Abstract
Background:
Hypopharyngeal squamous cell carcinoma (HSCC) is an aggressive malignancy with one of the worst prognoses among all head and neck cancers. Greater understanding of the pertinent molecular oncogenic pathways could help improve diagnosis, therapy, and prevention of this disease. The aim of this study was to identify tumour-suppressive microRNAs (miRNAs), based on miRNA expression signatures from clinical HSCC specimens, and to predict their biological target genes.
Methods:
Expression levels of 365 human mature miRNAs from 10 HSCC clinical samples were screened using stem-loop real-time quantitative PCR. Downregulated miRNAs were used in cell proliferation assays to identify a tumour-suppressive miRNA. Genome-wide gene expression analyses were then performed to identify the target genes of the tumour-suppressive miRNA.
Results:
Expression analysis identified 11 upregulated and 31 downregulated miRNAs. Gain-of-function analysis of the downregulated miRNAs revealed that miR-489 inhibited cell growth in all head and neck cancer cell lines examined. The gene PTPN11 coding for a cytoplasmic protein tyrosine phosphatase containing two Src Homology 2 domains was identified as a miR-489-targeted gene. Knockdown of PTPN11 resulted in the inhibition of cell proliferation in head and neck SCC cells.
Conclusion:
Identification of the tumour-suppressive miRNA miR-489 and its target, PTPN11, might provide new insights into the underlying molecular mechanisms of HSCC.
Keywords: hypopharyngeal squamous cell carcinoma (HSCC), microRNA, miR-489 , PTPN11
Hypopharyngeal squamous cell carcinoma (HSCC) is a relatively rare disease, with an incidence of about 10 cases per million people-years (Davies and Welch, 2006). Hypopharyngeal squamous cell carcinoma has a very poor prognosis compared with other head and neck squamous cell carcinomas (HNSCCs), with 5-year survival rates ranging from 30 to 35% (Hoffman et al, 1997; Bova et al, 2005). This poor prognosis is thought to result from advanced primary disease, a high rate of loco-regional recurrence, distant metastasis, and second primary tumours (Spector et al, 2001; Helliwell, 2003). Survival rates of HSCC patients have not markedly improved despite recent advances in various treatment modalities, including surgery, radiotherapy, and chemotherapy (Godballe et al, 2002). Understanding the molecular oncogenic pathways underlying HSCC could significantly improve diagnosis, therapy, and prevention of the disease.
MicroRNAs (miRNAs) are endogenous small non-coding RNAs that can control gene expression by targeting messenger RNAs (mRNAs) for cleavage or translational repression (Bartel, 2004). The miRNAs are involved in crucial biological processes, including development, differentiation, apoptosis, and proliferation (Bartel, 2004; Kloosterman and Plasterk, 2006). An important role for miRNAs in the development of cancer has emerged in recent years (Hwang and Mendell, 2006). The miRNAs are aberrantly expressed in many human cancers, and they may function as oncogenes and tumour suppressors. Upregulated miRNAs could function as oncogenes by negatively regulating tumour suppressor genes, while, downregulated miRNAs could act as tumour suppressors, inhibiting cancers by regulating oncogenes (Esquela-Kerscher and Slack, 2006; Hammond, 2006; Zhang et al, 2007).
A growing body of evidence indicates that unique miRNA expression profiles associated with particular cancers could serve as useful biomarkers for disease prognosis and diagnosis (Lu et al, 2005; Calin and Croce, 2006; Childs et al, 2009). Studies have been carried out for the purpose of identifying specific miRNA alterations in HNSCC (for review, see Liu et al, 2009). However, limited data are available on miRNA expression signatures in HSCC clinical specimens (Childs et al, 2009; Ramdas et al, 2009; Hui et al, 2010). Knockdown or overexpression of a specific miRNA allows functional investigation and validation of the specific role of the miRNAs in tumourigenesis. Analysis of the expression signature of laryngeal, oropharyngeal, or hypopharyngeal cancers showed that underexpression of miR-375 and overexpression of miR-106b-25 cluster might contribute to oncogenesis (Hui et al, 2010).
In this functional analysis of miRNA in HSCC, differentially expressed miRNAs were identified by evaluating 365 mature miRNAs from clinical specimens of HSCC. Cell proliferation assays were conducted to identify tumour-suppressive miRNAs, and genome-wide gene expression analysis was used to identify their targets. The identification of tumour-suppressive miRNAs, and their corresponding target genes, could provide new insights into HSCC carcinogenesis.
Materials and Methods
Clinical HSCC specimens
Tissue specimens of HSCC and adjacent non-cancerous hypopharynx tissue were obtained from patients undergoing surgical treatment for HSCC at Chiba University Hospital between 2004 and 2009. Tissues were immediately frozen in liquid nitrogen and stored at −80°C until further processing. Non-cancerous tissues were obtained far from the centre of the cancer in surgical specimens. No cancer cells were detected in neighbouring formalin-fixed paraffin-embedded specimens. Infection by human papillomavirus (HPV) types HPV16, HPV18, and HPV33 was investigated using genomic DNA from clinical specimens with the PCR Human Papillomavirus Detection Set (Takara, Tokyo, Japan) according to the manufacturer's instructions.
This study was approved by the Bioethics Committee of Chiba University. Prior written informed consent and approval were obtained from all patients.
Cell lines and cell culture
Four of the squamous cell carcinoma cell lines (FaDu, HSC2, HSC3, and D562) were maintained in Dulbecco's Modified Eagle's Medium/Nutrient Mixture F-12 Ham (Invitrogen, Carlsbad, CA, USA), supplemented with 10% foetal bovine serum (Invitrogen) in a humidified atmosphere containing 5% CO2 at 37°C. The FaDu cell line was derived from HSCC tissue (Rangan, 1972). The three remaining cell lines were derived from oral floor (HSC2), tongue (HSC3), and nasopharynx (D562) (Peterson et al, 1971; Momose et al, 1989).
RNA isolation
Total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. The concentrations of RNA were determined using a spectrophotometer, and molecule integrity was checked by gel electrophoresis. The quality of RNA was confirmed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
The miRNA expression signatures and data normalisation
The miRNA expression patterns were evaluated using the TaqMan Low Density Array Human MicroRNA Panel v1.0 (Applied Biosystems, Foster City, CA, USA). The assay was composed of two steps: generation of complementary DNA (cDNA) by reverse transcription, followed by a TaqMan real-time PCR assay. Briefly, miRNAs in the samples were converted to cDNA using 365 specific stem-loop reverse transcription primers. After cDNA conversion, the quantity of mature miRNAs was evaluated using specific TaqMan real-time PCR primers and probes. Real-time PCR was performed in duplicate using GeneAmp Fast PCR Master Mix (Applied Biosystems) and the ABI 7900HT Real-Time PCR System (Applied Biosystems). The Ct values were transformed using the following formula: expression score=2(40−Ct), and the calculated data were uploaded into GeneSpring GX version 7.3.1 software (Silicon Genetics, Redwood City, CA, USA) as described previously (Ichimi et al, 2009; Kano et al, 2010). Description of real-time PCR and the list of human miRNAs can be found on the Applied Biosystems website.
Three approaches were used to normalise the miRNA expression data: global normalisation and endogenous gene normalisation based on RNU44 and RNA48 (Ichimi et al, 2009; Kano et al, 2010). The miRNAs that were detected by all these three normalisation methods were chosen for further study. The fold change, normalisation ratio and P-values were calculated during global normalisation.
Mature miRNA transfection
Mature miRNA molecules, pre-miR miRNA precursors, and a negative control (Applied Biosystems) were incubated with Opti-MEM (Invitrogen) and Lipofectamine RNAiMax reagent (Invitrogen) as described previously (Ichimi et al, 2009). Transfection efficiency of pre-miR in the cell lines was confirmed on the basis of downregulation of PTK9 mRNA by transfection with miR-1 (as recommended by Applied Biosystems).
XTT (cell proliferation) assay
Cells were transfected with 10 nM miRNA by reverse transfection and plated into 96-well plates at 3 × 103 cells per well. After 72 h, cell viability was determined with the XTT assay, using Cell Proliferation Kit II (Roche Molecular Biochemicals, Mannheim, Germany) as described previously (Kano et al, 2010). Triplicate wells were assayed for cell viability in each treatment group.
Target gene search for miR-489
Expression profiles of FaDu cells transfected with miR-489 were screened and compared against miRNA-negative control transfectants using Oligo-microarray Human 44K arrays (Agilent Technologies; Chiyomaru et al, 2010; Kano et al, 2010). Hybridisation and washing steps were performed as described previously (Sugimoto et al, 2009). The arrays were scanned using a Packard GSI Lumonics ScanArray 4000 (Perkin Elmer, Boston, MA, USA). The data were analysed using DNASIS array software (Hitachi Software Engineering, Tokyo, Japan), which converted the signal intensity of each spot into text. The log2 ratios of the median subtracted background intensity were analysed. Data from each microarray study were subjected to a global normalisation (Sugimoto et al, 2009).
The predicted target genes and their conserved miRNA-binding site seed regions were investigated using TargetScan (release 5.1, http://www.targetscan.org/). The sequences of the predicted mature miRNAs were confirmed using miRBase release 13.0 (http://microrna.sanger.ac.uk/).
Real-time quantitative RT–PCR
First-strand cDNA was synthesised from 1 μg total RNA using random primers and the Reverse Transcription (RT) System (Promega, Tokyo, Japan). Gene-specific PCR products were assayed continuously using a 7900-HT Real-Time PCR System with TaqMan probes and primers for PTPN11 (P/N: Hs00818825_m1, Assay-On-Demand Gene Expression Products; Applied Biosystems), according to the manufacturer's protocol. The initial PCR step consisted of a 10-min hold at 95°C, followed by 40 cycles of 15-s denaturation at 95°C, and 1 min annealing/extension at 63°C. For cell lines and clinical samples, GAPDH (A/N: NM_002046) and 18S rRNA (P/N: 4333760F), respectively, were used as internal controls (Assay-On-Demand Gene Expression Products; Applied Biosystems). All reactions were performed in triplicate, and included negative control reactions that lacked cDNA.
Immunoblotting
Cells were collected 72 h after transfection and protein lysates were prepared. A total of 50 μg of lysate was separated by NuPAGE on a 4–12% bis–tris gel (Invitrogen) and transferred onto a polyvinylidene fluoride membrane. Immunoblotting was performed with diluted (1 : 100) monoclonal anti-PTPN11 antibody (ab76285, Abcam, Cambridge, UK), with β-actin serving as an internal control. The membrane was washed and incubated with goat anti-mouse IgG (H+L)–HRP conjugate (Bio-Rad, Hercules, CA, USA). Specific complexes were visualised by echochemiluminescence (GE Healthcare Bio-Sciences, Princeton, NJ, USA).
Plasmid construction and dual-luciferase assay
The miR-489 target sequences were chemically synthesised (Takara, Tokyo, Japan) and inserted between the XhoI and PmeI restriction sites in the 3′ UTR of the hRluc gene in the psiCHECK-2 vector (Promega). FaDu cells were then transfected with 5 ng vector, 10 nM mature miRNA molecules, pre-miRNA miR-489 (Applied Biosystems), and 1 μg Lipofectamine 2000 (Invitrogen) in 100 μl Opti-MEM. Firefly and Renilla luciferase activities in cell lysates were determined using a dual-luciferase assay system (Promega). Normalised data were calculated as the quotient of Renilla/firefly luciferase activities.
Small interfering RNA treatment
After co-transfection of 1 or 10 nM small interfering RNA PTPN11 (si-PTPN11; ID S11524, Ambion) or non-silencing small interfering RNA (si-control), FaDu cells were seeded into 96-well plates at a density of 3 × 103 cells per well. After 72 h, cell viability was determined using the XTT assay. Triplicate wells were measured for cell viability in each treatment group.
Statistical analysis
The relationships between two groups and the numerical values obtained by real-time RT–PCR were analysed using the non-parametric Mann–Whitney U test or the paired t-test. The relationship between miR-489 expression and PTPN11 expression was analysed using the Spearman rank correlation. Expert StatView (version 4, SAS Institute, Cary, NC, USA) was used for analyses, with statistical significance defined as P<0.05.
Results
Identification of differentially expressed miRNAs in clinical HSCC specimens
The expression of 365 mature miRNAs was evaluated in matched pairs of HSCC and their adjacent non-cancerous tissues from 10 patients (Table 1) after HPV infection was ruled out in all specimens. Following three normalisations (RNU44, RNU48 and global) of the raw data, 42 differentially expressed miRNAs were found using all three methods. Of these, 11 (3.0%) were upregulatedd and 31 (8.5%) were downregulated in cancerous tissues. The fold change, normalisation ratio, and P-values in Tables 2A and B were revealed by global normalisation.
Table 1. HSCC patients' characteristics for miRNA screening test.
| Patient | Age |
TNM stage
|
||||
|---|---|---|---|---|---|---|
| number | Gender | (years) | Differentiation | T | N | M |
| 1 | M | 58 | Well | 3 | 2c | 0 |
| 2 | M | 71 | Moderate | 1 | 0 | 0 |
| 3 | M | 60 | Moderate | 3 | 2c | 0 |
| 4 | M | 69 | Moderate | 3 | 2c | 0 |
| 5 | M | 60 | Moderate | 2 | 2c | 0 |
| 6 | F | 74 | Moderate | 4a | 2b | 0 |
| 7 | M | 57 | Moderate | 4a | 2c | 0 |
| 8 | M | 62 | Moderate | 2 | 1 | 0 |
| 9 | F | 52 | Well | 4a | 2b | 0 |
| 10 | M | 56 | Moderate | 4a | 2b | 0 |
Abbreviations: HSCC=hypopharyngeal squamous cell carcinoma; miRNA=microRNA.
Table 2. (a) Upregulated miRNAs in HSCC and (b) downregulated miRNAs in HSCC.
|
Normalized ratio
|
||||
|---|---|---|---|---|
| Gene/miRNA | Fold change | Non-cancer | Cancer | P-value |
| (A) | ||||
| miR-517c | 24.862 | 0.1511 | 3.7568 | 3.66E−05 |
| miR-196a | 10.073 | 0.7187 | 7.2388 | 1.40E−02 |
| miR-7 | 9.301 | 0.5490 | 5.1059 | 1.80E−04 |
| miR-196b | 6.698 | 0.4192 | 2.8074 | 8.24E−04 |
| miR-650 | 4.924 | 0.7011 | 3.4519 | 1.81E−02 |
| miR-18a | 3.518 | 0.6705 | 2.3590 | 2.76E−03 |
| miR-452 | 3.478 | 0.7385 | 2.5683 | 2.93E−02 |
| miR-183 | 3.063 | 0.6892 | 2.1110 | 2.93E−02 |
| miR-432 | 3.027 | 0.5053 | 1.5296 | 2.38E−02 |
| miR-301a | 2.822 | 0.7331 | 2.0691 | 1.37E−02 |
| miR-21 | 2.675 | 0.6324 | 1.6920 | 2.76E−03 |
| (B) | ||||
| miR-1 | 0.007 | 59.2640 | 0.4360 | 2.40E−02 |
| miR-375 | 0.033 | 4.0344 | 0.1322 | 7.25E−05 |
| miR-139-5p | 0.092 | 4.3646 | 0.4012 | 1.52E−04 |
| miR-504 | 0.147 | 2.5714 | 0.3793 | 1.20E−02 |
| miR-125b | 0.232 | 2.2884 | 0.5314 | 7.15E−04 |
| miR-199b | 0.268 | 1.5739 | 0.4217 | 6.53E−03 |
| miR-100 | 0.274 | 1.7713 | 0.4861 | 2.76E−03 |
| miR-497 | 0.278 | 2.0062 | 0.5575 | 7.15E−04 |
| let-7c | 0.282 | 1.8374 | 0.5182 | 3.66E−03 |
| miR-30a* | 0.318 | 1.6777 | 0.5330 | 1.20E−02 |
| miR-218 | 0.322 | 1.5021 | 0.4835 | 1.27E−02 |
| miR-10b | 0.328 | 1.9344 | 0.6353 | 5.89E−03 |
| miR-126* | 0.341 | 2.2839 | 0.7788 | 8.91E−03 |
| miR-378 | 0.342 | 2.0853 | 0.7125 | 2.93E−02 |
| miR-328 | 0.349 | 1.5447 | 0.5394 | 1.25E−03 |
| miR-204 | 0.356 | 2.1216 | 0.7556 | 3.36E−02 |
| miR-143 | 0.365 | 1.5665 | 0.5710 | 5.89E−03 |
| miR-126 | 0.372 | 1.6993 | 0.6324 | 5.64E−04 |
| miR-99a | 0.374 | 1.3994 | 0.5229 | 2.93E−02 |
| miR-195 | 0.393 | 1.7477 | 0.6864 | 1.29E−03 |
| miR-489 | 0.404 | 1.6276 | 0.6572 | 9.07E−03 |
| miR-203 | 0.446 | 1.4617 | 0.6512 | 3.36E−02 |
| miR-140-5p | 0.470 | 1.3766 | 0.6476 | 5.64E−04 |
| miR-29a | 0.484 | 1.4544 | 0.7046 | 5.23E−03 |
| miR-26a | 0.490 | 1.5074 | 0.7379 | 5.89E−03 |
| miR-214 | 0.490 | 1.3913 | 0.6815 | 3.70E−02 |
| miR-30a | 0.546 | 1.3110 | 0.7162 | 5.89E−03 |
| miR-26b | 0.550 | 1.2776 | 0.7030 | 1.27E−02 |
| miR-30e* | 0.571 | 1.3696 | 0.7814 | 4.71E−02 |
| miR-30b | 0.610 | 1.3094 | 0.7984 | 1.27E−02 |
| let-7b | 0.618 | 1.3212 | 0.8162 | 3.95E−02 |
Abbreviations: HSCC=hypopharyngeal squamous cell carcinoma; miRNA=microRNA.
Identification of tumour-suppressive miRNAs
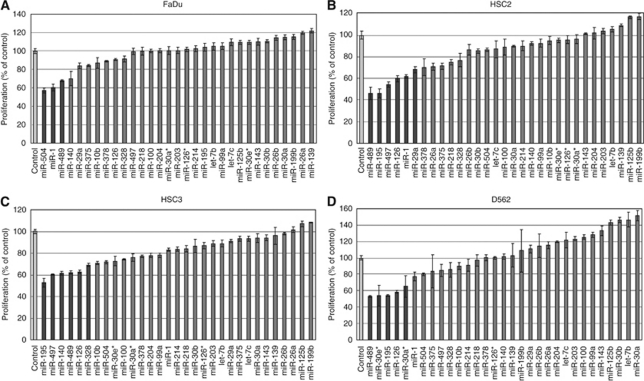
The effect of increasing levels of downregulated miRNAs on cancer cell proliferation was used to identify miRNAs with tumour suppression activity. The proliferation rates of HSCC transfectants are shown in Figures 1A–D. ‘Cell growth inhibiting miRNAs’ were defined as miRNA species that decreased cell proliferation by more than 30% compared with control transfectants. Three miRNAs (miR-504, miR-1, and miR-489) showed cell growth inhibition in FaDu cells (Figure 1A). Similarly, six miRNAs (miR-489, miR-195, miR-497, miR-126, miR-1, and miR-29a) were identified in HSC2 cells (Figure 1B), six miRNAs (miR-195, miR-497, miR-140, miR-489, miR-126, and miR-328) in HSC3 cells (Figure 1C), and five miRNAs (miR-489, miR-30e*, miR-195, miR-126, and miR-30a*) in D562 cells (Figure 1D). Of the 31 downregulated miRNAs (Table 2B), miR-489 inhibited cell growth in all four of the cancer cell lines tested and was, therefore, chosen for further study.
Figure 1.
Effect of transfection with 31 downregulated miRNAs on cancer cell proliferation. Cancer cells were transfected with 10 nM of the indicated mature miRNA. After incubation for 72 h, cell proliferation was determined using XTT assays. (A) FaDu cells; (B) HSC2 cells; (C) HSC3 cells; (D) D562 cells. The darkly shaded bars represent a decrease in cell proliferation of more than 30% compared with control transfections.
Screening of miR-489 target genes by genome-wide gene expression analysis
The molecular basis of miR-489 tumour suppression in HSCC was investigated by examining the effect of miR-489 on protein-coding genes. Mature miR-489 was transiently transfected into FaDu cells, with negative-miRNA transfection used as a control. Comprehensive gene expression analysis showed changes in gene expression patterns between miR-489 and negative-control transfectants. To identify candidate miR-489 target genes, a cut-off of values less than −2.00-fold was applied to the array data. This filtering resulted in the detection of 53 genes that were significantly downregulated upon miR-489 transfection (Table 3). Entries from the microarray data were approved by the Gene Expression Omnibus, and were assigned the Gene Expression Omnibus accession number GSE19718.
Table 3. Downregulated genes by miR-489 treatment in FaDu cells.
| No. | Symbol | Gene Name | Gene ID | Location | Fold | Target sites |
|---|---|---|---|---|---|---|
| 1 | CTDSPL2 | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 | NM_016396 | 15q15.3 | −3.59 | 3 |
| 2 | PTPN11 | Protein tyrosine phosphatase, non-receptor type 11 (Noonan syndrome 1) | NM_002834 | 12q24.13 | −3.40 | 2 |
| 3 | GPR110 | G protein-coupled receptor 110 | NM_025048 | 6p12.3 | −3.05 | — |
| 4 | CLIP4 | CAP-GLY domain containing linker protein family, member 4 | NM_024692 | 2p23.2 | −2.87 | 1 |
| 5 | VGF | VGF nerve growth factor inducible | NM_003378 | 7q22.1 | −2.85 | — |
| 6 | CD244 | CD244 molecule, natural killer cell receptor 2B4 | NM_016382 | 1q23.3 | −2.72 | — |
| 7 | SUZ12 | Suppressor of zeste 12 homologue (Drosophila) | NM_015355 | 17q11.2 | −2.68 | 3 |
| 8 | LIN28B | Lin-28 homologue B (Caenorhabditis elegans) | NM_001004317 | 6q21 | −2.68 | 1 |
| 9 | AP1S1 | Adaptor-related protein complex 1, sigma 1 subunit | NM_001283 | 7q22.1 | −2.62 | 1 |
| 10 | NF2 | Neurofibromin 2 (merlin) | NM_181831 | 22q12.2 | −2.55 | 1 |
| 11 | AP1M2 | Adaptor-related protein complex 1, mu 2 subunit | NM_005498 | 19p13.2 | −2.54 | 1 |
| 12 | A2ML1 | Alpha-2-macroglobulin-like 1 | NM_144670 | 12p13.31 | −2.52 | — |
| 13 | CRIPT | Cysteine-rich PDZ-binding protein | NM_014171 | 2p21 | −2.51 | 1 |
| 14 | EGR1 | Early growth response 1 | NM_001964 | 5q31.2 | −2.51 | — |
| 15 | CYP1B1 | Cytochrome P450, family 1, subfamily B, polypeptide 1 | NM_000104 | 2p22.2 | −2.49 | 2 |
| 16 | NAP1L1 | Nucleosome assembly protein 1-like 1 | NM_139207 | 12q21.2 | −2.48 | — |
| 17 | AHNAK | AHNAK nucleoprotein | NM_001620 | 11q12.3 | −2.48 | — |
| 18 | FAM26E | Family with sequence similarity 26, member E | NM_153711 | 6q22.1 | −2.43 | 1 |
| 19 | RAVER2 | Ribonucleoprotein, PTB-binding 2 | NM_018211 | 1p31.3 | −2.42 | — |
| 20 | RASL10A | RAS-like, family 10, member A | NM_001007279 | 22q12.2 | −2.40 | — |
| 21 | C14orf147 | Chromosome 14 open reading frame 147 | NM_138288 | 14q13.1 | −2.38 | 1 |
| 22 | C14orf143 | Chromosome 14 open reading frame 143 | NM_145231 | 14q32.11 | −2.37 | 1 |
| 23 | HTR2B | 5-hydroxytryptamine (serotonin) receptor 2B | NM_000867 | 2q37.1 | −2.37 | 1 |
| 24 | MYLK | Myosin light chain kinase | NM_053025 | 3q21.1 | −2.33 | — |
| 25 | TFAP4 | Transcription factor AP-4 (activating enhancer-binding protein 4) | NM_003223 | 16p13.3 | −2.33 | 1 |
| 26 | MYO3B | Myosin IIIB | NM_138995 | 2q31.1 | −2.32 | 1 |
| 27 | OSTM1 | Osteopetrosis-associated transmembrane protein 1 | NM_014028 | 6q21 | −2.32 | 1 |
| 28 | MARCKS | Myristoylated alanine-rich protein kinase C substrate | NM_002356 | 6q22.1 | −2.29 | 2 |
| 29 | KCTD4 | Potassium channel tetramerisation domain-containing 4 | NM_198404 | 13q14.12 | -2.26 | 1 |
| 30 | GCLC | Glutamate-cysteine ligase, catalytic subunit | NM_001498 | 6p12.1 | −2.26 | — |
| 31 | ERRFI1 | ERBB receptor feedback inhibitor 1 | NM_018948 | 1p36.23 | −2.26 | 1 |
| 32 | MDH1 | Malate dehydrogenase 1, NAD (soluble) | NM_005917 | 2p15 | −2.26 | — |
| 33 | IL15 | Interleukin 15 | NM_172174 | 4q31.21 | −2.26 | — |
| 34 | ZCCHC5 | Zinc finger, CCHC domain-containing 5 | NM_152694 | Xq21.1 | −2.26 | — |
| 35 | GRB10 | Growth factor receptor-bound protein 10 | NM_001001555 | 7p12.2 | −2.25 | 2 |
| 36 | KLHL5 | Kelch-like 5 (Drosophila) | NM_015990 | 4p14 | −2.21 | 1 |
| 37 | BLID | BH3-like motif containing, cell death inducer | NM_001001786 | 11q24.1 | −2.20 | — |
| 38 | CFL2 | Cofilin 2 (muscle) | NM_021914 | 14q13.2 | −2.19 | 3 |
| 39 | SLC24A1 | Solute carrier family 24 (sodium/potassium/calcium exchanger), member 1 | NM_004727 | 15q22.31 | −2.17 | 1 |
| 40 | CDIPT | CDP-diacylglycerol–inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) | NM_006319 | 16p11.2 | −2.14 | 1 |
| 41 | RTP4 | Receptor (chemosensory) transporter protein 4 | NM_022147 | 3q27.3 | −2.14 | — |
| 42 | ATP1B3 | ATPase, Na+/K+ transporting, beta 3 polypeptide | NM_001679 | 3q23 | −2.14 | 1 |
| 43 | NCOA3 | Nuclear receptor coactivator 3 | NM_181659 | 20q13.12 | −2.14 | 2 |
| 44 | CDK6 | Cyclin-dependent kinase 6 | NM_001259 | 7q21.2 | −2.12 | 1 |
| 45 | RP11-11C5.2 | Similar to RIKEN cDNA 2410129H14 | NM_001071775 | 13q22.1 | −2.11 | 2 |
| 46 | UNQ9438 | TIMM9 | NM_207377 | 14q23.1 | −2.08 | — |
| 47 | MAGEH1 | Melanoma antigen family H, 1 | NM_014061 | Xq11.21 | −2.07 | — |
| 48 | HPS3 | Hermansky–Pudlak syndrome 3 | NM_032383 | 3q24 | −2.05 | 1 |
| 49 | RNF149 | Ring finger protein 149 | NM_173647 | 2q11.2 | −2.04 | — |
| 50 | NUPL1 | Nucleoporin-like 1 | NM_014089 | 13q12.13 | −2.03 | — |
| 51 | SLC25A40 | Solute carrier family 25, member 40 | NM_018843 | 7q21.12 | −2.03 | — |
| 52 | ZCCHC4 | Zinc finger, CCHC domain-containing 4 | NM_024936 | 4p15.2 | −2.02 | 1 |
| 53 | TMEM64 | Transmembrane protein 64 | NM_001008495 | 8q21.3 | −2.01 | 1 |
The 3′ UTR regions of these downregulated genes were examined for miR-489 target sites using the TargetScan database. Of the 53 putative gene targets, 32 genes contained miR-489 target sites (Table 3).
Effect of miR-489 transfection on PTPN11 expression in cancer cells
One of the genes with miR-489 target sites in its 3′ UTR is PTPN11. This gene encodes a protein tyrosine phosphatase (PTP) that contains two Src Homology 2 domains. Although PTPs generally act as tumour suppressors, PTPN11 has been identified as the first PTP oncogene (Tonks, 2006). Therefore, this gene was investigated further as a target of miR-489.
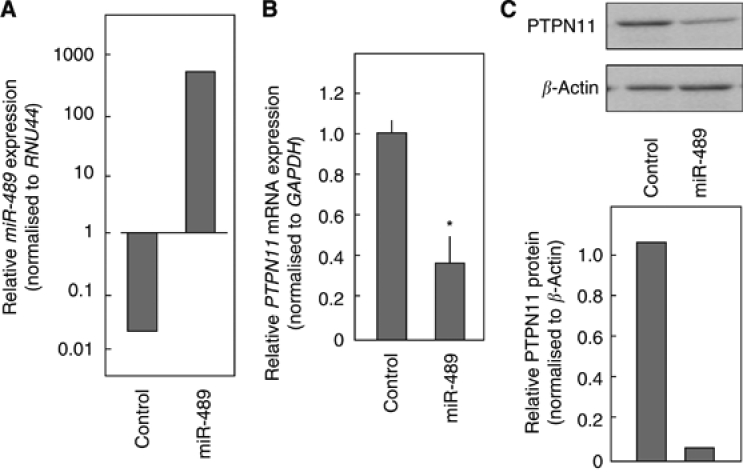
To determine whether miR-489 regulates PTPN11 expression, miR-489 was introduced into FaDu cells. Gain-of-function effects of miR-489 were investigated 72 h after transfection. The expression of miR-489 was elevated by >1000-fold in FaDu cells compared with the miR-negative control (Figure 2A). The mRNA levels for PTPN11 were significantly repressed (Figure 2B). Immunoblotting confirmed that PTPN11 protein expression was significantly decreased in miR-489 transfectants (Figure 2C).
Figure 2.
miR-489 negatively regulates PTPN11 expression. FaDu cells were transfected with miR-489. After incubation for 72 h, total RNA and proteins were isolated. (A) FaDu cells were treated with a miR-negative control (10 nM) or miR-489 (10 nM). After 72 h, miR-489 expression was measured by TaqMan quantitative real-time PCR. The results are normalised to RNU44 expression. (B) PTPN11 mRNA expression was analysed by TaqMan quantitative real-time PCR. The results are normalised to GAPDH expression and are presented relative to control expression. * P<0.05. (C) Cell lysates were analysed by immunoblotting. Membranes were incubated with anti-PTPN11 IgG and anti-β-actin IgG. The autoradiographic density of each protein band was quantified using NIH ImageJ software. The results are standardised against β-actin levels and are presented as the relative density.
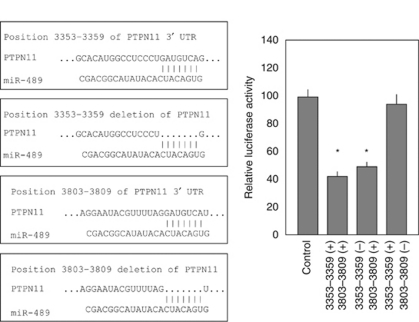
A luciferase reporter assay was performed to determine whether PTPN11 mRNA contains a miR-489 target site, as predicted by the TargetScan algorithm. A vector encoding the partial 3′ UTR of PTPN11 (position 3300–3850) exhibited significantly decreased luminescence intensity after miR-489 transfection (Figure 3). To determine the specific site targeted by miR-489, two vectors carrying deletions of candidate target sites were constructed (deleted positions 3353–3359 and 3803–3809). Luminescence intensity was significantly decreased for the vectors carrying the 3′ UTR and the deletion at position 3353–3359, but not in the vector with the deletion at position 3803–3809 (Figure 3), indicating that the region between positions 3803–3809 contains the miR-489 target site.
Figure 3.
miR-489 binds to the 3′ UTR of PTPN11 mRNA. A luciferase reporter assay used a vector encoding the partial PTPN11 3′ UTR (position 3300–3850). Renilla luciferase values are normalised against firefly luciferase values. Luciferase reporter assays were repeated using mutated vectors in which the candidate sites targeted by the miR-489 were deleted. *P<0.05
Effect of si-PTPN11 transfection
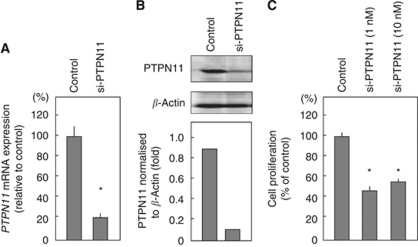
A loss-of-function assay using small interfering RNA analysis was performed to examine the oncogenic function of PTPN11, which is directly targeted by miR-489. The effect of si-PTPN11 on mRNA and protein expression levels was evaluated after transfection into FaDu cells. Both PTPN11 mRNA and protein levels had been reduced 72 h after transfection (Figures 4A and B). The contribution of PTPN11 to cell viability was assessed with si-PTPN11 loss-of-function assays in FaDu cells. Knockdown of PTPN11 significantly decreased cancer cell growth compared with si-control transfectants (Figure 4C).
Figure 4.
Proliferation is inhibited by transfection with si-PTPN11 in FaDu cells. FaDu cells were transfected with 10 nM si-PTPN11. Total RNA and proteins were isolated after 72-h incubation. (A) PTPN11 mRNA expression was analysed by TaqMan quantitative real-time PCR. The results are normalised to GAPDH expression and are presented as relative to control expression. *P<0.05. (B) Cell lysates were analysed by immunoblotting. Membranes were incubated with anti-PTPN11 IgG and anti-β-actin IgG. The autoradiographic density of each protein band was quantified using NIH ImageJ software. The results are standardised against β-actin levels and are presented the relative density. (C) FaDu cells were transfected with 1 or 10 nM si-PTPN11. After incubating for 72 h, cell proliferation was determined using an XTT assay. *P<0.05.
PTPN11 overexpression in HSCC clinical specimens
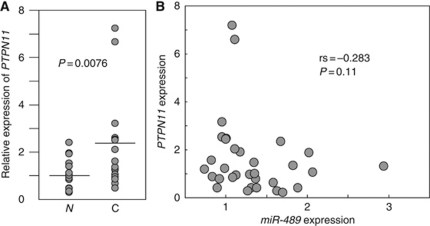
The mRNA expression levels of PTPN11 were significantly higher in 16 HSCC tissues than in adjacent non-cancerous hypopharyngeal tissues (Figure 5A). The possibility that the expression of PTPN11 and the miR-489 were correlated was tested using the Spearman rank correlation. However, the inverse correlation between PTPN11 and miR-489 expression levels was too low to be statistically significant (rs=−0.283 and P=0.11; Figure 5B).
Figure 5.
PTPN11 overexpression in clinical HSCC specimens. (A) PTPN11 mRNA expression levels were analysed by TaqMan quantitative real-time PCR and normalised to 18S rRNA expression. PTPN11 mRNA expression was compared between matched HSCC and non-cancerous tissues in 16 patients. Data were analysed using the paired t-test. N, non-cancerous tissues; C, cancer tissues. (B) Correlation between PTPN11 and miR-489 expression in HSCC clinical specimens.
Discussion
Unique miRNA expression profiles associated with particular cancers could serve as biomarkers for prognosis and diagnosis (Lu et al, 2005; Calin and Croce, 2006; Childs et al, 2009). This study of miRNA expression signatures in clinical HSCC specimens resulted in the identification of 42 differentially expressed miRNAs, of which 11 were upregulatedd (Table 2A) and 31 were downregulated (Table 2B). As HSCC has a very poor prognosis compared with other HNSCCs, these HSCC miRNA expression signatures could help elucidate the underlying molecular mechanisms of this disease.
The miRNA expression signatures of head and neck cancers have been reported by several laboratories (Chang et al, 2008; Wong et al, 2008; Avissar et al, 2009; Chen et al, 2009; Childs et al, 2009). A comparison of our data with these published expression signatures revealed that miR-21, miR-18a, and miR-196b are commonly upregulatedd in head and neck cancers. It was already known that miR-21, which functions as an oncogene (Chang et al, 2008), stands out as the miRNA most often overexpressed across a diverse range of malignancies (Esquela-Kerscher and Slack, 2006). Further studies are needed to clarify the functions of these upregulatedd miRNAs and their role in HSCC carcinogenesis.
A total of 17 of the 31 downregulated miRNAs identified in this study (miR-1, miR-375, miR-139–5p, miR-125b, miR-199b, miR-100, miR-497, miR-30a, miR-218, miR-10b, miR-204, miR-143, miR-99a, miR-195, miR-140–5p, miR-26b, and miR-30b) are previously reported head and neck cancer signatures. In HNSCC, miR-125b and miR-100 have tumour-suppressive functions (Henson et al, 2009). The miR-375 was the most downregulated miRNA in the HNSCC samples, including hypopharyngeal cancer, and its increased expression leads to a significant reduction in cell viability in cancer cells (Hui et al, 2010).
Tumour-suppressive miRNAs are usually underexpressed in cancer cells (Esquela-Kerscher and Slack, 2006; Hammond, 2006; Zhang et al, 2007). Therefore, we hypothesised that miRNAs with HSCC tumour-suppressive activity could be among the 31 downregulated miRNAs. In a screen for miRNAs that inhibited cancer cell proliferation, miR-489 inhibited cell growth in all cell lines examined (Figure 1), and was identified as a tumour-suppressive miRNA in HSCC. Although little is currently known regarding the function of miR-489, a recent report indicated that miR-489 may regulate early osteogenic differentiation in human mesenchymal stem cells, and that miR-489 has critical roles in osteogenesis (Schoolmeesters et al, 2009). However, the relationship between miR-489 and carcinogenesis remains unclear.
As miRNAs function by negatively regulating protein-coding genes, it is important to understand the miRNA-target gene network. Potential targets of miR-489 were observed in a genome-wide screen using FaDu (HSCC) cells. Of the 53 candidate genes, 32 contained miR-489 target sites, as predicted by the TargetScan database. More recently, we quickly and successfully screened miRNA target genes using microarray methods (Chiyomaru et al, 2010; Kano et al, 2010). Tumour-suppressive miRNAs usually prevent tumour development by inhibiting the activity of oncogenes (Esquela-Kerscher and Slack, 2006; Hammond, 2006; Zhang et al, 2007). Therefore, we expected that target genes of miR-489 would have oncogenic functions. One of the 53 candidates, PTPN11, is a cytoplasmic PTP that contains two Src Homology 2 domains. These PTPs are generally negative regulators because of their ability to oppose the effects of protein tyrosine kinases. Our data demonstrate that PTPN11 has an oncogenic role and is directly regulated by miR-489 in HSCC cells.
The PTPN11 gene is unusual in that it promotes the activation of RAS–MAPK signalling pathway in response to various growth factors and cytokines (Mohi and Neel, 2007; Matozaki et al, 2009). Interestingly, germline PTPN11 mutations have been identified in patients with Noonan syndrome, juvenile myelomonocytic leukaemia, and paediatric acute leukaemia (Aoki et al, 2008). Mutation of PTPN11 in Noonan syndrome and leukaemic cells resulted in gain-of-function enhanced phosphatase activity. Molecular and genetic studies have also shown that PTPN11 mediates cell signalling by epidermal growth factor (EGF), hepatocyte growth factor, and interleukin-6; specifically, PTPN11 has a role in the activation of ERK1/2 MAP kinase by EGF (Chen et al, 2006). The EGF signalling pathway is involved in a variety of cellular responses including cell growth and proliferation, and monoclonal antibodies and small-molecule inhibitors have been developed to inhibit EGF receptor (EGFR) pathways. These pathways, which include RAS–MAPK signalling, have been extensively studied in HNSCC, and seem to have a critical role in the survival and proliferation of cancer cells (Kalyankrishna and Grandis, 2006) and EGFR is overexpressed in more than 50% of HSCC specimens (Frank et al, 1993). Our data suggest that the silencing of miR-489 expression, and subsequent overexpression of PTPN11, leads to abnormal EGFR signalling. Future studies will clarify the mechanism by which deregulation of EGFR signalling networks contributes to HSCC carcinogenesis.
This study is to identify tumour-suppressive miRNAs based on clinical HSCC miRNA expression signature. We have specifically identified a tumour-suppressive miRNA (miR-489) and found its direct target (PTPN11). Disruption of this interaction may lead to the deregulation of miR-489-PTPN11 signalling in HSCC. The possibility of exploiting the therapeutic implications of these findings for future treatment of HSCC should be explored in future studies.
References
- Aoki Y, Niihori T, Narumi Y, Kure S, Matsubara Y (2008) The RAS/MAPK syndromes: novel roles of the RAS pathway in human genetic disorders. Hum Mutat 29: 992–1006 [DOI] [PubMed] [Google Scholar]
- Avissar M, Christensen BC, Kelsey KT, Marsit CJ (2009) MicroRNA expression ratio is predictive of head and neck squamous cell carcinoma. Clin Cancer Res 15: 2850–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Bova R, Goh R, Poulson M, Coman WB (2005) Total pharyngolaryngectomy for squamous cell carcinoma of hypopharynx: a review. Laryngoscope 115: 864–869 [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6: 857–866 [DOI] [PubMed] [Google Scholar]
- Chang SS, Jiang WW, Smith I, Poeta LM, Begum S, Glazer C, Shan S, Westra W, Sidransky D, Califano JA (2008) MicroRNA alterations in head and neck squamous cell carcinoma. Int J Cancer 123: 2791–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HC, Chen GH, Chen YH, Liao WL, Liu CY, Chang KP, Chang YS, Chen SJ (2009) MicroRNA deregulation and pathway alterations in nasopharyngeal carcinoma. Br J Cancer 100: 1002–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Sung SS, Yip ML, Lawrence HR, Ren Y, Guida WC, Sebit SM, Lawrence NJ, Wu J (2006) Discovery of a novel Shp2 protein tyrosine phosphatase inhibitor. Mol Pharmacol 70: 562–570 [DOI] [PubMed] [Google Scholar]
- Childs G, Fazzari M, Kung G, Kawachi N, Brandwein-Gensler M, McLemore M, Chen Q, Burk RD, Smith RV, Prystowsky MB, Belbin TJ, Schlecht NF (2009) Low-level expression of microRNAs let-7d and miR-205 are prognostic markers of head and neck squamous cell carcinoma. Am J Pathol 174: 736–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiyomaru T, Enokida H, Tatarano S, Kawahara K, Uchida Y, Nishiyama K, Fujimura L, Kikkawa N, Seki N, Nakagawa M (2010) miR-145 and miR-133a function as tumor suppressors and directly regulate FSCN1 expression in bladder cancer. Br J Cancer 102: 883–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies L, Welch HG (2006) Epidemiology of head and neck cancer in the United States. Otolaryngol Head Neck Surg 135: 451–457 [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ (2006) Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer 6: 259–269 [DOI] [PubMed] [Google Scholar]
- Frank JL, Garb JL, Banson BB, Peterman J, Neifeld JP, Kay S, Kornstein MJ, Sismanis A, Ware JL (1993) Epidermal growth factor receptor expression in squamous cell carcinoma of the hypopharynx. Surg Oncol 2: 161–167 [DOI] [PubMed] [Google Scholar]
- Godballe C, Jorgensen K, Hansen O, Bastholt L (2002) Hypopharyngeal cancer: results of treatment based on radiation therapy and salvage surgery. Laryngoscope 112: 834–838 [DOI] [PubMed] [Google Scholar]
- Hammond SM (2006) MicroRNAs as oncogenes. Curr Opin Genet Dev 16: 4–9 [DOI] [PubMed] [Google Scholar]
- Helliwell TR (2003) acp Best Practice No. 169. Evidence based pathology: squamous carcinoma of the hypopharynx. J Clin Pathol 56: 81–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson BJ, Bhattacharjee S, O'Dee DM, Feingold E, Gollin SM (2009) Decreased expression of miR-125b and miR-100 in oral cancer cells contributes to malignancy. Genes Chromosomes Cancer 48: 569–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman HT, Karnell LH, Shah JP, Ariyan S, Brown GS, Fee WE, Glass AG, Goepfert H, Ossoff RH, Fremgen AM (1997) Hypopharyngeal cancer patient care evaluation. Laryngoscope 107: 1005–1017 [DOI] [PubMed] [Google Scholar]
- Hui AB, Lenarduzzi M, Krushel T, Waldron L, Pintilie M, Shi W, Perez-Ordonez B, Jurisica I, O'Sullivan B, Waldron J, Gullane P, Cummings B, Liu FF (2010) Comprehensive microRNA profiling for head and neck squamous cell carcinomas. Clin Cancer Res 16: 1129–1139 [DOI] [PubMed] [Google Scholar]
- Hwang HW, Mendell JT (2006) MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer 94: 776–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimi T, Enokida H, Okuno Y, Kunimoto R, Chiyomayu T, Kawamoto K, Kawahara K, Toki K, Kawakami K, Nishiyama K, Tsujimoto G, Nakagawa M, Seki N (2009) Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer 125: 345–352 [DOI] [PubMed] [Google Scholar]
- Kano M, Seki N, Kikkawa N, Fujimura L, Hoshino I, Akutsu Y, Chiyomaru T, Enokida H, Nakagawa M, Matsubara H (2010) miR-145, miR-133a and miR-133b: tumor suppressive miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int J Cancer. E-pub ahead of print 2 March 2010 [DOI] [PubMed]
- Kalyankrishna S, Grandis JR (2006) Epidermal growth factor receptor biology in head and neck cancer. J Clin Oncol 24: 2666–2672 [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Plasterk RH (2006) The diverse functions of microRNAs in animal development and disease. Dev Cell 11: 441–450 [DOI] [PubMed] [Google Scholar]
- Liu X, Chen Z, Yu J, Xia J, Zhou X (2009) MicroRNA profiling and head and neck cancer. Comp Funct Genomics 837514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR (2005) MicroRNA expression profiles classify human cancers. Nature 435: 834–838 [DOI] [PubMed] [Google Scholar]
- Matozaki T, Murata Y, Saito Y, Okazawa H, Ohnishi H (2009) Protein tyrosine phosphatase SHP-2: a proto-oncogene product that promotes Ras activation. Cancer Sci 100: 1786–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohi MG, Neel BG (2007) The role of Shp2 (PTPN11) in cancer. Curr Opin Genet Dev 17: 23–30 [DOI] [PubMed] [Google Scholar]
- Momose F, Araida T, Negishi A, Ichijo H, Shinoda S, Sasaki S (1989) Variant sublines with different metastatic potentials selected in nude mice from human oral squamous cell carcinomas. J Oral Pathol Med 18: 391–395 [DOI] [PubMed] [Google Scholar]
- Peterson Jr WD, Stulberg CS, Simpson WF (1971) A permanent heteroploid human cell line with type B glucose-6-phosphate dehydrogenase. Proc Soc Exp Biol Med 136: 1187–1191 [DOI] [PubMed] [Google Scholar]
- Ramdas L, Giri U, Ashorn CL, Coombes KR, El-Nagger A, Ang KK, Story MD (2009) miRNA expression profiles in head and neck squamous cell carcinoma and adjacent normal tissue. Head Neck 31: 642–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan SR (1972) A new human cell line (FaDu) from a hypopharyngeal carcinoma. Cancer 29: 117–121 [DOI] [PubMed] [Google Scholar]
- Schoolmeesters A, Eklund T, Leake D, Vermeulen A, Smith Q, Force Aldred S, Fedorov Y (2009) Functional profiling reveals critical role for miRNA in differentiation of human mesenchymal stem cells. PloS One 4: e5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector JG, Sessions DG, Haughey BH, Chao KS, Simpson J, EL Mofty S, Perez CA (2001) Delayed regional metastases, distant metastases, and second primary malignancies in squamous cell carcinomas of the larynx and hypopharynx. Laryngoscope 111: 1079–1087 [DOI] [PubMed] [Google Scholar]
- Sugimoto T, Seki N, Shimizu S, Kikkawa N, Tsukada J, Shimada H, Sasaki K, Hanazawa T, Okamoto Y, Hata A (2009) The galanin signaling cascade is a candidate pathway regulating oncogenesis in human squamous cell carcinoma. Genes Chromosomes Cancer 48: 132–142 [DOI] [PubMed] [Google Scholar]
- Tonks NK (2006) Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol 7: 833–846 [DOI] [PubMed] [Google Scholar]
- Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI (2008) Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin Cancer Res 14: 2588–2592 [DOI] [PubMed] [Google Scholar]
- Zhang B, Pan X, Cobb GP, Anderson TA (2007) microRNAs as oncogenes and tumor suppressors. Dev Biol 302: 1–12 [DOI] [PubMed] [Google Scholar]