Abstract
T-cell immunoglobulin mucin-1 (Tim-1) has been proposed to be an important T-cell immunoregulatory molecule since its expression on activated T cells was discovered. To study the role of Tim-1 on T cells in vitro and in vivo we generated both Tim-1-deficient mice and several lines of Tim-1 transgenic mice with Tim-1 expression on either T cells, or B and T cells. We demonstrate that neither deficiency nor over-expression of Tim-1 on B and T cells results in modulation of their proliferation in vitro. More surprisingly, T helper type 2 cells generated either from Tim-1-deficient mice or Tim-1 transgenic mice did not show enhancement of interleukin-4 (IL-4), IL-5 and IL-10 production. Furthermore, using a Schistosoma mansoni egg challenge as a potent T helper type 2 response inducer we also show that Tim-1 is not essential for T- and B-cell responses in vivo. However, we observe induction of Tim-1 on B cells following B-cell receptor (BCR), but not Toll-like receptor 4 stimulation in vitro. We show that the induction of Tim-1 on B cells following BCR stimulation is phosphoinositide-3 kinase and nuclear factor-κB pathway dependent. More importantly, we conclude that Tim-1 is predominantly expressed on germinal centre B cells in vivo although the percentage of germinal centre B cells in wild-type and Tim-1-deficient mice is comparable. Identification of Tim-1 as a marker for germinal centre B cells will contribute to the interpretation and future analysis of the effects of the anti-Tim-1 antibodies in vivo.
Keywords: B-cell receptor, B cells, T cells, germinal centre
Introduction
The recently identified T-cell immunoglobulin and mucin domain-containing (Tim) molecules have been implicated in T-cell-mediated immune responses.1–4 This family of genes includes three members in humans and eight in mice.5 The human and rat homologues that most closely resemble mouse Tim-1, encoded by Havcr-1, were first identified as hepatitis A virus cellular receptor-1 and kidney injury molecule-1, respectively.6,7 Distinct genetic variants encoding Tim-1 were later proposed as being important in asthma and allergy susceptibility, both in mice and humans.4,8
Initial evidence for the role of Tim-1 in the regulation of asthma and allergic disease came from the observation that Tim-1 was expressed in CD4+ T cells and that its gene expression was increased during primary antigen stimulation.4 It was later shown that a low level of Tim-1 protein was detectable on CD4+ T cells, CD19+ B cells and CD11c+ cells, but not on CD8+ T cells using an anti-Tim-1 antibody clone 3B3.9 Furthermore, this antibody also detected increased expression of Tim-1 on CD4+ T cells activated in vitro and T helper type 2 (Th2) cells generated in vitro.9 Since then, many anti-Tim-1 antibodies of different clones have been generated by several groups to assess their potential therapeutic use in mouse models of allergy. However, results obtained from these studies using different anti-Tim-1 antibodies have proven to be complicated. First, multiple cell types including mast cells10 and thymocytes11 have been shown to express Tim-1 using different clones of anti-Tim-1 antibodies. Second, the binding patterns of different anti-Tim-1 antibodies on the same cell type have been shown to vary. Whereas anti-Tim-1 clone 3B3 showed induction of Tim-1 on almost all activated CD4 T cells after 48 hr stimulation in vitro, another clone of anti-Tim-1 showed minimal expression of Tim-1 on activated T cells.11 Third, anti-Tim-1 antibodies that recognize different epitopes on Tim-1 have been reported to mediate opposing biological effects.11 While anti-Tim-1 antibodies, distinguishing specific epitopes in both the immunoglobulin region and mucin domain, were shown to reduce lung eosinophilia and lung inflammation, another clone that binds at a different epitope in the mucin domain actually exacerbated the allergic response.11 Finally, even anti-Tim-1 antibodies (clones 3B3 and RMT1-10) recognizing the same epitope, but with different avidities for Tim-1 have been reported to elicit opposing functional effects.12 Furthermore, Tim family proteins have also been shown to bind to a diversity of ligands through their carbohydrate-binding capacity.13 Hence, the use of anti-Tim-1 antibodies has uncovered apparent complexity in the roles of Tim-1 in vivo.
To evaluate the roles of Tim-1 in vitro and in vivo, but avoid the potential complications of diverse anti-Tim-1 antibody recognition, we developed both Tim-1-deficient mice and transgenic mice over-expressing Tim-1 on T cells, or T and B cells. We confirmed the absence of Tim-1 or its transgenic expression using anti-Tim-1 antibody, clone RMT1-4, which we confirmed to be specific for Tim-1. Using this approach we were unable to detect Tim-1 expression on T cells stimulated in vitro or in vivo on T cells obtained from antigen-challenged mice. Furthermore, we found no deficit in the type 2 responses in vitro or in vivo. By contrast, we clearly observed Tim-1 induction on anti-immunoglobulin M (IgM) -stimulated B cells, and this staining of Tim-1 was completely abrogated on the anti-IgM-stimulated B cells from Tim-1-deficient mice. We also show that Tim-1 is predominantly expressed on germinal centre B cells following antigen challenge. However, Tim-1-deficient mice have normal levels of germinal centre B cells and antibody production, suggesting that Tim-1 is not essential or has a redundant role for the induction of germinal centre response. The identification of Tim-1 as a marker for germinal centre B cells will contribute to the interpretation and future analysis of the effects of the anti-Tim-1 antibodies in vivo.
Materials and methods
Mice
Wild-type BALB/c or C57BL/6 mice were obtained from Harlan, Oxford, UK. All mice were maintained in a specific pathogen-free environment according to UK Home Office regulations.
Generation of Tim-1 transgenic mice
The complementary DNAs (cDNAs) encoding either the BALB/c Tim-1 allotype or the C57BL/6 Tim-1 allotype were cloned into the pTEXCD2 vector (a kind gift from Michael Owen14), which contains regulatory elements from the human CD2 gene driving transgene expression in either T cells or T and B cells.14,15 The transgenes were purified and injected into fertilized mouse eggs. Transgenic mice (strain CBA × C57BL/6) were back-crossed to the C57BL/6 background for between three and six generations. Genotypes were screened by polymerase chain reaction (PCR) using primers ASEQ965 5′-ATATCTCAGGAATGGGATTGTGAC-3′ and ASEQ966 5′-CTACTGTATTTAACTGATTTGAAG-3′.
Generation of Tim-1-deficient mice by targeted disruption of the mouse havcr1−/− in embryonic stem cells
The Havcr1 replacement vector was constructed to insert the neomycin-resistance gene into exon 2 of the Havcr1 gene, deleting the nucleotides encoding amino acids Pro36–Asp118 (83 amino acids) of the 305-amino-acid Tim-1 translated sequence. The 4·5 kilobase 5′ arm of homology was generated using the RPCI21 PAC345-B13 as a template with PCR primers 5′-TGGGCATGGCGGCCGCTACCTGTAATCTTAGCATTCTGAACCTGG-3′ and 5′-CGTTGTGGATCCACGATATGTTGAGTAAGTACATGG-3′. The 4·1 kilobase 3′ homology arm was generated using the RPCI21 PAC345-B13 as a template with PCR primers 5′-TATTGTTGACTAGTGGAGATTCCTGGATGGTTTAATGATC-3′ and 5′-CTGGCTACTAGTGAATGCCCTGGGGATTTGATC-3′.
Havcr1 targeting vector was linearized and electroporated into E14·1 ES cells. Targeted embryonic stem (ES) cell clones were microinjected into 3·5-day C57BL/6 blastocysts to generate chimeras. These mice were mated with C57BL/6 mice and transmitted the ES cell genotype through the germline. Mice homozygous for the disrupted havcr1 gene were obtained by inter-breeding the heterozygotes (havcr1+/−). The havcr1−/− and corresponding wild-type animals used in the experiments reported below were derived from the original chimeras backcrossed for six generations to the BALB/c strain before inter-crossing to generate homozygous mice. Genotypes were screened by Southern blot analysis or PCR using primers 5′-GGGTCACCCTGTCACACTTCC-3′ and 5′-TTAACCATCCAGGAATCTCC-3′.
Antibodies and reagents
All antibodies used in enzyme-linked immunosorbent assay (ELISA) were purchased from BD Bioscience, Oxford, UK. These include affinity-purified capture antibodies anti-IgM (II/41), anti-IgG1 (A85-3), anti-IgG2a (R19-15), anti-IgE (R35-72), anti-interleukin-4 (IL-4; 11B11) and anti-IL-5 (TRFK5); as well as affinity-purified detection antibodies anti-IgM biotin (R6-60·2), anti-IgG1 biotin (A85-1), anti-IgG2a biotin (R19-15), anti-IgE biotin (R35-72), anti-IL-4 biotin (BVD6-24G2) and anti-IL-5 biotin (TRFK4). All antibodies used in flow cytometry, unless stated, were purchased from eBioscience, Hatfield, UK. These include anti-Tim-1 biotin (RMT1-4), anti-CD19 Pacific Blue (1D3), phycoerythrin-conjugated (PE) anti-CD19 (1D3), anti-CD3 PE Cy7 (145-2C11), anti-CD4 PE (GK1.5), anti-CD16/32 (clone93) and streptavidin PECy7. Anti-CD95 PE (Jo-2), IgG2b biotin, streptavidin PE and streptavidin allophycocyanin were obtained from BD Bioscience. Affinity-purified goat anti-mouse IgM F(ab′)2 was from Jackson ImmunoResearch (West Grove, PA). Fluorescein isothiocyanate-conjugated peanut agglutinin (PNA) and PNA biotin were from Vector Laboratories (Peterborough, UK). Phosphatidylinositol 3-kinase (PI3K) specific inhibitor (Ly294002) was from Calbiochem (Nottingham, UK). IκB-kinase (IKKε) -specific inhibitor (BX795) and p38 mitogen-activated protein kinase-specific inhibitor (BIRB796) were kind gifts from Professor Philip Cohen (University of Dundee, Dow Street, Dundee, Scotland, DD1 5EH). Schistosoma mansoni eggs (SE) and SE antigen (SEA) were kind gifts from Dr Padraic Fallon.
B-cell and CD4+ T-cell isolation using magnetic beads
Total splenocytes were isolated from the spleen and red blood cell lysis was performed using buffer containing ammonium chloride. B cells were purified by negative selection using a B-cell isolation kit (Miltenyi Biotec, Surrey, UK). CD4+ cells were isolated by positive selection using mouse CD4 (L3T4) MicroBeads (Miltenyi Biotec). Purified B or T cells were obtained using a magnetic antibody cell sorting (MACS) separation system following the standard manufacturer protocol. The purity of the isolated B and T cells was routinely checked by flow cytometry and determined to be > 98% and > 90%, respectively.
Cell culture
Total splenocytes or purified B cells were cultured either in standard RPMI media [with 10% fetal bovine serum (FBS), 1% penicillin and streptomycin, 0·1 mm 2-mercaptoethanol], or media containing 10 μg/ml goat anti-mouse IgM or 1 μg/ml pure lipopolysaccharide (LPS) for 1–5 days. For inhibitor studies, cells were pre-treated for 30 min with respective inhibitors before stimulation with anti-IgM. Th1 and Th2 cells were generated in vitro according to the previous described protocol.16
CFSE Proliferation assay
Total splenocytes in phosphate-buffered saline (PBS) were incubated with 2 μm carboxyfluorescein succinimidyl ester (CFSE; Invitrogen, Paisley, UK) at 37° for 10 min. Cells were then washed thrice with complete media. Cells were then plated at 5 × 105 cells per well in a 96-well plate with or without goat anti-mouse anti-IgM F(ab′)2 (10 μg/ml; Jackson ImmunoResearch) or LPS (1 μg/ml, 0127:B81; Sigma, Dorset, UK). Cells were cultured for 5 days. Cell proliferation was analysed as CFSE dilution.
[3H]Thymidine uptake cell proliferation assay
Total splenocytes were plated at 3 × 105 cells per well in a 96-well plate with or without stimulation for 48 hr. [3H]Thymidine (GE Healthcare, London, UK) was then added to each well at a final activity of 6·25 μCi. The cells were incubated for a further 18 hr before the thymidine incorporation was measured by scintillation counting.
Quantitative TaqMan PCR
Total RNA was prepared by phenol–chloroform extraction using RNAbee. Contaminating genomic DNA was removed using a DNAfree Turbo kit (Ambion, Warrington, UK) following the manufacturer's instructions. The cDNA was generated with Superscript III reverse transcriptase (Invitrogen) following manufacturer protocol. Relative expression of Tim-1 in each sample was analysed using TaqMan quantitative PCR using forward primer 5′-TCTATGTTGGCATCTGCATCG-3′, reverse primer 5′-GTACCTGGTGATAGCCACGGT-3′ and TaqMan probe 5′-6-FAM-AGCCCTGCTGCTACTGCTCCTTGTG-TAMRA-3′. An 18S ribosomal RNA primer-probe was used as an internal reference for normalization of well-to-well variability. Assay was performed on 7900HT Fast Real-Time PCR System (Applied Biosystems, Warrington, UK). Results were analysed using sds 2.2.2 software (Applied Biosystems). Standard procedures of analysis were followed to achieve relative expression values. Threshold cycles (CT) were normalized by subtraction of the average 18S ribosomal RNA CT value from the average Tim-1 CT value to achieve CT. Then ΔCT (CT sample – CT calibrator), which represents the relative n values in various samples compared with that of calibrator, was calculated. The quantitative data are means of triplicate experiments.
Flow cytometry analysis
All staining procedures were performed in PBS/2% FBS at 4°. A single-cell suspension was incubated with anti-CD16/32 antibodies for 20 min to block non-specific binding. The cells were then washed once and incubated with biotinylated or fluorochrome-conjugated antibodies for 30 min followed by two washing steps. When necessary, the cells were incubated with fluorochrome-conjugated streptavidin for 20 min followed by two washing steps. Cells were finally resuspended in PBS/2% FBS and analysed on FACS Caliber (BD Bioscience) or LSRII (BD Bioscience) following the manufacturer's standard operating procedures. The data obtained were analysed using flowjo software version 8.8.8 (Tree Star, Ashland, OR).
Immunization with sheep red blood cells
Sheep blood diluted 1 : 1 in Alsever's solution was purchased from Harlan, Oxford, UK and washed three times in sterile PBS. Mice were immunized with a single intraperitoneal injection of 2 × 107 sheep red blood cells (SRBC) in 200 μl PBS. Another group of mice with the same genotype were given 200 μl PBS as a control. Mice were killed on day 9 post-immunization with SRBC. Spleens were taken from each mouse group and used for flow cytometry analysis.
ELISA analysis
Flat-bottom 96-well ELISA microtitre plates (MAXISORP; Nunc, Roskilde, Denmark) were coated with 2 μg/ml of capture antibodies (refer to Antibodies and reagents) and held overnight at 4°. The plates were washed three times with PBS containing 0·05% Tween-20 (PBST) and blocked with PBS containing 10% fetal bovine serum (PBS/FBS) for 2 hr. For measuring antibody titre in the sera two-fold series dilution of the sera (starting at 1 in 20 dilution) was performed in PBS/FBS. Supernatant collected from the cell culture experiments was used without dilution. All the samples were added to the ELISA plate and incubated at room temperature for 2 hr. The plates were then washed three times with PBST; 1 μg/ml of biotinylated detection antibody was then added to their respective wells and incubated for 1 hr at room temperature before washing the plates three times with PBST and 1 μg/ml of streptavidin-horseradish peroxidase (VectaLab) was added and incubated at room temperature for 30 min before washing five times with PBST. A peroxidase substrate, 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulphonic acid) (Sigma) was then added, following the manufacturer's instructions. The signal was read spectrophotometrically at 405 nm.
Lung tissue collection and histology
Lungs were fixed in formalin (10% formaldehyde in 0·9% saline solution) and stained with Giemsa stain for inflammatory infiltrate and with periodic acid-Schiff reagent for goblet cells. Inflammation and mucus production were evaluated blindly by using numerical scoring expressed in arbitrary units.
Statistical analysis
Data shown in Fig. 3(a,b) were compared by linear and non-linear regression analysis. Other comparisons were analysed by unpaired Student's t-test. All analyses were performed using graphpad prism software (GraphPad Software, La Jolla, CA).
Figure 3.
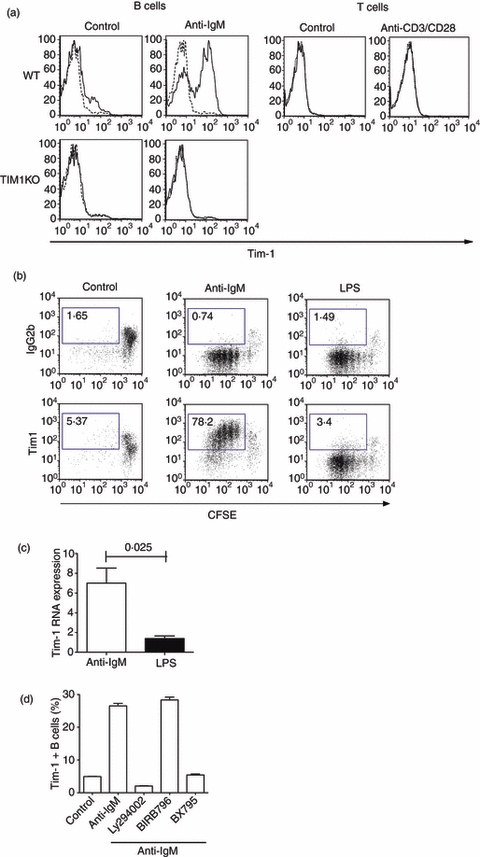
Tim-1 is induced on B cells through the B-cell receptor signalling pathway. (a) B and T cells were isolated from the spleen of either wild-type or Tim-1-deficient mice and cultured in media (control) or media containing anti-immunoglobulin M (IgM) or anti-CD3 and CD28. Expression of Tim-1 on each cell type was analysed by flow cytometry 48 hr later using anti-Tim-1 antibody (solid line) or isotype control (dash line). Data shown are representative from at least three independent experiments with three mice per group. (b) Total splenocytes from naive mice were labelled with carboxyfluorescein succinimidyl ester (CFSE) and cultured in media with or without anti-IgM or lipopolysaccharide (LPS). CFSE dilution and Tim-1 expression on CD19+ cells were analysed by flow cytometry on day 5. Data shown are representative from three independent experiments with three mice per group. (c) Purified B cells were cultured in media with or without anti-IgM or LPS for 24 hr. Tim-1 expression in each sample was analysed by real-time polymerase chain reaction, in triplicate, using 18S as an internal control. Tim-1 expression was then normalized to the unstimulated control (with value 1). Data shown are mean ± standard error of the mean from three independent experiments with two mice per group. (d) B cells were cultured in media or media containing anti-IgM with or without various inhibitors as shown. Percentage of Tim-1-expressing B cells was analysed by flow cytometry analysis. Data shown are derived from at least three independent experiments.
Results
Generation of Tim-1-deficient (Havcr-1−/−) mice and Tim-1-over-expressing transgenic mice
To study the role of Tim-1 in T-cell-mediated immune responses, we generated both Tim-1-deficient mice and Tim-1-over-expressing transgenic mice. Tim-1-deficient mice were generated by gene targeting in ES cells (Fig. 1a,b). Reverse transcription (RT) PCR analysis of cDNA preparations from naive tissue samples showed that the full length Havcr-1 transcript was highly expressed in the mesenteric lymph node and kidney but could not be detected in any of the samples taken from Tim-1-deficient mice, even following Tim-1 transcript-specific hybridization (Fig. 1c). Tim-1-over-expressing transgenic mice were generated by cloning the Tim-1 cDNA into the pTEXCD2 vector which contains regulatory elements from the human CD2 gene driving transgene expression in either T cells, or T and B cells (Fig. 1d,e). Tim-1 transgenic mice had highly elevated expression of Tim-1 messenger RNA compared to their wild-type littermates (Fig. 1f). We also confirmed Tim-1 cell surface expression using several transgenic lines, with some lines having Tim-1 expression restricted only to the T cells while other lines showed Tim-1 expression on both T and B cells (Fig. 1g). Both Tim-1-deficient and Tim-1-over-expressing transgenic mice were healthy and displayed no overt phenotypic abnormalities.
Figure 1.
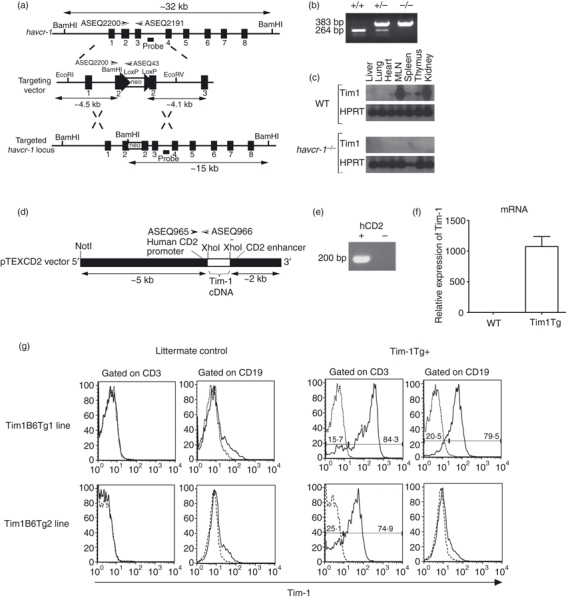
Generation of Tim-1-deficient mice and Tim-1-over-expressing transgenic mice. (a) The structure of the Havcr-1 locus, its targeting vector, and predicted homologous recombination event. neo, neomycin resistance cassette. (b) Genotyping of genomic DNA from wild-type (+/+), Havcr-1 gene-targeted heterozygous (+/−) and homozygous (−/−) mice by polymerase chain reaction (PCR) analysis. (c) Reverse transcription (RT-) PCR analysis of Tim-1 expression in various tissues collected from either wild-type (WT) or Havcr-1−/− mice. HPRT was used as a loading control for the analysis. Data shown are representative of three independent experiments performed. (d) Tim-1 complementary DNA was cloned into the XhoI site of the pTEXCD2 vector. The vector was then linearized by NotI digestion. (e) Genotyping of genomic DNA from Tim-1-over-expressing mouse and its littermate control by PCR analysis using primers ASEQ965 and ASEQ966. (f) RT-PCR analysis of Tim-1 expression on total splenocytes from either WT or Tim-1 transgenic (Tim1Tg) mice. (g) Flow cytometric analysis of Tim-1 expression on B and T cells from wild-type and Tim-1-over-expressing transgenic mice.
T-cell and B-cell proliferation and Th1 and Th2 differentiation are normal in Tim-1-deficient and Tim-1 transgenic mice
We initially assessed the proliferative capacity of naive T and B cells from the Tim-1-deficient mice and Tim-1-over-expressing transgenic mice. Total splenocytes from Tim-1-deficient mice or their wild-type controls were cultured in media in the absence (control) or presence of anti-CD3 or anti-IgM stimulation. This polyclonal stimulation induced strong T-cell and B-cell proliferation, respectively. However, there was no significant difference in proliferation between wild-type and Tim-1-deficient mice (Fig. 2a). Similar studies were performed using total splenocytes from Tim-1-over-expressing transgenic mice. Again, no differences were found between the wild-type mice and Tim-1 transgenic mice (Fig. 2b).
Figure 2.
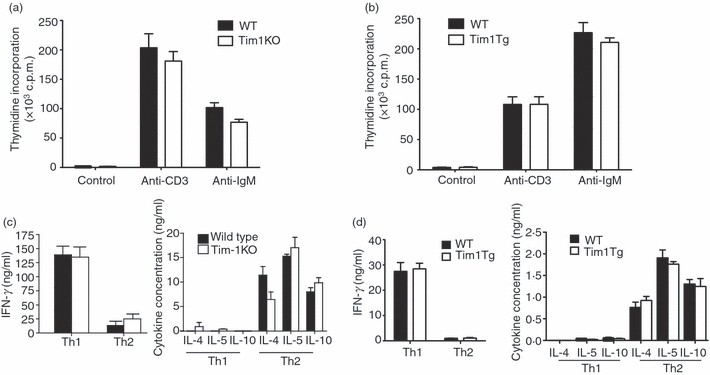
T-cell proliferation and differentiation is normal in Tim-1-deficient mice and Tim-1 transgenic mice. Proliferation of B and T cells from Tim-1-deficient mice (a) and Tim-1 transgenic mice (b) in media alone (control) or in media containing anti-CD3 or anti-immunoglobulin M (IgM). Thymidine incorporation assay for each sample was performed in triplicate. Data shown are from a single experiment with four or five mice per group but are representative of three repeat experiments. Cytokine production by total wild-type and Tim-1-deficient splenocytes (c), or by wild-type and Tim-1 transgenic CD4+ T cells (d) differentiated under T helper type 1 (Th1) or Th2 polarizing conditions and re-stimulated for 24 hr with anti-CD3ε antibody. Data shown are from a single experiment with four or five mice per group but are representative of three repeat experiments.
The original description of Tim-1 suggested a role for Tim-1 in regulating type-2 responses, so T-cell differentiation and proliferation were assessed in Tim-1-deficient mice by culturing splenocytes under Th1 and Th2 polarizing conditions in vitro. No differences were found in the production of the Th2 cytokines IL-4, IL-5 and IL-10 or the Th1 cytokine interferon-γ between wild-type and Tim-1-deficient mice (Fig. 2c). Proliferation of the wild-type and Tim-1-deficient Th1 or Th2 cultures was also normal (data not shown).
To complement this analysis we also undertook Th1 and Th2 differentiation assays using T cells derived from different lines of Tim-1 transgenic mice. We found no evidence for the modulation of in vitro Th1 and Th2 differentiation by over-expression of Tim-1 (Fig. 2d and data not shown). Furthermore, analysis of cytokine production by naive splenocytes from wild-type and Tim-1 transgenic mice was also similar (data not shown).
Induction of Tim-1 expression on B cells is B-cell receptor signalling dependent
Our data demonstrate that neither Tim-1-deficient mice nor Tim-1-over-expressing transgenic mice showed any modulation in B-cell and T-cell proliferation and differentiation. It has been demonstrated that activation of CD4+ T cells through their T-cell receptor induces expression of Tim-1 that can be detected using an anti-Tim-1 antibody, clone 3B3.9 We aimed to follow up this finding using a commercially available anti-Tim-1 antibody, clone RMT1-4. We purified mouse CD4+ T cells and B cells from naive mice and activated them in vitro using anti-CD3/CD28, and anti-IgM, respectively, for 48 hr to evaluate the expression of Tim-1 on activated cells. Tim-1 expression was detected on activated B cells, but not activated T cells (Fig. 3a). We were also unable to detect Tim-1 expression on in vitro differentiated Th2 cells, but reproducibly detected Tim-1 expression on HEK293 cells transfected with Tim-1 cDNA (data not shown). To ensure that this anti-Tim-1 antibody did not cross-react with other Tim-1 family members, especially Tim-2, we also purified B cells from Tim-1-deficient mice and activated them in vitro using anti-IgM. As expected, the binding of this antibody on activated Tim-1-deficient B cells was totally ablated in the absence of Tim-1, confirming the specificity of this antibody, and verifying the regulation of Tim-1 on B cells (Fig. 3a).
Both B-cell receptor (BCR) cross-linking17 and LPS18 have been shown to induce B-cell proliferation in vitro. We next determined if anti-IgM and LPS have similar effects on Tim-1 expression on B cells. Total splenocytes from naive mice were labelled with CFSE and cultured in the presence or absence of anti-IgM or LPS. After 5 days of culture, we evaluated the proliferation of B cells as well as the induction of Tim-1 using flow cytometric analysis. Interestingly, although both anti-IgM and LPS induced B-cell proliferation, only anti-IgM induced Tim-1 expression on B cells (Fig. 3b). This was further substantiated by analysis of Tim-1 messenger RNA expression by B cells, demonstrating that anti-IgM but not LPS up-regulated Tim-1 gene expression (Fig. 3c). These data demonstrate that the induction of Tim-1 expression on B cells is dependent on the BCR signalling pathway.
B-cell antigen receptor signalling has been shown to involve the PI3K and nuclear factor-κB pathways,19–21 but also activation of extracellular signal-regulated kinase 2 (ERK2) and more weakly ERK1, Jun N-terminal kinase and p38.22 To evaluate the role of these signalling molecules in the induction of Tim-1 by BCR signalling, we used PI3K (Ly294002), IKKε (BX795)23 and p38α/β mitogen-activated protein kinase (MAPK; BIRB796)24 inhibitors to interfere with these signalling molecules. Purified B cells were pre-treated with or without inhibitors for 15 min before being cultured with anti-IgM. Expression of Tim-1 was then analysed 24 hr later using flow cytometry. Interestingly, although PI3K and IKKε inhibitors completely abrogated the induction of Tim-1 on B cells, the MAPK inhibitor had no effect on Tim-1 induction (Fig. 3d).
B-cell and T-cell responses are unaffected during a model of type-2 immune response when Tim-1 is over-expressed
Our in vitro characterization of Tim-1 transgenic T and B cells failed to show any overt phenotype compared to their wild-type controls. We next evaluated whether the over-expression of Tim-1 on B and T cells would affect type-2 immune responses in vivo. Because Tim-1 has been proposed to regulate Th2 responses, we used Schistosoma mansoni eggs (SE) as inducers of a potent Th2 response. This in vivo model has been shown to induce IL-4 and IL-5 production as well as eosinophilia, increased mucus production and elevated IgE.25 We injected SE intraperitoneally into Tim-1 transgenic mice and their littermate controls on day 0 and re-challenged the mice with intravenous injection of SE on day 14. Control groups only received PBS. Four days after the final challenge the mice were killed and tissues were subjected to various immunological analyses.
As we had identified that Tim-1 is expressed on BCR-triggered B cells in vitro, we assessed whether the over-expression of Tim-1 on B cells would alter their antibody response in vivo by determining the level of the various SE-specific immunoglobulin isotypes in the sera of PBS or SE wild-type and transgenic mouse groups. Although levels of SE-specific IgM, IgE and IgG1 increased substantially in the mouse groups that received SE challenge compared with the PBS-treated controls the magnitude of this was similar in the Tim-1 transgenic mice and their wild-type littermates (Fig. 4a). These data show that over-expression of Tim-1 on B and T cells has no detectable effect on B-cell antibody responses in vivo.
Figure 4.
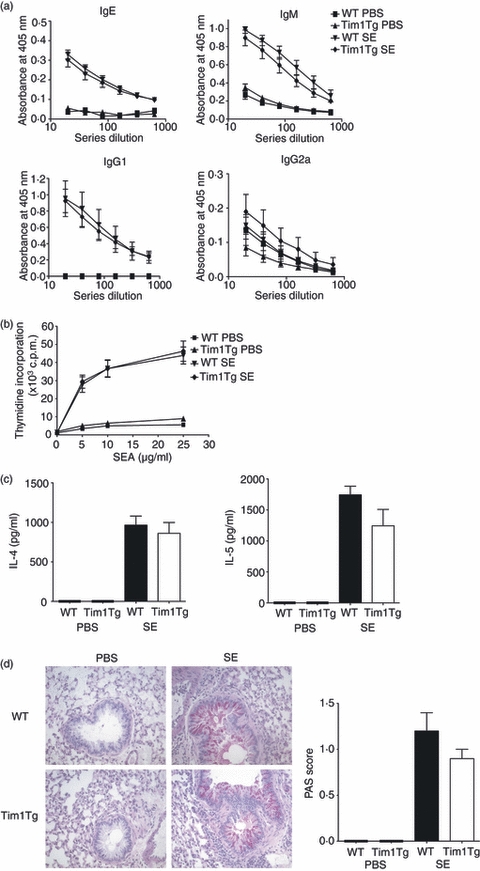
Tim-1 over-expressing mice have a normal in vivo type 2 immune response. Wild-type or Tim-1 transgenic mice received intraperitoneal injection of phosphate-buffered saline (PBS) or Schistosoma mansoni eggs. Two weeks later the mice received PBS or Schistosoma mansoni eggs intravenously. Mice were killed 3 days later. (a) Relative amount of various isotypes of Schistosoma mansoni egg-specific antibodies in each mouse group were analysed by direct enzyme-linked immunosorbent assay (ELISA). (b) Splenocytes from each mouse group were re-stimulated in vitro with various concentrations of Schistosoma mansoni egg antigen (SEA). Proliferation was measured by thymidine incorporation assay, in triplicate, after 48 hr. (c) Interelukin-4 (IL-4) and IL-5 production from the day 2 SEA-stimulated total splenocytes from each mouse group was measured by sandwich ELISA. (d) Lungs from each mouse group were fixed in formalin. Paraffin sections were prepared and stained with Giemsa for cell infiltrate and periodic acid-Schiff (PAS) for goblet cells (pictures). Mucus production was evaluated blindly by using numerical scoring expressed in arbitrary units (histogram). Data shown are from a single experiment with five or six mice per group but are representative of three repeat experiments.
We also assessed T-cell responses from the SE-challenged mice. Total splenocytes were isolated from all the mouse groups and re-stimulated in vitro with SE antigen for 48 hr. We then measured cell proliferation by thymidine incorporation. As shown in Fig. 4(b), while splenocytes from PBS-treated mouse groups had a minimal proliferative response, SE-challenged mice responded strongly to the SE antigen restimulation in a dose-dependent manner (Fig. 4b). However, there was no significant difference in cell proliferation between the Tim-1 transgenic mice and their littermate controls that received SE challenge (Fig. 4b). We also measured the IL-4 and IL-5 production from the splenocytes that were re-stimulated with SE antigen. As expected, splenocytes from SE mouse groups, but not PBS-treated mouse groups, produced IL-4 and IL-5 in response to SE antigen re-stimulation. However, once again the cytokine levels from the Tim-1 transgenic mice and their littermate controls were very similar (Fig. 4c).
Intravenous injection of SE into the mice results in the circulation of the SE to the lung where they become trapped and provoke T-cell-mediated granulomatous inflammation.26 Paraffin lung sections were prepared from all the mouse groups and stained with Giemsa for inflammatory infiltrate and periodic acid-Schiff (PAS) for goblet cells. As shown in Fig. 4(d), both PBS-treated Tim-1 transgenic mice and their littermate controls showed healthy lung airways with no cell infiltrate or mucus production. Challenge with SE resulted in inflammatory cell infiltration into the lungs and increased mucus production (Fig. 4d). The over-expression of Tim-1 did not alter the magnitude of these responses (Fig. 4d,e).
We also performed similar studies using Tim-1-deficient mice. Again, there was no difference in the immune response between the wild-type and the Tim-1-deficient mice (data not shown). In addition we also infected Tim-1-deficient mice with helminth Nippostrongylus brasiliensis and observed no significant differences in the responses (such as worm burden) of Tim-1-deficient mice compared with wild-type mice (data not shown).
Tim-1 is expressed predominantly on germinal centre B cells in vivo
Although our in vitro studies showed that Tim-1 could be induced on B cells, our in vivo analysis using Tim-1 transgenic mice failed to demonstrate a functional effect for this expression. This led us to ask if Tim-1 is indeed expressed on B cells in vivo. As Tim-1 expression on B cells is dependent on BCR cross-linking, we hypothesized that Tim-1 would be expressed on BCR-triggered B cells in vivo. During a T-cell-dependent B-cell response, activated B cells migrate to the B-cell follicle and initiate germinal centres where somatic hypermutation and isotype switching occur.27 These B cells are subjected to selection through BCR signalling.28 Therefore, we hypothesized that Tim-1 would be predominantly expressed on germinal centre B cells. We used SRBC immunization to induce germinal centre responses in wild-type mice and then compared the expression of Tim-1 on germinal centre B cells and non-germinal centre B cells. Using PNA and CD95 as markers for germinal centre B cells we detected the induction of germinal centre B cells in the spleens of mice that received SRBC injection (Fig. 5a). Strikingly, all of the germinal centre B cells had high-level cell surface expression of Tim-1 when compared with non-germinal centre B cells (Fig. 5b). This observation led us to ask if Tim-1 expression is essential for germinal centre induction. To test this we injected SRBC into wild-type or Tim-1-deficient mice and assessed the induction of germinal centre B cells on day 9 after challenge. As shown in Fig. 5(c), comparable proportions of germinal centre B cells were observed in wild-type and Tim-1-deficient mice. Hence, although Tim-1 is highly expressed on the surface of germinal centre B cells it is not essential for their development in vivo.
Figure 5.
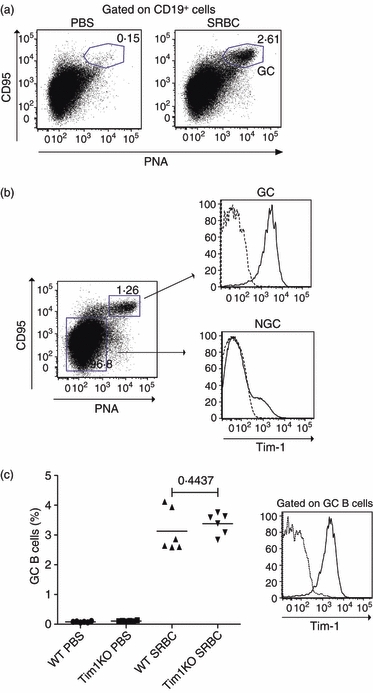
Tim-1 is predominantly expressed on germinal centre B cells. (a) Wild-type BALB/c mice were given intraperitoneal injections of phosphate-buffered saline (PBS) or 2 × 107 sheep red blood cells (SRBCs). On day 9, induction of germinal centre (GC) B cells was analysed by flow cytometry using anti-Tim-1 (solid line) or isotype control (dashed line). CD19, peanut agglutinin (PNA) and CD95 were used as markers for GC B cells. (b) Expression of Tim-1 on GC B cells and non-GC (NGC) B cells on day 9 for SRBC-challenged mice was analysed by flow cytometry using anti-Tim-1 (solid line) or isotype control (dashed line). (c) Wild-type (WT) and Tim-1-deficient (Tim1KO) mice were given intraperitoneal injections of PBS or 2 × 107 SRBCs. On day 9, the percentage of GC B cells within the B-cell population was analysed (left). Expression of Tim-1 on GC B cells from day 9 SRBC challenged WT (solid line) and Tim1KO (dashed line) mice was analysed by flow cytometry using anti-Tim-1 antibody (right). All data shown are derived from a single experiment with five or six mice per group but are representative of three repeat experiments.
Discussion
Our studies convincingly show that Tim-1 is induced on germinal centre B cells during an immune response in vivo. Although previous studies reported the expression of Tim-1 on activated T cells,9 we have found that high-level expression of Tim-1 is predominantly on activated B cells rather than T cells. As explained in the Introduction, a number of studies of Tim-1 have relied on a limited number of anti-Tim-1 antibodies that show highly variable staining and biological properties. During our studies we tested a number of anti-Tim-1 antibodies and to resolve the issue of specificity, we used three criteria to evaluate the various antibodies: an anti-Tim-1-specific antibody must (i) bind a cell line transfected with the Tim-1 transgene; (ii) bind cells from our Tim-1 over-expressing transgenic mice; and (iii) not bind to cells from Tim-1-deficient mice. We found that anti-Tim-1 clone RMT1-4 was the only antibody, that we tested, that met these criteria. However, using this anti-Tim-1 antibody we failed to detect Tim-1 expression on in vitro activated T cells or in vitro differentiated Th2 cells. Although this could be a result of the level of Tim-1 expression on these cells being below the detection limit of this antibody we were able to detect Tim-1 expression on anti-IgM-activated B cells.
Since the identification of Tapr (an airway hyper-reactivity regulatory locus)4 and the report of preferential Tim-1 expression on Th2 cells9, a number of studies have examined the expression and function of murine Tim-1 using an anti-Tim-1 antibody9,11,12,29 or a TIM-1-immunoglobulin fusion protein.30 However, these studies suggest that the proteins of this family function in a much broader manner than just on T cells. Mast cells have been shown to express Tim-1 and its activation led to enhancement of Th2 cytokine production.10 Tim-1 has also been shown to be a phosphatidylserine receptor and it plays an important role in controlling phagocytosis.31–33 In fact it has recently been suggested that the definition of Tim should be adjusted to ‘transmembrane immunoglobulin and mucin’.34 Furthermore, the V-type immunoglobulin domain and glycosylated mucin-like domain of Tim proteins have been proposed to bind a diversity of ligands, suggesting a degree of promiscuity in their molecular interactions.13 These data suggest that there may be limitations in using anti-Tim-1 antibodies and fusion proteins to understand the function of Tim-1 on T cells. To address this, we generated transgenic mice either deficient in Tim-1 expression or with Tim-1 over-expression. We successfully generated several lines of Tim-1 transgenic mice in which the regulatory elements from the human CD2 gene were used to drive Tim-1 transgene expression in either T cells, or T and B cells. In this report, we have presented the results obtained from one of the lines with Tim-1 over-expressed on both T and B cells, but this line mirrored the results with transgenic Tim-1 solely in the T-cell compartment. Previously it has been reported that ectopic expression of Tim-1 using retroviral transduction during T-cell differentiation resulted in a significant increase in the number of IL-4-producing cells.35 Tim-1 protein has also been proposed to influence the activation and differentiation of T cells.36 However, results from our transgenic mouse studies showed that high-level expression of Tim-1 on T cells had no significant effect on T-cell proliferation or cytokine production in vitro. Furthermore, ablation of Tim-1 in gene-targeted mice also failed to show any effect on T-cell proliferation or cytokine production in vitro. This was surprising given the reports of Tim-1 correlating with type 2 immune responses. However, when we assessed a potent in vivo inducer of type 2 immunity, using Tim-1 transgenic and Tim-1-deficient mice, we also failed to demonstrate any critical requirement for Tim-1 in the regulation of type 2 cytokines, or pathological features such as inflammatory cell infiltration, eosinophilia, mucus production or IgE expression. We have previously used SE challenge in vivo and assays of T-cell function in vitro to address the roles of several molecules that have been implicated in type 2 immune responses and they have proven robust measures of involvement in type 2 immunity. Our results suggest that Tim-1 plays only a minor role in type 2 responses.
Despite these unexpected results, we did identify high levels of Tim-1 expression on germinal centre B cells from antigen-challenged animals. This expression was absent on the germinal centre B cells derived from the Tim-1-deficient mice, confirming the specificity of the observed staining. However, we have been unable to show an essential role for Tim-1 expression for the induction of the germinal response because the levels of germinal centre B cells in the Tim-1-deficient mice were comparable to controls. In addition, antibody responses were also unaffected by the constitutive presence of Tim-1 on B cells or its ablation. This may result from redundancy between Tim family molecules. Murine Tim-1 and Tim-2 have been shown to have highly similar sequences.5 Furthermore, Tim-2 had also been shown to be expressed on T cells and to regulate Th2 immune responses.37 Expression of Tim-2 on B cells has also been reported recently.38 It is currently not known if Tim-1 and Tim-2 have redundant roles in germinal centre B-cell activities. However, Tim-2 has been found on all splenic B cells and is a receptor for H-ferritin endocytosis.38 In contrast to the reported ubiquitous expression of Tim-2 on splenic B cells, our studies demonstrate a low level of Tim-1 expression on a small subset (5–10%) of non-germinal centre B cells in the spleen. However, we do not have any evidence that these B cells are functionally different from the Tim-1-negative B cells.
Our results conclude that Tim-1 is induced on B cells through BCR signalling. We observed that although both anti-IgM and LPS induced B-cell proliferation in vitro, only anti-IgM stimulation induced Tim-1 expression on activated B cells. This result suggests a requirement for BCR-induced but not Toll-like receptor-4-induced signalling pathways for the induction of Tim-1. We also showed the potential role for PI3K and nuclear factor-κB pathways in the induction of Tim-1 expression because both PI3K and IKKε-specific inhibitors totally abrogated Tim-1 induction following BCR activation. This result was supported by previous studies showing a critical role for PI3K and nuclear factor-κB in BCR-mediated signalling.20,21 However, LPS has also been shown previously to activate IKKε in human macrophages,39 and to induce B-cell proliferation in a PI3K-dependent manner,40 suggesting that there are other signalling components specific to BCR signalling involved in the induction of Tim-1.
In summary, we have generated both Tim-1-deficient and Tim-1-over-expressing transgenic mice and assessed their type-2 immune responses both in vitro and in vivo. Notably, neither of these mouse lines showed any deficit or gain of function in these responses. Interestingly, using this approach we discovered that B-cell signalling leads to the up-regulation of Tim-1 on activated B cells and robust high-level expression of Tim-1 on germinal centre B cells in vivo. Although we were unable to identify a critical role for Tim-1 in the development of germinal centre reactions or antibody responses, Tim-1 in combination with other cell surface receptors represents a useful marker of this population.
Acknowledgments
We would like to thank the animal facility staff for their technical assistance. See Heng Wong and Stephen Nabbaro are supported by a grant from Asthma, UK.
Disclosures
The authors have no financial conflict of interest.
References
- 1.de Souza AJ, Kane LP. Immune regulation by the TIM gene family. Immunol Res. 2006;36:147–55. doi: 10.1385/IR:36:1:147. [DOI] [PubMed] [Google Scholar]
- 2.Hafler DA, Kuchroo V. TIMs: central regulators of immune responses. J Exp Med. 2008;205:2699–701. doi: 10.1084/jem.20082429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McIntire JJ, Umetsu DT, DeKruyff RH. TIM-1, a novel allergy and asthma susceptibility gene. Springer Semin Immunopathol. 2004;25:335–48. doi: 10.1007/s00281-003-0141-3. [DOI] [PubMed] [Google Scholar]
- 4.McIntire JJ, Umetsu SE, Akbari O, et al. Identification of Tapr (an airway hyperreactivity regulatory locus) and the linked Tim gene family. Nat Immunol. 2001;2:1109–16. doi: 10.1038/ni739. [DOI] [PubMed] [Google Scholar]
- 5.Meyers JH, Sabatos CA, Chakravarti S, Kuchroo VK. The TIM gene family regulates autoimmune and allergic diseases. Trends Mol Med. 2005;11:362–9. doi: 10.1016/j.molmed.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 6.Ichimura T, Bonventre JV, Bailly V, Wei H, Hession CA, Cate RL, Sanicola M. Kidney injury molecule-1 (KIM-1), a putative epithelial cell adhesion molecule containing a novel immunoglobulin domain, is up-regulated in renal cells after injury. J Biol Chem. 1998;273:4135–42. doi: 10.1074/jbc.273.7.4135. [DOI] [PubMed] [Google Scholar]
- 7.Kaplan G, Totsuka A, Thompson P, Akatsuka T, Moritsugu Y, Feinstone SM. Identification of a surface glycoprotein on African green monkey kidney cells as a receptor for hepatitis A virus. EMBO J. 1996;15:4282–96. [PMC free article] [PubMed] [Google Scholar]
- 8.McIntire JJ, Umetsu SE, Macaubas C, et al. Immunology: hepatitis A virus link to atopic disease. Nature. 2003;425:576. doi: 10.1038/425576a. [DOI] [PubMed] [Google Scholar]
- 9.Umetsu SE, Lee WL, McIntire JJ, et al. TIM-1 induces T cell activation and inhibits the development of peripheral tolerance. Nat Immunol. 2005;6:447–54. doi: 10.1038/ni1186. [DOI] [PubMed] [Google Scholar]
- 10.Nakae S, Iikura M, Suto H, Akiba H, Umetsu DT, Dekruyff RH, Saito H, Galli SJ. TIM-1 and TIM-3 enhancement of Th2 cytokine production by mast cells. Blood. 2007;110:2565–8. doi: 10.1182/blood-2006-11-058800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sizing ID, Bailly V, McCoon P, et al. Epitope-dependent effect of anti-murine TIM-1 monoclonal antibodies on T cell activity and lung immune responses. J Immunol. 2007;178:2249–61. doi: 10.4049/jimmunol.178.4.2249. [DOI] [PubMed] [Google Scholar]
- 12.Xiao S, Najafian N, Reddy J, et al. Differential engagement of Tim-1 during activation can positively or negatively costimulate T cell expansion and effector function. J Exp Med. 2007;204:1691–702. doi: 10.1084/jem.20062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilker PR, Sedy JR, Grigura V, Murphy TL, Murphy KM. Evidence for carbohydrate recognition and homotypic and heterotypic binding by the TIM family. Int Immunol. 2007;19:763–73. doi: 10.1093/intimm/dxm044. [DOI] [PubMed] [Google Scholar]
- 14.Lake RA, Wotton D, Owen MJ. A 3′-transcriptional enhancer regulates tissue-specific expression of the human CD2 gene. EMBO J. 1990;9:3129. doi: 10.1002/j.1460-2075.1990.tb07510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lang G, Wotton D, Owen MJ, Sewell WA, Brown MH, Mason DY, Crumpton MJ, Kioussis D. The structure of the human CD2 gene and its expression in transgenic mice. EMBO J. 1988;7:1675. doi: 10.1002/j.1460-2075.1988.tb02995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cousins DJ, Lee TH, Staynov DZ. Cytokine coexpression during human Th1/Th2 cell differentiation: direct evidence for coordinated expression of Th2 cytokines. J Immunol. 2002;169:2498. doi: 10.4049/jimmunol.169.5.2498. [DOI] [PubMed] [Google Scholar]
- 17.Mongini P, Friedman S, Wortis H. Accessory cell requirement for anti-IgM-induced proliferation of B lymphocytes. Nature. 1978;276:709–11. doi: 10.1038/276709a0. [DOI] [PubMed] [Google Scholar]
- 18.Marcelletti JF. IL-10 inhibits lipopolysaccharide-induced murine B cell proliferation and cross-linking of surface antigen receptors or ligation of CD40 restores the response. J Immunol. 1996;157:3323–33. [PubMed] [Google Scholar]
- 19.Jou ST, Carpino N, Takahashi Y, Piekorz R, Chao JR, Carpino N, Wang D, Ihle JN. Essential, nonredundant role for the phosphoinositide 3-kinase p110δ in signaling by the B-cell receptor complex. Mol Cell Biol. 2002;22:8580. doi: 10.1128/MCB.22.24.8580-8591.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okkenhaug K, Bilancio A, Farjot G, et al. Impaired B and T cell antigen receptor signaling in p110δ PI 3-kinase mutant mice. Sci STKE. 2002;297:1031. doi: 10.1126/science.1073560. [DOI] [PubMed] [Google Scholar]
- 21.Su TT, Guo B, Kawakami Y, et al. PKC- controls I B kinase lipid raft recruitment and activation in response to BCR signaling. Nat Immun. 2002;3:780–6. doi: 10.1038/ni823. [DOI] [PubMed] [Google Scholar]
- 22.Sutherland CL, Heath AW, Pelech SL, Young PR, Gold MR. Differential activation of the ERK, JNK, and p38 mitogen-activated protein kinases by CD40 and the B cell antigen receptor. J Immunol. 1996;157:3381. [PubMed] [Google Scholar]
- 23.Clark K, Plater L, Peggie M, Cohen P. Use of the pharmacological inhibitor BX795 to study the regulation and physiological roles of TBK1 and IkappaB kinase epsilon: a distinct upstream kinase mediates Ser-172 phosphorylation and activation. J Biol Chem. 2009;284:14136–46. doi: 10.1074/jbc.M109.000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuma Y, Sabio G, Bain J, Shpiro N, Marquez R, Cuenda A. BIRB796 inhibits all p38 MAPK isoforms in vitro and in vivo. J Biol Chem. 2005;280:19472–9. doi: 10.1074/jbc.M414221200. [DOI] [PubMed] [Google Scholar]
- 25.Sabin EA, Kopf MA, Pearce EJ. Schistosoma mansoni egg-induced early IL-4 production is dependent upon IL-5 and eosinophils. J Exp Med. 1996;184:1871–8. doi: 10.1084/jem.184.5.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chensue SW, Terebuh PD, Warmington KS, Hershey SD, Evanoff HL, Kunkel SL, Higashi GI. Role of IL-4 and IFN-γ in Schistosoma mansoni egg-induced hypersensitivity granuloma formation. Orchestration, relative contribution, and relationship to macrophage function. J Immunol. 1992;148:900–6. [PubMed] [Google Scholar]
- 27.Liu YJ, Johnson GD, Gordon J, MacLennan IC. Germinal centres in T-cell-dependent antibody responses. Immunol Today. 1992;13:17–21. doi: 10.1016/0167-5699(92)90199-H. [DOI] [PubMed] [Google Scholar]
- 28.Liu YJ, Joshua DE, Williams GT, Smith CA, Gordon J, MacLennan IC. Mechanism of antigen-driven selection in germinal centres. Nature. 1989;342:929–31. doi: 10.1038/342929a0. [DOI] [PubMed] [Google Scholar]
- 29.Encinas JA, Janssen EM, Weiner DB, Calarota SA, Nieto D, Moll T, Carlo DJ, Moss RB. Anti-T-cell Ig and mucin domain-containing protein 1 antibody decreases TH2 airway inflammation in a mouse model of asthma. J Allergy Clin Immunol. 2005;116:1343–9. doi: 10.1016/j.jaci.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 30.Meyers JH, Chakravarti S, Schlesinger D, et al. TIM-4 is the ligand for TIM-1, and the TIM-1–TIM-4 interaction regulates T cell proliferation. Nat Immunol. 2005;6:455–64. doi: 10.1038/ni1185. [DOI] [PubMed] [Google Scholar]
- 31.Ichimura T, Asseldonk EJ, Humphreys BD, Gunaratnam L, Duffield JS, Bonventre JV. Kidney injury molecule-1 is a phosphatidylserine receptor that confers a phagocytic phenotype on epithelial cells. J Clin Invest. 2008;118:1657–68. doi: 10.1172/JCI34487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kobayashi N, Karisola P, Pena-Cruz V, et al. TIM-1 and TIM-4 glycoproteins bind phosphatidylserine and mediate uptake of apoptotic cells. Immunity. 2007;27:927–40. doi: 10.1016/j.immuni.2007.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyanishi M, Tada K, Koike M, Uchiyama Y, Kitamura T, Nagata S. Identification of Tim4 as a phosphatidylserine receptor. Nature. 2007;450:435–9. doi: 10.1038/nature06307. [DOI] [PubMed] [Google Scholar]
- 34.Su EW, Lin JY, Kane LP. TIM-1 and TIM-3 proteins in immune regulation. Cytokine. 2008;44:9–13. doi: 10.1016/j.cyto.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Souza AJ, Oriss TB, O’malley KJ, Ray A, Kane LP. T cell Ig and mucin 1 (TIM-1) is expressed on in vivo-activated T cells and provides a costimulatory signal for T cell activation. Proc Natl Acad Sci U S A. 2005;102:17113–8. doi: 10.1073/pnas.0508643102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mariat C, Degauque N, Balasubramanian S, et al. Tim-1 signaling substitutes for conventional signal 1 and requires costimulation to induce T cell proliferation. J Immunol. 2009;182:1379–85. doi: 10.4049/jimmunol.182.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rennert PD, Ichimura T, Sizing ID, et al. T cell, Ig domain, mucin domain-2 gene-deficient mice reveal a novel mechanism for the regulation of Th2 immune responses and airway inflammation. J Immunol. 2006;177:4311–21. doi: 10.4049/jimmunol.177.7.4311. [DOI] [PubMed] [Google Scholar]
- 38.Chen TT, Li L, Chung DH, et al. TIM-2 is expressed on B cells and in liver and kidney and is a receptor for H-ferritin endocytosis. J Exp Med. 2005;202:955–65. doi: 10.1084/jem.20042433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Solis M, Romieu-Mourez R, Goubau D, et al. Involvement of TBK1 and IKKε in lipopolysaccharide-induced activation of the interferon response in primary human macrophages. Eur J Immunol. 2007;37:528–39. doi: 10.1002/eji.200636090. [DOI] [PubMed] [Google Scholar]
- 40.Venkataraman C, Shankar G, Sen G, Bondada S. Bacterial lipopolysaccharide induced B cell activation is mediated via a phosphatidylinositol 3-kinase dependent signaling pathway. Immunol Lett. 1999;69:233–8. doi: 10.1016/s0165-2478(99)00068-1. [DOI] [PubMed] [Google Scholar]


