All of these samples came from men, with ages of 22, 68, 72, 75 and 84 years, with four of five being smokers. For the analyses presented here, the case group was restricted to Caucasian transitional cell carcinomas having smoking status data and promoter methylation data and excluded cases that were diagnosed as carcinoma in situ due to small numbers; this included a total of 310 cases (n = 53 from series 1 of 459 cases and n = 257 from series 2 of 398 cases), whose characteristics are presented in Table I. Ninety-five percent of cases and controls in this study were Caucasian, giving us limited power to detect differences in bladder cancer risk factors or prognosis in other racial/ethnic groups; therefore, we restricted our analyses to only Caucasians with race/ethnicity being obtained through self-report. For efficiency purposes for phase 1, the same control group used in a study of non-melanoma skin cancer conducted from 1 July 1993 to 30 June 1995 was used (24). Additional controls were selected afterward up to 2002 frequency matched to cases by age (25–34, 35–44, 45–54, 55–64, 65–69 and 70+) and gender and randomly assigned a reference date comparable with the cases' diagnosis date as described previously (22) In both series, controls <65 years of age were selected from records obtained from the New Hampshire Department of Transportation and controls >65 years of age were chosen from records obtained from the Health Care Financing Administration's Medicare Program. Approximately 70% controls eligible for this study agreed to participate and the same methods were used to collect controls in all phases of the study. For the analyses presented here, the control group was restricted to 1546 Caucasian controls (n = 637 from series 1 and n = 909 from series 2) with complete smoking data, whose characteristics are presented in Table I. No significant differences in the demographic or risk factor variables examined were found between subjects with and without promoter methylation and smoking status data. All procedures and study materials were approved by the appropriate Institutional Review Boards.
Table I.
Selected characteristics of bladder cancer cases stratified by RPMMs class and controls with smoking status data
| Covariates (N = 1856) | All cases (N = 310), n (%) | Controls (N = 1546), n (%) | P-value |
| Smoke status (N = 1856) | |||
| Never | 53 (17.1) | 537 (34.73) | <0.0001 |
| Former | 161 (51.94) | 751 (48.58) | |
| Current | 96 (30.97) | 258 (16.69) | |
| Gender (N = 1856) | |||
| Female | 75 (24.19) | 561 (36.29) | <0.0001 |
| Male | 235 (75.81) | 985 (63.71) | |
| Age (N = 1856) | |||
| <50 years | 27 (8.71) | 233 (15.07) | <0.0004 |
| 50–59 years | 60 (19.35) | 296 (19.15) | |
| 60–69 years | 100 (32.26) | 569 (36.8) | |
| 70–79 years | 123 (39.68) | 448 (28.98) | |
| Water Arsenic (N = 1483) | |||
| 0.002 to <0.104 μg/l | 56 (18.79) | 297 (25.06) | 0.02 |
| 0.104–160.50 μg/l | 242 (81.21) | 888 (74.94) | |
Personal interview and arsenic measurement
Consenting subjects underwent a detailed in-person interview, usually at their home, which assessed sociodemographic information, occupational history, detailed information about the use of tobacco products (such as history of cigarette smoking) and medical history prior to the reference or diagnosis date. In addition, environmental exposures such as average water arsenic levels were measured from participants' drinking water. More specifically, water samples from household drinking water were placed into mineral-free high-density polyethylene bottles (I-Chem vials; Fisher Scientific, Pittsburgh, PA) with strict precautions taken to avoid contamination. Within 6 h, cooled samples were taken to the laboratory and kept frozen at −80°C until the analysis of total arsenic as described previously (25). Samples of drinking water were analyzed for arsenic concentration using an Agilent 7500c Octopole inductively coupled plasma mass spectrometer (Agilent Technologies, Palo Alto, CA) in the Dartmouth Trace Element Analysis Core Facility. Intraclass correlations between replicate (masked) samples of 0.98 down to concentrations of ≤0.010 μg/l has been achieved using this approach (26–28).
DNA methylation analysis
DNA was extracted from the formalin-fixed paraffin-embedded tumor samples as described previously (29). One microgram of DNA was then subjected to sodium bisulfite modification using the EZ DNA Methylation Kit (Zymo Research, Orange, CA) following the manufacturer's protocol; this allowed the conversion of unmethylated cytosines to uracil, whereas methylated cytosines remained unchanged. Methylation profiling of this DNA was performed using the Illumina GoldenGate Methylation Bead Array at the UCSF Institute for Human Genetics Genomic Core Facility as described previously (30,31). All array data points were represented by fluorescent signals from both methylated (Cy5) and unmethylated (Cy3) alleles to create the average methylation (β) value derived from ∼30 replicate methylation measurements. The data was then assembled with BeadStudio methylation software from Illumina. Quality assurance and quality control (QA/QC) was performed to remove poor performing loci and samples as determined by each sample and each loci's detection P-value. More specifically, at each locus for each sample, the detection P-value is defined as 1-P-value computed from the background model characterizing the chance that the signal was distinguishable from negative controls (21). Using this as a metric for quality control for sample performance, eight CpG loci (0.5%) had a median detection P-value >0.05 and were subsequently dropped from the analysis; leaving 1497 loci for analysis (21). In addition, all CpG loci on the X chromosome were excluded from the analysis, leaving a final 1413 CpG loci associated with 773 genes.
Statistical methods
A model-based form of unsupervised clustering known as recursively partitioned mixture models (RPMMs, available through the CRAN website, http://cran.r-project.org/) was used to model the methylation profiles defining subgroups of tumors. To restrict the RPMM analysis to the most informative loci that differed the most between normal and tumor tissues, we computed the mean methylation at each of the 1413 CpG loci among the normal tissue samples (n = 5). We then computed the difference in methylation (delta beta) between each of the raw beta values for the tumor samples and the mean methylation of normal tissues. We subsequently calculated the median value of the delta betas for each of the 1413 loci and selected only those loci with an absolute median delta beta value >0.2, as this value represent, approximately, the minimum detectable difference of the methylation value between two samples detectable using the GoldenGate platform (32). To assess the clinical relevance between the classes obtained from the RPMM analysis, unconditional logistic regression was used to assess the association between each methylation class and invasive disease status. We then looked at univariate associations of exposures and demographics by tumor class using permutation tests (running 10 000 permutations), specifically Kruskal–Wallis for continuous covariates and Chi-squared for categorical covariates. We found significant associations at the P < 0.05 level for age, smoking status (never, former and current), gender and average water arsenic levels. Including only the significant covariates in our model, namely smoking status, gender, average water arsenic levels and age, we used unconditional logistic regression and multinomial (nominal) logistic regression to examine the association between risk factors of bladder cancer and tumor subclass, using controls (as RPMM Class 0) to model the risk of belonging to a class conditional on being a case. We also controlled for study series in our multinomial logistic regression models. Age was coded as a categorical variable into <50 (used as a referent category), 50–59, 60–69 and 70+ years of age as shown in our previous work (9). We categorized smoking as a categorical variable defined as never, former or current smokers (9,33). Based on the standardized histopathologic review conducted by our single study pathologist, we coded our cases as being carcinoma in situ (which were excluded), non-invasive low grade, non-invasive high grade or invasive. We then collapsed the non-invasive low- and high-grade cases into one non-invasive category for our analyses. Finally, average water arsenic levels were classified into quartiles and as the effects were similar among the upper three quartiles and to improve power, the lowest quartile was compared with the higher three (0.002 to <0.104 μg versus 0.104–160.50 μg, respectively). Data were analyzed by use of SAS statistical software, version 9.1 and R, version 2.10.
Results
The characteristics of controls and cases used in these analyses are shown in Table I. We initially examined the difference in the profiles of methylation between the non-diseased bladder epithelium and bladder tumors (supplementary Figure 1A is available at Carcinogenesis Online). An RPMM utilizing all autosomal loci comparing tumor to non-diseased tissue found that all of the non-diseased tissue clustered into a single class, and thus the profiles of DNA methylation are significantly different between tumor and non-diseased tissue (supplementary Figure 1B is available at Carcinogenesis Online; P < 0.00001).
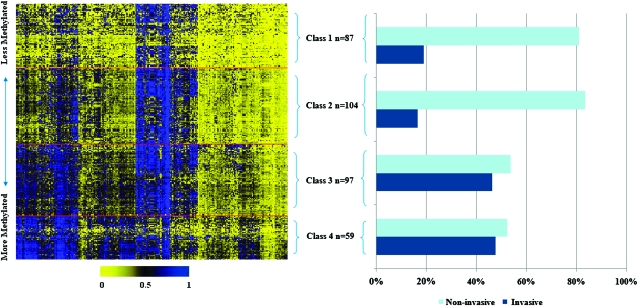
Figure 1 depicts the DNA methylation data for four distinct classes of bladder tumors resulting from RPMM. We found 267 loci that had an absolute median delta–beta value that met the >0.2 threshold so we then fit a beta distributed RPMM to the tumor samples including only these loci. The automated RPMM solution resulted in nine classes; however, due to the small number of participants in some classes, we combined the smallest classes with their RPMM siblings, effectively pruning the RPMM dendrogram further than the automated solution to four final RPMM classes, resulting in four classes. The intensity of methylation is shown in the heatmap with yellow indicating unmethylated and blue indicating fully methylated. Overall, class-specific mean methylation across all loci was lowest in class 1 (mean average beta = 0.25), followed by class 2 (mean average beta = 0.35), then class 3 (mean average beta = 0.44) and class 4 was the most highly methylated (mean average beta = 0.48). The proportion of non-invasive and invasive cases by RPMM class is also shown in Figure 1. The association between having invasive bladder cancer and RPMM class was found to be significant in a permutation chi-square test at the P < 0.05 level.
Fig. 1.
Heatmap and mean methylation by RPMMs class. The heatmap in Panel A shows the intensity of methylation with 0 (yellow) being unmethylated and 1 (blue) being fully methylated. The columns represent the 267 CpG loci and the rows represent individual subjects. The red lines within the heatmap denote the four classes derived from the model. Panel B shows a chart of the percentages of the non-invasive and invasive cases within each RPMM class. Samples sizes are included by RPMM class. The association between having invasive bladder cancer and RPMM class was found to be significant in a permutation chi-square test, at the P < 0.05 level.
Table II presents the odds of invasive bladder cancer by methylation class, adjusted for gender, age and smoking status using unconditional logistic regression. Compared with class 1, a significant 3.93-fold increased risk of being invasive [95% confidence interval (CI): 1.96–7.89] was observed among subjects in class 3, and a significant 4.89-fold increased risk of being invasive (95% CI: 2.15–11.09) was found in subjects in class 4.
Table II.
Risk of invasiveness for each RPMMs class adjusting for smoke status, age, gender and average water arsenic levels
| RPMM Class (N = 310 cases)a | Non-invasive (n = 90), n (%) | Invasive (n = 211), n (%) | OR for invasive disease (95% CI) |
| Class 1 (n = 84) | 68 (81.0) | 16 (19.0) | Ref |
| Class 2 (n = 91) | 76 (83.5) | 15 (16.5) | 0.97 (0.44–2.10) |
| Class 3 (n = 82) | 44 (53.7) | 38 (46.3) | 3.93 (1.96–7.89) |
| Class 4 (n = 53)b | 23 (52.3) | 21 (47.73) | 4.89 (2.15–11.09) |
Italicized values represent statistically significant associations.
The model is adjusted for smoke status, age, gender and average water arsenic levels.
Class 4 is missing nine stage or grade values.
To look at the potential associations between bladder cancer risk factors and specific methylation class, we used controls as our referent group and ran a multinomial logistic regression on all four classes against controls (Table III) and as a stratified analysis using unconditional logistic regression on each class versus controls (supplementary Table 1 is available at Carcinogenesis Online). We only included, in our final model, covariates found to be significant at the P < 0.05 level in previously run univariate analyses as well as the matching factors of age and gender. Current smokers had similar, significant odds of membership in any methylation class compared with controls (Table III; P < 0.05), whereas former-smokers were significantly associated with membership in class 2 and 3. Being male significantly predicted class membership only in class 3 [odds ratio (OR) 1.75, 95% CI: 1.04–2.95] along with being >70 years of age (OR 2.28, 95% CI: 1.02–5.08). Finally, cases in the highest quartile of average water arsenic levels significantly predicted membership in classes 2 and 3 with an OR of 2.02 (95% CI: 1.12–3.63) and 1.98 (95% CI: 1.09–3.60), respectively. Further, the stratified analysis presented in supplementary Table 1 (available at Carcinogenesis Online) is consistent with the results of the multinomial regression.
Table III.
Nominal logistic regression of association of covariates and risk of being in a specific RPMMs class using controls as the referent group
| Nominal logistic regression of covariates (N = 1856) with controls as the referent group | Controls, N = 1546 (referent group), n (%) | Class 1 = 84 cases, n (%) | RPMM class 1, OR (95% CI) | Class 2 = 91 cases, n (%) | RPMM class 2, OR (95% CI) | Class 3 = 82 cases, n (%) | RPMM class 3, OR (95% CI) | Class 4 = 53 cases, n (%) | RPMM class 4, OR (95% CI) |
| Age (N = 1856) | |||||||||
| <50 years | 233 (15.1) | 10 (11.9) | Ref | 7 (7.7) | Ref | 4 (4.9) | Ref | 6 (11.3) | Ref |
| 50–59 years | 296 (19.1) | 13 (15.5) | 0.84 (0.35–2.01) | 23 (25.3) | 2.05 (0.89–4.72) | 14 (17.1) | 1.17 (0.48–2.86) | 10 (18.9) | 0.96 (0.33–2.79) |
| 60–69 years | 569 (36.8) | 25 (29.7) | 0.86 (0.39–1.90) | 34 (37.4) | 1.53 (0.69–3.42) | 24 (29.3) | 1.02 (0.45–2.35) | 17 (32.1) | 0.92 (0.35–2.42) |
| 70+ years | 448 (29.0) | 36 (42.9) | 1.61 (0.75–3.45) | 27 (29.7) | 1.65 (0.73–3.77) | 40 (48.7) | 2.28 (1.02–5.08) | 20 (37.7) | 1.49 (0.58–3.83) |
| Gender (N = 1856) | |||||||||
| Female | 561 (36.3) | 24 (28.6) | Ref | 23 (25.3) | Ref | 18 (22.0) | Ref | 10 (18.9) | Ref |
| Male | 985 (63.7) | 60 (71.4) | 1.29 (0.77–2.16) | 68 (74.7) | 1.32 (0.82–2.11) | 64 (78.0) | 1.75 (1.04–2.95) | 43 (81.1) | 1.62 (0.85–3.10) |
| Smoke status (N = 1856) | |||||||||
| Never | 537 (34.7) | 14 (16.7) | Ref | 15 (16.5) | Ref | 13 (15.9) | Ref | 11 (20.8) | Ref |
| Former | 751 (48.6) | 39 (46.4) | 1.76 (0.94–3.28) | 48 (52.7) | 1.98 (1.12–3.52) | 45 (54.9) | 2.28 (1.23–4.22) | 29 (54.7) | 1.59 (0.79–3.21) |
| Current | 258 (16.7) | 31 (36.9) | 5.26 (2.73–10.12) | 28 (30.8) | 3.91 (2.10–7.30) | 24 (29.2) | 4.46 (2.26–8.79) | 13 (24.5) | 2.75 (1.23–6.11) |
| Water Arsenic (N = 1483) | |||||||||
| 0.002 to <0.104 μg/l | 297 (25.1) | 17 (21.0) | Ref | 13 (14.9) | Ref | 13 (16.3) | Ref | 13 (26.0) | Ref |
| 0.104–160.50 μg/l | 888 (74.9) | 64 (79.0) | 1.01 (0.58–1.76) | 74 (85.1) | 2.02 (1.12–3.63) | 67 (83.7) | 1.98 (1.09–3.60) | 37 (74.0) | 0.84 (0.45–1.55) |
Note. controlling for series in all models.
Italicized values represent statistically significant associations.
Discussion
This study utilized methylation profiles to define subtypes of bladder cancer and associated these subtypes with clinical disease presentation and carcinogen exposure histories. As expected, our initial analyses demonstrate that the profiles identified in tumors are significantly distinct from those identified in non-diseased bladder epithelium. Among tumors, we demonstrated that the mean methylation level differs among methylation profile classes, suggesting that there are distinct phenotypes associated with the methylation profiles, and that membership in the most methylated classes is associated with ORs for invasive bladder cancer of ≥4. This is consistent with our initial analyses using a highly selected reduced number of loci in a smaller series of tumors, which demonstrated that a greater propensity for DNA methylation was associated with more aggressive forms of bladder cancer (15). Our previous work also suggested that the propensity identified by a small number of genes may in fact have been exemplifying a more widespread process of epigenetic dysregulation across the genome (15).
Again, consistent with this previous report and previously published work (14–17,34,35), we also have demonstrated associations between male gender, age and former smoking status with specific subgroups of bladder tumors defined by methylation profile. Compared with non-smokers and controls, current-smokers demonstrated relatively similar odds of membership in all methylation-based subgroups of bladder cancer. This suggests that the specificity of class membership is based on additional exposures, beyond current smoking. For example, high water arsenic levels were associated with cases that had a class 3 methylation profile, suggesting that arsenic exposure has a distinct phenotype represented by a highly specific epigenetic profile. Arsenic exposure has been associated with epigenetic effects in animal models (36–39) and we have demonstrated that specific gene methylation events are associated with arsenic exposure in bladder cancer (14). This class was also almost four times more probably to be an invasive tumor compared with class 1, consistent with our findings that arsenic exposure is associated with more aggressive disease and poorer patient survival (28). Although there is controversy over the levels at which arsenic exposure is carcinogenic in humans, our data suggest that levels found commonly in the USA (40,41) give rise to a detectable specific molecular subgroup of this disease. Former smoking cases were more probably to be class 2 or 3 molecular subtypes, similarly suggesting that this exposure leads to an intermediate overall hypermethylation status. We cannot tell from this type of cross-sectional analysis if this represents a state attained by smoking and then quitting or a return to a lesser state following quitting, but model-system studies should be initiated to better understand the mechanisms by which smoking leads to these effects. Finally, we also observed that males were more probably to be in the class 3 molecular subtype as compared with females. Men generally are three to four times more at risk of developing bladder cancer as compared with females and previous authors have shown that after accounting for exposures such as cigarette smoking, urinary infections and occupational hazards, men still had an excessive risk of bladder cancer as compared with women (42,43). This excessive risk may be related to anatomical differences between men and women, and may be related, especially in older men, with an inability to completely void their bladder (due to prostate enlargement or other conditions) allowing exposures present in residual urine to persist for a longer time. As greater frequency of urination decreases the risk of bladder cancer, this persistence may provide different selective pressures by gender driving these methylation profiles (44–46). However, further research is needed to further examine this potential mechanism.
Strengths of this study included the large size and population-based nature of the study, as well as the use of the Illumina GoldenGate Methylation Bead Array for methylation profiling and detailed exposure assessment including the use of average water arsenic concentrations measured by an inductively coupled plasma mass spectrometer. Limitations of this study include the retrospective nature of the study and thus, the inability to determine true causality to the associations described. Another limitation is the use of only five non-diseased bladder epithelium obtained from individuals without cancer as these individuals may not be representative of normal bladder tissue. The methylation of these normal tissues is relatively homogenous and is significantly distinct from those of the bladder tumors (supplementary Figure 1 is available at Carcinogenesis Online). Therefore, we believe that these are relatively representative and can serve as an appropriate comparator. Larger examinations of non-diseased tissue would be necessary to determine how representative these samples are and how much the demographics of the individual and their exposure history can influence the pattern of DNA methylation in non-diseased bladder epithelium. A final limitation is the time reference for arsenic exposure as the measures of water arsenic are taken at the time of enrollment, which for cases, is following diagnosis, and thus it is possible that the exposure levels at that time do not reflect those which may have been related to bladder tumorigenesis. At the same time, long-term reproducibility of these measures is particularly probably for stable populations, and those for which remediation efforts have not occurred, such as the New Hampshire population under study, and thus these measures are probably reflective of exposures over some period of time (47). We have previously demonstrated reproducible measures of arsenic in tap-water over a 3–5 year period in this population and found that our population used their tap water system for >15 years on average (9,48).
In summary, this study demonstrates that profiles of DNA methylation can be used to distinguish phenotypically and clinically important subgroups of bladder cancer. Smoking history as well as arsenic exposure, age and gender are not only risks of bladder cancer in general but also predispose individuals to specific molecular subtypes of disease. The novelty of these results lies in the use of array-based methodologies to examine CpG methylation of a large number of CpG loci instead of examining only specific promoter regions of certain genes, thereby allowing for a more comprehensive understanding of the epigenetic landscape of bladder tumors. These findings indicate that the methylation profiles of CpG loci can be used as a potential diagnostic marker of bladder cancer and can help further identify novel molecular subtypes of bladder cancer. Future work should examine if these subtypes can be used to create more individualized, targeted regiments of therapy for bladder cancer and aid in the prognosis of this disease.
Supplementary material
Supplementary Figure 1 and Tables 1 and 2 can be found at http://carcin.oxfordjournals.org/
Funding
Flight Attendant Medical Research Institute (YCSA 052341 to C.J.M.); National Institutes of Health (R01CA121147, P42ES007373, R01CA057494).
Conflict of Interest Statement: None declared.
Supplementary Material
Glossary
Abbreviations
- CI
confidence interval
- OR
odds ratio
- RPMM
recursively partitioned mixture model
References
- 1.Schilsky RL. Personalized medicine in oncology: the future is now. Nat. Rev. Drug Discov. 2010;9:363–366. doi: 10.1038/nrd3181. [DOI] [PubMed] [Google Scholar]
- 2.Genomic Health, Inc. Oncotype Dx Breast Cancer Assay Website. Redwood City, CA: Genomic Health, Inc; 2010. [Google Scholar]
- 3.Wikman H, et al. Gene expression and copy number profiling suggests the importance of allelic imbalance in 19p in asbestos-associated lung cancer. Oncogene. 2007;26:4730–4737. doi: 10.1038/sj.onc.1210270. [DOI] [PubMed] [Google Scholar]
- 4.Tan MH, et al. Genomic expression and single-nucleotide polymorphism profiling discriminates chromophobe renal cell carcinoma and oncocytoma. BMC Cancer. 2010;10:196. doi: 10.1186/1471-2407-10-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.American Cancer Society. Cancer Facts and Figures 2009. Atlanta, GA: American Cancer Society; 2009. [Google Scholar]
- 6.Kaufman DS, et al. Bladder cancer. Lancet. 2009;374:239–249. doi: 10.1016/S0140-6736(09)60491-8. [DOI] [PubMed] [Google Scholar]
- 7.Dinney CP, et al. Focus on bladder cancer. Cancer Cell. 2004;6:111–116. doi: 10.1016/j.ccr.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 8.Parkin MP. The global burden of urinary bladder cancer. Scand. J. Urol. Nephrol. Suppl. 2008;42:12–20. doi: 10.1080/03008880802285032. [DOI] [PubMed] [Google Scholar]
- 9.Karagas MR, et al. Incidence of transitional cell carcinoma of the bladder and arsenic exposure in New Hampshire. Cancer Causes Control. 2004;15:465–472. doi: 10.1023/B:CACO.0000036452.55199.a3. [DOI] [PubMed] [Google Scholar]
- 10.Boffetta P. Human cancer from environmental pollutants: the epidemiological evidence. Mutat. Res. 2006;608:157–162. doi: 10.1016/j.mrgentox.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 11.Baylin SB, et al. Epigenetic gene silencing in cancer—a mechanism for early oncogenic pathway addiction. Nat. Rev. Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 12.Jones PA, et al. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 13.Wolff EM, et al. Mechanisms of disease: genetic and epigenetic alterations that drive bladder cancer. Nat. Clin. Pract. Urol. 2005;2:502–510. doi: 10.1038/ncpuro0318. [DOI] [PubMed] [Google Scholar]
- 14.Marsit CJ, et al. Carcinogen exposure and gene promoter hypermethylation in bladder cancer. Carcinogenesis. 2006;27:112–116. doi: 10.1093/carcin/bgi172. [DOI] [PubMed] [Google Scholar]
- 15.Marsit CJ, et al. Promoter hypermethylation is associated with current smoking, age, gender and survival in bladder cancer. Carcinogenesis. 2007;28:1745–1751. doi: 10.1093/carcin/bgm116. [DOI] [PubMed] [Google Scholar]
- 16.Zhu Y, et al. Methyl-CpG-binding domain 2: a protective role in bladder carcinoma. Cancer. 2004;100:1853–1858. doi: 10.1002/cncr.20199. [DOI] [PubMed] [Google Scholar]
- 17.Malekzadeh K, et al. Methylation patterns of Rb1 and Casp-8 promoters and their impact on their expression in bladder cancer. Cancer Invest. 2009;27:70–80. doi: 10.1080/07357900802172085. [DOI] [PubMed] [Google Scholar]
- 18.Marsit CJ, et al. Identification of methylated genes associated with aggressive bladder cancer. PLoS One. 2010;5:e12334. doi: 10.1371/journal.pone.0012334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tao MH, et al. DNA hypermethylation and clinicopathological features in breast cancer: the Western New York Exposures and Breast Cancer (WEB) Study. Breast Cancer Res. Treat. 2009;114:559–568. doi: 10.1007/s10549-008-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mokarram P, et al. Distinct high-profile methylated genes in colorectal cancer. PLoS One. 2009;4:e7012. doi: 10.1371/journal.pone.0007012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marsit CJ, et al. Epigenetic profiling reveals etiologically distinct patterns of DNA methylation in head and neck squamous cell carcinoma. Carcinogenesis. 2009;30:416–422. doi: 10.1093/carcin/bgp006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baris D, et al. A case-control study of smoking and bladder cancer risk: emergent patterns over time. J. Natl Cancer Inst. 2009;101:1553–1561. doi: 10.1093/jnci/djp361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karagas MR, et al. Design of an epidemiologic study of drinking water arsenic exposure and skin and bladder cancer risk in a U.S. population. Environ. Health. Perspect. 1998;106:1047–1050. doi: 10.1289/ehp.98106s41047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wallace K, et al. Selenium and risk of bladder cancer: a population-based case-control study. Cancer Prev. Res. (Phila. Pa) 2009;2:70–73. doi: 10.1158/1940-6207.CAPR-08-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meza MM, et al. Developmentally restricted genetic determinants of human arsenic metabolism: association between urinary methylated arsenic and CYT19 polymorphisms in children. Environ. Health. Perspect. 2005;113:775–781. doi: 10.1289/ehp.7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwong RC, et al. Arsenic exposure predicts bladder cancer survival in a US population. World J. Urol. 2009;28:487–492. doi: 10.1007/s00345-009-0477-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karagas MR, et al. Measurement of low levels of arsenic exposure: a comparison of water and toenail concentrations. Am. J. Epidemiol. 2000;152:84–90. doi: 10.1093/aje/152.1.84. [DOI] [PubMed] [Google Scholar]
- 28.Andrew AS, et al. DNA repair genotype interacts with arsenic exposure to increase bladder cancer risk. Toxicol. Lett. 2009;187:10–14. doi: 10.1016/j.toxlet.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marsit CJ, et al. Epigenetic inactivation of SFRP genes and TP53 alteration act jointly as markers of invasive bladder cancer. Cancer Res. 2005;65:7081–7085. doi: 10.1158/0008-5472.CAN-05-0267. [DOI] [PubMed] [Google Scholar]
- 30.Bibikova M, et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006;16:383–393. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Christensen BC, et al. Breast cancer DNA methylation profiles are associated with tumor size and alcohol and folate intake. PLoS Genet. 2010;6:e1001043. doi: 10.1371/journal.pgen.1001043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bibikova M, et al. GoldenGate assay for DNA methylation profiling. Methods Mol. Biol. 2009;507:149–163. doi: 10.1007/978-1-59745-522-0_12. [DOI] [PubMed] [Google Scholar]
- 33.Wilhelm CS, et al. Implications of LINE1 methylation for bladder cancer risk in women. Clin. Cancer Res. 2010;16:1682–1689. doi: 10.1158/1078-0432.CCR-09-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sobti RC, et al. Hypermethylation-mediated partial transcriptional silencing of DAP-kinase gene in bladder cancer. Biomarkers. 2010;15:167–174. doi: 10.3109/13547500903395124. [DOI] [PubMed] [Google Scholar]
- 35.Brait M, et al. Aberrant promoter methylation of multiple genes during pathogenesis of bladder cancer. Cancer Epidemiol. Biomarkers Prev. 2008;17:2786–2794. doi: 10.1158/1055-9965.EPI-08-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rossman TG. Mechanism of arsenic carcinogenesis: an integrated approach. Mutat. Res. 2003;533:37–65. doi: 10.1016/j.mrfmmm.2003.07.009. [DOI] [PubMed] [Google Scholar]
- 37.Zhao CQ, et al. Association of arsenic-induced malignant transformation with DNA hypomethylation and aberrant gene expression. Proc. Natl Acad. Sci. USA. 1997;94:10907–10912. doi: 10.1073/pnas.94.20.10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhong CX, et al. Both hypomethylation and hypermethylation of DNA associated with arsenite exposure in cultures of human cells identified by methylation-sensitive arbitrarily-primed PCR. Toxicol. Lett. 2001;122:223–234. doi: 10.1016/s0378-4274(01)00365-4. [DOI] [PubMed] [Google Scholar]
- 39.Mass MJ, et al. Arsenic alters cytosine methylation patterns of the promoter of the tumor suppressor gene p53 in human lung cells: a model for a mechanism of carcinogenesis. Mutat. Res. 1997;386:263–277. doi: 10.1016/s1383-5742(97)00008-2. [DOI] [PubMed] [Google Scholar]
- 40.Meliker JR, et al. Lifetime exposure to arsenic in drinking water and bladder cancer: a population-based case-control study in Michigan, USA. Cancer Causes Control. 2010;21:745–757. doi: 10.1007/s10552-010-9503-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mink PJ, et al. Low-level arsenic exposure in drinking water and bladder cancer: a review and meta-analysis. Regul. Toxicol. Pharmacol. 2008;52:299–310. doi: 10.1016/j.yrtph.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 42.Puente D, et al. Gender-related differences in clinical and pathological characteristics and therapy of bladder cancer. Eur. Urol. 2003;43:53–62. doi: 10.1016/s0302-2838(02)00496-7. [DOI] [PubMed] [Google Scholar]
- 43.Hartge P, et al. Unexplained excess risk of bladder cancer in men. J. Natl Cancer Inst. 1990;82:1636–1640. doi: 10.1093/jnci/82.20.1636. [DOI] [PubMed] [Google Scholar]
- 44.Radosavljević V, et al. Non-occupational risk factors for bladder cancer: a case-control study. Tumori. 2004;90:175–180. doi: 10.1177/030089160409000203. [DOI] [PubMed] [Google Scholar]
- 45.Michaud DS, et al. Fluid intake and the risk of bladder cancer in men. N. Engl. J. Med. 1999;340:1390–1397. doi: 10.1056/NEJM199905063401803. [DOI] [PubMed] [Google Scholar]
- 46.Villanueva CM, et al. Total and specific fluid consumption as determinants of bladder cancer risk. Int. J. Cancer. 2006;118:2040–2047. doi: 10.1002/ijc.21587. [DOI] [PubMed] [Google Scholar]
- 47.Karagas MR, et al. Assessment of cancer risk and environmental levels of arsenic in New Hampshire. Int. J. Hyg. Environ. Health. 2002;205:85–94. doi: 10.1078/1438-4639-00133. [DOI] [PubMed] [Google Scholar]
- 48.Karagas MR, et al. Markers of low level arsenic exposure for evaluating human cancer risks in a US population. Int. J. Occup. Med. Environ. Health. 2001;14:171–175. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.