Abstract
Ligase-mediated gene detection has proven valuable for detection and precise distinction of DNA sequence variants. We have recently shown that T4 DNA ligase can also be used to distinguish single nucleotide variants of RNA sequences. Here we describe parameters that influence RNA-templated DNA ligation by T4 DNA ligase. The reaction proceeds much more slowly, requiring more enzyme, compared to ligation of the same oligonucleotides hybridized to the corresponding DNA sequence. The reaction is inhibited at high concentrations of ATP and NaCl and both magnesium and manganese ions can support the reaction. We define reaction conditions where 80% of RNA target molecules can template a diagnostic ligation reaction. Ligase-mediated RNA detection should provide a useful mechanism for sensitive and accurate detection and distinction of RNA sequence variants.
INTRODUCTION
Specific RNA sequences are measured to gauge the level of gene expression, either averaged over a tissue sample or according to the distribution of transcripts in tissue sections or within individual cells. Related RNA sequences can be distinguished by taking advantage of the reduced hybridization stability of imperfectly matched hybridization probes. This can be problematical, however, when many sequences are investigated under one set of hybridization conditions and when closely similar variants must be resolved. This is a significant, perhaps underrated, problem in expression profiling, since many genes are members of gene families and are expressed at widely different levels. Improved sequence distinction is also required for in situ analyses of related genes or allelic variants of single genes.
DNA ligases can be used to distinguish single nucleotide variation among DNA sequences by taking advantage of the inefficient ligation of terminally mismatched oligonucleotides. This mechanism has been used for gene detection in the oligonucleotide ligation assay (1) and the ligase chain reaction (2). Early studies of the enzyme T4 DNA ligase showed that the enzyme could join DNA oligonucleotides hybridized to RNA templates, albeit with a substantially lower efficiency compared to DNA-templated reactions (3,4). More recently, RNA-templated ligation of DNA and RNA probes has been used to generate molecules that can be amplified by PCR (5–7) and the Qβ replicase, respectively (8). However, the mechanism of RNA-templated DNA ligation has not been carefully studied, as required to define optimal reaction conditions. The reaction mechanism of the T4 DNA ligase-catalyzed sealing of nicked DNA substrates is known in some detail (9–11). The enzyme is first activated through ATP hydrolysis, resulting in covalent addition of an AMP group to the enzyme. After binding to a nicked site in a DNA duplex, the ligase transfers this AMP to the phosphorylated 5′-end at the nick, forming a 5′–5′ pyrophosphate bond. Finally, the ligase catalyzes an attack on this pyrophosphate bond by the OH group at the 3′-end of the nick, thereby sealing it, whereafter ligase and AMP are released. If the ligase detaches from the substrate before the 3′ attack, e.g. because of premature AMP reloading of the enzyme, then the 5′ AMP is left at the 5′-end, blocking further ligation attempts.
Here we investigate the conditions for efficient T4 DNA ligase-mediated joining of oligonucleotides hybridizing to in vitro transcribed RNA target molecules. Both the 5′-adenylation and subsequent joining step of the ligation reaction proceed considerably more slowly than on the corresponding DNA targets, and the overall reaction is inhibited by elevated concentrations of NaCl and ATP. However, under the appropriate conditions RNA targets are efficient templates for probe ligation.
MATERIALS AND METHODS
Oligonucleotides and transcription templates
Ligation probes were synthesized on an ABI 374 oligonucleotide synthesizer and purified by reversed phase chromatography (RP-18; Pharmacia Biotech, Uppsala, Sweden) before and after detritylation. The 5′ probes, 5′-TGACCGCTGAAGGGCTX-3′, had additions of 12, 8, 4 or no T residues at the 5′-end of the sequence, size-coding for G, C, A or T as the 3′-most residue, respectively. The variable 3′ ultimate position in the probes is identified by an X in the sequence above and in Figure 1. The 3′ ligation probe, 5′-TTGAACTCTGCTTAAATCCAGTGGCT-3′, was chemically 5′-phosphorylated (12). This oligonucleotide also contained a C residue modified with a primary amine (13), shown in bold in the sequence. A Cy5 fluorophore was conjugated to the primary amine using a Cy5-N-hydroxysuccinimide ester (Pharmacia Biotech). Oligonucleotides used as PCR primers or as templates for DNA ligation were purchased from Interactiva (Ulm, Germany). Templates for in vitro transcription and purification of transcribed RNA were prepared as described (14).
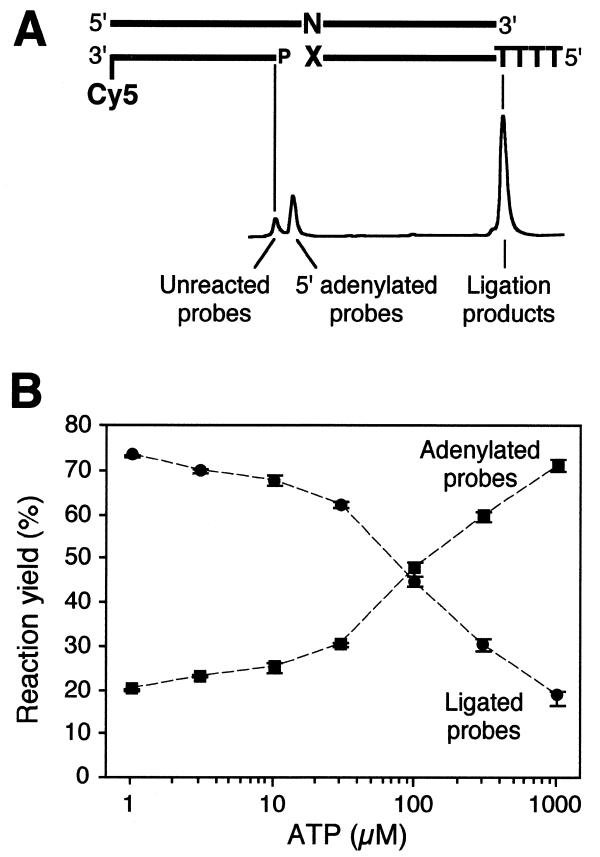
Figure 1.
RNA-templated probe ligation. (A) Schematic illustration of the experimental set-up. N denotes the variable position in the RNA target, whereas X is the corresponding variable 3′ position in the 5′ ligation probe. Different numbers of T residues were added at the 5′-end of these probe variants in order to distinguish the four different ligation probes according to size upon gel electrophoresis. Below the ligation substrate the three classes of reaction products recorded by a fluorescence sequencer are identified. (B) ATP dependence of probe adenylation and ligation reactions in RNA-templated DNA ligation. Averages and ranges of adenylated and ligated products (squares and circles, respectively) from triplicate reactions are shown after a 60 min reaction with a ligation probe having a A at the 3′-most position and an RNA target with an U at the variable site of the DNA probe.
Ligation reactions on RNA templates
Forty nanomolar ligation substrates were formed in 10 mM TrisOAc pH 7.5, 10 mM MgOAc2, 10 µM ATP, by combining the 3′ oligonucleotide labeled with Cy5, the in vitro transcribed RNA target and the 5′ oligonucleotides at a molar ratio of 1:2:4. The ligation mixes were incubated at 65°C for 3 min, slowly cooled to room temperature and then placed on ice, whereafter T4 DNA ligase (Amersham Pharmacia Biotech, Uppsala, Sweden) was added to the reactions to a final concentration of 0.5 U/µl. The reactions were incubated at 37°C for up to 2 h. The magnesium/manganese experiment was performed as above but in 10 mM Tris–HCl pH 7.5, with 10 mM MgCl2 or MnCl2. Chloride anions were used in this experiment because MnOAc solution was not stable and failed to support efficient RNA-templated DNA ligation. No difference in efficiency was observed between RNA-templated DNA ligation reactions in buffers containing MgCl2 or MgOAc. Ligation reactions were terminated by adding an equal volume of stop buffer (95% formamide, 25 mM EDTA and dextran blue) or, in time-course reactions, 5 µl aliquots of the reactions were added to 5 µl of stop buffer. One microliter of 1 M NaOH was added to the terminated reactions, followed by 15 min incubation at 65°C to degrade the RNA target, in order to avoid renaturation of probes and targets which could result in extra bands during electrophoresis. Products of the ligation reactions were analyzed using an ALF Express DNA sequencer (Amersham Pharmacia Biotech) with 10% polyacrylamide, 7 M urea gels in 0.6× TBE buffer. Fluorescent products could be measured over a 1000-fold concentration range. The recorded fluorescence was processed using software developed for this instrument (AlleleLinks). All experiments have been reproduced at least once in order to verify the results.
RESULTS AND DISCUSSION
Experimental set-up
We monitored the course of events in ligation reactions, where a 3′-labeled oligonucleotide was ligated to an unlabeled oligonucleotide hybridizing immediately 5′ to it on a RNA target sequence (see Fig. 1A), by removing samples from the reactions at several time points. The RNA target sequences were produced by in vitro transcription. To ensure that all labeled probe oligonucleotides participated in ligation substrates, the template strands and the unlabeled size-coded 5′ probe oligonucleotides were added in 2- and 4-fold molar excess, respectively, over the fluorescence-labeled probe oligonucleotides. In experiments where the ability to discriminate single nucleotide variations were assessed, the ligation rates of size-coded 5′-oligonucleotides with terminally mismatched 3′-ends were measured in the presence of an equimolar amount of the perfectly matched oligonucleotides. The products of the two reactions could be distinguished because of different length additions to the 5′-ends of the mismatched or perfectly matched oligonucleotides. Ligation reaction products were analyzed by gel electrophoresis in a fluorescence sequencing instrument, to identify and quantify the different reaction products. One of the observed reaction products is the 5′-adenylated form of the labeled probe oligonucleotide, which is formed as an intermediary product during the ligation reaction.
Reaction mechanism and kinetics
Rossi et al. proposed a model for ligation reactions that involves two different DNA-binding complexes (11). According to their model, the adenylated enzyme scans the duplex for substrates and binds at nicks as a transient complex. Once the AMP has been transferred from the enzyme to the 5′-phosphate of the substrate a stable complex is formed which facilitates the joining reaction. The model predicts that the joining of ‘difficult’ substrates, e.g. blunt end ligation, may be inhibited by premature AMP reloading of the ligase, resulting in dissociation of the enzyme from the substrate after the 5′-adenylation step (11). Due to the slow kinetics of the DNA joining reaction on RNA targets, the reaction could be expected from Rossi’s model to be inhibited by ATP concentrations exceeding the Km for ATP binding (14 µM) (11). An ATP titration experiment in RNA-templated ligation reactions indeed supports this model, since the yield of adenylated end products increased with increasing ATP concentration, while less ligated end products were obtained (Fig. 1B). This was the case for all four RNA sequence variants studied below (data not shown). At ATP concentrations above the Km of ATP binding adenylation predominates over completed ligation reactions. DNA-templated DNA ligation is usually performed in 1 mM ATP, well above the Km. This ATP concentration is clearly sub-optimal for RNA-templated DNA ligation reactions.
DNA ligation on DNA substrates follows first order kinetics and can be divided into a series of discrete reaction steps (9–11). Under conditions where the enzyme is turned over and the ATP concentration is above the Km for ATP binding, substrate binding is the rate limiting reaction step (11). In contrast, DNA probe ligation reactions on RNA targets in the presence of a molar excess of ligase follow quite different kinetics (Fig. 2). The accumulation of probes, processed by T4 DNA ligase to form either adenylated or ligated reaction products, follows first order kinetics, but completed ligation alone does not. The accumulation of 5′-adenylated reaction intermediates suggests that the subsequent joining step is rate limiting. Moreover, the ligation rates differ substantially between the four templates used in this study.
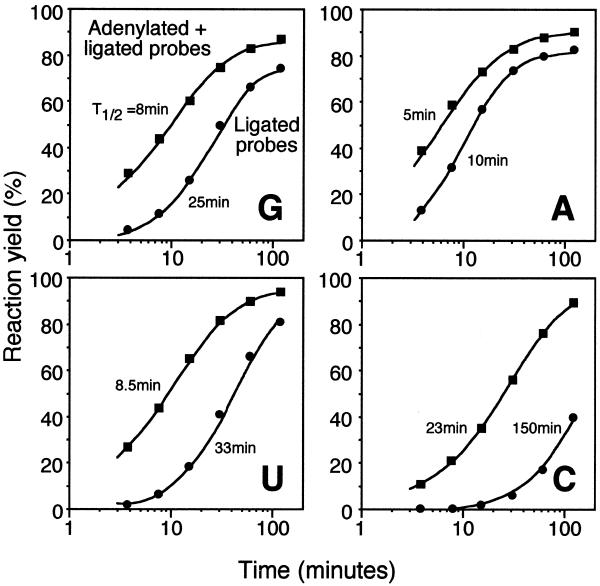
Figure 2.
Time-course of ligation of DNA probes, correctly base paired to four RNA targets, that differed in a single nucleotide position. Aliquots of the four different reactions, each including one of the four RNA targets and a matched probe pair, were withdrawn at different time points. The RNA target used in each experiment is identified by a capital letter in the lower right corner of the diagram. Squares represent formation of adenylated reaction intermediates deduced from the sum of adenylated and ligated probes, while circles represent ligated probes. The curves were obtained by iteratively fitting the functions Rad = Rmax × [1 – e–kad × t/(1 + 0.17 × kad × t)] and Rlig = Rad × Rmax × (1 – e–klig × t) to the data points in the adenylation and ligation time series, respectively. R is the reaction yield at time point t and k is the apparent initial rate of the reaction. The t1/2 estimates of the reactions in minutes are shown in the figure and were determined at the time points when R = Rmax/2.
The initial rate (≈Vmax) of DNA ligation on DNA targets using T4 DNA ligase has been estimated at 5 turnovers/s (15). A ligase titration experiment with DNA probes on an RNA template suggests that the enzyme is not turned over in the reaction, since it reaches saturation only if an excess of enzyme over substrate is added (data not shown). It is therefore not meaningful to measure the DNA ligation rate on RNA in turnover numbers. Instead, the time required to process half the substrates (t1/2) can be used as a measure of the reaction rate. Under our conditions the t1/2 for the complete ligation reaction ranged from 10 to 150 min for the four different targets, indicating that the ligation reaction was at least several thousand-fold less efficient on RNA targets compared to DNA targets.
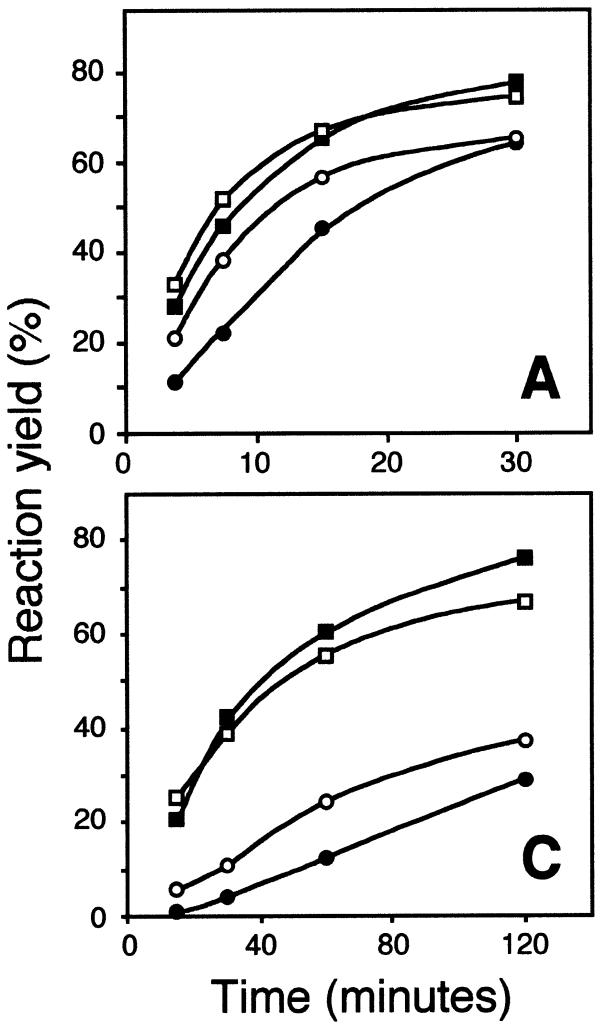
The abilities of magnesium and manganese ions to support the RNA-templated ligation reaction were compared (Fig. 3), both ions having previously been shown to support the reaction (16). In this experiment the two target RNA sequences were used that resulted in the fastest (the A target) and slowest (the C target) reaction kinetics in Tris–HCl buffer, in order to assess whether there is a sequence dependency with regard to the ability of divalent cations to support ligation. Adenylation proceeded at a similar rate in the presence of either of the two divalent cations. However, the joining reaction was about twice as fast for both target sequences in the presence of 10 mM MnCl2 compared to 10 mM MgCl2. Manganese ions therefore seem to be more efficient than magnesium ions as cofactors for the joining reaction, while the two ions are equally efficient in the 5′-adenylation reaction.
Figure 3.
Time–course of ligation of correctly matched DNA probes on RNA targets having A or C in the diagnostic position, respectively, in the presence of 10 mM MgCl2 (filled symbols) or MnCl2 (open symbols). The RNA targets are identified by capital letters in the lower right corners of the diagrams. Squares represent probes that were either adenylated or ligated and circles denote probes that had been ligated.
Mismatch inhibition of DNA ligation on RNA targets
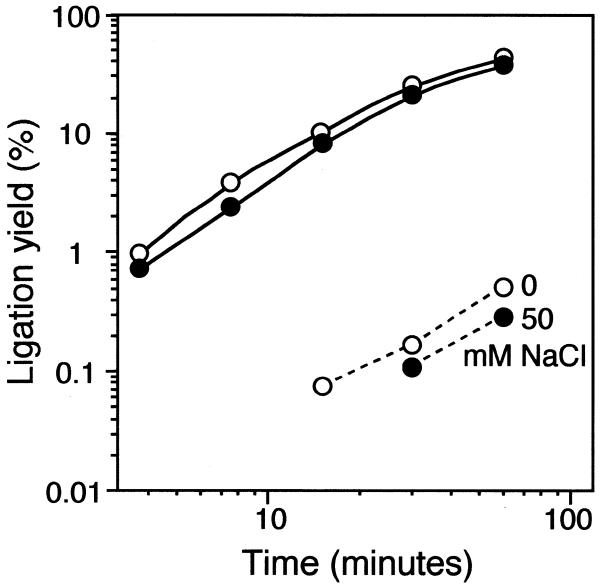
It is well established that the ability of T4 DNA ligase to discriminate mismatches in DNA substrates is increased at elevated concentrations of NaCl (1,17). We investigated the effect of NaCl concentration on ligation efficiency and distinction of RNA target sequence variants, by probing U target sequences with a mix of ligation probes having a 3′ A or G, at four NaCl concentrations (Fig. 4). After a 30 min ligation reaction at 0 mM NaCl the G-U mismatch was discriminated from the corresponding matched probe and target pair by a factor of 150. Ligase fidelity was only modestly increased by addition of 50 mM NaCl, while the ligation rate was somewhat reduced. Addition of 150 or 250 mM NaCl completely inhibited both the 5′-adenylation and joining reactions (data not shown). Similar results were obtained when the G target sequence was probed with the same mix of ligation probes (data not shown). Addition of NaCl therefore seems to be of little value to enhance sequence discrimination of RNA sequence variants.
Figure 4.
Time–course of ligation reactions of probes with a 3′-most A or G residue on an RNA target with U at the diagnostic position, at 0 and 50 mM NaCl concentrations (open and filled symbols). Solid and broken lines denote matched and mismatched reactions, respectively.
CONCLUSION
Our results demonstrate that RNA target molecules can be efficiently detected via ligation of oligonucleotides by T4 DNA ligase, provided that the concentrations of both NaCl and ATP are kept low, a molar excess of ligase over substrate is used and the reaction is given sufficient time. In most biological samples the concentration of any specific RNA sequence is low enough that sufficient T4 DNA ligase can be added to detection reactions. A potentially greater problem is manifested in the considerable differences in ligation kinetics among the four closely similar RNA target sequences used in this study. The reason for this difference is not known. Nonetheless, we have recently shown that all mismatched RNA targets can be clearly distinguished from the corresponding matched ones under the reaction conditions reported here (14).
Direct analysis of RNA sequences without a preceding cDNA synthesis step may more faithfully report the relative abundance of specific mRNAs in cellular extracts. Moreover, ligase-assisted probe ligation could be used to more accurately distinguish members of gene families and to identify splice variants, compared to traditional hybridization-based analyses. By using ligase-mediated probe circularization (padlock probes) (18), reacted probes can be replicated through rolling circle replication (19–23) or by PCR (16), enabling quantitative and sensitive detection of RNA sequence variants in solution, on arrays or in situ. This strategy should be particularly well suited for highly parallel quantitation of RNA expression, because under the appropriate conditions only intramolecular probe circularization events generate an amplification product, allowing large probe sets to be applied with minimal risk of cross-reactions between probes. RNA-templated probe ligation should thus be of value for detection and quantitation of numerous closely related transcripts over a wide dynamic range and from small tissue samples.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by grants from the Beijer Foundation, the Swedish Medical and Technological Research Councils and the Swedish Cancer Foundation. M.N. is supported by a long-term EMBO fellowship.
References
- 1.Landegren U., Kaiser,R., Sanders,J. and Hood,L. (1988) A ligase-mediated gene detection technique. Science, 241, 1077–1080. [DOI] [PubMed] [Google Scholar]
- 2.Barany F. (1991) Genetic disease detection and DNA amplification using cloned thermostabile ligase. Proc. Natl Acad. Sci. USA, 88, 189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kleppe K., van de Sande,J.H. and Khorana,H.G. (1970) Polynucleotide ligase-catalyzed joining of deoxyribo-oligonucleotides on ribopolynucleotide templates and of ribo-oligonucleotides on deoxyribopolynucleotide templates. Proc. Natl Acad. Sci. USA, 67, 68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fareed G.C., Wilt,E.M. and Richardson,C.C. (1971) Enzymatic breakage and joining of deoxyribonucleic acid. VIII. Hybrids of ribo- and deoxyribonucleotide homopolymers as substrates for polynucleotide ligase of bacteriophage T4. J. Biol. Chem., 246, 925–932. [PubMed] [Google Scholar]
- 5.Hsuih T.C.H., Park,Y.N., Zaretsky,C., Wu,F., Tyagi,S., Kramer,F.R., Sperling,R. and Zhang,D.Y. (1996) Novel, ligation-dependent PCR assay for detection of hepatitis C virus in serum. J. Clin. Microbiol., 34, 501–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park Y.N., Abe,K., Li,H., Hsuih,T., Thung,S.N. and Zhang,D.Y. (1996) Detection of hepatitis C virus RNA using ligation-dependent polymerase chain reaction in formalin-fixed, paraffin-embedded liver tissues. Am. J. Pathol., 149, 1485–1491. [PMC free article] [PubMed] [Google Scholar]
- 7.Miyauchi I., Moriyama,M., Zhang,D.Y. and Abe,K. (1998) Further study of hepatitis C virus RNA detection in formalin-fixed, paraffin-embedded liver tissues by ligation-dependent polymerase chain reaction. Pathol. Int., 48, 428–432. [DOI] [PubMed] [Google Scholar]
- 8.Tyagi S., Landegren,U., Tazi,M., Lizardi,P.M. and Kramer,F.R. (1996) Extremely sensitive, background-free gene detection using binary probes and Qb replicase. Proc. Natl Acad. Sci. USA, 93, 5395–5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higgins N.P. and Cozzarelli,N.R. (1979) DNA-joining enzymes: a review. Methods Enzymol., 68, 50–71. [DOI] [PubMed] [Google Scholar]
- 10.Engler M.J. and Richardson,C.C. (1982) DNA ligases. The Enzymes, XV, 3–29. [Google Scholar]
- 11.Rossi R., Montecucco,A., Ciarrocchi,G. and Biamonti,G. (1997) Functional characterization of the T4 DNA ligase: a new insight into the mechanism of action. Nucleic Acids Res., 25, 2106–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Connolly B.A. (1987) Solid phase 5′-phosphorylation of oligonucleotides. Tetrahedron Lett., 28, 463–466. [Google Scholar]
- 13.Sund C., Ylikoski,J., Hurskainen,P. and Kwiatkowski,M. (1988) Construction of europium (Eu3+)-labelled oligo DNA hybridization probes. Nucl. Nucl., 7, 655–659. [Google Scholar]
- 14.Nilsson M., Barbany,G., Antson,D., Gertow,K. and Landegren,U. (2000) Enhanced detection and distinction of RNA by enzymatic probe ligation. Nature Biotechnol., 18, 791–793. [DOI] [PubMed] [Google Scholar]
- 15.Tong J., Cao,W. and Barany,F. (1999) Biochemical properties of a high fidelity DNA ligase from Thermus species AK16D. Nucleic Acids Res., 27, 788–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang D.Y., Brandwein,M., Hsuih,T.C.H. and Li,H. (1998) Amplification of target-specific, ligation dependent circular probe. Gene, 211, 277–285. [DOI] [PubMed] [Google Scholar]
- 17.Wu D.Y. and Wallace,R.B. (1989) Specificity of the nick-closing activity of bacteriophage T4 DNA ligase. Gene, 76, 245–254. [DOI] [PubMed] [Google Scholar]
- 18.Nilsson M., Malmgren,H., Samiotaki,M., Kwiatkowski,M., Chowdhary,B.P. and Landegren,U. (1994) Padlock probes: circularizing oligonucleotides for localized DNA detection. Science, 265, 2085–2088. [DOI] [PubMed] [Google Scholar]
- 19.Fire A. and Xu,S.-Q. (1995) Rolling replication of short DNA circles. Proc. Natl Acad. Sci. USA, 92, 4641–4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu D., Daubendiek,S.L., Zillman,M.A., Ryan,K. and Kool,E.T. (1996) Rolling circle DNA synthesis: small circular oligonucleotides as efficient templates for DNA polymerases. J. Am. Chem. Soc., 118, 1587–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banér J., Nilsson,M., Mendel-Hartvig,M. and Landegren,U. (1998) Signal amplification of padlock probes by rolling circle replication. Nucleic Acids Res., 22, 5073–5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lizardi P.M., Huang,X., Zhu,Z., Bray-Ward,P., Thomas,D.C. and Ward,D.C. (1998) Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nature Genet., 19, 225–232. [DOI] [PubMed] [Google Scholar]
- 23.Thomas D.C., Nardone,G.A. and Randall,S.K. (1999) Amplification of padlock probes for DNA diagnostics by cascade rolling circle amplification or the polymerase chain reaction. Arch. Pathol. Lab. Med., 123, 1170–1176. [DOI] [PubMed] [Google Scholar]