Abstract
DM catalyses class II-associated invariant chain peptide (CLIP) release, edits the repertoire of peptides bound to major histocompatibility complex (MHC) class II molecules, affects class II structure, and thereby modulates binding of conformation-sensitive anti-class II antibodies. Here, we investigate the ability of DM to enhance the cell surface binding of monomorphic antibodies. We show that this enhancement reflects increases in cell surface class II expression and total cellular abundance, but notably these effects are selective for particular alleles. Evidence from analysis of cellular class II levels after cycloheximide treatment and from pulse-chase experiments indicates that DM increases the half-life of affected alleles. Unexpectedly, the pulse-chase experiments also revealed an early effect of DM on assembly of these alleles. The allelically variant feature that correlates with susceptibility to these DM effects is low affinity for CLIP; DM-dependent changes in abundance are reduced by invariant chain (CLIP) mutants that enhance CLIP binding to class II. We found evidence that DM mediates rescue of peptide-receptive DR0404 molecules from inactive forms in vitro and evidence suggesting that a similar process occurs in cells. Thus, multiple mechanisms, operating along the biosynthetic pathway of class II molecules, contribute to DM-mediated increases in the abundance of low-CLIP-affinity alleles.
Keywords: antigen presentation/processing, class II-associated invariant chain peptides (CLIP), human leucocyte antigen (HLA)-DM, major histocompatibility complex (MHC) class II
Introduction
DM [human leucocyte antigen (HLA)-DM in humans or H-2M in mice] is known to play several critical roles in the major histocompatibility complex (MHC) class II antigen presentation pathway.1 In this pathway, MHC class II α and β chains are synthesized in the endoplasmic reticulum (ER) and assembled onto invariant chain (Ii) trimers. Nonameric class II/Ii complexes are transported to endosomes, where Ii is degraded. The terminal Ii fragments, a nested set of peptides termed class II-associated Ii peptides (CLIP), occupy the peptide-binding groove of MHC class II molecules and are replaced during antigen presentation. For most alleles, DM is required for efficient removal of CLIP.2,3 DM also serves to edit peptide/class II complexes in favour of those with increased stability.4,5In vitro, DM specifically recognizes and binds to unstable conformations of peptide/MHC complexes6 and to empty MHC class II proteins7–9 and preserves their peptide-binding capacity.8,9 DM also has been shown to extinguish expression of certain class II/peptide conformers.10 Investigation of the molecular mechanisms of the DM peptide exchange function has provided evidence for a ‘hit-and-run’ mechanism11 and a ‘compare-exchange’ mechanism.12 In the ‘hit-and-run’ mechanism, DM interacts transiently and repetitively with MHC class II to induce conformational changes that lead to peptide dissociation. In the ‘compare-exchange’ mechanism, an unbound peptide makes a transient tetramolecular complex with MHC class II, bound peptide and DM; a peptide with better fit replaces the pre-bound peptide via a DM-facilitated process. There is also evidence for a direct role of DM in peptide association with empty DR molecules.9,13
Prior work demonstrated that DM promotes binding of certain class II allele-specific antibodies, such as monoclonal antibody (mAb) 16.23.14,15 Where investigated, these DM-dependent antibodies have been found, not surprisingly, to recognize the peptide-binding domain of class II.16 The DM dependence of such antibodies may derive from the requirement for particular peptides, for example mAb YAe,17,18 or particular conformations, for example mAb 16.23.19
For certain alleles, we noted that DM also augments the binding of class II isotype-specific (monomorphic) antibodies, suggesting an increase in class II abundance. DM has been found to enhance surface class II levels in some previous (mostly murine) reports,20–24 but the DM-mediated cellular effects that contribute to this enhancement have not been fully elucidated. In addition, common features of alleles that are susceptible to this effect have not been highlighted in prior studies. Here, we address these aspects of the chaperoning activity of DM.
Materials and methods
Cell lines and antibodies
The B lymphoblastoid cell lines (B-LCLs) 2.2.93,25 9.5.3,14 8.1.614 and 5.2.426 were used (see Table 1). DM transfectants of 2.2.93 and 9.5.3 (2.2.93-DM and 9.5.3-DM, respectively) have been described previously.28 DM-, DO-, and DM/DO-transfectants of 5.2.4 were generated using retroviral infection as previously described.29 cDNA for a DO-GFP fusion protein (a gift from S. M. van Ham, Netherlands Cancer Institute, Amsterdam, the Netherlands)30 was cloned into the pBMN vector and used for transfection of 5.2.4. DR transfectants of 5.2.4 and 5.2.4-DM have been described.29 Using the same retroviral transfection system, the H-2M-null mouse B cell lines 3A531 and 3A5.g7 (C. H. Rinderknecht, unpublished data) were stably transfected with 6xHis-tagged or 3xFLAG-tagged wild-type (wt) or high-CLIP-affinity mutant Ii (32 and C. H. Rinderknecht, unpublished data). The cDNA for murine DMα was obtained by RNA extraction and reverse transcription–polymerase chain reaction (RT-PCR) from A20 cells,33 the DM-expressing parent cell line of 3A5, and was also cloned into a pBMN vector. All B-cell transfectants made in our laboratory for this study were polyclonal populations, excluding the possibility of selection of atypical clonal variants. T2 cells ± DR0401 ± DM (T2 cells with or without transfected DR0401 and with or without transfected DM) have been previously described,34,35 and the cells were provided by P. A. Roche [National Cancer Institute, National Institutes of Health (NIH), Bethesda, MD]. Schneider-2 (S2) Drosophila melanogaster transfectants expressing soluble DM and DR molecules have been described.3,36,37
Table 1.
Expression of class II molecules in B-cell lines
| 2.2.93 | 9.5.3 | 8.1.6 | 5.2.4 | T2-DR4 | 3A5 | 3A5.g7 | |
|---|---|---|---|---|---|---|---|
| Class II | DR1, DR3, DQ1, DP4 | DR3, DQ2, DP4 | DR3, DQ2, DP4 | DP4 | DR0401 | I-Ed, I-Ad | I-Ed, I-Ad, I-Ag7 |
| DM1 | DMB only | DMA only | DMA, DMB | DMA only | – | DMB only | DMB only |
| DO2 | DOA, DOB | DOA, DOB | DOA, DOB | DOA only | – | DOA, DOB | DOA, DOB |
Mature DM protein requires both DMA (alpha) and DMB (beta) expression. For cell lines lacking DM protein, the DM genes that are intact are shown in italics.
DO protein cannot exit the endoplasmic reticulum without interaction with DM.27 The DO genes that are intact in cell lines lacking DM protein are shown in italics.
Antibodies used in this study were L243 [anti-DR, immunoglobulin G2a (IgG2a)],38 B7/21.2 (anti-DP, IgG3),39 SPVL-3 (anti-DQ, IgG2a),40 DA6.231 (anti-DR and DP β, IgG1),41 DA6.147 (anti-DR α, IgG1),42 IA3 (anti-DQ, IgG2a; Biodesign, Saco, ME), XD5.a11 (anti-class II β chain, IgG1),43 anti-HLA-DR (clone TÜ36, IgG2b; CALTAG/Invitrogen, Carlsbad, CA), anti-human-CD19 (clone HIB19, IgG1; BD Pharmingen, San Jose, CA), W6/32 (anti-HLA class I, IgG2a),44,45 anti-β-actin (IgG1; Sigma, St Louis, MO), ISCR3 (anti-DR, IgG2b),46 5C1 (anti-DM α, IgG1),47 DOB.L1 (anti-DO β, IgG2b; BD Pharmingen), CHAMP (anti-DR rabbit serum),48 MEM-264 (anti-empty DR, IgG2b)48 (CHAMP and MEM-264 provided by L. Stern, University of Massachusetts, Worcester, MA), K455 (rabbit anti-serum to denatured HLA class I, provided by L. Karlsson, R.W. Johnson Pharmaceutical Research Institute, La Jolla, CA),49 AF8 (anti-human-calnexin, IgG1; ascites provided by M. Brenner, Brigham & Women's Hospital, Boston, MA),50 Pin1.1 (anti-human-Ii, IgG1; ascites provided by P. Cresswell, Yale University School of Medicine, New Haven, CT),51 14-4-4S (anti-I-Eα, IgG2a; Southern Biotech, Birmingham, AL),52 rabbit-anti-I-Edα cytoplasmic tail antiserum (provided by R. N. Germain, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, MD),53 OX-6 (anti-rat RT1B mAb that cross-reacts with I-Ag7, IgG1; Serotec, Oxford, UK) and anti-H2-M (rat IgG1; BD Pharmingen).
Flow cytometry
Cells were stained on ice with directly fluorophore-conjugated antibodies, or, for indirect staining, with unlabelled primary antibodies followed by detection of bound antibody using appropriate fluorophore-conjugated secondary antibodies. For combined cell surface and intracellular staining, surface staining was performed first, followed by fixation and permeabilization using the Cytofix/Cytoperm kit (BD Pharmingen) and intracellular staining. Cells were analysed using a FACSscan flow cytometer (Becton Dickinson, Mountain View, CA) and data were analysed using CellQuest (Becton Dickinson) or FlowJo (Tree Star, Inc, Ashland, OR) software.
Pulse-chase and immunoprecipitation
Cells were washed and starved for 1–2 hr in Cys/Met-free RPMI containing 10% dialysed fetal bovine serum (FBS) (Invitrogen). Cells were pulsed with 100–150 μCi/ml ExpreSS [35S] labelling mix (Perkin Elmer, Boston, MA) for the indicated times, then washed and chased in complete RPMI containing 10% FBS and 2 mm l-glutamine (at 37° and 5% CO2). Aliquots of cells were collected and washed at the indicated time-points and lysed in lysis buffer [Tris-HCl, pH 8.0, with MgCl2, 1% NP-40 and complete protease inhibitors (Roche Diagnostics, Mannheim, Germany)] at 4°. Lysates were pre-cleared with normal mouse serum, Pansorbin (Calbiochem, La Jolla, CA), and protein A or protein G sepharose beads (formerly Amersham Pharmacia Biotech, now GE Healthcare, Piscataway, NJ), and then normalized based on starting cell number at time 0 or total radioactivity, measured by beta-counter (Wallac, Turku, Finland), as indicated in figure legends. Immunoprecipitations were performed by incubating the normalized lysates with protein A or protein G sepharose beads and class II-specific antibodies (≥ 1 hr at 4°). Proteins were eluted by boiling the precipitates in reducing sodium dodecyl sulphate (SDS) sample buffer (containing 62.5 mm Tris-HCl, pH 6.8, 1% SDS, 3% glycerol, 0.007% bromophenol blue and 1% 2-mercaptoethanol) and then separated by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE). Bands were visualized by exposing dried gels to radiography films (Kodak, Rochester, NY). Densitometry was performed using a Bio-Rad GS-710 densitometer and QuantityOne software (BioRad, Hercules, CA).
For immunoprecipitation of denatured class II chains, metabolic labelling and preparation of cell lysates were performed as described above with a few modifications. For detection of molecular half-life, excess unlabelled Cys/Met (1 mm) was added during the chase period. Cell lysis and centrifugation for clearing of nuclear and cellular debris were accomplished in a small volume of lysis buffer. To denature proteins, concentrated SDS was added to 1% final concentration and samples were boiled for 10 min. The lysates were then diluted at least tenfold in NP-40 lysis buffer to a final concentration of ≤ 0.1% SDS, and pre-cleared with Pansorbin, protein G sepharose, normal mouse serum, normal rat serum, and rabbit-anti-mouse/rat IgG (Zymed Laboratories, now Invitrogen), as appropriate. Subsequent immunoprecipation of individual denatured chains, SDS-PAGE analysis of precipitates, and image analysis were performed as described above.
Immunoblotting
For analysis of total cellular class II abundance, untreated cells were lysed in 1% NP-40 lysis buffer, protein concentration was measured using the Bradford assay, and samples were normalized for total protein. For the observation of class II turnover, cells treated with 100 μg/ml cycloheximide (Sigma) for the indicated times were lysed in 1% NP-40 lysis buffer, and samples were normalized for live cell equivalents (as described in 32). Cell lysate samples were mixed with SDS sample buffer, analysed by SDS-PAGE and transferred onto Immobilon PVDF membrane (Millipore, Bedford, MA). After blocking, membranes were incubated with antibody, and antibody binding was detected with horseradish peroxidase-conjugated secondary antibody followed by an enhanced chemiluminescence (ECL) substrate (Amersham). Densitometry was performed using a Bio-Rad GS-710 densitometer and QuantityOne software (Bio-Rad).
Expression and purification of recombinant MHC class II molecules
Soluble HLA-DR0404 (sDR0404) and soluble DM (sDM) molecules were produced in stably transfected S2 cells and purified by immunoaffinity chromatography and (for sDM) size exclusion chromatography, as described previously 3,36,37.
Synthesis and purification of peptide
A myelin basic protein peptide (MBP 84-99, DENPVVHFFKNIVTPR) was synthesized with standard 9H-fluoren-9-ylmethoxycarbonyl (FMOC) chemistry and labelled at the N-terminus with 5-(and 6-) carboxyfluorescein succinimidyl ester (Molecular Probes, Eugene, OR).54 After high-performance liquid chromatography (HPLC) purification, the peptide was analysed by mass spectrometry and the purity was > 90%.
Peptide association assays
sDR0404 was thermally equilibrated overnight at 37° in citrate/phosphate-buffered saline (PBS), pH 5.3 (nine parts by volume of PBS, 10 mm sodium phosphate, 150 mm NaCl and 0.02% NaN3, pH 7.0, plus one part 1 m sodium citrate, pH 4.8). During the thermal equilibration period, the concentration of DR was 1.25–5 times the ultimate target concentration. Peptide association was initiated by diluting the equilibrated DR solution to the desired concentration with a solution of fluoresceinated peptide (f-peptide). When used, sDM was added together with peptide. Mixtures of sDR and peptide with or without sDM were incubated at 37°. Peptide binding to sDR was carried out for 10 min or less without sDM, and 3 min or less with sDM. These times are generally < 20% of the dissociation half-lives of the complexes.55 After various times, reactions were quenched, and aliquots (45 μl) were injected into a 5-μm particle high-performance size exclusion chromatography (HPSEC) column (dimensions 7.8 mm × 30 cm; G3000 SWXL TSK-GEL; TosoHaas, Montgomery, PA) coupled to a fluorescence detector. The excitation and emission wavelengths were set to 492 and 522 nm, respectively, for detection of fluorescein. The size exclusion column separates the f-peptide/sDR complex (elution time 9 min) from the excess f-peptide (elution time 11 min) under standard separation conditions (mobile phase, PBS pH 7.0; flow rate, 1 ml/min; 25°). The concentration of complex was calculated by comparing the size of the complex peak to that of a carboxyfluorescein standard.
Detection of empty sDR0404 molecules
sDR0404 was thermally equilibrated overnight, as described above. Comparable amounts of untreated or equilibrated sDR0404 (visualized using SDS-PAGE and silver staining; SilverQuest Silver Stain Kit; Invitrogen) were incubated for 2 hr at pH 7 with protein A sepharose beads conjugated with MEM-264 antibody. After washes using PBS, beads were mixed with SDS sample buffer and boiled. Immunoprecipitates were analysed by SDS-PAGE and immunoblotted with CHAMP anti-DR antiserum, detected with horseradish peroxidase-conjugated donkey anti-rabbit immunoglobulin and ECL, as described above.
Quantification of total class II peptide binding sites
‘Bindable class II’ concentration was the concentration of f-peptide/sDR formed after 4 days at pH 5.3 and 37° at saturating peptide concentrations (≥ 2 μm). The concentration of the f-peptide/sDR complex was quantified by HPSEC as described above. The concentration of ‘bindable’ DR was typically 10–15% of the total protein concentration, based on the absorbance at 280 nm.
Calculations
P-values were calculated using Student's t-test. In peptide association assays, association rates were determined by measuring the amount of complex formed after short incubation times during the linear part of the binding curve. Apparent first-order or pseudo first-order association rate constants (with peptide in large excess) were calculated from the relationship rate = kapp [DR], where kapp is the apparent rate constant and [DR] is the concentration of soluble DR, and converted to half-lives using the relationship k = ln(2)/t1/2, where t1/2 is the half-life. Linear and non-linear regression analyses were performed using KaleidaGraph software (Synergy Software, Reading, PA).
Results
For a subset of class II alleles, DM increases cell surface staining by monomorphic antibodies
Although there is some previous evidence that DM increases cell surface levels of some class II molecules,20–24 this property of DM has not been extensively investigated. We evaluated the effect of DM on surface staining by monomorphic anti-class II antibodies. We used several DM-negative mutant lymphoblastoid cell lines (Table 1) and paired transfectants in which DM expression is reconstituted. This approach was chosen to ensure that the only difference between the paired lines was expression of DM. 2.2.93 and 2.2.93-DM express the HLA class II alleles DR1, DR3, DQ1 and DP4; 5.2.4 and 5.2.4-DM express only DP4; 9.5.3 and 9.5.3-DM express DR3, DQ2 and DP4; T2-DR0401 (T2-DR4) and T2-DR0401-DM (T2-DR4-DM) express only DR0401; 3A5 and 3A5-DM express I-Ed and I-Ad; 3A5.g7 and 3A5.g7-DM express I-Ed, I-Ad and I-Ag7.
We surface-stained 2.2.93 and 2.2.93-DM cells with five different monomorphic, anti-class II antibodies: L243 (anti-DR), CALTAG clone TÜ36 (anti-DR), B7/21.2 (anti-DP), SPVL-3 and IA3 (both anti-DQ). The DM-positive cells consistently showed higher levels of antibody binding to DP4 and DQ1 [mean fold change of 1.723 (B7/21.2), 2.092 (SPVL-3) and 1.85 (IA3); Fig. 1a,b], while the effect on DR was limited [mean fold change of 1.145 (L243); Fig. 1a,b]. Compared with 5.2.4 cells, 5.2.4-DM cells showed increased binding of mAb B7/21.2 (anti-DP; Fig. 1e) and DA6.231 (anti-DR and -DP; Fig. 1b), both of which detect DP4 molecules on these DR-negative cells. With 9.5.3 and 9.5.3-DM cells, we found DM-dependent increases in monomorphic antibody binding to DP4, but not to DQ2 or DR3 (not shown and 56). To assess other DR alleles, we examined mAb ISCR3 (anti-DR) binding to DR0401, DR0402, and DR0404 expressed in 5.2.4 cells, in the presence and absence of DM (Fig. 1c). DR0401 and DR0404, but not DR0402, showed DM-dependent increases in mAb ISCR3 binding. Binding of two different anti-DR mAbs (TÜ36 and L243) to DR0401 was also increased in DM transfected T2-DR0401 cells compared with the DM-null parental line (Fig. 1c, and data not shown), consistent with previously reported results for mAbs L24357 and 3.5.9-13F10.22 We also observed the same effect in a murine system: DMA mutant, DM protein-null 3A5 or 3A5.g7 cells (Table 1) transfected with murine DMA showed enhanced binding of the mAb 14-4-4S to cell surface I-Ed and of mAb OX-6 to I-Ag7 (Fig. 1b). These murine antibodies recognize multiple alleles.
Figure 1.
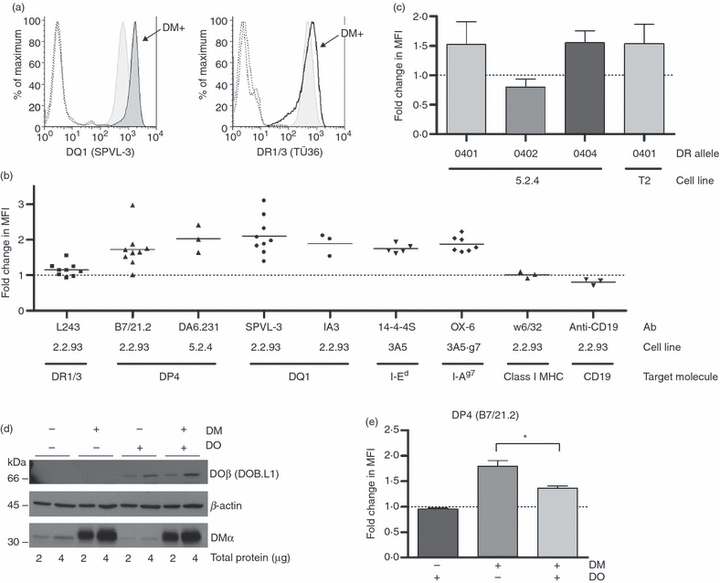
Cell surface staining of a subset of class II alleles with monomorphic antibodies is increased in the presence of DM. (a) Left: representative histograms of staining of 2.2.93 (shaded, light) and 2.2.93-DM (shaded, dark) with SPVL-3 (anti-DQ). Right: representative histograms of staining of 2.2.93 (shaded) and 2.2.93-DM (open) with TÜ36 [anti-human leucocyte antigen (HLA)-DR]. Dotted line histograms represent staining with isotype controls. (b) Fold change in mean fluorescence intensities (MFIs) of staining of 2.2.93-DM compared with 2.2.93 (L243, B7/21.2, SPVL-3, IA3, w6/32 and anti-CD19 staining), of 5.2.4-DM compared with 5.2.4 (DA6.231 staining), of 3A5-DM compared with 3A5 (14-4-4S staining), or of 3A5.g7-DM compared with 3A5.g7 (OX-6 staining). 2.2.93-DM and 5.2.4-DM were stable, drug-selected, polyclonal cell lines. The 3A5-DM and 3A5.g7-DM cells were transient transfectants, assessed in the days immediately following retroviral infection with H-2MA. Fold-change in cell surface I-Ed or I-Ag7 is expressed as class II level (MFI) on cells in the H-2M-positive gate relative to the class II level on cells in the H-2M-negative gate within the same culture. Data are from three independent transient transfections of H-2MA into 3A5 [specifically, 3A5 + 6xHis-wt invariant chain (Ii)], and two independent transfections into 3A5.g7 (specifically, 3A5.g7 + 3xFLAG-wt Ii). Mean values from multiple experiments are shown as horizontal bars. P-values were calculated using one-sample Student's t-test comparing mean values of each column to a hypothetical value of 1: P = 0.0473 (L243), 0.0035 (B7/21.2), 0.0447 (DA6.231), 0.0003 (SPVL-3), 0.0387 (IA3), 0.0002 (14-4-4S), < 0.0001 (OX-6), 0.8527 (w6/32; not significant) and 0.0663 (anti-CD19; not significant). Some of these data have been published 56. (c) 5.2.4 and 5.2.4-DM cells were transfected with DR0401, DR0402 or DR0404 and stained with ISCR3. T2-DR0401 and T2-DR0401-DM cells were stained with TÜ36 (anti-DR). Relative DR expression levels are shown as fold change of MFI of DM-positive cells compared with DM-negative cells. P-values were calculated using one-sample Student's t-test comparing mean values of each column to a hypothetical value of 1: P = 0.0219 (DR0401 in 5.2.4; n = 6), 0.0082 (DR0402 in 5.2.4; n = 7), 0.0116 (DR0404 in 5.2.4; n = 4) and 0.0051 (DR0401 in T2; n = 7). Dotted lines in (b) and (c) indicate no change (MFI ratio = 1). (d) Expression of DM and DO in 5.2.4, 5.2.4-DM, 5.2.4-DO and 5.2.4-DM/DO was analysed by western blot using antibodies 5C1 (anti-DMα) and DOB.L1 (anti-DOβ). The DO β chain appears at ∼66 kD because of the green fluorescent protein (GFP) conjugation (see Materials and methods). (e) Cell surface DP4 levels on indicated cell lines were detected using B7/21.2. Median MFIs from multiple experiments were: 218 (5.2.4), 217 (5.2.4-DO), 395 (5.2.4-DM) and 319 (5.2.4-DM/DO). MFIs were compared with that for parental 5.2.4 cells and represented as fold change. The difference between DP4 levels on 5.2.4-DM and 5.2.4-DM/DO was statistically significant (P = 0.04; n = 4) in a paired t-test.
In the experiments on 3A5 cells, a transient transfection system [i.e. fluorescence-activated cell sorter (FACS) staining was performed after retroviral infection, but before drug selection] was used. 14-4-4S staining of untransfected cells, control vector (expressing lacZ) transfected cells, and DM-negative cells within the retrovirally infected culture was comparable and expression of transfected DM (by intracellular FACS) was equivalent to or lower than DM levels in A20 (parent line of 3A5, with endogenous DM expression) (data not shown). These observations argue against transfection, drug selection, or over-expression of DM as artifactual causes of shifts in mAb binding to class II. Similarly, we observed lower DP4 staining in 9.5.3 compared with its DM-expressing but non-transfected parent line, 8.1.6 (Table 1; data not shown). The effects of DM on class II surface expression were observed even in 5.2.4-DM cells, where DM expression was relatively inefficient (∼1/3 the endogenous levels observed in the parent line, 8.1.6; data not shown). The argument for allelic variation in susceptibility to DM effects on surface class II abundance and against the results being attributable to superphysiological DM levels is especially clear in 2.2.93 and 2.2.93-DM, where DR1/3 alleles showed little or no change in cell surface expression, despite being exposed to the same conditions and DM levels as the susceptible alleles DQ1 and DP4 (Fig. 1a,b). In addition, surface staining of other proteins [MHC class I or CD19 on 2.2.93 (Fig. 1b) and MHC class I on 5.2.4-DR0404 (not shown)] was comparable on DM− and DM+ cells. (5.2.4 cells do not express CD19.) All B-cell transfectants used, except T2-DR4 and T2-DR4-DM, were polyclonal populations, excluding the possible selection of atypical clonal variants.
HLA-DO inhibits the effect of DM on monomorphic antibody binding
HLA-DO (DO), a direct modulator of DM activity, is expressed in B cells, thymic medullary epithelial cells, and subsets of dendritic cells.1,58–61 DO inhibits DM catalysis of peptide exchange in vitro and influences the peptide repertoire in cells.30,62–65 As an additional test that the effects on monomorphic antibody binding were specifically caused by DM expression, we tested whether they were inhibited by DO. 5.2.4 cell lines were transfected with either DM or DO, or both (Fig. 1d), and tested for surface DP4 levels (Fig. 1e). 5.2.4-DM cells were chosen for this experiment as their lower DM levels facilitate detection of inhibition. DP4 surface staining was ∼1.8-fold higher in 5.2.4-DM than in DM/DO-null 5.2.4 cells (Fig. 1b,e). 5.2.4-DM/DO showed a lower level of DP4 than 5.2.4-DM, but a higher level (∼1.4-fold) than 5.2.4 cells; transfection of 5.2.4 cells with DO alone did not affect the DP4 level (Fig. 1e). Taken as a whole, the evidence shown in Figure 1 strongly supports the conclusion that DM augments binding of monomorphic antibodies to a subset of HLA class II alleles.
DM increases the cell surface abundance of DP4, DQ1, DR0404 and I-Ed
The increased binding of monomorphic antibodies to certain class II molecules on DM-expressing cells could result from conformational changes in the class II molecules that increase the affinity of the class II–antibody interaction. An alternate, but not mutually exclusive, explanation is that HLA-DM affects the abundance and/or distribution of these proteins. To test this, we measured levels of binding to cell surface DP4, DQ1, DR0404 and DR0402 with titrated amounts of antibody. If the class II surface abundance is different between DM-expressing and non-expressing cells, the staining levels at saturating concentrations of antibody also will be different. If there is only an affinity difference, the staining on DM-positive and DM-null cells will be comparable at saturating concentrations of antibody. Plateau levels of antibody staining were higher in DM-positive cells than in DM-null cells for DP4, DQ1 and DR0404, consistent with higher surface abundance (Fig. 2). In contrast, DR0402 showed a small reduction in surface levels in the presence of DM. The amounts of 14-4-4S used in cell surface I-Ed staining (Fig. 1b) are saturating,32 implying that the increased 14-4-4S staining in DM-transfected 3A5 also reflects increased I-Ed abundance.
Figure 2.
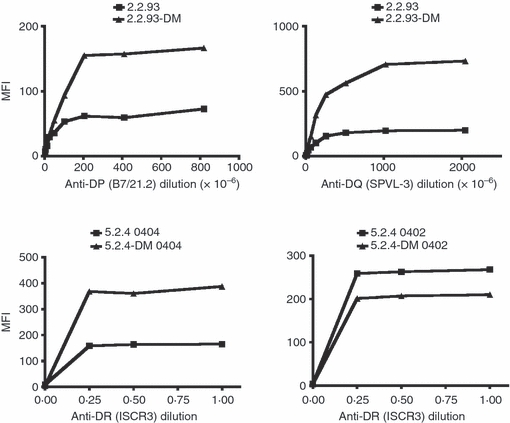
Cell surface abundance of a subset of class II molecules is increased in the presence of DM. Cell surface fluorescence-activated cell sorter (FACS) staining for class II alleles was carried out in the presence or absence of DM. Serial dilutions of B7/21.2 (top left), SPVL-3 (top right, both ascites), or ISCR3 (bottom, supernatant from hybridoma cell line) were used to stain for cell surface DP4, DQ1, DR0404 or DR0402. Mean fluorescence intensity (MFI) is plotted against antibody concentration (1 = undiluted supernatant or ascites; 0.1 = 10-fold dilution, etc.). For each allele, data are from one of two independent experiments.
DM increases total cellular DP4, DQ1, DR0404 and I-Ed
Increased surface class II could reflect increased total class II levels or altered distribution of class II between intracellular compartments and the cell surface. To investigate the effect of DM on the total amount of class II, we measured class II levels using immunoblot analyses of titrated amounts of reduced and boiled whole cell lysates from DM-positive and DM-negative cells. Figure 3a shows that the total cellular levels of DP4, DQ1 and DR0404, but not DR0402, were higher in DM-positive cells. Densitometric analysis (Fig. 3b) confirmed a greater than twofold increase for all three susceptible alleles in DM-positive cells. Total cellular levels of I-Ed detected by western blot were dramatically increased in stable 3A5-DM transfectants (Fig. 6b; wt Ii). FACS staining of total class II showed increased DQ1 in permeabilized DM-expressing 2.2.93 cells (Fig. 3c) and increased DR0401 in permeabilized T2-DR4-DM cells (not shown). Together, these data show that DM acts on certain class II alleles to increase their total cellular abundance.
Figure 3.
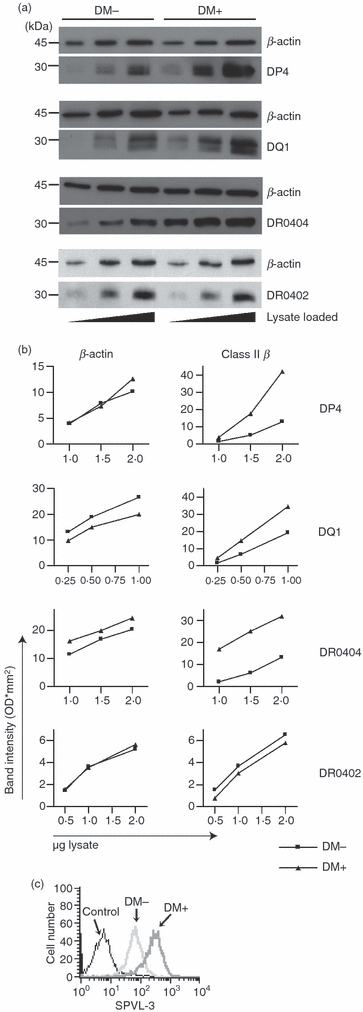
Total abundance of a subset of class II molecules is increased in the presence of DM. (a) Total cellular abundance of class II in the presence or absence of DM was analysed by western blot. Top: cell lysates from 2.2.93 and 2.2.93-DM, pre-cleared for DR in three sequential immunoprecipitations with DA6.147, were reduced, boiled and analysed for DP4 using DA6.231. Second from top: Cell lysates from 2.2.93 and 2.2.93-DM, pre-cleared for DR and DP in three sequential immunoprecipitations with DA6.231, were analysed for DQ1 using XD5.a11. Second from bottom: 5.2.4 DR0404 and 5.2.4-DM DR0404 were analysed for DR0404 using DA6.147. Bottom: 5.2.4 DR0402 and 5.2.4-DM DR0402 were analysed for DR0402 using DA6.147. Each membrane was re-blotted for β-actin as a loading control. (b) Graphs show band intensities (densitometry) for β-actin and class II β chain bands shown in (a). In (a) and (b), one of three to four experiments with similar results is shown. (c) Total DQ1 was measured by fluorescence-activated cell sorter (FACS) staining of permeablized 2.2.93 and 2.2.93-DM using SPVL-3. 5.2.4 cells (DQ-null) served as a (genetic) control for the background level of staining with SPVL-3. 5.2.4 staining was comparable to an isotype control + secondary stain on 2.2.93 cells (not shown). A representative histogram from one of three experiments is shown.
Figure 6.
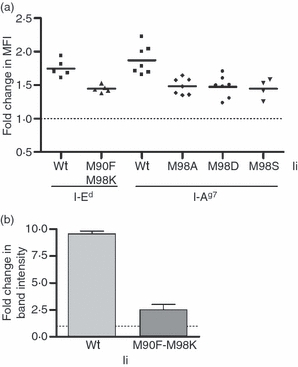
Effect of DM chaperoning on I-Ed and I-Ag7 abundance is reduced in the presence of high class II-associated invariant chain peptide (CLIP) affinity invariant chain (Ii). (a) 3A5 cells stably expressing 6xHis-tagged wild-type (wt) or M90F-M98K CLIP mutant Ii and 3A5.g7 cells stably expressing 3xFLAG-tagged wt, M98A, M98D or M98S CLIP mutant Ii were transiently transfected with H-2MA. The fold change in cell surface I-Ed or I-Ag7 is expressed as class II level [mean fluorescence intensity (MFI)] on cells in the DM-positive gate relative to the class II level on cells in the DM-negative gate within the same short-term culture. Data are from three independent transient transfections in 3A5, two independent transfections in 3A5.g7 with wt, M98A and M98D Ii, and one transfection in 3A5.g7 with M98S Ii. Significance of reduction in fold increase in I-Ed or I-Ag7: M90F-M98K, P = 0.0013; M98A, P = 0.0008; M98D, P = 0.0002; M98S, P = 0.0036 by paired Student's t-test. (b) Total cellular levels of I-Ed in stable 3A5 H-2MA transfectants, drug-selected from cultures in (a), were analysed in quantitative immunoblotting for I-Edα. The indicated fold change was calculated by densitometry analysis from three experiments. The difference between wild-type and mutant Ii-expressing cells was significant (P = 0.011 by paired Student's t-test).
The half-life of alleles susceptible to DM-mediated abundance changes is greater in DM-expressing than non-expressing cells
We predicted that increased total abundance of affected alleles would reflect, at least in part, increases in their half-life in the presence of DM. In an initial experiment to test this prediction, we treated 2.2.93 and 2.2.93-DM cells with cycloheximide (CHX) to block de novo protein synthesis and measured total cellular DP4 and DQ1 levels by immunoblot of denatured molecules at sequential time-points after initiating CHX treatment. In CHX-treated DM-null cells, levels of DP4 and DQ1 began to fall by 17 and 8 hrs, respectively, whereas in CHX-treated, DM+ cells, levels of these proteins appeared constant over this time frame (Fig. 4a). To assess DM effects on turnover in cells without pharmacological treatment, we used T2-DR4 and T2-DR4-DM; cell surface DR0401 levels were increased 1.5–2-fold in the presence of DM (Fig. 1c, and 22,57). We compared turnover of total DR0401 in these cells using pulse-chase and conformation-independent immunoprecipitation. After a short pulse and various chase times, DR4 α or β chains were precipitated from SDS-denatured NP-40 whole cell lysates of metabolically labelled cells, using conformation-independent antibodies DA6.147 (anti-DRα cytoplasmic tail) or XD5.a11 (anti-class II β chain, non-conformational), and analysed by SDS-PAGE. When normalized for an initial difference in level, the disappearance of DR0401 was faster in the absence of DM, with the most marked difference in stability detected in DRα in the 2–12-hr post-synthesis window (Fig. 4b and data not shown). This method did not reveal similar differences for class II proteins in 2.2.93 or 5.2.4 cells compared with their respective DM-transfectants (not shown), perhaps because of the need for higher levels of DM (DM levels in T2-DM > 2.2.93-DM > 5.2.4-DM) to observe a difference in this assay, as discussed below.
Figure 4.
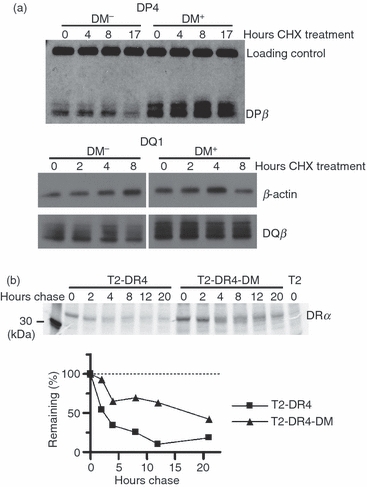
The lifetime of an affected allele is greater in DM-expressing than non-expressing cells. (a) 2.2.93 and 2.2.93-DM were treated with cycloheximide (CHX) for the indicated time periods. DP4: cell lysates from 2.2.93 and 2.2.93-DM, pre-cleared for DR in three sequential immunoprecipitations with DA6.147, were reduced, boiled, and analysed for DP4 levels using DA6.231. DQ1: cell lysates from 2.2.93 and 2.2.93-DM, pre-cleared for DR and DP in five sequential immunoprecipitations (three with DA6.147 and two with DA6.231), were reduced, boiled, and analysed for DQ1 levels using XD5.a11. Loading controls were β-actin (re-blotting of DQ1 membrane) and non-class II bands in the DP4 blots. Representative images from one of two experiments are shown. (b) T2-DR4 and T2-DR4-DM cells (normalized for 5 × 106 cells at time zero per lane) were pulsed for 12 min with S35-Cys/Met and then chased for the indicated time periods. Individual DR α and β chains were immunoprecipitated from clarified and denatured NP-40 cell lysates and analysed by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE). DRα precipitated with DA6.147 is shown (one of four experiments). Densitometric analysis of per cent remaining, compared with band intensity at time 0, is shown.
DM also influences abundance of selected class II alleles early after synthesis
As observed for DRα in T2-DR4-DM, we noted modest but reproducible increases in initial amounts of precipitable class II (DQ1, DR0404, DR0401 and DP4), even after short pulse times (10–15 min), in all DM+ cells tested (Fig. 4b, time 0, and not shown; summarized in Fig. 5c). To examine this more carefully, we assessed levels of several proteins expressed by these cells, including DR alleles that are relatively insensitive to the DM effect on abundance (DR1/3 in 2.2.93), and MHC class I, invariant chain, and calnexin as additional controls. To measure the abundance of these proteins without selectivity for molecular subsets recognized by conformationally sensitive antibodies, we used SDS-denatured cell lysates and non-conformational antibodies (except for calnexin). We observed a DM-dependent effect on the abundance of DQ1(beta), DP4(beta) and DR0404, but not on control proteins (Fig. 5 and data not shown). These results suggest an unexpected effect of HLA-DM on the abundance of selected class II alleles during or immediately after synthesis. The degree of change in precipitated class II chains did not correlate with length of pulse, arguing against a contribution of later events to this observation (not shown).
Figure 5.
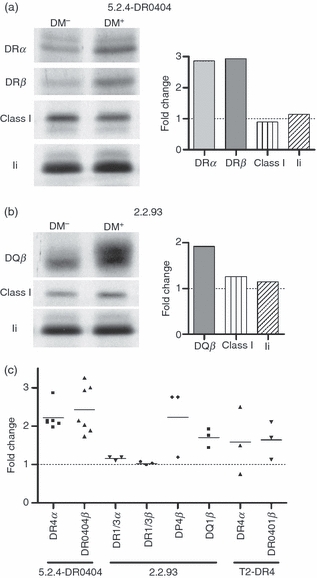
Increased abundance of selected class II alleles early after synthesis in the presence of DM. (a) 5.2.4-DR0404 and 5.2.4-DR0404-DM cells (107 cells per lane) were pulsed for 10 min with S35-Cys/Met and immediately collected for lysis (no chase). Individual chains were immunoprecipitated from denatured and pre-cleared NP-40 cell lysates and analysed by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE): DRα (DA6.147), DRβ (XD5.a11), class I heavy chain (K455), and invariant chain (Ii) (Pin1.1). (b) 2.2.93 and 2.2.93-DM cells (107 cells per lane) were pulsed for 10 min with S35-Cys/Met and immediately collected for lysis. Native DR1/3 was cleared in three sequential immunoprecipitations with DA6.147 (not shown). Individual chains were then immunoprecipitated from denatured, pre-cleared lysates and analysed by SDS-PAGE: DP4β (DA6.231, repeated immunoprecipitations to clear all DP; not shown), DQ1β (XD5.a11), class I heavy chain (K455) and Ii (Pin1.1). Control immunoprecipitations from both cell line pairs under native conditions also showed no DM effect on class I (W6/32), Ii (Pin1.1) and calnexin (AF8) levels (not shown). Densitometry data are shown as fold change in band intensity for the DM+ sample compared with the DM− sample. Representative data from two experiments are shown. (c) Summary of fold change in class II band intensity in DM+ versus DM− cell lines from these experiments and from 0-hr time-points in similar experiments (e.g. Fig. 4b, and data not shown). All pulse times were 10–20 min, except for two experiments (35- and 45-min pulse for 5.2.4-DR0404 ± DM). All data represent immunoprecipitation of individual denatured class II chains except DR1/3 in 2.2.93.
Allelic differences in susceptibility to DM-mediated abundance change: the role of CLIP affinity
DQ1, DP4, DR0401, DR0404, I-Ed and I-Ag7 showed DM-dependent increases in abundance, whereas DR1, DR3 and DR0402 did not. We have shown that low affinity for CLIP peptides is a feature of DR0401 and 0404 compared with DR0402.29,66 The other alleles susceptible to the DM effect on abundance also had low CLIP affinity: in DM-deficient cells, co-precipitation of CLIP with DP4 and DQ1 was substantially lower than with DR1 and/or DR3 (Fig. 8), and I-Ed and I-Ag7 both have high IC50 (high half maximal inhibitory concentration, or low affinity) for wt CLIP.66–68
Figure 8.
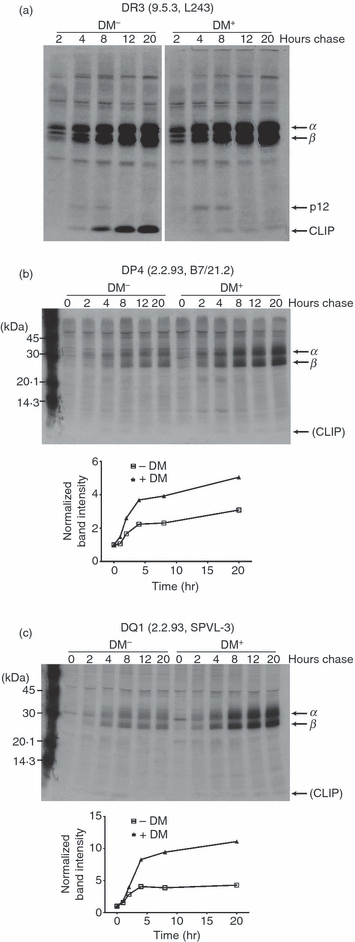
DM effects on levels of class II with mature conformation. 9.5.3 and 9.5.3-DM (a) and 2.2.93 and 2.2.93-DM (b and c) cells were pulsed for 1 hr with S35-Cys/Met and chased for the indicated time periods. Aliquots of cell lysates, normalized for counts, were immunoprecipitated with L243 for DR3 (a, parts of the same gel), B7/21.2 for DP4 (b) and SPVL-3 for DQ1 (c) and then analysed by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE). Graphs show densitometric analysis of band intensity at each time-point normalized to the time zero band. Representative images from one of two to three experiments with similar results are shown. Similar results were obtained for DP4 immunoprecipitated from 9.5.3-DM compared with 9.5.3 (similar to b; not shown), and for DR1/3 immunoprecipitated from 2.2.93 compared with 2.2.93-DM (similar to a; not shown).
The correlation between CLIP affinity and susceptibility to DM effects on abundance is strong, but comparison of distinct alleles introduces confounding variables related to intrinsic stability, pH sensitivity, etc. To isolate the influence of CLIP affinity, we used Ii mutants (32 and C. H. Rinderknecht, unpublished data) that have increased CLIP affinity for I-Ed or I-Ag7. 3A5 (an H-2Mα-deficient B-cell line expressing I-Ed) and 3A5.g7 (a 3A5 transfectant expressing I-Ag7) cells were stably transfected with wt Ii or with the relevant mutant Ii. We have shown that transfected Ii of higher affinity competes successfully with endogenous wt Ii for assembly with I-Ed.32 H-2Mα was then introduced into the 3A5 Ii transfectants to reconstitute H2-M (murine DM) expression. Cell surface (Fig. 6a) and total cellular (Fig. 6b) class II levels were increased by H2-M transfection (fold change > 1). Importantly, the fold change between the DM-positive and DM-negative cells was significantly lower in the cells with the high-CLIP-affinity mutant Ii compared with the cells with wt Ii. These data directly establish the importance of CLIP affinity in class II allelic susceptibility to a DM-mediated abundance change.
DM promotes MHC class II activation
Another potential source of DM effects on class II abundance is rescue of inactivated class II molecules. Whereas in alleles with sufficient CLIP affinity, DM-mediated CLIP removal and the stabilization of the resulting empty class II molecule in a peptide-receptive form may be rapidly sequential, if not concerted, events, alleles that spontaneously lose CLIP, like empty molecules at endosomal pH in vitro, likely inactivate.69,70 In the loading compartment(s), DM is present at substoichiometric levels compared with class II (1 : 4–5) (71 and Elizabeth D. Mellins, unpublished data) and endogenous peptide is limiting,8,72 both of which favour inactivation of empty class II. Studies with recombinant class II molecules have shown that most empty, inactive class II molecules require a slow, rate-limiting transition to an active conformation before they can bind peptide.70,73 To efficiently rescue molecules, DM may be required to catalyse reactivation of class II molecules that have undergone inactivation following spontaneous CLIP release.
To explore this possibility in vitro, we used recombinant soluble DR0404 and DM molecules, which cannot form stable complexes with each other because of a lack of membrane tethering.36,74 We generated empty sDR0404 molecules by overnight incubation of insect-cell derived sDR0404 at acidic pH (5.3) and 37° (‘equilibration’). In this way, we could preclude the possibility of DM generating active class II molecules simply by releasing pre-bound peptide. To confirm that empty molecules were generated in the equilibration step, we used an antibody, MEM-264, that specifically recognizes empty DR molecules.48 Untreated or equilibrated sDR0404 molecules were immunoprecipitated using MEM-264 (Fig. 7a). The amount of immunoprecipitated material was higher in the equilibrated sample, indicating more ‘empty’ molecules.
Figure 7.
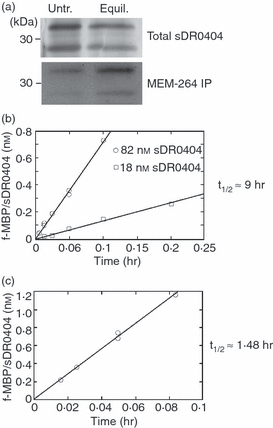
DM affects the peptide association rate of empty DR0404. (a) Empty DR molecules were immunoprecipitated with the monoclonal antibody (mAb) MEM-264 from untreated sDR0404 (untr.) or sDR0404 incubated overnight at 37° and pH 5.3 (equilibrated: equil.), analysed by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotted with CHAMP antiserum (bottom, MEM-264 IP). CHAMP stained DRα more intensely than DR0404β as it was raised against DR1. Aliquots of starting material were silver-stained as controls for initial input (top, total sDR0404). (b) Initial peptide binding to sDR0404. f-MBP peptide (2 μm) was allowed to react with 18 nm (circle) or 82 nM (square) of bindable sDR0404 (quantified as described in Materials and methods) at 37° and pH 5.3. After the indicated times, aliquots were removed, and the amount of f-MBP/sDR0404 complex was measured by high-performance size exclusion chromatography (HPSEC) analysis. Best-fitting regression lines are shown. Assuming a burst phase > 3× the standard error of the mean (SEM) would be detectable at P < 0.01, we estimate that the concentration of active molecules at the beginning of the reaction did not exceed 0.04–0.05% of total DR binding sites (see Methods). Equilibration brings the activation/inactivation reaction to or near equilibrium; the steady-state fraction of active DR molecules at initiation of peptide binding should approximate the ratio of activation and inactivation rate constants, Kai = ka/ki < 5 × 10−4. Thus, the estimated half-life of inactivation of sDR0404 cannot exceed 10–20 seconds. (c) Peptide binding to sDR0404 in the presence of DM. Soluble DM (0.25 μm) and sDR0404 (30 nm) were preincubated overnight before reaction with a saturating concentration of f-MBP peptide (2 μm).
We measured the rate of fluorescence-labelled MBP peptide (f-MBP) binding to two different concentrations of equilibrated sDR0404 (Fig. 7b). The amount of peptide/DR complex formed during the initial binding reaction increased linearly with incubation time. The measured on-rates were slow and were higher at the higher DR concentration. As peptide concentration was held in excess, we used pseudo first-order kinetics to calculate an apparent association half-life (t1/2) of about 9 hr (9.8 hr at 18 nm DR and 7.8 hr at 82 nm DR). Dissociation during the initial rate measurements was negligible, as the dissociation half-life of f-MBP/sDR0404 complexes is ∼21 hr under these conditions.55 Regression analysis indicated that the best-fit straight lines describing the data extrapolated to zero complex at time zero within experimental error (0.0057 ± 0.003 and −0.019 ± 0.011 nm for 18 and 82 nm DR, respectively). The lack of an early burst of binding (positive Y-intercept) at time zero indicates the absence of accumulated active DR. There was also no evidence for initial accumulation of unstable binding intermediates that dissociated during HPSEC analysis, which would have produced a lag phase, and a negative Y-intercept. Thus, we interpret the peptide association half-life as a reflection of the activation half-life (t1/2,a) of sDR0404 in this system.
To measure the effect of DM on sDR0404/peptide complex formation, we performed initial rate measurements in the presence of 0.25 μm sDM and excess peptide (Fig. 7c). Under these conditions, dissociation of f-MBP/sDR0404 complexes is accelerated about 10-fold,55 but not enough to interfere with initial rate measurements. The apparent t1/2,a was about 1.48 hr, which is ∼6 times faster than in the absence of DM. Linear curve-fitting again provided no evidence for an initial burst of peptide binding that would reflect an accumulated pool of active DR. We also measured initial binding rates at two different DM concentrations. With 0.25 μm DM, the estimated activation half-life (t1/2,a) was ∼1.2 hr; at a fourfold higher DM concentration, DM-dependent activation was fourfold faster (t1/2,a = 0.30 hr), consistent with the participation of a single sDM molecule in the activation reaction (not shown). The most likely interpretation of these findings is that sDM can shorten the activation half-life by acting catalytically on inactive sDR 0404 molecules.
DM enhances the abundance of mature class II conformations
We sought evidence compatible with the model that DM rescues inactive forms of class II in cells and that this process is of particular consequence for the level of expression of low-CLIP-affinity alleles. Invariant chain processing in endosomes occurs at 2–4 hr time-points in pulse-chase experiments in the B-LCL studied here (Fig. 8). Using antibodies whose epitopes are preferentially expressed on mature class II molecules, we assessed the generation of mature conformers in the absence and presence of DM. For DR3, an allele that retains CLIP in the absence of DM, immunoprecipitation of mature (L243+) molecules was enhanced coincident with Ii degradation in DM− cells (9.5.3), and CLIP-bound mature molecules were precipitated throughout the later points of chase (Fig. 8a, left). In the presence of DM (9.5.3-DM), CLIP was exchanged for other peptides (detected as an absence of co-precipitated CLIP), and the level of precipitated mature forms was comparable to that of DM-null cells, suggesting that CLIP preserves mature molecules in the absence of DM for DR3 (Fig. 8a). For DP4 and DQ1, the generation of mature molecules (as defined by B7/21.2 and SPVL3 recognition, respectively) that had spontaneously lost CLIP was detected in DM-negative cells in the expected time frame (Fig. 8b,c, left). However, in contrast to findings with DR3, a significantly larger cohort of these molecules (especially for DQ1) was found in DM-expressing (compared with non-expressing) cells (Fig. 8b,c, right). Although various explanations for this finding are plausible, the rescue/reactivation of molecules that have inactivated is attractive, because the levels of DM are substoichiometric, probably prohibiting immediate stabilization of all molecules that release CLIP without DM catalysis.
Discussion
Initial studies of DM effects on class II molecules demonstrated that DM enhanced expression of allele-specific DR epitopes, such as those recognized by mAbs 16.23 and NFLD.D2.75,76 These epitopes map to the allelically variant, peptide-binding domains of DR3 and DR4 molecules, respectively. In studies of DM effects on DR3 molecules, no significant changes in binding of monomorphic anti-DR antibodies were observed, indicating that DR3 levels were not detectably modulated by DM expression.14 In contrast, one finding of this report is the DM-dependent increase in cell surface binding of monomorphic antibodies to certain alleles (such as DP4, DQ1, DR0401, DR0404, I-Ed and I-Ag7), but not to others (such as DR1, DR3, DR0402 and DQ2). Total cellular levels of affected alleles were also increased in DM-positive as compared with DM-negative cells. Quantification by titrated lysates on western blots is sufficient to observe this increase, but is not precise enough to compare the extent of change in total versus surface class II levels and to determine if the ratio of intracellular to surface class II levels also is altered by DM. Although the changes in surface abundance are modest, they are likely to have physiological consequences, as T cells detecting class II/peptide signals demonstrate exquisite sensitivity to quantitative changes.77–79
Notably, the allelic variation in susceptibility to the DM-mediated increases in class II abundance correlates with CLIP affinity for multiple alleles (human and murine) studied here. Studies of other murine alleles also revealed allelic differences in DM-mediated effects.80,81 I-Ak (a low-CLIP-affinity allele), but not I-Ab (a high-CLIP-affinity allele), is dependent on DM for optimal cell surface expression.21,66,82 Using I-Ed, I-Ag7 and CLIP mutant Ii constructs, we explicitly demonstrated that increased CLIP affinity reduced DM effects on both surface and total protein for susceptible alleles. Honey et al.23 demonstrated the converse: when expressed in association with a mutant Ii that reduces binding of CLIP to class II, optimal I-Ab surface expression becomes dependent on DM. Together, these data establish the generality of this conditional phenotype.
Other allele-specific characteristics, such as intrinsic molecular stability and affinity for DM, also probably modulate susceptibility to DM chaperone activity. For example, the I-Ek molecule has a fast CLIP peptide (murine a.a. 85–99) off-rate, with a t½ of 4.4 min,67 but after peptide dissociation, the empty form inactivates slowly with a t½ of 13 min and empty soluble I-Ek is not prone to aggregation.70 Unlike other low-CLIP-affinity alleles, the level of I-Ek protein expression is nearly equivalent in the presence and absence of DM expression.21 In another example, the IC50 values for human CLIP (aa 80–103) are intermediate and fairly comparable for DR3 and DR0404 (118 and 141 nm, respectively).66 However, DR3, an allele whose abundance is not substantially affected by the DM, is quite resistant to functional inactivation,8 while DR0404, a susceptible allele, has an inactivation half-life of ∼20 seconds (Fig. 7 legend). One consequence of a slower inactivation rate would be the opportunity to bind available endosomal peptides and resist aggregation and eventual proteolysis, even after spontaneous CLIP release.
Reductions in total cellular levels of DP4 and DQ1 in DM-null cells were not detectable until after 17 or 8 hr, respectively, of CHX treatment, suggesting that CLIP-free molecules might not be immediately degraded (although CHX could affect these kinetics). These molecules might aggregate after spontaneous loss of CLIP: a lag time between class II aggregation and degradation in cells is consistent with a detectable steady-state level of aggregated DR0401 in DM-negative T2-DR4 cells.8 Alternatively, in the absence of DM, empty DP4 and DQ1 molecules may bind endogenous peptides, but form shorter-lived complexes, resulting in detection of their loss after 8 hr.
For a subset of alleles with reduced interaction with Ii, we showed that DM increased class II levels during or shortly after synthesis (within 10 min metabolic labelling). Our result is surprising, as the time frame suggests a DM effect in the ER (or other early compartments) and DM-mediated peptide exchange on most alleles of class II is suboptimal at neutral pH.3 However, another group has reported an early chaperoning effect of DM in transfectants lacking Ii.83 This early DM effect may reflect enhanced assembly of heterodimers, which protects against degradation of nascent class II chains.84 We have found evidence for reduced assembly of low-CLIP-affinity alleles with Ii (C. H. Rinderknecht, unpublished data), possibly explaining the compensatory increase in their dependence on DM at this stage. In addition, DM may act in the most traditional sense of a chaperone: incompletely folded segments of nascent proteins, even while still ribosome-bound, can be protected from co-translational ubiquitination and degradation by association with chaperones.85,86
In endosomal compartments, DM is present in substoichiometric amounts relative to DR.71 For CLIP-bound class II molecules that encounter DM, a peptide-receptive conformation is probably maintained, first by CLIP and then by DM, after catalysis of CLIP release. For those molecules that release CLIP spontaneously, CLIP release probably generates sufficient empty class II molecules in endosomes to exceed the limiting amounts of DM available to stabilize them through stoichiometric interaction. For alleles that inactivate rapidly, such as DR0404, DM could act directly on inactive class II molecules and increase their rate of reactivation to a peptide receptive form, or could stabilize a spontaneous but transient conversion of inactive to active conformations. Similar to our findings, Marin-Esteban et al.87 showed that soluble DM enhanced the rate of HA peptide binding to soluble, insect cell-derived HLA-DR1, an observation they interpreted as catalysis of formation of active molecules, although they did not show directly that the DR1 was initially empty or inactive. Grotenbreg et al.9 used peptide photolysis to generate empty soluble DR2 molecules and obtained results similar to ours, indicating that the ‘reactivation’ capacity of DM is not limited to DR0404. In contrast, Kropshofer et al.8 found that inactivation of full-length DR1 was irreversible and resistant to reactivation by purified DM. This discrepancy may be attributable to the Zwittergent-12 detergent in these assays, which may enhance denaturation/chain dissociation of the empty molecules. Whereas the capacity of DM to reactivate empty class II molecules in vitro will probably be observed for any allele, this property is most relevant physiologically for those alleles that have low CLIP affinity and unstable peptide-receptive forms.
The class II half-life in cells reflects a combination of DM effects on molecular survival in endosomes through stabilization and reactivation (chaperoning) and DM effects on peptide occupancy (editing), which is known to influence class II half-life.88 The DM chaperone function seems to require a higher molar ratio of DM to substrate than that needed for peptide exchange. High molar ratio to substrate is common for chaperones, such as Ii and heat-shock protein 70 (hsp-70). Observation of different class II turnover rates in pulse-chase experiments with T2-DR0401 ± DM, but not with lower DM expressing lines 5.2.4-DR0404 or 2.2.93, might reflect a threshold level needed for detection of this process in these assays. The molar ratio of functional DM to class II differs in different antigen-presenting cell (APC) types and is affected by APC activation state.58,59 This ratio will probably influence the extent of the chaperone effect on susceptible alleles. Non-transfected cells achieve sufficient DM:class II ratios to increase surface class II, as observed in wild-type versus DM knockout mice.21,23,82
There may be subtle mechanistic differences between the peptide exchange and chaperone functions of DM; for example, class II/peptide complexes that are resistant to conformational editing can still be substrates for the DM-mediated peptide-release reaction.89 We show DO inhibition of the chaperone function of DM, adding to prior data on DO inhibition of DM-mediated peptide exchange. Our findings corroborate results in transgenic mice, where ectopic expression of human DO in dendritic cells led to decreased cell surface and total levels of I-Ak, but not I-Ab (although the mechanism was not defined).24
Our observation that DM effects on abundance are important for certain alleles with low affinity for CLIP is of special interest, because of the relationship between low-CLIP-affinity alleles and autoimmune disease susceptibility. Examples of this correlation (reviewed in 90) include disease associations with DR4 subtypes, DQ1 and DP4.29, 91–94 Our results predict that alterations in expression of functional DM (e.g. by DO inhibition, distinct cellular localization of DM and class II, DM promoter polymorphism, or cytokine milieu) will especially impact abundance of low-CLIP-affinity alleles, supporting our current models for the roles of chaperones in class II association with autoimmunity.90
Acknowledgments
We thank Ms Stacey Chu and Dr Henriette Macmillan for assistance with experiments, Drs Larry Stern, Paul Roche, Janice Blum, Lars Karlsson, Peter Cresswell and Michael Brenner for kind gifts of antibodies and cells, and Dr Phillip Lavori for advice on statistical analyses of our data. This work was supported by financial support from the National Institutes of Health [to E.D.M., and Immunology Training Grant (T32 AI07290) to M.P.B. and C.H.R.], the Arthritis Foundation (to E.D.M.), the Cancer Research Institute (to A.P.), the National Science Foundation (to C.H.R.), and the NRSA [postdoctoral fellowship (1 F32 AI 10298-01) to M.P.B].
Glossary
Abbreviations:
- CHX
cycloheximide
- CLIP
class II-associated invariant chain peptides
- ER
endoplasmic reticulum
- Ii
invariant chain
- IP
immunoprecipitation
- MFI
mean fluorescence intensity
Disclosures
The authors have no financial conflict of interest.
References
- 1.Alfonso C, Karlsson L. Nonclassical MHC class II molecules. Annu Rev Immunol. 2000;18:113–42. doi: 10.1146/annurev.immunol.18.1.113. [DOI] [PubMed] [Google Scholar]
- 2.Denzin LK, Cresswell P. HLA-DM induces CLIP dissociation from MHC class II alpha beta dimers and facilitates peptide loading. Cell. 1995;82:155–65. doi: 10.1016/0092-8674(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 3.Sloan VS, Cameron P, Porter G, et al. Mediation by HLA-DM of dissociation of peptides from HLA-DR. Nature. 1995;375:802–6. doi: 10.1038/375802a0. [DOI] [PubMed] [Google Scholar]
- 4.Weber DA, Evavold BD, Jensen PE. Enhanced dissociation of HLA-DR-bound peptides in the presence of HLA-DM. Science. 1996;274:618–20. doi: 10.1126/science.274.5287.618. [DOI] [PubMed] [Google Scholar]
- 5.Kropshofer H, Vogt AB, Moldenhauer G, Hammer J, Blum JS, Hammerling GJ. Editing of the HLA-DR-peptide repertoire by HLA-DM. EMBO J. 1996;15:6144–54. [PMC free article] [PubMed] [Google Scholar]
- 6.Chou CL, Sadegh-Nasseri S. HLA-DM recognizes the flexible conformation of major histocompatibility complex class II. J Exp Med. 2000;192:1697–706. doi: 10.1084/jem.192.12.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denzin LK, Hammond C, Cresswell P. HLA–DM interactions with intermediates in HLA-DR maturation and a role for HLA-DM in stabilizing empty HLA-DR molecules. J Exp Med. 1996;184:2153–65. doi: 10.1084/jem.184.6.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kropshofer H, Arndt SO, Moldenhauer G, Hammerling GJ, Vogt AB. HLA-DM acts as a molecular chaperone and rescues empty HLA-DR molecules at lysosomal pH. Immunity. 1997;6:293–302. doi: 10.1016/s1074-7613(00)80332-5. [DOI] [PubMed] [Google Scholar]
- 9.Grotenbreg GM, Nicholson MJ, Fowler KD, et al. Empty class II major histocompatibility complex created by peptide photolysis establishes the role of DM in peptide association. J Biol Chem. 2007;282:21425–36. doi: 10.1074/jbc.M702844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pu Z, Lovitch SB, Bikoff EK, Unanue ER. T cells distinguish MHC-peptide complexes formed in separate vesicles and edited by H2-DM. Immunity. 2004;20:467–76. doi: 10.1016/s1074-7613(04)00073-1. [DOI] [PubMed] [Google Scholar]
- 11.Narayan K, Chou CL, Kim A, et al. HLA-DM targets the hydrogen bond between the histidine at position beta81 and peptide to dissociate HLA-DR-peptide complexes. Nat Immunol. 2007;8:92–100. doi: 10.1038/ni1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrante A, Anderson MW, Klug CS, Gorski J. HLA-DM mediates epitope selection by a “compare-exchange” mechanism when a potential peptide pool is available. PLoS ONE. 2008;3:e3722. doi: 10.1371/journal.pone.0003722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belmares MP, Busch R, Mellins ED, McConnell HM. Formation of two peptide/MHC II isomers is catalyzed differentially by HLA-DM. Biochemistry. 2003;42:838–47. doi: 10.1021/bi020466p. [DOI] [PubMed] [Google Scholar]
- 14.Pious D, Dixon L, Levine F, Cotner T, Johnson R. HLA class II regulation and structure. Analysis with HLA-DR3 and HLA-DP point mutants. J Exp Med. 1985;162:1193–207. doi: 10.1084/jem.162.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mellins E, Smith L, Arp B, Cotner T, Celis E, Pious D. Defective processing and presentation of exogenous antigens in mutants with normal HLA class II genes. Nature. 1990;343:71–4. doi: 10.1038/343071a0. [DOI] [PubMed] [Google Scholar]
- 16.Drover S, Kovats S, Masewicz S, Blum JS, Nepom GT. Modulation of peptide-dependent allospecific epitopes on HLA-DR4 molecules by HLA-DM. Hum Immunol. 1998;59:77–86. doi: 10.1016/s0198-8859(97)00263-2. [DOI] [PubMed] [Google Scholar]
- 17.Rudensky A, Rath S, Preston-Hurlburt P, Murphy DB, Janeway CA., Jr On the complexity of self. Nature. 1991;353:660–2. doi: 10.1038/353660a0. [DOI] [PubMed] [Google Scholar]
- 18.Murphy DB, Rath S, Pizzo E, et al. Monoclonal antibody detection of a major self peptide. MHC class II complex. J Immunol. 1992;148:3483–91. [PubMed] [Google Scholar]
- 19.Verreck FA, Fargeas CA, Hammerling GJ. Conformational alterations during biosynthesis of HLA-DR3 molecules controlled by invariant chain and HLA-DM. Eur J Immunol. 2001;31:1029–36. doi: 10.1002/1521-4141(200104)31:4<1029::aid-immu1029>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 20.Albert L, Ghumman B, Watts T. Effect of HLA-DM transfection on hen egg lysozyme presentation by T2.Ak cells. J Immunol. 1996;157:2247–55. [PubMed] [Google Scholar]
- 21.Koonce CH, Wutz G, Robertson EJ, Vogt AB, Kropshofer H, Bikoff EK. DM loss in k haplotype mice reveals isotype-specific chaperone requirements. J Immunol. 2003;170:3751–61. doi: 10.4049/jimmunol.170.7.3751. [DOI] [PubMed] [Google Scholar]
- 22.Lich JD, Jayne JA, Zhou D, Elliott JF, Blum JS. Editing of an immunodominant epitope of glutamate decarboxylase by HLA-DM. J Immunol. 2003;171:853–9. doi: 10.4049/jimmunol.171.2.853. [DOI] [PubMed] [Google Scholar]
- 23.Honey K, Forbush K, Jensen PE, Rudensky AY. Effect of decreasing the affinity of the class II-associated invariant chain peptide on the MHC class II peptide repertoire in the presence or absence of H-2M. J Immunol. 2004;172:4142–50. doi: 10.4049/jimmunol.172.7.4142. [DOI] [PubMed] [Google Scholar]
- 24.Fallas JL, Tobin HM, Lou O, Guo D, Sant’Angelo DB, Denzin LK. Ectopic expression of HLA-DO in mouse dendritic cells diminishes MHC class II antigen presentation. J Immunol. 2004;173:1549–60. doi: 10.4049/jimmunol.173.3.1549. [DOI] [PubMed] [Google Scholar]
- 25.Fling SP, Arp B, Pious D. HLA-DMA and -DMB genes are both required for MHC class II/peptide complex formation in antigen-presenting cells. Nature. 1994;368:554–8. doi: 10.1038/368554a0. [DOI] [PubMed] [Google Scholar]
- 26.Mellins E, Kempin S, Smith L, Monji T, Pious D. A gene required for class II-restricted antigen presentation maps to the major histocompatibility complex. J Exp Med. 1991;174:1607–15. doi: 10.1084/jem.174.6.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liljedahl M, Kuwana T, Fung-Leung WP, Jackson MR, Peterson PA, Karlsson L. HLA-DO is a lysosomal resident which requires association with HLA-DM for efficient intracellular transport. EMBO J. 1996;15:4817–24. [PMC free article] [PubMed] [Google Scholar]
- 28.Pashine A, Busch R, Belmares MP, et al. Interaction of HLA–DR with an acidic face of HLA-DM disrupts sequence–dependent interactions with peptides. Immunity. 2003;19:183–92. doi: 10.1016/s1074-7613(03)00200-0. [DOI] [PubMed] [Google Scholar]
- 29.Patil NS, Pashine A, Belmares MP, et al. Rheumatoid arthritis (RA)-associated HLA-DR alleles form less stable complexes with class II-associated invariant chain peptide than non-RA-associated HLA-DR alleles. J Immunol. 2001;167:7157–68. doi: 10.4049/jimmunol.167.12.7157. [DOI] [PubMed] [Google Scholar]
- 30.Van Ham SM, Tjin EP, Lillemeier BF, et al. HLA-DO is a negative modulator of HLA-DM-mediated MHC class II peptide loading. Curr Biol. 1997;7:950–7. doi: 10.1016/s0960-9822(06)00414-3. [DOI] [PubMed] [Google Scholar]
- 31.Dang LH, Lien LL, Benacerraf B, Rock KL. A mutant antigen-presenting cell defective in antigen presentation expresses class II MHC molecules with an altered conformation. J Immunol. 1993;150:4206–17. [PubMed] [Google Scholar]
- 32.Rinderknecht CH, Belmares MP, Catanzarite TL, et al. Posttranslational regulation of I-Ed by affinity for CLIP. J Immunol. 2007;179:5907–15. doi: 10.4049/jimmunol.179.9.5907. [DOI] [PubMed] [Google Scholar]
- 33.Kim KJ, Kanellopoulos-Langevin C, Merwin RM, Sachs DH, Asofsky R. Establishment and characterization of BALB/c lymphoma lines with B cell properties. J Immunol. 1979;122:549–54. [PubMed] [Google Scholar]
- 34.Riberdy JM, Cresswell P. The antigen-processing mutant T2 suggests a role for MHC-linked genes in class II antigen presentation. J Immunol. 1992;148:2586–90. [PubMed] [Google Scholar]
- 35.Denzin LK, Robbins NF, Carboy-Newcomb C, Cresswell P. Assembly and intracellular transport of HLA-DM and correction of the class II antigen-processing defect in T2 cells. Immunity. 1994;1:595–606. doi: 10.1016/1074-7613(94)90049-3. [DOI] [PubMed] [Google Scholar]
- 36.Busch R, Reich Z, Zaller DM, Sloan V, Mellins ED. Secondary structure composition and pH-dependent conformational changes of soluble recombinant HLA-DM. J Biol Chem. 1998;273:27557–64. doi: 10.1074/jbc.273.42.27557. [DOI] [PubMed] [Google Scholar]
- 37.Hall FC, Rabinowitz JD, Busch R, et al. Relationship between kinetic stability and immunogenicity of HLA-DR4/peptide complexes. Eur J Immunol. 2002;32:662–70. doi: 10.1002/1521-4141(200203)32:3<662::AID-IMMU662>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 38.Lampson LA, Levy R. Two populations of Ia-like molecules on a human B cell line. J Immunol. 1980;125:293–9. [PubMed] [Google Scholar]
- 39.Robbins PA, Evans EL, Ding AH, Warner NL, Brodsky FM. Monoclonal antibodies that distinguish between class II antigens (HLA-DP, DQ, and DR) in 14 haplotypes. Hum Immunol. 1987;18:301–13. doi: 10.1016/0198-8859(87)90077-2. [DOI] [PubMed] [Google Scholar]
- 40.Spits H, Borst J, Giphart M, Coligan J, Terhorst C, De Vries JE. HLA-DC antigens can serve as recognition elements for human cytotoxic T lymphocytes. Eur J Immunol. 1984;14:299–304. doi: 10.1002/eji.1830140404. [DOI] [PubMed] [Google Scholar]
- 41.Van Heyningen V, Guy K, Newman R, Steel CM. Human MHC class II molecules as differentiation markers. Immunogenetics. 1982;16:459–69. doi: 10.1007/BF00372104. [DOI] [PubMed] [Google Scholar]
- 42.Guy K, Van Heyningen V, Cohen BB, Deane DL, Steel CM. Differential expression and serologically distinct subpopulations of human Ia antigens detected with monoclonal antibodies to Ia alpha and beta chains. Eur J Immunol. 1982;12:942–8. doi: 10.1002/eji.1830121109. [DOI] [PubMed] [Google Scholar]
- 43.Radka SF, Machamer CE, Cresswell P. Analysis of monoclonal antibodies reactive with human class II beta chains by two-dimensional electrophoresis and Western blotting. Hum Immunol. 1984;10:177–86. doi: 10.1016/0198-8859(84)90038-7. [DOI] [PubMed] [Google Scholar]
- 44.Barnstable CJ, Bodmer WF, Brown G, et al. Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens-new tools for genetic analysis. Cell. 1978;14:9–20. doi: 10.1016/0092-8674(78)90296-9. [DOI] [PubMed] [Google Scholar]
- 45.Parham P, Barnstable CJ, Bodmer WF. Use of a monoclonal antibody (W6/32) in structural studies of HLA-A,B,C, antigens. J Immunol. 1979;123:342–9. [PubMed] [Google Scholar]
- 46.Watanabe M, Suzuki T, Taniguchi M, Shinohara N. Monoclonal anti-Ia murine alloantibodies crossreactive with the Ia-homologues of other mammalian species including humans. Transplantation. 1983;36:712–8. doi: 10.1097/00007890-198336060-00025. [DOI] [PubMed] [Google Scholar]
- 47.Sanderson F, Thomas C, Neefjes J, Trowsdale J. Association between HLA-DM and HLA-DR in vivo. Immunity. 1996;4:87–96. doi: 10.1016/s1074-7613(00)80301-5. [DOI] [PubMed] [Google Scholar]
- 48.Carven GJ, Chitta S, Hilgert I, et al. Monoclonal antibodies specific for the empty conformation of HLA-DR1 reveal aspects of the conformational change associated with peptide binding. J Biol Chem. 2004;279:16561–70. doi: 10.1074/jbc.M314315200. [DOI] [PubMed] [Google Scholar]
- 49.Rask L, Lindblom JB, Peterson PA. Structural and immunological similarities between HLA antigens from three loci. Eur J Immunol. 1976;6:93–100. doi: 10.1002/eji.1830060205. [DOI] [PubMed] [Google Scholar]
- 50.Hochstenbach F, David V, Watkins S, Brenner MB. Endoplasmic reticulum resident protein of 90 kilodaltons associates with the T- and B-cell antigen receptors and major histocompatibility complex antigens during their assembly. Proc Natl Acad Sci USA. 1992;89:4734–8. doi: 10.1073/pnas.89.10.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roche PA, Marks MS, Cresswell P. Formation of a nine-subunit complex by HLA class II glycoproteins and the invariant chain. Nature. 1991;354:392–4. doi: 10.1038/354392a0. [DOI] [PubMed] [Google Scholar]
- 52.Ozato K, Mayer N, Sachs DH. Hybridoma cell lines secreting monoclonal antibodies to mouse H-2 and Ia antigens. J Immunol. 1980;124:533–40. [PubMed] [Google Scholar]
- 53.Sant AJ, Hendrix LR, Coligan JE, Maloy WL, Germain RN. Defective intracellular transport as a common mechanism limiting expression of inappropriately paired class II major histocompatibility complex alpha/beta chains. J Exp Med. 1991;174:799–808. doi: 10.1084/jem.174.4.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Belmares MP, Rabinowitz JD, Liu W, Mellins ED, McConnell HM. pH stability of HLA-DR4 complexes with antigenic peptides. Biochemistry. 2000;39:14558–66. doi: 10.1021/bi001544g. [DOI] [PubMed] [Google Scholar]
- 55.Belmares MP, Busch R, Wucherpfennig KW, McConnell HM, Mellins ED. Structural factors contributing to DM susceptibility of MHC class II/peptide complexes. J Immunol. 2002;169:5109–17. doi: 10.4049/jimmunol.169.9.5109. [DOI] [PubMed] [Google Scholar]
- 56.Fallang LE, Roh S, Holm A, et al. Complexes of two cohorts of CLIP peptides and HLA-DQ2 of the autoimmune DR3-DQ2 haplotype are poor substrates for HLA-DM. J Immunol. 2008;181:5451–61. doi: 10.4049/jimmunol.181.8.5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Poloso NJ, Denzin LK, Roche PA. CDw78 defines MHC class II-peptide complexes that require Ii chain-dependent lysosomal trafficking, not localization to a specific tetraspanin membrane microdomain. J Immunol. 2006;177:5451–8. doi: 10.4049/jimmunol.177.8.5451. [DOI] [PubMed] [Google Scholar]
- 58.Denzin LK, Fallas JL, Prendes M, Yi W. Right place, right time, right peptide: DO keeps DM focused. Immunol Rev. 2005;207:279–92. doi: 10.1111/j.0105-2896.2005.00302.x. [DOI] [PubMed] [Google Scholar]
- 59.Hornell TM, Burster T, Jahnsen FL, et al. Human dendritic cell expression of HLA-DO is subset specific and regulated by maturation. J Immunol. 2006;176:3536–47. doi: 10.4049/jimmunol.176.6.3536. [DOI] [PubMed] [Google Scholar]
- 60.Douek DC, Altmann DM. HLA-DO is an intracellular class II molecule with distinctive thymic expression. Int Immunol. 1997;9:355–64. doi: 10.1093/intimm/9.3.355. [DOI] [PubMed] [Google Scholar]
- 61.Fallas JL, Yi W, Draghi NA, O’Rourke HM, Denzin LK. Expression patterns of H2-O in mouse B cells and dendritic cells correlate with cell function. J Immunol. 2007;178:1488–97. doi: 10.4049/jimmunol.178.3.1488. [DOI] [PubMed] [Google Scholar]
- 62.Karlsson L. DM and DO shape the repertoire of peptide-MHC-class-II complexes. Curr Opin Immunol. 2005;17:65–70. doi: 10.1016/j.coi.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 63.Denzin LK, Sant’Angelo DB, Hammond C, Surman MJ, Cresswell P. Negative regulation by HLA-DO of MHC class II-restricted antigen processing. Science. 1997;278:106–9. doi: 10.1126/science.278.5335.106. [DOI] [PubMed] [Google Scholar]
- 64.Liljedahl M, Winqvist O, Surh CD, et al. Altered antigen presentation in mice lacking H2-O. Immunity. 1998;8:233–43. doi: 10.1016/s1074-7613(00)80475-6. [DOI] [PubMed] [Google Scholar]
- 65.Van Ham M, Van Lith M, Lillemeier B, et al. Modulation of the major histocompatibility complex class II-associated peptide repertoire by human histocompatibility leukocyte antigen (HLA)-DO. J Exp Med. 2000;191:1127–36. doi: 10.1084/jem.191.7.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sette A, Southwood S, Miller J, Appella E. Binding of major histocompatibility complex class II to the invariant chain-derived peptide, CLIP, is regulated by allelic polymorphism in class II. J Exp Med. 1995;181:677–83. doi: 10.1084/jem.181.2.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liang MN, Beeson C, Mason K, McConnell HM. Kinetics of the reactions between the invariant chain (85–99) peptide and proteins of the murine class II MHC. Int Immunol. 1995;7:1397–404. doi: 10.1093/intimm/7.9.1397. [DOI] [PubMed] [Google Scholar]
- 68.Hausmann DH, Yu B, Hausmann S, Wucherpfennig KW. pH-dependent peptide binding properties of the type I diabetes-associated I-Ag7 molecule: rapid release of CLIP at an endosomal pH. J Exp Med. 1999;189:1723–34. doi: 10.1084/jem.189.11.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Germain RN, Rinker AG., Jr Peptide binding inhibits protein aggregation of invariant-chain free class II dimers and promotes surface expression of occupied molecules. Nature. 1993;363:725–8. doi: 10.1038/363725a0. [DOI] [PubMed] [Google Scholar]
- 70.Rabinowitz JD, Vrljic M, Kasson PM, et al. Formation of a highly peptide-receptive state of class II MHC. Immunity. 1998;9:699–709. doi: 10.1016/s1074-7613(00)80667-6. [DOI] [PubMed] [Google Scholar]
- 71.Schafer PH, Green JM, Malapati S, Gu L, Pierce SK. HLA-DM is present in one-fifth the amount of HLA-DR in the class II peptide-loading compartment where it associates with leupeptin-induced peptide (LIP)-HLA-DR complexes. J Immunol. 1996;157:5487–95. [PubMed] [Google Scholar]
- 72.Germain RN, Hendrix LR. MHC class II structure, occupancy and surface expression determined by post-endoplasmic reticulum antigen binding. Nature. 1991;353:134–9. doi: 10.1038/353134a0. [DOI] [PubMed] [Google Scholar]
- 73.Natarajan SK, Assadi M, Sadegh-Nasseri S. Stable peptide binding to MHC class II molecule is rapid and is determined by a receptive conformation shaped by prior association with low affinity peptides. J Immunol. 1999;162:4030–6. [PubMed] [Google Scholar]
- 74.Weber DA, Dao CT, Jun J, Wigal JL, Jensen PE. Transmembrane domain-mediated colocalization of HLA-DM and HLA-DR is required for optimal HLA-DM catalytic activity. J Immunol. 2001;167:5167–74. doi: 10.4049/jimmunol.167.9.5167. [DOI] [PubMed] [Google Scholar]
- 75.Morris P, Shaman J, Attaya M, et al. An essential role for HLA-DM in antigen presentation by class II major histocompatibility molecules. Nature. 1994;368:551–4. doi: 10.1038/368551a0. [DOI] [PubMed] [Google Scholar]
- 76.Drover S, Karr RW, Fu XT, Marshall WH. Analysis of monoclonal antibodies specific for unique and shared determinants on HLA-DR4 molecules. Hum Immunol. 1994;40:51–60. doi: 10.1016/0198-8859(94)90021-3. [DOI] [PubMed] [Google Scholar]
- 77.Krogsgaard M, Juang J, Davis MM. A role for “self” in T-cell activation. Semin Immunol. 2007;19:236–44. doi: 10.1016/j.smim.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Constant SL, Bottomly K. Induction of Th1 and Th2 CD4+ T cell responses: the alternative approaches. Annu Rev Immunol. 1997;15:297–322. doi: 10.1146/annurev.immunol.15.1.297. [DOI] [PubMed] [Google Scholar]
- 79.Ridgway WM, Fasso M, Fathman CG. A new look at MHC and autoimmune disease. Science. 1999;284:749. doi: 10.1126/science.284.5415.749. [DOI] [PubMed] [Google Scholar]
- 80.Miyazaki T, Wolf P, Tourne S, et al. Mice lacking H2-M complexes, enigmatic elements of the MHC class II peptide-loading pathway. Cell. 1996;84:531–41. doi: 10.1016/s0092-8674(00)81029-6. [DOI] [PubMed] [Google Scholar]
- 81.Martin WD, Hicks GG, Mendiratta SK, Leva HI, Ruley HE, Van Kaer L. H2-M mutant mice are defective in the peptide loading of class II molecules, antigen presentation, and T cell repertoire selection. Cell. 1996;84:543–50. doi: 10.1016/s0092-8674(00)81030-2. [DOI] [PubMed] [Google Scholar]
- 82.Wolf PR, Tourne S, Miyazaki T, Benoist C, Mathis D, Ploegh HL. The phenotype of H-2M-deficient mice is dependent on the MHC class II molecules expressed. Eur J Immunol. 1998;28:2605–18. doi: 10.1002/(SICI)1521-4141(199809)28:09<2605::AID-IMMU2605>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 83.Serradell L, Muntasell A, Catalfamo M, et al. HLA-DM can partially replace the invariant chain for HLA-DR transport and surface expression in transfected endocrine epithelial cells. Tissue Antigens. 1999;53:447–58. doi: 10.1034/j.1399-0039.1999.530501.x. [DOI] [PubMed] [Google Scholar]
- 84.Cotner T, Pious D. HLA-DR beta chains enter into an aggregated complex containing GRP-78/BiP prior to their degradation by the pre-Golgi degradative pathway. J Biol Chem. 1995;270:2379–86. doi: 10.1074/jbc.270.5.2379. [DOI] [PubMed] [Google Scholar]
- 85.Turner GC, Varshavsky A. Detecting and measuring cotranslational protein degradation in vivo. Science. 2000;289:2117–20. doi: 10.1126/science.289.5487.2117. [DOI] [PubMed] [Google Scholar]
- 86.Frydman J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem. 2001;70:603–47. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- 87.Marin-Esteban V, Falk K, Rotzschke O. “Chemical analogues” of HLA-DM can induce a peptide-receptive state in HLA-DR molecules. J Biol Chem. 2004;279:50684–90. doi: 10.1074/jbc.M407598200. [DOI] [PubMed] [Google Scholar]
- 88.Nelson CA, Petzold SJ, Unanue ER. Peptides determine the lifespan of MHC class II molecules in the antigen-presenting cell. Nature. 1994;371:250–2. doi: 10.1038/371250a0. [DOI] [PubMed] [Google Scholar]
- 89.Lovitch SB, Pu Z, Unanue ER. Amino-terminal flanking residues determine the conformation of a peptide-class II MHC complex. J Immunol. 2006;176:2958–68. doi: 10.4049/jimmunol.176.5.2958. [DOI] [PubMed] [Google Scholar]
- 90.Busch R, Rinderknecht CH, Roh S, et al. Achieving stability through editing and chaperoning: regulation of MHC class II peptide binding and expression. Immunol Rev. 2005;207:242–60. doi: 10.1111/j.0105-2896.2005.00306.x. [DOI] [PubMed] [Google Scholar]
- 91.Van Kerckhove C, Luyrink L, Taylor J, et al. HLA-DQA1*0101 haplotypes and disease outcome in early onset pauciarticular juvenile rheumatoid arthritis. J Rheumatol. 1991;18:874–9. [PubMed] [Google Scholar]
- 92.Kavak A, Baykal C, Ozarmagan G, Akar U. HLA in alopecia areata. Int J Dermatol. 2000;39:589–92. doi: 10.1046/j.1365-4362.2000.00921.x. [DOI] [PubMed] [Google Scholar]
- 93.Barahmani N, De Andrade M, Slusser JP, Zhang Q, Duvic M. Major histocompatibility complex class I chain-related gene A polymorphisms and extended haplotypes are associated with familial alopecia areata. J Invest Dermatol. 2006;126:74–8. doi: 10.1038/sj.jid.5700009. [DOI] [PubMed] [Google Scholar]
- 94.Bugawan TL, Angelini G, Larrick J, Auricchio S, Ferrara GB, Erlich HA. A combination of a particular HLA-DP beta allele and an HLA-DQ heterodimer confers susceptibility to coeliac disease. Nature. 1989;339:470–3. doi: 10.1038/339470a0. [DOI] [PubMed] [Google Scholar]


