Abstract
Toll-like receptor-3 (TLR-3) recognizes double-stranded RNA and induces multiple intracellular events responsible for innate anti-viral immunity against a number of viral infections. Activation of TLR-3 inhibits human immunodeficiency virus (HIV) replication, but the mechanism(s) underlying the action of TLR-3 activation on HIV are largely unknown. Here we demonstrate that treatment of monocyte-derived macrophages with poly I:C, a synthetic ligand for TLR-3, significantly inhibited HIV infection and replication. Investigation of the mechanisms showed that TLR-3 activation resulted in the induction of type I interferon inducible antiviral factors, including APOBEC3G and tetherin, the newly identified anti-HIV cellular proteins. In addition, poly I:C-treated macrophages expressed increased levels of CC chemokines, the ligands for CCR5. Furthermore, TLR-3 activation in macrophages induced the expression of cellular microRNAs (miRNA-28, -125b, -150, -223 and -382), the newly identified intracellular HIV restriction factors. These findings indicate that TLR-3-mediated induction of multiple anti-HIV factors should be beneficial for the treatment of HIV disease where innate immune responses are compromised by the virus.
Keywords: CC chemokine, microRNA, poly I:C, tetherin, type I interferon
Introduction
Innate immunity plays an important role in the control of viral infections including those with human immunodeficiency virus (HIV).1 Macrophages, one of the major components in the innate immune system, have the abilities to activate both innate and adaptive immune responses. The importance of macrophage in the pathogenesis of HIV infection is highlighted by its dual roles in HIV infection, where macrophages on the one hand participate in the host anti-HIV immune response, and on the other hand, macrophages are the target for HIV. The HIV can persist in macrophages, and therefore, it is believed that macrophages are an important virus reservoir and contribute to viral latency. The biology of macrophages and their anti-viral activity have been intensively studied, showing that macrophages mount broad anti-viral responses through producing chemokines and cytokines, including macrophage inflammatory proteins (MIPs) and type I interferons (IFNs), and thus are able to inhibit HIV infection at multiple levels.2,3 It has been shown that exposure of macrophages to IFNs results in the restriction of HIV-1 replication at several steps of a viral replication cycle.4,5 However, the precise mechanism of IFN-mediated intracellular antiviral response in macrophages remains to be determined.
As HIV latency is the major obstacle in preventing the eradication of HIV, it is important to identify innate immune factors that suppress and eliminate HIV in its reservoir, such as monocytes/macrophages. Induction of the anti-viral innate immune response depends on a family of innate immune receptors, Toll-like receptors (TLRs). These are a family of pattern recognition receptors expressed by immune cells, including macrophages.6–8 Engagement of TLRs activates signalling cascades that culminate in inflammatory and immune defence responses.9,10 Among the 11 identified human TLRs,11,12 TLR-3 has been recognized as a major receptor in virus-mediated innate immune responses.13,14 The TLR-3 specifically senses double-stranded RNA (dsRNA), an almost universal viral intermediate generated during most viral replications. A synthetic ligand, poly I:C, can also mediate immune responses through activation of TLR-3. The TLR-3 activation induces multiple cellular antiviral responses, including production of IFN-α/β and up-regulation of the anti-HIV cellular factor APOBEC3G.15–17 Activation of TLR-3 has been shown to inhibit a number of viral infections, including those involving herpes simplex virus-1,18 West Nile virus,19 hepatitis C virus20 and influenza virus.21 However, it is still unclear about the role of TLR-3 activation in protecting macrophages from HIV infection. The present study was undertaken to determine whether the treatment of macrophage with the TLR-3 ligand, poly I:C, can inhibit HIV infection of macrophages. We also examined the mechanisms involved in TLR-3-mediated anti-HIV activity in macrophages.
Materials and methods
Macrophage preparation
Purified human monocytes obtained from Human Immunology Core at the University of Pennsylvania were plated in 96-well plates (5 × 105 cells/well) in complete Dulbecco's modified Eagle's medium (DMEM) with 10% fetal calf serum. Monocytes differentiated into macrophages during in vitro cultures (5–7 days).
Poly I:C treatment and HIV infection
Cultured macrophages were incubated with or without poly I:C (0·5 or 1 μg/ml) for 4 or 12 hr before HIV infection. The cells were then infected with an equal amount (p24 protein content) of cell-free HIV strains (Bal or Jago) for 2 hr at 37° in the presence or absence of poly I:C. The cells were washed three times with DMEM to remove input viruses, and fresh medium without poly I:C was added to the cultures. The final wash was tested for reverse transcriptase activity and shown to be free of residual viruses. The cells were incubated for 12 days, and culture supernatants were harvested for HIV reverse transcriptase activity assay.
HIV strains and reverse transcriptase assay
Based on the differential usage of co-receptors (CCR5 and CXCR4), HIV isolates have been referred to as R5, X4 or dual strains.22 The HIV R5 strains (Bal and Jago) were obtained from the AIDS Research and Reference Program (National Institutes of Health, Bethesda, MD).
The HIV reverse transcriptase activity was determined based on the technique of Willey et al.23 with modifications. In brief, 10 μl of culture supernatants was added to a cocktail containing poly A, oligo-dT and (32P) dTTP and incubated for 20 hr at 37°. Then, 20 μl of the cocktail was spotted onto DE81 paper, dried, and washed five times with 2× saline-sodium citrate buffer and once with 95% ethanol. The filter paper was then air-dried. Radioactivity was counted in a liquid scintillation counter.
Real-time reverse transcription–polymerase chain reaction
Total cellular RNA was extracted from cells using Tri-Reagent (Molecular Research Center, Cincinnati, OH) as described previously.24 Total RNA (1 μg) was subjected to the reverse transcription using reagents obtained from Promega (Madison, WI). The real-time reverse transcription–polymerase chain reaction (RT-PCR) for the quantification of messenger RNAs (mRNAs) for IFN-α, IFN-β, APOBEC3G (A3G), myxovirus resistance protein A (MxA), the interferon-stimulated gene 56 (ISG56), obstructive sleep apnoea-1 (OSA-1), protein kinase R (PKR), MIP-α, MIP1-β, IFN regulatory factor-1 (IRF-1), IRF-3, IRF-5, IRF-7, IRF-9, TLR-1, TLR-2, TLR-3, TLR-4, TLR-5, TLR-6, TLR-7, TLR-8, TLR-9, TLR-10 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were performed with the iQ SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA) as described previously.24 The levels of GAPDH mRNA were used as an endogenous reference to normalize the quantities of target mRNA. To conduct micro RNA (miRNA) detection, total cellular RNA, including miRNA, was extracted from cells using a miRNeasy Mini Kit from Qiagen (Valencia, CA). Total RNA (1 μg) was reverse-transcribed with a miScript Reverse Transcription Kit from Qiagen. The real-time RT-PCR for the quantification of a subset of miRNAs (miRNA-28, miRNA-125b, miRNA-150, miRNA-223 and miRNA-382) was carried out as described elsewhere,24 with miScript Primer Assays and miScript SYBR Green PCR Kit from Qiagen. The special oligonucleotide primers used in this study are listed in Table 1. The oligonucleotide primers were synthesized by Integrated DNA Technologies Inc. (Coralville, IA).
Table 1.
Primer sets for real-time reverse transcription–polymerase chain reaction
| Primer | Accession no. | Orientation | Sequences |
|---|---|---|---|
| GAPDH | NM_002046 | Sense: | 5′-GGTGGTCTCCTCTGACTTCAACA-3′ |
| Antisense: | 5′-GTTGCTGTAGCCAAATTCGTTGT-3′ | ||
| IFN-α | NM_002175 | Sense: | 5′-TTTCTCCTGCCTGAAGGACAG-3′ |
| Antisense: | 5′-GCTCATGATTTCTGCTCTGACA-3′ | ||
| IFN-β | NM_002176 | Sense: | 5′-GCCGCATTGACCATCTATGAGA-3′ |
| Antisense: | 5′-GAGATCTTCAGTTTCGGAGGTAAC-3′ | ||
| IRF-1 | NM_002198 | Sense: | 5′-TGAAGCTACAACAGATGAGG-3′ |
| Antisense: | 5′-AGTAGGTACCCCTTCCCATC-3′ | ||
| IRF-3 | NM_001571 | Sense: | 5′-ACCAGCCGTGGACCAAGAG-3′ |
| Antisense: | 5′-TACCAAGGCCCTGAGGCAC-3′ | ||
| IRF-5 | NM_001098631 | Sense: | 5′-AAGCCGATCCGGCCAA-3′ |
| Antisense: | 5′-GGAAGTCCCGGCTCTTGTTAA-3′ | ||
| IRF-7 | NM_001572 | Sense: | 5′-TGGTCCTGGTGAAGCTGGAA-3′ |
| Antisense: | 5′-GATGTCGTCATAGAGGCTGTTGG-3′ | ||
| IRF-9 | NM_006084 | Sense: | 5′-GCATCAGGCAGGGCACGCTGCACC-3′ |
| Antisense: | 5′-GCCTGCATGTTTCCAGGGAATCCG-3′ | ||
| TLR-1 | NM_003263 | Sense: | 5′-GCCTATATGCAAAGAGTTTGGC-3′ |
| Antisense: | 5′-CTCTCCTAAGACCAGCAAGACC-3′ | ||
| TLR-2 | NM_003264 | Sense: | 5′-GGCTTCTCTGTCTTGTGACC-3′ |
| Antisense: | 5′-GGGCTTGAACCAGGAAGACG-3′ | ||
| TLR-3 | NM_003265 | Sense: | 5′-AGCCACCTGAAGTTGACTCAGG-3′ |
| Antisense: | 5′-CAGTCAAATTCGTGCAGAAGGC-3′ | ||
| TLR-4 | NM_138554 | Sense: | 5′-CAGAGTTTCCTGCAATGGATCA-3′ |
| Antisense: | 5′-GCTTATCTGAAGGTGTTGCACAT-3′ | ||
| TLR-5 | NM_003268 | Sense: | 5′-AGCCATCTGACTGCATTAAGG-3′ |
| Antisense: | 5′-GACTTCCTCTTCATCACAACC-3′ | ||
| TLR-6 | NM_006068 | Sense: | 5′-ATTGAAAGCATTCGTGAAGAAG-3′ |
| Antisense: | 5′-ACGGTGTACAAAGCTGTCTGTG-3′ | ||
| TLR-7 | NM_016562 | Sense: | 5′-AAAATGGTGTTTCCAATGTGG-3′ |
| Antisense: | 5′-GGCAGAGTTTTAGGAAACCATC-3′ | ||
| TLR-8 | NM_138636 | Sense: | 5′-TTATGTGTTCCAGGAACTCAGAGAA-3′ |
| Antisense: | 5′-TAATACCCAAGTTGATAGTCGATAAGTTTG-3′ | ||
| TLR-9 | NM_017442 | Sense: | 5′-TACCAACATCCTGATGCTAGACTC-3′ |
| Antisense: | 5′-TAGGACAACAGCAGATACTCCAGG-3′ | ||
| TLR-10 | NM_001017388 | Sense: | 5′-GGCCAGAAACTGTGGTCAAT-3′ |
| Antisense: | 5′-AAATGACTGCATCCAGGGAG-3′ | ||
| MIP1-α | NM_002983 | Sense: | 5′-GCTGACTACTTTGAGACGAGC-3′ |
| Antisense: | 5′-CCAGTCCATAGAAGAGGTAGC-3′ | ||
| MIP1-β | NM_002984 | Sense: | 5′-CCAAACCAAAAGAAGCAAGC-3′ |
| Antisense: | 5′-AGAAACAGTGACAGTGGACC-3′ | ||
| A3G | NM_021822 | Sense: | 5′-TCAGAGGACGGCATGAGACTTAC-3′ |
| Antisense: | 5′-AGCAGGACCCAGGTGTCATTG-3′ | ||
| MxA | M_30817 | Sense: | 5′-GCCGGCTGTGGATATGCTA-3′ |
| Antisense: | 5′-TTTATCGAAACATCTGTGAAAGCAA-3′ | ||
| ISG56 | X_03557 | Sense: | 5′-TTCGGAGAAAGGCATTAGA-3′ |
| Antisense: | 5′-TCCAGGGCTTCATTCATAT-3′ | ||
| OSA-1 | NM_016816 | Sense: | 5′-AGAAGGCAGCTCACGAAACC-3′ |
| Antisense: | 5′-CCACCACCCAAGTTTCCTGTA-3′ | ||
| PKR | NM_003690 | Sense: | 5′-AGAGTAACCGTTGGTGACATAACCT-3′ |
| Antisense: | 5′-GCAGCCTCTGCAGCTCTATGTT-3′ | ||
| Tetherin | BC_033873 | Sense: | 5′-AAGAAAGTGGAGGAGCTTGAGG-3′ |
| Antisense: | 5′-CCTGGTTTTCTCTTCTCAGTCG-3′ |
Flow cytometric analysis
Cultured macrophages (5 × 105 cells/well in 48-well plates) were incubated with or without poly I:C (1 μg/ml) for 12 hr. Cells were then harvested, washed twice with phosphate-buffered saline containing 1% fetal bovine serum, incubated with Alexo-Fluoro-488-conjugated anti-human tetherin (CD317; eBiosciences, San Diego, CA) on ice for 30 min. Unstained or isotype-matched mouse immunoglobulin G-stained cells were included as a negative control. Stained cells were acquired by fluorescence-activated cell sorting (FACSCalibur; BD Bioscience, San Jose, CA) and analysed using Flow-Jo software (Tree Star InC, Ashland, OR).
Enzyme-linked immunosorbent assay for IFN-α/β and CC chemokines
Enzyme-linked immunosorbent assays (ELISA) for analysis of IFN-α and IFN-β proteins were performed as described in the protocol provided by the manufacturers (PBL Biomedical Laboratories, Piscataway, NJ; Fujirebio Inc., Tokyo, Japan). Both MIP-1α and MIP-1β proteins were analysed by ELISA with the specific kits produced by R&D Systems Inc. (Minneapolis, MN). The plate was read on a microplate reader (ELX800; Bio-Tek Instruments, Inc., Winooski, VT).
Statistical analysis
Where appropriate, data were expressed as mean ± SD of triplicate cultures. For comparison of the mean of two groups (treated versus untreated), statistical significance was assessed by Student's t-test. If there were more than two groups, one-way repeated measures of analysis of variance were used. Statistical analyses were performed with Graphpad Instat Statistical Software (GraphPad Software Inc., San Diego, CA). Statistical significance was defined as P < 0·05.
Results
TLR expression and regulation in macrophages
Toll-like receptors recognize pathogen-associated molecular patterns (PAMPs) and are expressed on immune cells including macrophages.11 Among the 11 identified human TLRs, TLR-1 to -10 have been well established functionally.11,12 In addition, the ligands for these TLRs have been identified.25 We showed that macrophages expressed all TLR-1 to TLR-10 at mRNA levels (Fig. 1a). We next investigated whether TLRs expressed by macrophages were biologically functional. We were particularly interested in TLR-3, because the ligand (poly I:C) for TLR-3 mimics a viral replication intermediate, dsRNA. Analysis by RT-PCR of poly I:C-treated macrophages showed an up-regulation of TLR-1, TLR-2, TLR-3 and TLR-7 by three-, four-, seven- and two-fold, respectively (Fig. 1b).
Figure 1.
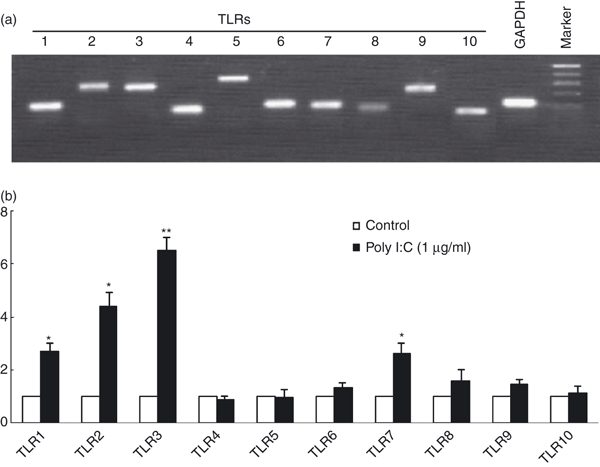
Toll-like receptor (TLR) expression and regulation in macrophages. (a) TLR expressions in macrophages. Total RNA extracted from blood monocyte-derived macrophages (7 day-cultured macrophages) was subjected to the reverse transcription–polymerase chain reaction (RT-PCR) using the primers specific for human TLR-1 to TLR-10. Amplified PCR products were displayed on 2% agarose gel. Sizes were estimated from the DNA ladder (100-base-pair fragments) co-electrophoresed with gyceraldehyde 3-phosphate dehydrogenase (GAPDH). (b) Effect of TLR-3 activation on TLR expression. Seven-day-cultured macrophages were treated with or without poly I:C at the indicated concentrations for 12 hr. Total RNA extracted from cells were then subjected to the real-time RT-PCR for the messenger RNA (mRNA) levels of TLR-1 to TLR-10 and GAPDH. The data are expressed as mRNA levels for TLR-1 to TLR-10 relative (fold) to the control (without poly I:C treatment, which is defined as 1).
TLR-3 activation inhibits HIV-1 infection of macrophages
To evaluate the effect of TLR-3 activation on HIV replication in macrophages, macrophages derived from monocytes of the healthy donors were treated with/without poly I:C before or after infection with different strains (Bal and Jago) of HIV. As shown in Fig. 2, a single pre-treatment of macrophages with poly I:C for 12 hr completely protected cells from infection with both Bal (Fig. 2a) and Jago (Fig. 2b) HIV strains. The duration of this protective effect on macrophages was up to 12 days (Fig. 2). We next examined whether the treatment with poly I:C after HIV infection could inhibit the virus replicated in macrophages. As shown in Fig. 2, macrophages infected first with Bal or Jago HIV strains and then treated with poly I:C once (12 hr) showed a significant decrease (70–80%) in HIV reverse transcriptase activity at day 12 post-infection (Fig. 2). Morphologically, HIV-1 Bal-infected macrophage cultures without poly I:C pre-treatment demonstrated characteristic giant syncytium formation (Fig. 2c), whereas poly I:C-treated macrophages failed to develop HIV-induced giant syncytia (Fig. 2c).
Figure 2.
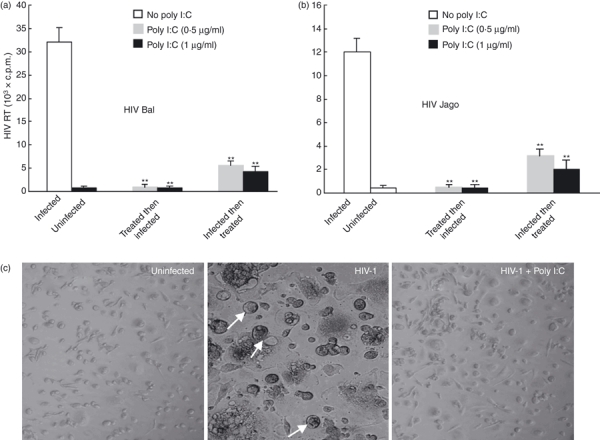
Toll-like receptor-3 (TLR-3) activation suppresses human immunodeficiency virus (HIV) infection of macrophages. (a and b) Effect of poly I:C treatment on HIV Bal (a) or Jago (b) infection of macrophages. Seven-day-cultured macrophages were treated with or without poly I:C at the indicated doses, either 12 hr before or 72 hr after HIV infection for 12 hr. Culture supernatants collected at day 12 after HIV infection were subjected to reverse transcriptase assay. (c) Effect of poly I:C treatment on HIV-induced syncytium formation in macrophages. The morphology of untreated and uninfected, untreated and HIV-infected (Bal strain), and poly I:C-pretreated (1 μg/ml) and HIV-infected macrophages was observed and photographed under a light microscope (magnification, ×200) at day 8 post-infection. The arrows indicate giant syncytium formation.
TLR-3 activation induces type I IFN expression
Since TLR-3 activation triggers intracellular signalling, resulting in the production of chemokines and antiviral cytokines, including type I IFNs,9,26 we next examined whether endogenous IFN-α/β expression was induced in macrophages. We showed that poly I:C treatment induced the expression of IFN-α/β at both mRNA (Fig. 3a) and protein (Fig. 3b) levels in macrophages. This effect was dose dependent (Fig. 3). To determine the mechanism for the effect of TLR-3 activation on IFN-α/β expression, we then examined whether the TLR-3 activation could induce the expression of IRFs. We showed that poly I:C treatment selectively enhanced the expression of IRF-1, -5, -7 and -9 in macrophages (Fig. 3c).
Figure 3.
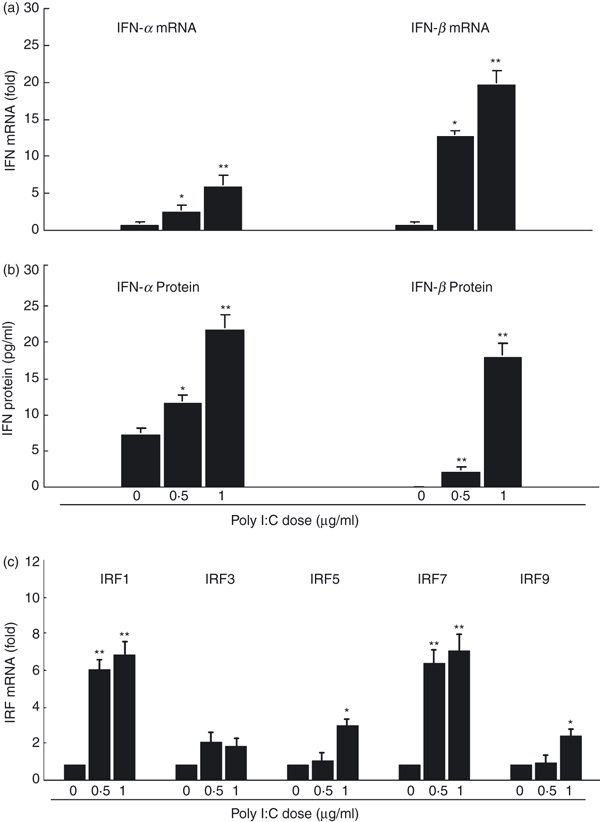
Toll-like receptor-3 (TLR-3) activation induces interferon-α/β (IFN-α/β) and interferon regulation factor (IRF) expressions. (a) IFN-α/β messenger RNA (mRNA) expression. Seven-day-cultured macrophages were treated with or without poly I:C at the indicated concentrations for 12 hr. Total RNA extracted from cells was then subjected to the real-time reverse transcription–polymerase chain reaction (RT-PCR) for the mRNA levels of IFN-α/β and glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The data are expressed as IFN mRNA levels relative (fold) to the control (without poly I:C treatment, which is defined as 1). (b) IFN-α/β protein expression. Seven day-cultured macrophages were treated with or without poly I:C at the indicated concentrations for 24 hr. Supernatants collected from cultures were then assayed by enzyme-linked immunosorbent assay to measure the IFN-α/β proteins. (c) IRF mRNA expression. Seven-day-cultured macrophages were treated with or without poly I:C at the indicated concentrations for 12 hr. Total RNA extracted from cells was then subjected to the real-time RT-PCR for the mRNA levels of IRF-1, IRF-3, IRF-5, IRF-7, IRF-9 and GAPDH. The data are expressed as IRF mRNA levels relative (fold) to the control. The results shown are the mean ± SD of triplicate wells, representing three independent experiments (**P < 0.01; *P < 0.05).
TLR-3 activation induces the expression of IFN-signalling responsive antiviral elements
To provide information on the molecular mechanism(s) by which TLR-3 activation can inhibit HIV infection of macrophages, we examined the expression of several viral restriction factors in macrophages. These cellular restriction factors, including A3G, OSA-1, PKR, MxA and ISG-56, serving as IFN signalling responsive elements, have been reported to be able to inhibit viral replication as part of the cellular innate antiviral machinery. More importantly, these factors have been shown to be regulated by type I IFNs.15,27,28 It is therefore important to determine whether poly I:C treatment of macrophages can induce the expression of these factors. We observed that poly I:C treatment selectively enhanced the expression of A3G, MXA and ISG-56 in macrophages in a dose-dependent manner (Fig. 4a). In addition to these cellular restriction factors, we also examined whether TLR-3 activation in macrophages could induce the expression of the antiviral miRNAs. Five anti-HIV miRNAs (miR-28, -125b, -150, -223 and -382) have been identified to be able to inhibit HIV replication in human CD4+ T cells29 and monocytes/macrophages.30 We demonstrated that poly I:C treatment, in a dose-dependent and time-dependent fashion, induced the expression of all five anti-HIV miRNAs in macrophages by two- to four-fold (Fig. 4b). In addition, we showed that both IFN-α and IFN-β could induce the expression of the anti-HIV miRNAs in macrophages (Fig. 4c).
Figure 4.
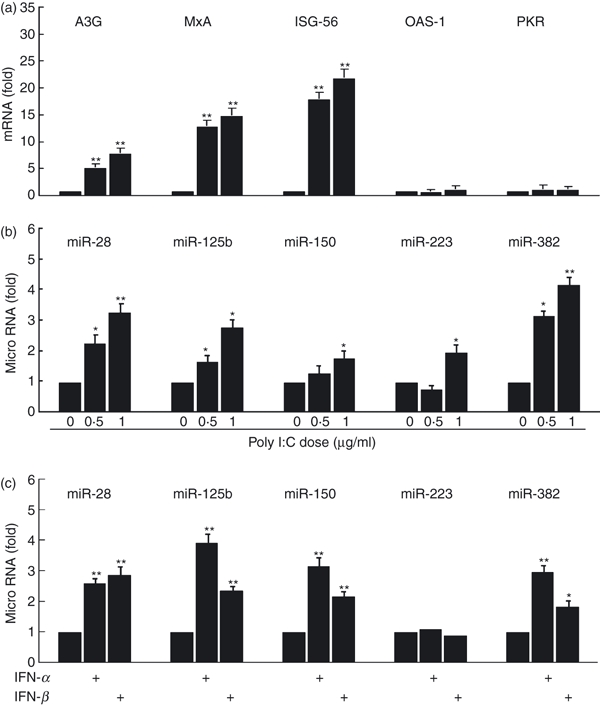
Toll-like receptor (TLR) activation modulates interferon (IFN) -signalling responsive antiviral elements. (a) Anti-viral factors expression. Seven-day-cultured macrophages were treated with poly I:C at the indicated doses for 12 hr. Total RNA extracted from cells was then subjected to the real-time reverse transcription–polymerase chain reaction (RT-PCR) for the messenger RNA (mRNA) levels of A3G, MxA, ISG-56, OAS-1, PKR and glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The data are expressed as A3G, MxA, ISG-56, OAS-1 or PKR mRNA levels relative (fold) to the control (without poly I:C treatment, which is defined as 1). (b) Anti-human immunodeficiency virus (HIV) microRNA (miRNA) expression. Total RNA extracted from cells was then subjected to the real-time RT-PCR for cellular miRNA28, miRNA125b, miRNA150, miRNA223, miRNA382 and GAPDH mRNA quantification. (c) IFN-α/β induces anti-HIV miRNAs in macrophages. Seven-day-cultured macrophages were cultured in the presence or absence of IFN-α/β (100 μg/ml) for 6 hr. Cells were collected and subjected to RNA extraction for miRNA expression by real-time RT-PCR. The data are expressed as the miRNA levels relative (fold) to the control (without IFN-α/β treatment, which is defined as 1). The results shown are the mean ± SD of triplicate wells, representing three independent experiments (**P < 0.01; *P < 0.05).
TLR-3 activation induces CC chemokine expression
Both MIP1-α and MIP1-β, the natural ligands for CCR5 co-receptor, have been reported to be the HIV suppressive factors for virus entry.31 We therefore investigated whether TLR-3 activation had an impact on MIP1-α and MIP1-β expression in macrophages. As demonstrated in Fig. 5, poly I:C treatment of macrophages significantly increased the mRNA levels of both MIP1-α and MIP1-β (Fig. 5a). In addition, poly I:C-treated macrophages produced significantly higher levels of MIP1-α/β proteins than untreated cells (Fig. 5b).
Figure 5.
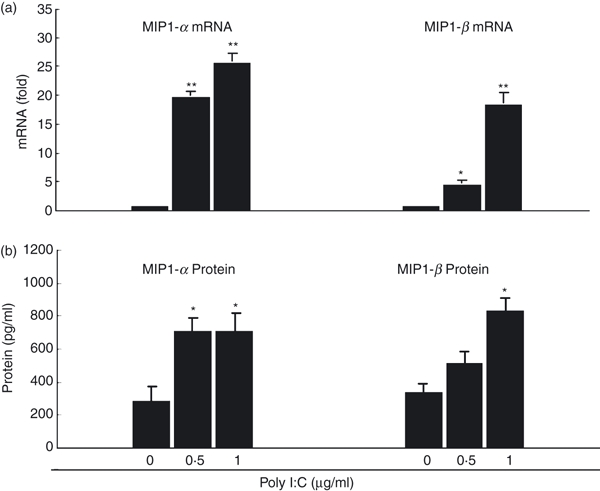
Toll-like receptor-3 (TLR-3) activation up-regulates macrophage inflammatory protein 1-α/β (MIP-1α/β) expression. (a) MIP1-α/β messenger RNA (mRNA) expression. Seven-day-cultured macrophages were treated with or without poly I:C at the indicated concentrations for 12 hr. Total RNA extracted from cells was then subjected to the real-time polymerase chain reaction for the mRNA levels of MIP1-α, MIP1-β and glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The data are expressed as MIP1-α or MIP1-β mRNA levels relative (fold) to the control (without poly I:C treatment, which is defined as 1). (b) MIP1-α/β protein expression. Seven day-cultured macrophages were treated with or without poly I:C at the indicated concentrations for 24 hr. Supernatants were then collected from cell cultures for the protein levels of MIP1-α/β. The results shown are the mean ± SD of triplicate wells, representing three independent experiments (**P < 0.01; *P < 0.05).
TLR-3 activation induces tetherin expression
Tetherin has been recently identified as an important IFN-α inducible cellular restriction factor that inhibits HIV infection of host cells by preventing release of virus from an infected cell.32,33 We therefore examined whether poly I:C treatment modulated tetherin expression in macrophages. Compared with untreated cells, poly I:C-treated macrophages expressed significantly higher levels (more than four-fold) of tetherin mRNA (Fig. 6a). In addition, poly I:C treatment up-regulated the tetherin protein expression in macrophages as demonstrated by flow cytometry analysis (Fig. 6b,c).
Figure 6.
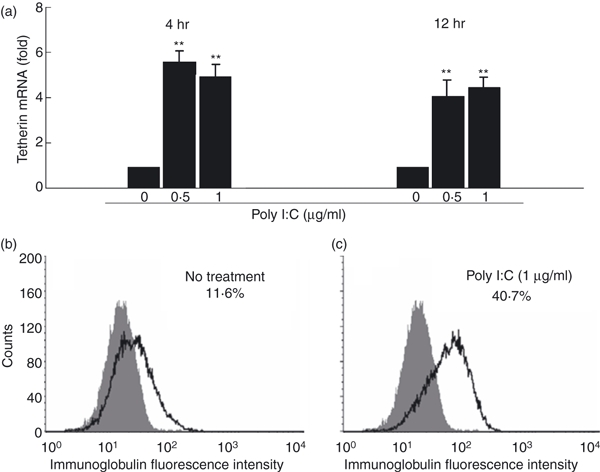
Toll-like receptor-3 (TLR-3) activation up-regulates tetherin expression. (a) Tetherin messenger RNA (mRNA) expression. Seven-day-cultured macrophages were treated with or without poly I:C at the indicated concentrations for 4 or 12 hr. Total RNA extracted from cells was then subjected to the real-time reverse transcription–polymerase chain reaction (RT-PCR) for the mRNA levels of tetherin and glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The data are expressed as tetherin mRNA levels relative (fold) to the control (without poly I:C treatment, which is defined as 1). The results shown are the mean ± SD of triplicate wells, representing three independent experiments (**P < 0.01; *P < 0.05). (b and c) Tetherin protein expression. Seven day-cultured macrophages were treated with (c) or without (b) poly I:C (1 μg/ml) for 12 hr. Cells were stained with fluorescence-conjugated anti-human tetherin (CD317) antibody and analysed for tetherin expression by flow cytometry. Shaded histogram, control staining with isotope-matched antibody (immunoglobulin G2b); open histogram, tetherin staining with monoclonal antibody CD317. A representative histogram graph was shown.
Discussion
Toll-like receptors are crucial in the innate immune response to pathogens because they recognize and respond to PAMPs, which leads to activation of intracellular signalling pathways. Immune cells, including macrophages, mount anti-microbial responses by the recognition of PAMPs by diverse TLRs.6 Among the 11 identified human TLRs, TLR-3 plays a crucial role in virus-mediated innate immune responses, because TLR-3 specifically recognizes dsRNA (a universal viral molecular pattern) and initiates anti-viral signalling pathways in macrophages.25,34 It is known that dsRNA is formed in the HIV replication cycle,35,36 which raises the question whether activation of TLR-3 is able to inhibit HIV replication in macrophages. In this study, we have provided the experimental evidence that TLR-3 activation by a dsRNA analogue, poly I:C, resulted in the inhibition of HIV infection of macrophages. This poly I:C action is TLR-3-mediated, as treatment with the ligands (Pam3CSK4 and HKLM) to TLR-1 and TLR-2 could not inhibit HIV infection of macrophages (data not shown). This poly I:C-mediated antiviral effect is highly effective and durable. When macrophages were pretreated once with poly I:C for 12 hr, a nearly complete inhibition of HIV infection of macrophages was observed (Fig. 2). This inhibitory effect of poly I:C treatment was seen even after HIV infection had been initiated in macrophages (Fig. 2). The anti-HIV activity of poly I:C treatment has also been documented in other cell types.15,37 A recent study using human astrocytes showed that HIV replication was inhibited by poly I:C treatment.37 More recently, Trapp et al.15 reported that poly I:C inhibits HIV in dendritic cells. Taken together, these findings suggest that TLR-3 activation-induced HIV inhibition is a natural and innate immune response possessed by various cell types.
Activation of TLR-3 triggers potent antiviral activity against HIV through multiple mechanisms at both cellular and molecular levels. We first investigated whether TLR-3 activation results in the production of type I IFNs. Type I IFNs (IFN-α/β) have been recognized as the first line of the TLR-3 activation-mediated antiviral response.6 Interferon-α/β has the ability to inhibit HIV infection and replication in macrophages. Our data showed that following poly I:C treatment, IFN-α/β expression was remarkably up-regulated in macrophages. In contrast, the treatment with the ligands to TLR-1 or TLR-2 had little impact on IFN expression (data not shown), suggesting that TLR-1 and TLR-2 that could be induced by poly I:C are not involved in poly I:C-mediated action on IFN-α/β. This induction of IFN-α/β was the result of TLR-3 activation-mediated IRF expression in macrophages. It has been documented that IRFs such as IRF7 have a key role in activating type I IFN expression. We showed that several IRFs, particularly IRF7, were induced in macrophages by poly I:C treatment (Fig. 3c), which provides a sound mechanism for the induction of IFN-α/β by poly I:C. A strong type I IFN response to TLR-3 activation is critical for the production of down-stream antiviral mediators, because IFN-α/β induced by poly I:C treatment can be released from macrophages, and bind to IFN receptors on the cell membrane, triggering a secondary response: activating the JAK/STAT pathway, which results in the induction of the expression of IFN-inducible genes such as MxA and ISG56 (Fig. 4a). We also demonstrated that APOBEC3G (A3G), a specific anti-HIV cellular factor, was induced in macrophages treated with poly I:C (Fig. 4a). This finding is in agreement with the recent report that poly I:C, through type I IFN-mediated activation of APOBEC3G, inhibits HIV amplification in dendritic cells.15 A3G is a cytidine deaminase that has the ability to induce c to u mutation in the negative strand of HIV DNA, resulting in inhibition of HIV replication in both CD4+ T cells and macrophages.38 A3G can either edit the newly synthesized viral DNA or have an inhibitory effect at another site(s) of the HIV life cycle.39,40 In addition, tetherin, which has also been identified as an IFN-α inducible cellular factor,32 was induced in macrophages treated with poly I:C. Tetherin is a transmembrane protein that specifically inhibits HIV infection by preventing its release from infected cells.33 Furthermore, we demonstrated that poly I:C treatment enhanced CC chemokine (MIP-1α/β) expression by macrophages. The CC chemokines block the entry of HIV strains that use CCR5 co-receptor by competitive inhibition.
In addition to cellular restriction proteins against viral infection, there is increasing interest in the role of miRNA in regulating HIV replication. Several lines of evidence support the regulatory role of cellular miRNAs in viral infections.41 Bioinformatic analyses indicate that several human miRNAs can target several viruses,42,43 including HIV.44 Huang et al.29 reported that five human miRNAs (miR-28, -125b, -150, -223 and -382) can target the 3′-untranslated region of HIV-1 transcript and so be responsible for viral latency in resting CD4+ T cells. Our recent study30 showed that these anti-HIV miRNAs also play a crucial role in control of HIV replication in monocytes and macrophages. Therefore, we investigated whether TLR-3 activation could modulate the expression of the anti-HIV miRNAs in macrophages. We demonstrated that poly I:C treatment induced the expression of all five anti-HIV miRNAs in macrophages (Fig. 4c). This effect appeared to be TLR-3-specific, as the ligands to TLR-1 and TLR-2 had little impact on miRNA expression (data not shown). These data provide an additional mechanism involved in TLR-3 activation-induced anti-HIV action in macrophages. This poly I:C treatment-mediated induction of the anti-HIV miRNA could be the result of the activation of IFNs, as both IFN-α and IFN-β up-regulated the expression of anti-HIV miRNAs (Fig. 4c). This finding supports the study by others showing that type I IFNs modulate cellular miRNAs as an antiviral mechanism.45
Taken together, our study provides compelling experimental evidence that TLR-3 activation by poly I:C in macrophages significantly suppresses HIV infection and replication through multiple antiviral mechanisms at both cellular and molecular levels. Although additional mechanism(s) might also be involved, the induction of type I IFNs and IFN-dependent antiviral mechanisms accounts for much of the TLR-3-mediated anti-HIV activity. There are at least two distinct components involved in type I IFN-dependent anti-HIV activities: the induction of extracellular factors, CC chemokines that block HIV entry into macrophages; the activation of intracellular viral restriction factors such as A3G, MxA, ISG, miRNAs and tetherin. Each of these antiviral factors plays an important role in restriction of HIV replication, as they directly inhibit HIV at different steps of the viral replication cycle. These findings are clinically important, as the activation of the TLR-3 signalling pathway may represent a promising novel strategy for treatment of people with HIV infection. Because this approach activates the intracellular innate immune system, leading to enhance and/or restore type I IFN production as well as multiple IFN-inducible antiviral gene expression in HIV-infected host cells, it is less likely for HIV to develop resistance. Currently, the therapeutic TLR agonists are being developed for the treatment of cancer allergies and viral infections. Agonists for TLRs, particularly TLR-3, TLR-7 and TLR-9, have been shown to have promise as treatment for infectious diseases, especially viral infections including HIV.46,47 A number of TLR agonists are now in clinical or preclinical trails such as the anti-HIV TLR-3 agonist (poly I:C 12U).46,47 Obviously, the findings of this study support the notion for further developing a TLR-3 agonist-based therapy for HIV disease in which host cell innate immune responses are significantly compromised by the virus. It is hopeful that ongoing and future studies will yield promising data that TLR-3 agonists will be therapeutically useful for the treatment of people infected with HIV.
Acknowledgments
The study was supported by grants NIDA012815, NIDA027550, NIDA025477 and NIDA022177 (to W.Z. Ho), and the grant A0901 from W. W. Smith Charitable Trust to W. Z. Ho from the National Institutes of Health.
References
- 1.Deeks SG, Walker BD. Human immunodeficiency virus controllers: mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity. 2007;27:406–16. doi: 10.1016/j.immuni.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 2.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5:953–64. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 3.Stout RD, Suttles J. Functional plasticity of macrophages: reversible adaptation to changing microenvironments. J Leukoc Biol. 2004;76:509–13. doi: 10.1189/jlb.0504272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang FX, Huang J, Zhang H, Ma X, Zhang H. APOBEC3G upregulation by alpha interferon restricts human immunodeficiency virus type 1 infection in human peripheral plasmacytoid dendritic cells. J Gen Virol. 2008;89:722–30. doi: 10.1099/vir.0.83530-0. [DOI] [PubMed] [Google Scholar]
- 5.Adalid-Peralta L, Godot V, Colin C, et al. Stimulation of the primary anti-HIV antibody response by IFN-alpha in patients with acute HIV-1 infection. J Leukoc Biol. 2008;83:1060–7. doi: 10.1189/jlb.1007675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 7.Delneste Y, Beauvillain C, Jeannin P. Innate immunity: structure and function of TLRs. Med Sci (Paris) 2007;23:67–73. doi: 10.1051/medsci/200723167. [DOI] [PubMed] [Google Scholar]
- 8.Hasko G, Pacher P, Deitch EA, Vizi ES. Shaping of monocyte and macrophage function by adenosine receptors. Pharmacol Ther. 2007;113:264–75. doi: 10.1016/j.pharmthera.2006.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 10.Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–7. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 11.Kabelitz D, Medzhitov R. Innate immunity – cross-talk with adaptive immunity through pattern recognition receptors and cytokines. Curr Opin Immunol. 2007;19:1–3. doi: 10.1016/j.coi.2006.11.018. [DOI] [PubMed] [Google Scholar]
- 12.Lauw FN, Caffrey DR, Golenbock DT. Of mice and man: TLR11 (finally) finds profilin. Trends Immunol. 2005;26:509–11. doi: 10.1016/j.it.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 13.Lafon M, Megret F, Lafage M, Prehaud C. The innate immune facet of brain: human neurons express TLR-3 and sense viral dsRNA. J Mol Neurosci. 2006;29:185–94. doi: 10.1385/JMN:29:3:185. [DOI] [PubMed] [Google Scholar]
- 14.Jackson AC, Rossiter JP, Lafon M. Expression of Toll-like receptor 3 in the human cerebellar cortex in rabies, herpes simplex encephalitis, and other neurological diseases. J Neurovirol. 2006;12:229–34. doi: 10.1080/13550280600848399. [DOI] [PubMed] [Google Scholar]
- 15.Trapp S, Derby NR, Singer R, et al. Double-stranded RNA analog poly(I:C) inhibits human immunodeficiency virus amplification in dendritic cells via type I interferon-mediated activation of APOBEC3G. J Virol. 2009;83:884–95. doi: 10.1128/JVI.00023-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang YJ, Wang X, Zhang H, et al. Expression and regulation of antiviral protein APOBEC3G in human neuronal cells. J Neuroimmunol. 2009;206:14–21. doi: 10.1016/j.jneuroim.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abrahams VM, Schaefer TM, Fahey JV, et al. Expression and secretion of antiviral factors by trophoblast cells following stimulation by the TLR-3 agonist, Poly(I : C) Hum Reprod. 2006;21:2432–9. doi: 10.1093/humrep/del178. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Y, Ye L, Wan Q, et al. Activation of Toll-like receptors inhibits herpes simplex virus-1 infection of human neuronal cells. J Neurosci Res. 2009;87:2916–25. doi: 10.1002/jnr.22110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daffis S, Samuel MA, Suthar MS, Gale M, Jr, Diamond MS. Toll-like receptor 3 has a protective role against West Nile virus infection. J Virol. 2008;82:10349–58. doi: 10.1128/JVI.00935-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Broering R, Wu J, Meng Z, et al. Toll-like receptor-stimulated non-parenchymal liver cells can regulate hepatitis C virus replication. J Hepatol. 2008;48:914–22. doi: 10.1016/j.jhep.2008.01.028. [DOI] [PubMed] [Google Scholar]
- 21.Lau YF, Tang LH, Ooi EE. A TLR3 ligand that exhibits potent inhibition of influenza virus replication and has strong adjuvant activity has the potential for dual applications in an influenza pandemic. Vaccine. 2009;27:1354–64. doi: 10.1016/j.vaccine.2008.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berger EA, Doms RW, Fenyo EM, et al. A new classification for HIV-1. Nature. 1998;391:240. doi: 10.1038/34571. [DOI] [PubMed] [Google Scholar]
- 23.Willey RL, Smith DH, Lasky LA, Theodore TS, Earl PL, Moss B, Capon DJ, Martin MA. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J Virol. 1988;62:139–47. doi: 10.1128/jvi.62.1.139-147.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo CJ, Douglas SD, Gao Z, Wolf BA, Grinspan J, Lai JP, Riedel E, Ho WZ. Interleukin-1beta upregulates functional expression of neurokinin-1 receptor (NK-1R) via NF-kappaB in astrocytes. Glia. 2004;48:259–66. doi: 10.1002/glia.20079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 26.Alsharifi M, Mullbacher A, Regner M. Interferon type I responses in primary and secondary infections. Immunol Cell Biol. 2008;86:239–45. doi: 10.1038/sj.icb.7100159. [DOI] [PubMed] [Google Scholar]
- 27.Cunningham S, Graham C, Hutchinson M, et al. Pharmacogenomics of responsiveness to interferon IFN-beta treatment in multiple sclerosis: a genetic screen of 100 type I interferon-inducible genes. Clin Pharmacol Ther. 2005;78:635–46. doi: 10.1016/j.clpt.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 28.Hamano E, Hijikata M, Itoyama S, et al. Polymorphisms of interferon-inducible genes OAS-1 and MxA associated with SARS in the Vietnamese population. Biochem Biophys Res Commun. 2005;329:1234–9. doi: 10.1016/j.bbrc.2005.02.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang J, Wang F, Argyris E, et al. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat Med. 2007;13:1241–7. doi: 10.1038/nm1639. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, Ye L, Hou W, Zhou Y, Wang YJ, Metzger DS, Ho WZ. Cellular microRNA expression correlates with susceptibility of monocytes/macrophages to HIV-1 infection. Blood. 2009;113:671–4. doi: 10.1182/blood-2008-09-175000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Proost P, Schols D. Role of chemokines in the HIV infection process. Verh K Acad Geneeskd Belg. 2002;64:403–20. [PubMed] [Google Scholar]
- 32.Kawai S, Azuma Y, Fujii E, Furugaki K, Ozaki S, Matsumoto T, Kosaka M, Yamada-Okabe H. Interferon-alpha enhances CD317 expression and the antitumor activity of anti-CD317 monoclonal antibody in renal cell carcinoma xenograft models. Cancer Sci. 2008;99:2461–6. doi: 10.1111/j.1349-7006.2008.00968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–30. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 34.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–8. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 35.Brule F, Marquet R, Rong L, Wainberg MA, Roques BP, Le Grice SF, Ehresmann B, Ehresmann C. Structural and functional properties of the HIV-1 RNA-tRNA(Lys)3 primer complex annealed by the nucleocapsid protein: comparison with the heat-annealed complex. Rna. 2002;8:8–15. doi: 10.1017/s1355838202010981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fedoroff OY, Ge Y, Reid BR. Solution structure of r(gaggacug):d(CAGTCCTC) hybrid: implications for the initiation of HIV-1+-strand synthesis. J Mol Biol. 1997;269:225–39. doi: 10.1006/jmbi.1997.1024. [DOI] [PubMed] [Google Scholar]
- 37.Suh HS, Zhao ML, Rivieccio M, et al. Astrocyte indoleamine 2,3-dioxygenase is induced by the TLR3 ligand poly(I:C): mechanism of induction and role in antiviral response. J Virol. 2007;81:9838–50. doi: 10.1128/JVI.00792-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–50. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 39.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 40.Mariani R, Chen D, Schrofelbauer B, et al. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell. 2003;114:21–31. doi: 10.1016/s0092-8674(03)00515-4. [DOI] [PubMed] [Google Scholar]
- 41.Kumar A. The silent defense: micro-RNA directed defense against HIV-1 replication. Retrovirology. 2007;4:26. doi: 10.1186/1742-4690-4-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghosh Z, Mallick B, Chakrabarti J. Cellular versus viral microRNAs in host–virus interaction. Nucleic Acids Res. 2009;37:1035–48. doi: 10.1093/nar/gkn1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saib A, Voinnet O. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–60. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 44.Hariharan M, Scaria V, Pillai B, Brahmachari SK. Targets for human encoded microRNAs in HIV genes. Biochem Biophys Res Commun. 2005;337:1214–8. doi: 10.1016/j.bbrc.2005.09.183. [DOI] [PubMed] [Google Scholar]
- 45.Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–22. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Averett DR, Fletcher SP, Li W, Webber SE, Appleman JR. The pharmacology of endosomal TLR agonists in viral disease. Biochem Soc Trans. 2007;35:1468–72. doi: 10.1042/BST0351468. [DOI] [PubMed] [Google Scholar]
- 47.Kanzler H, Barrat FJ, Hessel EM, Coffman RL. Therapeutic targeting of innate immunity with Toll-like receptor agonists and antagonists. Nat Med. 2007;13:552–9. doi: 10.1038/nm1589. [DOI] [PubMed] [Google Scholar]


