Abstract
A better understanding of similarities and differences in the composition of the cellular immune system in non-human primates (NHPs) compared with human subjects will improve the interpretation of preclinical studies. It will also aid in addressing the usefulness of NHPs as subjects for studying chronic diseases, vaccine development and immune reconstitution. We employed high content colour flow cytometry and analysed simultaneously the expression of CD3, CD4, CD8α, CD8β, CD16/CD56, CD45RA, CCR7, CD27, CD28, CD107a and the interleukin-7 receptor α-chain (IL-7Rα) in peripheral blood mononuclear cells (PBMCs) of 27 rhesus macaques and 16 healthy human subjects. Regulatory T cells (Tregs) were identified using anti-CD3, -CD4, -CD25, -FoxP3, and -IL-7Rα monoclonal antibodies. Responsiveness to IL-7 was gauged in a signal transducer and activation of transcription 5 (STAT-5) phosphorylation assay. Human and NHP PBMCs showed a similar T-cell composition pattern with some remarkable differences. Similarities: human and NHP CD4+ and CD8+ cells showed a similar STAT-5 phosphorylation pattern in response to IL-7. Multicolour flow cytometric analysis identified a CD4+ CD8αα+ CD8αβ+ T-cell population in NHPs as well as in human subjects that expressed the degranulation marker CD107a and may represent a unique CD4+ T-cell subset endowed with cytotoxic capacity. Differences: we identified in PBMCs from NHPs a higher proportion (5·16% in CD3+ T cells) of CD8αα+ T cells when compared with human donors (1·22% in CD3+ T cells). NHP CD8αα+ T cells produced tumour necrosis factor-α / interferon-γ (TNF-α/IFN-γ) or TNF-α, whereas human CD8αα+ T cells produced simultaneously TNF-α/IFN-γ and IL-2. A minor percentage of human CD8+ T cells expressed CD25bright and FoxP3 (0·01%). In contrast, 0·07% of NHP CD8+ T cells exhibited the CD25bright FoxP3+ phenotype. PBMCs from NHPs showed less IL-7Rα-positive events in all T-cell subsets including CD4+ Tregs (median 5%) as compared with human (median 12%). The data visualize commonalities and differences in immune cell subsets in humans and NHPs, most of them in long-lived memory cells and cells with suppressive functions. This provides a matrix to assess future efforts to study diseases and vaccines in NHPs.
Keywords: flow cytometry, immune markers, immune profiling, non-human primate model, T cells, vaccination
Introduction
Non-human primates (NHPs) provide an indispensable model to study human diseases, including chronic infections and human immunodeficiency virus and tuberculosis vaccine development.1,2 They have been instrumental in the study of aging and immune reconstitution.3–6 Despite general differences in T-cell immunology between species, other factors play an important role in gauging immune responses. Animals live in a protected environment and are not exposed to the same pathogens that affect humans. This may impact on the breadth of the cellular immune repertoire and on the maintenance of immune cell memory.7–12 Other observations underline the need to study the differences between human and NHP immune responses: a humanized anti-CD28 monoclonal superagonist antibody caused severe side-effects in a phase 1 clinical trial;13 it induced a delayed and sustained Ca2+-influx in human CD4+ T cells, but not in CD4+ T cells from NHPs.14 Any experimental study of cellular, adaptive immune responses addresses also T-cell homeostasis, the active and dynamic process by which immune cells mature traffic and produce cytokines upon activation. Key elements of the analysis of adaptive cellular immune responses are (i) T-cell subsets (CD4/CD8, CD8αα+ memory T cells) in concert with differentiation and homing markers (CD45RA, CCR7, CD28, CD27, CD62L),15 cytotoxicity (measure of CD107a) and cytokine production (polyfunctionality);16 (ii) regulatory T cells (Tregs);17 and (iii) the response to interleukin-7 (IL-7), a key cytokine for T-cell survival, homeostasis and T-cell memory.18 T-cell compartment composition and phenotype has been studied previously in rhesus macaques19,20 with a limited panel of immune markers. Different combinations of immune markers were used in these studies to define memory and effector T-cell compartments in rhesus monkey.21 To our knowledge, the current report analyses for the first time the simultaneous expression of CD45RA, CCR7, CD27 and CD28 in T-cell subsets in healthy rhesus monkeys. We took advantage of a high-content, multicolour flow cytometry to assess the distribution of immune cells in peripheral blood mononuclear cells (PBMCs) from female rhesus monkeys (defined by expression of CD45RA, CCR7, CD28, CD27, CD107a, IL-7 receptor α-chain), to compare cytokine [IL-2, interferon-γ (IFN-γ) and tumour necrosis factor-α (TNF-α)] production in T-cell subsets, IL-7-induced signal transducer and activator of transcription 5 (STAT-5) phosphorylation, and Treg frequencies.
Materials and methods
Blood samples
Peripheral blood was obtained from 16 healthy human donors (19–66 years, median 31 years) from the Blood Bank (Ethical Permit DNR 00-097). Peripheral blood was obtained from 27 female rhesus macaques (Macaca mulatta) of Chinese origin with an age range between 3 and 4 years housed in the Astrid Fagraeus laboratory at the Swedish Institute for Infectious Disease Control. Housing of the animals and care procedures were in compliance with the provisions and general guidelines of the Swedish Animal Welfare Agency, the Local Ethical Committee responsible for Animal Experiments approved all procedures (protocol DNR238/2006-54). The PBMCs were isolated from freshly obtained, heparinized peripheral blood by Ficoll–Hypaque density gradient centrifugation.
Multicolour flow cytometric analysis
Immune marker analysis was performed on freshly isolated PBMCs by a standard Ficoll procedure from heparinized blood samples. Cells (1 × 106) were incubated at 4°C for 15 min with the following antibodies: phycoerythrin (PE) -Cy7-conjugated anti-CCR7 (3D12), peridinin chlorophyll protein (PerCP) -conjugated anti-CD3 (SP-34-2), PE-conjugated anti-CD16 (3G8), PE-conjugated anti-CD56 (B159), allophycocyanin (APC) -Cy7-conjugated anti-CD8α chain (SK1), Amcyan-conjugated anti-CD28 (CD28.2), purchased from BD Biosciences (Stockholm, Sweden), APC-Alexa Fluor 700-conjugated anti-CD107a (H4A3), APC-conjugated anti-IL-7 receptor α-chain (IL-7Rα; R34.34), PE-Texas Red-conjugated anti-CD45RA (2H4), fluorescein isothiocyanate (FITC) -conjugated anti-CD8β chain (2ST8.5H7), purchased from Beckman Coulter (Marseille, France), and Pacific Blue-conjugated anti-CD4 (S3.5) purchased from Caltag Laboratories (Burlingame, CA). The lymphocytes were then washed in phosphate-buffered saline (PBS) with 0·1% fetal bovine serum, and incubated at 4° for 15 min with the anti-CD27 (1A4CD27) antibody (Beckman Coulter) labelled with Pacific Orange using the Zenon Pacific Orange Mouse IgG1 Labeling Kit obtained from Invitrogen (Stockholm, Sweden). Human samples were processed the same day, and NHP samples were processed on a different occasion, but also the same day. The median fluorescence intensity (MFI) of IL-7Rα expression therefore allows a comparison of the intensity of IL-7Rα expression on T cells within each species but not between humans and NHPs. Data acquisition was performed using a FACSAria Flow cytometer (BD Biosciences) and results were analysed with FlowJo software (Tree Star Inc., Ashland, OR).
Intracellular cytokine staining
Cytokine production was analysed in frozen PBMCs, which were thawed, rested overnight and stimulated for 6 hr in the presence of brefeldin A (10 mg/ml) purchased from Sigma-Aldrich (Sweden AB, Stockholm, Sweden) either with medium: RPMI-1640 containing l-glutamine (2 mm), penicillin (100 IU/ml) and streptomycin (10 mg/ml), 10% heat-inactivated fetal bovine serum (Gibco, Invitrogen), or medium and phorbol 12-myristate 13-acetate (PMA)/ionomycin (25 ng/ml and 1 mg/ml, respectively; Sigma-Aldrich). Cells were then washed in PBS, and stained with cell surface marker antibodies: Pacific Blue-conjugated anti-CD3 (SP34-2), PerCP-Cy5.5 conjugated anti-CD4 (L200; BD Biosciences), APC-Cy7-conjugated anti-CD8α chain (SK1), and FITC-conjugated anti-CD8β chain (2ST8.5H7), in the presence of the live/dead fixable dead cell marker (Aqua LIVE/DEAD; Invitrogen), for 30 min at 4°. After washing with PBS, cells were fixed and permeabilized using the IntraPrep Fix/Perm Kit (Beckman Coulter) and incubated with antibodies specific for intracellular cytokines for 30 min at 4°: PE-conjugated anti-IL-2 (MQ1-17H12), PE-Cy7-conjugated anti-IFN-γ (B27), and APC-conjugated anti-TNF-α, all obtained from BD Biosciences. Cells were analysed using a BD FACSCanto flow cytometer (BD Biosciences) and data analysis was performed using FlowJo software.
T helper type 17 induction and detection of IL-17-producing cells
Human and NHP frozen PBMCs were thawed, rested overnight and distributed into 96-well plates (0·4 × 106 cells/well) coated with 50 μl anti-CD3 (OKT3, 1 μg/ml) and anti-CD28 (CD28.2; Beckman Coulter, 1 μg/ml) antibodies. Cells were cultured in AIM-V/Dulbecco's modified Eagle's medium (Gibco, Invitrogen) containing 1% heat-inactivated human serum (Biowest, Nuaillé, France), either (i) in medium alone, (ii) in the presence of human IL-23 (10 ng/ml) (Humanzyme, Chicago, IL) or (iii) in a combination of IL-23 with human IL1-β (10 ng/ml) (R&D Systems, Abingdon, UK). After 6 days, cells were stimulated with PMA/ionomycin for 6 hr, and IL-17, IFN-γ and TNF-α production was detected in CD4+, CD8αα+ and CD8αβ+ T cells as described above, using a PE-conjugated anti-IL-17 antibody (eBio64DEC17) purchased from eBioscience (San Diego, CA) simultaneously with PE-Cy7-conjugated anti-IFN-γ (B27) and APC-conjugated anti-TNF-α antibodies.
IL-7-induced STAT-5 phosphorylation assay
Constitutive and IL-7-induced phosphorylated STAT-5 (P-STAT-5) expression was evaluated in frozen PBMCs as described previously.71 Briefly, overnight starved, thawed PBMCs were incubated with recombinant human IL-7 (rhIL-7; 100 ng for 105 cells, provided by Dr Michel Morre, Cytheris, Issy-les-Moulineaux, France) for 15 min at 37°. The cells were then incubated for 15 min at 4° with the following cell surface antibodies: APC-conjugated anti-CD4 (SK3; BD Biosciences), and APC-Cy7-conjugated anti-CD8α chain, and immediately after fixed with 2% paraformaldehyde. The cells were washed with Stain Buffer (BD Biosciences) and permeabilized with 90% methanol for 30 min on ice, followed by two washes with Stain Buffer. The cells were incubated with Alexa-Fluor 488-conjugated anti-P-STAT-5a antibody (Y694) (BD Biosciences) for 1 hr at room temperature and analysed immediately using a FACSAria flow cytometer and data analysis was performed using FlowJo software. Because of the fixation procedure, we could not include the anti-CD3 monoclonal antibody as it did not exhibit sufficient stability in the fixation procedure required for intracellular staining, so the data are obtained by gating on CD8+ and CD4+ cells for STAT-5 phosphorylation analysis. The anti-CD8β chain antibody could not be used in this panel (also because of the fixation procedure). The CD8+ subset encompasses therefore the CD8αα+ and CD8αβ+ cell subsets. Human IL-7 shows similar activity to NHP IL-7 (personal communication, Dr Michel Morre, Cytheris, Issy-les-Moulineaux, France).
Regulatory T cells
Frozen PBMCs were thawed and incubated at 4° for 15 min with the following antibodies: PerCP-conjugated anti-CD3 (SP34-2), PerCP Cy5.5-conjugated anti-CD4 (L200), APC-Cy7-conjugated anti-CD8α chain (SK1), APC-conjugated anti-IL-7Rα (R34.34), PE-Cy7-conjugated anti-CD25 (2A3; BD Biosciences). The PBMCs were then washed with Stain Buffer (BD Biosciences) and fixed with FOXP3 Fix/Perm Buffer (BioLegend, San Diego, CA) at room temperature for 20 min followed by one washing with Stain Buffer and one washing with FOXP3 Perm Buffer (BioLegend). The PBMCs were resuspended in FOXP3 Perm Buffer and incubated at room temperature for 15 min. After washing, cells were resuspended in FOXP3 Perm Buffer and incubated at room temperature for 30 min with the Alexa-Fluor 488-conjugated anti-FoxP3 (259D) antibody purchased from BioLegend. Analysis was performed using a FACSAria flow cytometer and data were analysed using FlowJo software. CD8β-expressing cells could not be measured because the monoclonal antibody anti-CD8β chain did now exhibit sufficient stability in the fixation procedure required for FoxP3 protein analysis.
Statistics
Data are presented as median ± SD, and P-values were derived using a Mann–Whitney U-test.
Results
NHPs and humans share a similar T-cell compartment
The phenotype of the T-cell compartment in the peripheral blood of 16 healthy human donors (HDs) and 27 rhesus monkeys was assessed by multicolour flow cytometric analysis. CD3− lymphocytes, which express CD56 and CD16, identify natural killer (NK) cells in humans. CD56 identifies mainly monocytes and CD16+ NK cells in rhesus macaques.22 T lymphocytes were determined by CD3 expression and after exclusion of CD16+ and CD56+ cells (gating strategy see Fig. 1a). The (co)expression of CD4, CD8α and CD8β in the T-cell (CD56 CD16− CD3+) compartment was determined in HDs and NHPs. CD8αβ+ T cells and CD8αα+ T cells represented 23·8% and 1·2% in HDs, and 28% and 5·2% in NHPs. In PBMCs from HDs and NHPs, γδ T-cell receptor (TCR)+/− cells exhibited the CD8αα+/− phenotype. Yet the majority (> 70%) of CD8αα+/− T cells were present in the TCR-αβ T-cell compartment (data not shown). CD4+ T cells represented the prevalent T-cell subset: 74·3% and 63·6% of T cells in HDs and NHPs, respectively. Two other less frequent cell subsets could be identified: CD4+ T cells expressing either the CD8αα homodimer or the CD8αβ heterodimer (0·2% and 0·1% in HDs; 1·3% and 1·4% in NHPs) (see Fig. 1b). CD8αα+, CD4+ CD8αα+ and CD4+ CD8αβ+ T cells showed a statistically higher frequency in NHPs than in HDs.
Figure 1.
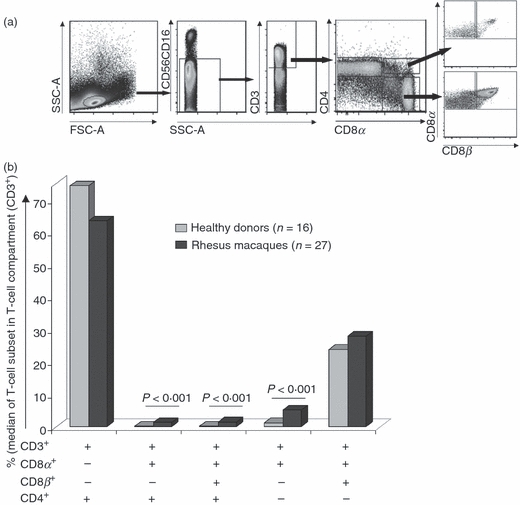
Identification of T-cell subsets. (a) Gating strategy to identify T cells, based on expression of CD3, CD8α, CD8β and CD4. (b) T-cell frequencies. CD8αα+, CD4+ CD8αα+ and CD4+ CD8αβ+ T-cell frequencies were statistically higher in non-human primates. P <0·001 (Mann–Whitney U-test).
Four functional T-cell compartments are defined in humans by the expression of CD45RA and CCR7: precursor (CD45RA+ CCR7+), central memory (CD45RA− CCR7+), effector memory (CD45RA− CCR7−) and differentiated effector (CD45RA+ CCR7−) T-cell subsets.15,23 The distribution of the T-cell subsets defined by CD45RA and CCR7 expression within the different T-cell populations was statistically different in PBMCs between HDs and NHPs (Table 1).
Table 1.
T-cell compartments CD45RA/CCR7 expression in healthy human donors and non-human primates
| CD45RA+ CCR7+ | CD45RA+ CCR7+ | CD45RA+ CCR7+ | CD45RA+ CCR7+ | |
|---|---|---|---|---|
| CD8αβ+ | ||||
| Healthy donors | 47·6 ± 10·7** | 29 ± 7·1* | 10·3 ± 6·8** | 13·1 ± 6·8** |
| Rhesus macaques | 70 ± 9·2 | 25·7 ± 9·3 | 2·7 ± 2·1 | 1·5 ± 1·3 |
| CD8αα+ | ||||
| Healthy donors | 19·4 ± 9·1** | 27·8 ± 10·1** | 19·4 ± 8·5** | 33·4 ± 10·2** |
| Rhesus macaques | 46·3 ± 7·8 | 49·7 ± 7·7 | 1·6 ± 1·6 | 2·3 ± 2·4 |
| CD4+ | ||||
| Healthy donors | 35·4 ± 7·9** | 20 ± 5·8** | 23·9 ± 5·3** | 20·7 ± 6·5** |
| Rhesus macaques | 73·4 ± 9·7 | 12·9 ± 5·6 | 11 ± 7·6 | 2·5 ± 2·2 |
| CD4+ CD8αα+ | ||||
| Healthy donors | 24·8 ± 6·7** | 19·6 ± 6·9* | 26·7 ± 7·7** | 28·8 ± 7·7** |
| Rhesus macaques | 59·2 ± 10·6 | 28·3 ± 9·2 | 7·4 ± 6·2 | 5 ± 3·4 |
| CD4+ CD8αβ+ | ||||
| Healthy donors | 39·1 ± 14·1** | 17·1 ± 10·3* | 27·2 ± 13·4** | 16·5 ± 7·1** |
| Rhesus macaques | 83·7 ± 6·5 | 10·8 ± 5·2* | 4·8 ± 3·3 | 0·6 ± 0·6 |
Healthy donors n = 16, rhesus macaques n = 27. (%) Mean values and standard deviation.
P <0·05 (Mann–Whitney U-test).
P <0·0001
We assessed the CD28 and/or CD27 expression within the CD45RA/CCR7 subsets. The median value of the expression frequency of CD45RA+/− CCR7+/− CD28+/− CD27+/− subsets in the parental T-cell population from the PBMC of HDs and NHPs is displayed as heat-maps (Fig. 2). In PBMCs from HDs, precursor, effector memory and central memory CD8αβ+ T-cells co-expressed CD28 and CD27 (CD28− CD27+ and CD28+ CD27− subsets were also found). In contrast, differentiated effector CD8αβ+ T cells were enriched in cells expressing only CD27. In NHPs, CD45RA+ CCR7+ and CD45RA+ CCR7− cells represented the dominant T-cell subsets in the CD8αβ+ T-cell compartment, and the expression of CD28 and CD27 differed from that by HDs within these T-cell compartments. In NHPs, CD8αβ+ T cells expressed predominantly either CD28, or only CD27. We observed also an enrichment of CD28− CD27− (and a parallel decrease of CD28+ CD27+) T cells in PBMCs from NHPs compared with HDs.
Figure 2.
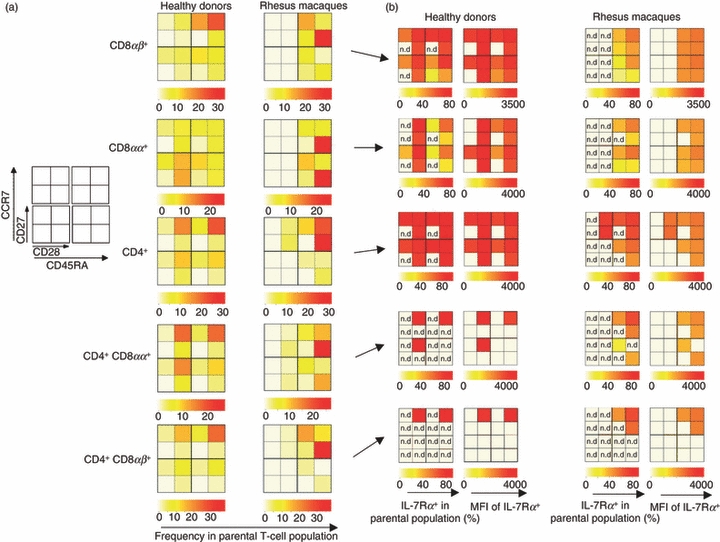
Overview of T-cell subsets defined by CD45RA/CCR7 and CD27/CD28 expression using heat-map analysis. (a) Frequency of immune cell subsets in human donors and non-human primates. (b) Interleukin-7 receptor α (IL-7Rα) expression and relative IL-7Rα density [as measured by mean fluorescence intensity (MFI)] in these T-cell subsets. The percentage of IL-7Rα (and MFI) in T-cell subsets displaying low number of events (< 100 events) was not determined (n.d.) for quality control reasons.
The CD8αα+ T-cell subset displayed a different profile as compared to CD8αβ+ T cells. In HDs, CD8αα+ T cells were enriched in differentiated T-cells (particularly CD45RA+/− CCR7−) as compared to CD8αβ+ T cells. Effector memory CD8αα+ T cells expressed CD28 alone or in combination with CD27, and differentiated CD8αα+ T cells CD27 or CD28. In NHPs, CD8αα+ T cells displayed either a CD45RA+ CCR7+ or a CD45RA+ CCR7− profile. Most of the CD45RA+ CCR7± CD8αα+ T cells stained positive only for CD28.
CD4+ T cells were observed within the four CD45RA+/− CCR7+/− compartments in HDs, whereas 75·5% of CD4+/− T cells from NHPs stained positive for CD45RA+ CCR7+. Similar to the phenotype of CD8+ T cells, NHP CD4+ T cells were enriched in cells expressing only CD28 and not CD27. Interestingly, CD4+/− CD8αβ+/− T cells displayed a phenotype, based on CD45RA and CCR7 expression, comparable (not statistically different) to CD4± T cells in PBMCs from HDs. Of note, CD4+ CD8αα+ and CD4+ CD8αβ+ T cells represented the only immune cell subsets that stained positive for CD107a+ (particularly in CD45RA+ CCR7 cells expressing CD28 and or CD27): 5·5% and 3·7% of total CD4+ CD8αα+ and CD4+ CD8αβ+ T cells in HDs, and 1·3% and 1·7% in NHPs (data not shown).
In HDs, most CD8αβ+ T cells and approximately 50% of CD8αα+ T cells expressed the IL-7Rα. CD4+ T cells and CD4+ CD8αα+ CD8αβ+ T cells showed an increased frequency of IL-7Rα+ T cells and higher levels of IL-7Rα expression/cell (measured by MFI) compared with CD8+ T cells. The PBMCs obtained from NHPs showed a similar trend for IL-7Rα expression to HDs: more CD4+ T cells expressed more IL-7Rα compared with the CD8+ T-cell subsets, but the frequency of IL-7Rα+ in all T-cell subsets was decreased in PBMCs obtained from NHPs compared with the frequency observed in HDs (e.g. in 86% of CD4+ T cells in HDs and 67% in NHPs were IL-7Rα+, Fig. 2b).
Differential cytokine production in T cells from humans compared with NHPs
The cytokine profile of CD4+, CD4+ CD8+, CD8αα+, CD8αβ+ and CD4− CD8− T cells upon PMA/ionomycin stimulation (used to induce maximal cytokine production) in NHPs (n = 27) and HDs (n = 5) was assessed. The frequency of different T-cell subsets in the medium control and upon PMA/ionomycin stimulation (Fig. 3a) was similar in PBMCs from NHPs. In HDs, the frequency of CD4− CD8− T cells upon PMA/ionomycin stimulation was increased (from 3·6% to 10%) as a result of the down-regulation of CD4 and CD8 co-receptors in the CD4+ and CD8αβ+ T-cell subsets24 (and concomitant decreased frequency of those subsets upon PMA/ionomycin stimulation as seen in some HDs). In PBMCS from NHPs and from HDs, CD4+ and CD8αα+ T cells showed similar frequencies of cytokine-producing cells in response to PMA/ionomycin stimulation. In contrast, higher frequencies of cytokine-producing cells were detected in the CD8αβ+, CD4+ CD8+, and CD4− CD8− T-cell compartments in HDs compared with NHPs (Fig. 3b). CD4− CD8− T cells were sorted by fluorescence-activated cell sorting, followed by intracellular staining with anti-cytokine (IL-2, TNF-α, IFN-γ), -CD4 and -CD8 monoclonal antibodies to decipher whether the increased frequency of cytokine producing CD4− CD8− T cells after PMA/ionomycin stimulation in PBMCs from HDs as compared to NHPs was the result of ‘bona fide’ CD4− CD8− T cells or to T cells that down-regulated the cell surface expression of the CD4 or CD8 co-receptors. The CD4− CD8− T cells from HDs that do not express (at the cell surface or intracellularly) CD4 or CD8 showed a higher frequency of cytokine-producing cells than the NHPs CD4− CD8− T cells (data not shown).
Figure 3.
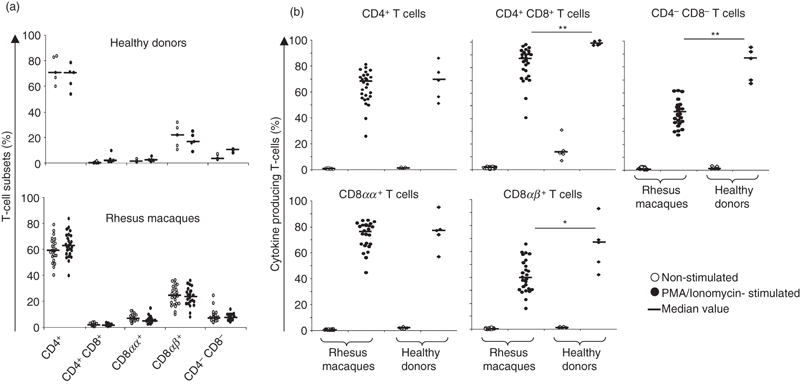
Cytokine producing T-cell subsets in human donors and non-human primates. (a) Presence of immune cell subsets defined by CD3, CD4, CD8α and CD8β expression with no stimulation (medium control) and phorbol 12-myristate 13-acetate (PMA)/ionomycin stimulation. (b) Percentage of cytokine producing T cells. **P <0·001, *P <0·05 (Mann–Whitney U-test).
The production of IL-2, TNF-α and IFN-γ was measured simultaneously on the single cell level to assess the presence of polyfunctional T cells. The profile of two representative PBMC samples from monkeys and from two HDs is shown in Fig. 4. In NHPs, CD4+ T cells produced TNF-α and IL-2, either in combination or alone, CD8αβ+ T cells produced mainly IFN-γ and TNF-α, either in combination or alone, and to a lesser extent IL-2. The CD8αα+ T-cell subset showed a cytokine production profile very similar to that of the CD8αβ+ T-cell subset. CD4+ CD8+ T cells displayed a polyfunctional profile (the vast majority of CD4+ CD8+ T cells produced two or three cytokines simultaneously). CD4− CD8− T cells displayed a profile similar to CD4+ T cells, they produced IL-2 and TNF-α, but also IL-2 or TNF-α alone. The cytokine profile in the different T-cell compartments from HDs was very similar to the profile identified in NHPs, but they exhibited a higher frequency of polyfunctional T cells (e.g. 18·8% of CD8αβ+ T cells in NHPs produced three cytokines compared with 27·2% in HDs). To further characterize the different T-cell subsets, we assessed the presence of IL-17+ producing T cells. The PBMCs from four HDs were either cultured without cytokines, or in Th17 differentiation conditions (in the presence of IL-23 either alone or in combination with IL-1β). The combination of IL-23 and IL-1β was found to induce the highest frequency of IL-17+ producing cells. CD4+ CD8+ T cells showed, after PMA/ionomycin stimulation, an enrichment in IL-17+ producing cells compared with CD4+ T cells (Fig. S1). In the presence of IL-23 and IL-1β, IL-17 production was detected in 20% (median value) of CD4+ CD8+ T cells, and in 10% of CD4+ T cells. Interleukin-17 was produced in combination with TNF-α in CD4+ CD8+ and CD4+Τ cells and to a lesser extent also with IFN-γ. Higher frequencies of IL-17+ producing cells were detected in CD8αα+ than in CD8αβ+ T cells. The NHP PBMCs from five animals were cultured using identical conditions, yet we could not study the nature of IL-17+ T cells because of the low number of IL-17-positive events.
Figure 4.
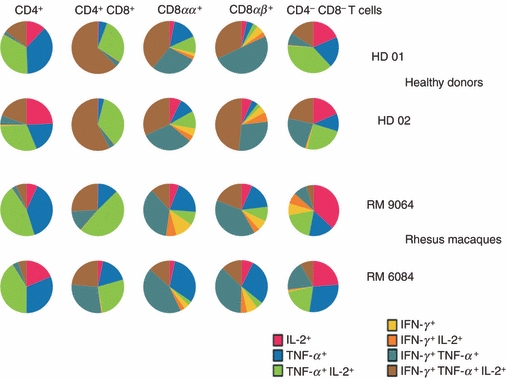
Analysis of polyfunctional T cells. Interleukin-2 (IL-2), interferon-γ (IFN-γ) and tumor necrosis factor-α (TNF-α) production were measured by intracellular cytokine staining on the single-cell level. Two representative individuals from human donors and non-human primates are shown as a paradigm. Note the increased frequency of polyfunctional T-cells in human donors compared with non-human primates.
IL-7-induced STAT-5 phosphorylation in NHPs and humans
The binding of IL-7 to the IL-7Rα induces the activation by phosphorylation of the transcription factor STAT-5. IL-7 and IL-7 T-cell responsiveness play a pivotal role in T-cell homeostasis and in immune formation.18 A STAT-5 phosphorylation assay was used to gauge IL-7 responsiveness in CD4+ and CD8+ cells. The increase of the percentage of P-STAT-5 cells, and an example of constitutive P-STAT-5 and IL-7-induced P-STAT-5, in HD and NHP are shown in Fig. 5(a,b). In NHPs, (n = 15) 84·4 ± 10·8% and 60·6 ± 12% of CD4+ and CD8+ cells showed an increase of P-STAT-5 cells in response to IL-7 stimulation. Similar numbers were obtained in PBMCs from HDs (n = 10): 87·6 ± 7·6% and 62·3 ± 15·4% in CD4+ and CD8+ cells, respectively. CD4+ and CD8+ subsets showed comparable responses to IL-7 stimulation as measured by STAT-5 phosphorylation in NHPs and HDs. In HDs and NHPs more CD4+ cells than CD8+ cells showed STAT-5 phosphorylation (consistent with higher levels of IL-7Rα expression on CD4+ T cells) but the amplitude (measured by MFI) was not statistically different between CD4+ and CD8+ cells.
Figure 5.
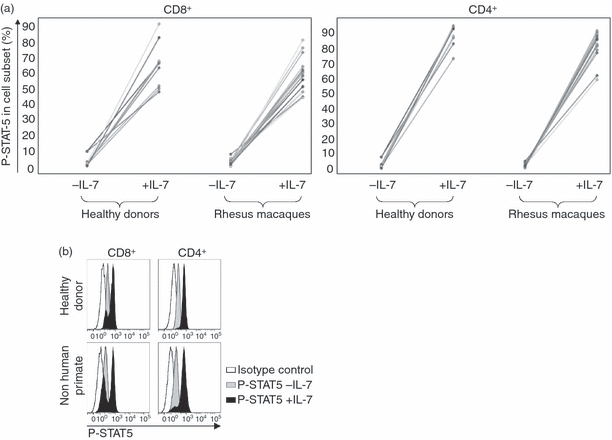
Interleukin-7 (IL-7) -induced signal transducer and activator of transcription 5 (STAT-5) phosphorylation. Phosphorylated (P-) STAT-5 was determined in CD4+ and CD8+ cells before and after exposure to IL-7. (a) Percentage of P-STAT-5 positive cells. Similar levels of constitutive and IL-7-induced STAT-5 phosphorylation in peripheral blood mononuclear cells from human donors and non-human primates. (b) Example of constitutive and IL-7-induced P-STAT-5 in human and non-human primate peripheral blood mononuclear cells determined by flow cytometry.
Differences in Tregs defined by IL-7Rα expression
The presence of regulatory cells was assessed by expression analysis of CD25 and FoxP3 in the CD4+, CD8+ and CD4+ CD8+ T-cell compartments (gating strategy see Supplementary Fig. S2). In NHPs, the CD4+ T-cell compartment showed the following frequencies: 16·5% (median values) were CD25intermediate (CD25interm.) and 0·5% stained for CD25bright; in CD4+ CD8+ T cells: 19·6% cells were CD25interm. and 1·4% were CD25bright; in CD8+ T cells: 1% were CD25interm. and 0·07% CD25bright (Table 2). Compared with HDs, higher frequencies of CD4+ CD25interm. T cells and CD4+ CD8+ CD25interm./bright, and CD8+ CD25bright T cells (21%) were detected in PBMCs from NHPs. Analysis of FoxP3 expression in the different CD25+/− T-cell compartments showed that the majority of CD25bright T cells in NHPs were FoxP3+ (87·5% of CD4+ and 76% of CD4+ CD8+ and CD8+ T cells), whereas only 10–20% of CD25interm. T cells showed FoxP3 expression (Fig. 6a). In contrast, almost no FoxP3 expression could be observed in human CD4+ CD8+ CD8+ T cells that exhibited the CD25interm. phenotype. Analysis of PBMCs from four of eight HDs showed that CD4+ CD8+ and CD8+ T cells, which displayed a CD25bright phenotype, stained also positive for FoxP3. In summary, statistically higher frequencies (P≤0·05) of CD4+/− CD25interm.FoxP3+/−, CD4± CD8± CD25interm./high FoxP3+/− and CD8± CD25interm./high FoxP3+/− were detected in NHPs than in HDs.
Table 2.
CD25 expression in CD4+, CD4+ CD8+ and CD8+ T-cell compartments in healthy human donors and non-human primates
| CD3+ CD4+ | CD3+ CD4+ CD8+ | CD3+ CD8+ | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Median | CD25− | CD25interm | CD25bright | CD25− | CD25interm. | CD25bright | CD25− | CD25interm | CD25bright |
| Rhesus macaques | 82** | 16·5** | 0·5 | 79·4** | 19·6** | 1·4* | 99 | 1 | 0·07* |
| Healthy donors | 95·9 | 3·4 | 0·5 | 96·5 | 2·3 | 0·4 | 98·8 | 1 | 0·01 |
(%) Median values.
P <0·05 (Mann–Whitney U-test).
P <0·001
Figure 6.
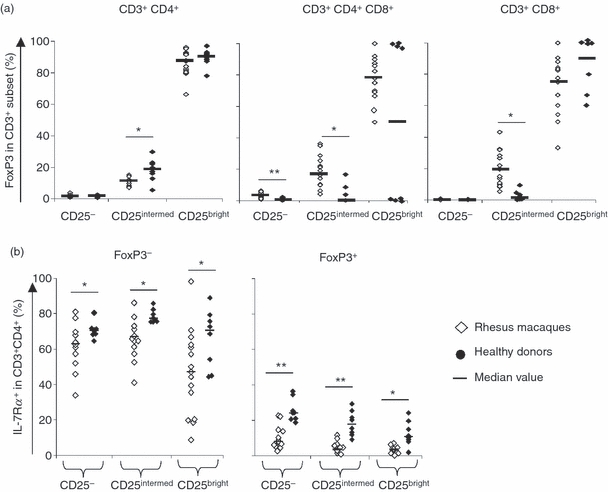
Determination of regulatory T cells based on marker analysis. (a) CD4+ CD8– (left), CD4+ CD8+ (middle panel) and CD8+ CD4− (right) T cells were segregated based on CD25 expression and FoxP3 analysis was performed as shown in the supplementary Fig. S2. Note that some CD8+ (CD4−) CD25bright T-cells showed low FoxP3 expression in non-human primates. (b) Interleukin-7 receptor α (IL-7Rα) -expressing T cells in CD4+ CD8− regulatory T cells (Tregs) based on FoxP3 analysis. Higher percentage of IL-7Rα+ T cells in Foxp3− CD25bright Tregs in peripheral blood mononuclear cells from humans than in those from non-human primates. **P <0·001, *P <0·05 (Mann–Whitney U-test).
Expression of the IL-7Rα on NHP CD25bright T cells was inversely correlated with expression of FoxP3, which is similar to the situation described in humans.25 More than 90% of NHP CD4+ CD8+ CD25interm./bright FoxP3+ T-cell subsets did not express the IL-7Rα, whereas the majority of CD4+ CD8+ CD25+/− FoxP3− (33–67%) were positive for IL-7Rα (% of IL-7Rα expression is shown for CD4+ T cells in Fig. 6b). The same trend was observed in HDs. However, 9% of human CD4+ CD25bright FoxP3+ T cells expressed the IL-7Rα (Fig. 6b), this was not true for the same T-cell subset in NHPs (3·8%). The MFI of IL-7Rα in the percentage of IL-7Rα expressing FoxP3+ cells was decreased compared with FoxP3− T cells, which indicates a lower number of IL-7Rα molecules/cell compared with FoxP3− T cells (data not shown).
Discussion
Limitations of any immune composition analysis may be related to the origin of the animals, previous exposure to environmental pathogens and age.26–29 It has been shown that gender may affect lymphocyte frequencies. In humans, females were found to show higher CD4+ T-cell frequencies,30 whereas others showed similar20 or different31 CD4+/− and CD8+/− T-cell compositions in PBMCs from female and male Chinese rhesus monkeys. The presence of steroid receptors on immune cells32 may account for differences in lymphocytes in females compared with males. A variety of other factors also impacts on PBMC composition. An increase of peripheral blood lymphocytes can be induced by exercise or stress with preferential mobilization of certain lymphocyte subsets during exercice.33 The aim of this study was to characterize the immune compartment, compare the phenotype of different T-cell subsets in a large cohort (27 animals) of young female Chinese rhesus macaques using reagents cross-reacting with human and NHP CD markers. The results were compared with a cohort of younger to older (19–66 years) female and male HDs with the limitations discussed above.
The NHP cohort, like in the human population, is outbred and individual variation is to be expected; to match exactly the age of the HDs and NHPs is not easily feasible because of the limited access to blood from appropriately age-matched individuals. Yet despite gender and age differences between HDs and NHPs, our study provides useful insights into some of the commonalities and differences between human and NHP immune cell compartments.
Differences in T-cell differentiation/homing markers
In this report we describe the composition of different T-cell compartments based on the simultaneous detection of CD45RA, CCR7, CD28 and CD27 in humans, and for the first time in rhesus monkeys. In NHPs and HDs CD8αβ+ T cells showed a preferential distribution within the precursor CD45RA+ CCR7+ compartment. In PBMCs from HDs, precursor T cells predominantly co-expressed CD28 and CD27 as shown previously by Romero et al.34 We identified CD45RA+ CCR7+ T cells that expressed only CD28 or CD27 and this was not described in their report. Human CD4+ T cells exhibited precursor phenotype and co-expressed CD28 and CD27 as described by Okada et al.35 Differentiated effector (CD45RA+ CCR7−) T cells represented 19% of the CD4+ T-cell compartment. This frequency is higher than reported by previous studies;35,36 differences in age,37 sex, or ethnic composition of the human cohorts may account for these differences. The majority of the CD8αβ+ and CD4+ T cells (> 70%) in PBMCs from rhesus monkeys co-expressed CD45RA and CCR7. However, the expression of CD28 and CD27 differed from that in HDs: fewer T cells co-expressed CD28 and CD27, most T cells expressed only CD28 (e.g. 40% of CD8αβ+ T cells). Pitcher et al.3 have characterized T-cell subsets in rhesus macaques based on the expression of markers such as CD45RA, CD95 and CD28, and concluded that CD95 and CD28 expression represented valid markers for the definition of precursor/effector/memory T-cell subsets, whereas the use of CD45RA and CD62L expression failed to identify precursor T cells. CD45RA expression was found on putative memory T cells and cytomegalovirus antigen-experienced cells. In humans, central memory T cells display a CD45RA+ CCR7− phenotype, and antigen-specific T cells have been found in different T-cell memory compartments.38 Furthermore, in the report by Pitcher et al. the marker CCR7 was not used so it does not exclude the use of CD45RA in combination with other markers (including CCR7) to delineate T-cell subsets.39,40 Our results show that more CD45RA+ CCR7+ CD28+ CD27+ cells (putative precursor cells) were present in the CD4+ than in the CD8αβ+ T-cell compartment in NHPs. This observation is consistent with the report by Pitcher et al. that the frequency of memory cells increases faster in CD8αβ+ T cells than in CD4+ T cells. Furthermore, CD45RA+ CCR7+ CD28+ CD27+ CD4+ and CD8αβ+ T cells were enriched for IL7-Rα+ T cells (77·4% and 55%, respectively are IL-7Rα+), suggesting that these cells may indeed represent precursor T cells.18 The biology of CD45RA+ CCR7+ CD28+ CD27− T cells in NHPs remains to be defined, they could represent T cells that entered differentiation. Alternatively, they could represent antigen-experienced T cells that regained CD45RA+ CCR7+ expression.41 A different area in NHP research attempts to reveal why natural simian immunodeficiency virus (SIV)-infection of African NHPs does not lead to disease.42 A key difference is that NHPs may develop an anti-inflammatory response that prevents chronic activation, and T-cell proliferation.43,44 Our observation that lower frequencies in NHPs of cytokine-producing cells in CD4+ CD8+, CD4− CD8− and CD8αβ+ T cells after PMA/ionomycin stimulation may indicate intrinsic differences in the levels of activation and T-cell responses between humans and NHPs. Lower levels on T cells of IL-7Rα expression were observed in NHPs, T-cell homeostasis in NHPs may have a lower requirement for IL-7. Interestingly, it was recently described that higher levels of plasmatic soluble IL-7Rα are detected in rhesus monkeys than in humans,45 suggesting that IL-7Rα shedding could also explain the lower detection of cell surface IL-7Rα in NHPs.
Differences in CD8αα+ T cells
CD3+ T cells that express the CD8αα homodimer have been described in mice46 and man.47,48 The CD8αα homodimer was transiently expressed in antigen lymphocytic choriomeningitis virus (LCMV) specific T cells along with markers for increased T-cell survival, i.e. IL-7Rα and Bcl-2.46 Mice defective in expressing CD8αα homodimers (E8I−/−) showed impaired CD8+ T-cell memory formation. However, other reports could not identify impaired CD8+ T-cell memory in E8I−/− mice in response to LCMV49 and influenza virus A50 (yet in the latter report a correlation between CD8αα expression and elevated levels of IL-7Rα and Bcl-2 was observed). The CD8αα homodimer, a ligand for the non-classical major histocompatibility complex (MHC) molecule thymic leukaemia antigen,51 is transiently expressed on CD8αβ+50 T cells that down-regulated the CD8β chain. Studies performed on human blood samples identified CD8αα+ T cells as a particular memory T-cell subset47,48 which is stable over time52 and enriched in antigen-specific T cells. Our data showed that CD8αα+ T cells are not only present in NHPs, but are also present at higher frequency, in the peripheral circulation of NHPs, and that in HDs and NHPs CD8αα+ T cells were enriched in differentiated T cells compared with CD8αβ+ T cells.
The NHP CD8αα+ T cells may therefore also represent a memory T-cell subsets for long-lived antigen-specific immune responses:53 we have previously shown that NHP CD8αα+ T cells, and not CD8αβ+ T cells specifically proliferate in response to molecularly defined Mycobacterium tuberculosis antigens.53 Down-regulation of the CD8β chain may represent a mechanism that lowers the avidity of the TCR to its MHC–peptide ligand to secure long-term immune cell memory limiting T-cell activation54 and the risk of activation-induced apoptosis.55,56.
Identification of CD4+ CD8+ T cells
Two additional T-cell compartments were present in HDs and at a higher frequency in NHPs: CD4+ CD8αα+ and CD4+ CD8αβ+ T cells as reported previously.57–59 Their frequency appeared to be higher in female rhesus monkeys.20 CD4+ CD8+ T cells stained positive for the degranulation marker CD107a. In contrast to a previous report,59 CD4+ CD8αα+ and CD4+ CD8αβ+ T cells in NHPs showed similar frequencies and their maturation/differentiation marker profile reflected the phenotype of the ‘conventional’ CD4+ CD8– T cells. We postulate that CD4+ CD8+ T cells represent a specialized compartment of CD4+ T cells formed during the different stages of T-cell differentiation, characterized by CD8 expression. Because the CD4+ CD8+ T cells were endowed with effector capacity (CD107a expression) (model Fig. 7); it could be that CD4+ CD8− T cells represent a CD4+ T-cell compartment capable of lysing target cells, the co-expression of CD8 enables intracellular calcium levels to be increased, enhances cytotoxicity and may prevent apoptosis60 upon binding to MHC class I molecules.
Figure 7.
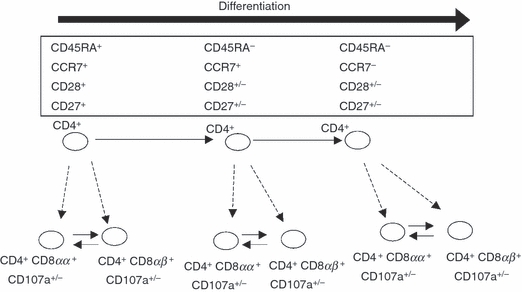
Model of CD3+ CD4+ CD8+ T-cell differentiation. Human donor and non-human primate CD3+ CD4+ CD8αα+ and CD3+ CD4+ CD8αβ+ T cells expressed CD107a and displayed a very similar phenotype to CD3+ CD4+, suggesting that CD3+ CD4+ CD8+ arise from CD3+ CD4+ and represent a ‘back-up’ compartment endowed with cytotoxic functions. CD4+ CD8αα+ may arise from CD4+ CD8αβ+ (or vice versa).
IL-17 production in CD4+ CD8+ and CD8αα+ T cells
To examine the role of CD4+ CD8+ T cells, we evaluated IL-17 production in PBMCs from HDs and NHPs in the presence IL-23 and IL-1β.61 Only data from HDs could be analysed because of the low number of IL-17-positive events in NHP PBMCs. CD4+ CD8+ T cells showed a higher, and CD8αα+ T cells a comparable, frequency of IL-17 production, yet a different profile (more polyfunctional IL-17+ TNF-α+ IFN-γ+) as compared with CD4+ (CD8−) T cells. These data support the notion that CD4+ CD8+ T cells appear to represent a distinct CD4+ T-cell memory compartment, in part characterized by IL-17 production. Our results also showed that the differentiation of IL-17+ producing CD8+ T cells occurred preferentially within the CD8αα+ T-cell compartment. Interleukin-17 production by memory CD8+ T cells, displaying a CD27+ CD28+/− CD45RA− phenotype in humans, was described by Kondo et al.62
Differences in T cells with putative regulatory or suppressive functions
CD4+ Tregs are characterized by co-expression of FoxP3 and high levels of CD25.63 We observed comparable frequencies of CD4+ (CD25high FoxP3+) Tregs in PBMCs from HD and NHPs. CD8+ Tregs (CD8+ CD25+ FoxP3+) have been described in humans,64,65 and in rhesus monkeys.66 We show that CD8+ Tregs (CD8+ CD25interm./high FoxP3+) were present in PBMCs from NHPs in higher frequencies compared with HDs. The same was true for other T-cell subsets co-expressing FoxP3 and CD25 with putative regulatory functions, i.e. CD4+ CD25interm FoxP3+, CD4+ CD8+ CD25interm./high FoxP3+. The FoxP3 and CD25 can be induced upon T-cell activation, it is exclusively expressed by Tregs.
The observation that NHPs showed a decreased number of bona fide IL-7Rα+ in CD4+ Tregs underlines the fact that differential suppressive functions may be present in NHPs compared with HDs. FoxP3 interacts with the IL-7Rα promoter and facilitates the down-regulation of IL-7Rα in CD4+ CD25bright Tregs;67 negative staining for IL-7Rα was postulated as a marker for human Tregs in concert with CD4, CD25 and FoxP3 analysis.68,69 A low percentage of human Tregs express IL-7Rα and these cells are important in diseases: a recent study showed that human CD3+ CD4+ CD25+ Tregs, which stain positive for IL-7Rα, exhibit an aberrant functional capacity in patients with autoimmune diseases: they exhibit increased proliferation and more IFN-γ/IL-2 production compared with the same cells from healthy individuals.70 The number of IL-7Rα+ expressing CD4+ Tregs was lower in NHPs than in HDs and this may also provide the cellular basis for differential suppressive networks in NHPs.
In summary, we showed, using high content flow cytometry, that the cellular immune system in humans and NHPs exhibited high level of communalities, including a unique CD4+ CD8αα/αβ+ T-cell population with cytotoxic potential. Differences between humans and NHPs reside in immune cell subsets with long-term memory, i.e. in CD8αα+ T cells and in cells with regulatory functions. This may be biologically important in chronic disease models where inflammatory patterns contribute to immune pathology.
Acknowledgments
We would like to thank Meryl Forman, Beckman Coulter (Miami, FL) for her valuable advice concerning antibody selection and the choice of fluorochromes on custom-labelled reagents. The project was funded in part by the AERAS foundation, from Karolinska Institutet, from SIDA, Vetenskaprådet and from the Söderberg Foundation, Sweden. The study was in part financed by the Aeras foundation, by a Marie-Curie Host Fellowship for Early Stage Researchers Training grant to I.M., from Cancerfonden, the Söderberg foundation, SIDA, Vetenskapsrådet and Karolinska Institutet to M.M.
Disclosures
The authors declare that there is no conflict of interest.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1. Identification of IL-17 producing cells.
Figure S2. Gating strategy to identify Tregs.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- 1.Watkins DI, Burton DR, Kallas EG, Moore JP, Koff WC. Nonhuman primate models and the failure of the Merck HIV-1 vaccine in humans. Nat Med. 2008;14:617–21. doi: 10.1038/nm.f.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Langermans JA, Andersen P, Van Soolingen D, et al. Divergent effect of bacillus Calmette–Guérin (BCG) vaccination on Mycobacterium tuberculosis infection in highly related macaque species: implications for primate models in tuberculosis vaccine research. Proc Natl Acad Sci USA. 2001;98:11497–502. doi: 10.1073/pnas.201404898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pitcher CJ, Hagen SI, Walker JM, Lum R, Mitchell BL, Maino VC, Axthelm MK, Picker LJ. Development and homeostasis of T cell memory in rhesus macaque. J Immunol. 2002;168:29–43. doi: 10.4049/jimmunol.168.1.29. [DOI] [PubMed] [Google Scholar]
- 4.Jankovic V, Messaoudi I, Nikolich-Zugich J. Phenotypic and functional T-cell aging in rhesus macaques (Macaca mulatta): differential behavior of CD4 and CD8 subsets. Blood. 2003;102:3244–51. doi: 10.1182/blood-2003-03-0927. [DOI] [PubMed] [Google Scholar]
- 5.Messaoudi I, Warner J, Fischer M, et al. Delay of T cell senescence by caloric restriction in aged long-lived nonhuman primates. Proc Natl Acad Sci USA. 2006;103:19448–53. doi: 10.1073/pnas.0606661103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fry TJ, Mackall CL. Current concepts of thymic aging. Springer Semin Immunopathol. 2002;24:7–22. doi: 10.1007/s00281-001-0092-5. [DOI] [PubMed] [Google Scholar]
- 7.Welsh RM, Kim SK, Cornberg M, Clute SC, Selin LK, Naumov YN. The privacy of T cell memory to viruses. Curr Top Microbiol Immunol. 2006;311:117–53. doi: 10.1007/3-540-32636-7_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welsh RM, Selin LK. No one is naive: the significance of heterologous T-cell immunity. Nat Rev Immunol. 2002;2:417–26. doi: 10.1038/nri820. [DOI] [PubMed] [Google Scholar]
- 9.Welsh RM, Selin LK, Szomolanyi-Tsuda E. Immunological memory to viral infections. Annu Rev Immunol. 2004;22:711–43. doi: 10.1146/annurev.immunol.22.012703.104527. [DOI] [PubMed] [Google Scholar]
- 10.Hohn H, Kortsik C, Nilges K, Necker A, Freitag K, Tully G, Neukirch C, Maeurer MJ. Human leucocyte antigen-A2 restricted and Mycobacterium tuberculosis 19-kDa antigen-specific CD8+ T-cell responses are oligoclonal and exhibit a T-cell cytotoxic type 2 response cytokine-secretion pattern. Immunology. 2001;104:278–88. doi: 10.1046/j.1365-2567.2001.01307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hohn H, Kortsik C, Tully G, et al. Longitudinal analysis of Mycobacterium tuberculosis 19-kDa antigen-specific T cells in patients with pulmonary tuberculosis: association with disease activity and cross-reactivity to a peptide from HIVenv gp120. Eur J Immunol. 2003;33:1613–23. doi: 10.1002/eji.200323480. [DOI] [PubMed] [Google Scholar]
- 12.Nilges K, Hohn H, Pilch H, Neukirch C, Freitag K, Talbot PJ, Maeurer MJ. Human papillomavirus type 16 E7 peptide-directed CD8+ T cells from patients with cervical cancer are cross-reactive with the coronavirus NS2 protein. J Virol. 2003;77:5464–74. doi: 10.1128/JVI.77.9.5464-5474.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suntharalingam G, Perry MR, Ward S, Brett SJ, Castello-Cortes A, Brunner MD, Panoskaltsis N. Cytokine storm in a phase 1 trial of the anti-CD28 monoclonal antibody TGN1412. N Engl J Med. 2006;355:1018–28. doi: 10.1056/NEJMoa063842. [DOI] [PubMed] [Google Scholar]
- 14.Waibler Z, Sender LY, Merten C, et al. Signaling signatures and functional properties of anti-human CD28 superagonistic antibodies. PLoS ONE. 2008;3:e1708. doi: 10.1371/journal.pone.0001708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu Rev Immunol. 2004;22:745–63. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- 16.Appay V, Douek DC, Price DA. CD8+ T cell efficacy in vaccination and disease. Nat Med. 2008;14:623–8. doi: 10.1038/nm.f.1774. [DOI] [PubMed] [Google Scholar]
- 17.Kursar M, Bonhagen K, Fensterle J, Kohler A, Hurwitz R, Kamradt T, Kaufmann SH, Mittrucker HW. Regulatory CD4+ CD25+ T cells restrict memory CD8+ T cell responses. J Exp Med. 2002;196:1585–92. doi: 10.1084/jem.20011347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fry TJ, Mackall CL. The many faces of IL-7: from lymphopoiesis to peripheral T cell maintenance. J Immunol. 2005;174:6571–6. doi: 10.4049/jimmunol.174.11.6571. [DOI] [PubMed] [Google Scholar]
- 19.Sopper S, Stahl-Hennig C, Demuth M, Johnston IC, Dorries R, Ter Meulen V. Lymphocyte subsets and expression of differentiation markers in blood and lymphoid organs of rhesus monkeys. Cytometry. 1997;29:351–62. [PubMed] [Google Scholar]
- 20.Qiu CL, Zhao H, Yang GB, Liu Q, Shao Y. Flow cytometric characterization of T lymphocyte subsets in the peripheral blood of Chinese rhesus macaques: normal range, age- and sex-related differences. Vet Immunol Immunopathol. 2008;4:313–21. doi: 10.1016/j.vetimm.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 21.Mattapallil JJ, Douek DC, Buckler-White A, Montefiori D, Letvin NL, Nabel GJ, Roederer M. Vaccination preserves CD4 memory T cells during acute simian immunodeficiency virus challenge. J Exp Med. 2006;203:1533–41. doi: 10.1084/jem.20060657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carter DL, Shieh TM, Blosser RL, Chadwick KR, Margolick JB, Hildreth JE, Clements JE, Zink MC. CD56 identifies monocytes and not natural killer cells in rhesus macaques. Cytometry. 1999;37:41–50. [PubMed] [Google Scholar]
- 23.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–12. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 24.DiSanto JP, Klein JS, Flomenberg N. Phosphorylation and down-regulation of CD4 and CD8 in human CTLs and mouse L cells. Immunogenetics. 1989;30:494–501. doi: 10.1007/BF02421181. [DOI] [PubMed] [Google Scholar]
- 25.Liu W, Putnam AL, Xu-Yu Z, et al. CD127 expression inversely correlates with FoxP3 and suppressive function of human CD4+ T reg cells. J Exp Med. 2006;203:1701–11. doi: 10.1084/jem.20060772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia Vela JA, Delgado I, Bornstein R, Alvarez B, Auray MC, Martin I, Ona F, Gilsanz F. Comparative intracellular cytokine production by in vitro stimulated T lymphocytes from human umbilical cord blood (HUCB) and adult peripheral blood (APB) Anal Cell Pathol. 2000;3:93–8. doi: 10.1155/2000/572952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcondes MC, Penedo MC, Lanigan C, Hall D, Watry DD, Zandonatti M, Fox HS. Simian immunodeficiency virus-induced CD4+ T cell deficits in cytokine secretion profile are dependent on monkey origin. Viral Immunol. 2006;19:679–89. doi: 10.1089/vim.2006.19.679. [DOI] [PubMed] [Google Scholar]
- 28.Ibegbu C, Brodie-Hill A, Kourtis AP, Carter A, McClure H, Chen ZW, Nahmias AJ. Use of human CD3 monoclonal antibody for accurate CD4+ and CD8+ lymphocyte determinations in macaques: phenotypic characterization of the CD3− CD8+ cell subset. J Med Primatol. 2001;30:291–8. doi: 10.1034/j.1600-0684.2001.300601.x. [DOI] [PubMed] [Google Scholar]
- 29.DeMaria MA, Casto M, O’Connell M, Johnson RP, Rosenzweig M. Characterization of lymphocyte subsets in rhesus macaques during the first year of life. Eur J Haematol. 2000;65:245–57. doi: 10.1034/j.1600-0609.2000.065004245.x. [DOI] [PubMed] [Google Scholar]
- 30.Tollerud DJ, Clark JW, Brown LM, Neuland CY, Pankiw-Trost LK, Blattner WA, Hoover RN. The influence of age, race, and gender on peripheral blood mononuclear-cell subsets in healthy nonsmokers. J Clin Immunol. 1989;9:214–22. doi: 10.1007/BF00916817. [DOI] [PubMed] [Google Scholar]
- 31.Xia HJ, Zhang GH, Wang RR, Zheng YT. The influence of age and sex on the cell counts of peripheral blood leukocyte subpopulations in Chinese rhesus macaques. Cell Mol Immunol. 2009;6:433–40. doi: 10.1038/cmi.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Z, Shackleton CH, Pahwa S, White PC, Speiser PW. Prominent sex steroid metabolism in human lymphocytes. Mol Cell Endocrinol. 1998;2:61–9. doi: 10.1016/s0303-7207(98)00052-5. [DOI] [PubMed] [Google Scholar]
- 33.Campbell JP, Riddell NE, Burns VE, Turner M, Van Zanten JJ, Drayson MT, Bosch JA. Acute exercise mobilises CD8+ T lymphocytes exhibiting an effector-memory phenotype. Brain Behav Immun. 2009;23:767–75. doi: 10.1016/j.bbi.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 34.Romero P, Zippelius A, Kurth I, et al. Four functionally distinct populations of human effector-memory CD8+ T lymphocytes. J Immunol. 2007;178:4112–9. doi: 10.4049/jimmunol.178.7.4112. [DOI] [PubMed] [Google Scholar]
- 35.Okada R, Kondo T, Matsuki F, Takata H, Takiguchi M. Phenotypic classification of human CD4+ T cell subsets and their differentiation. Int Immunol. 2008;20:1189–99. doi: 10.1093/intimm/dxn075. [DOI] [PubMed] [Google Scholar]
- 36.Harari A, Vallelian F, Pantaleo G. Phenotypic heterogeneity of antigen-specific CD4 T cells under different conditions of antigen persistence and antigen load. Eur J Immunol. 2004;34:3525–33. doi: 10.1002/eji.200425324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koch S, Larbi A, Derhovanessian E, Ozcelik D, Naumova E, Pawelec G. Multiparameter flow cytometric analysis of CD4 and CD8 T cell subsets in young and old people. Immun Ageing. 2008;5:6. doi: 10.1186/1742-4933-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Appay V, Dunbar PR, Callan M, et al. Memory CD8+ T cells vary in differentiation phenotype in different persistent virus infections. Nat Med. 2002;8:379–85. doi: 10.1038/nm0402-379. [DOI] [PubMed] [Google Scholar]
- 39.Gauduin MC, Yu Y, Barabasz A, Carville A, Piatak M, Lifson JD, Desrosiers RC, Johnson RP. Induction of a virus-specific effector-memory CD4+ T cell response by attenuated SIV infection. J Exp Med. 2006;203:2661–72. doi: 10.1084/jem.20060134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monceaux V, Viollet L, Petit F, Ho Tsong Fang R, Cumont MC, Zaunders J, Hurtrel B, Estaquier J. CD8+ T cell dynamics during primary simian immunodeficiency virus infection in macaques: relationship of effector cell differentiation with the extent of viral replication. J Immunol. 2005;174:6898–908. doi: 10.4049/jimmunol.174.11.6898. [DOI] [PubMed] [Google Scholar]
- 41.Bell EB, Sparshott SM, Bunce C. CD4+ T-cell memory, CD45R subsets and the persistence of antigen – a unifying concept. Immunol Today. 1998;19:60–4. doi: 10.1016/s0167-5699(97)01211-5. [DOI] [PubMed] [Google Scholar]
- 42.Liovat AS, Jacquelin B, Ploquin MJ, Barre-Sinoussi F, Muller-Trutwin MC. African non human primates infected by SIV – why don't they get sick? Lessons from studies on the early phase of non-pathogenic SIV infection. Curr HIV Res. 2009;7:39–50. doi: 10.2174/157016209787048546. [DOI] [PubMed] [Google Scholar]
- 43.Kornfeld C, Ploquin MJ, Pandrea I, et al. Antiinflammatory profiles during primary SIV infection in African green monkeys are associated with protection against AIDS. J Clin Invest. 2005;115:1082–91. doi: 10.1172/JCI23006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pandrea I, Sodora DL, Silvestri G, Apetrei C. Into the wild: simian immunodeficiency virus (SIV) infection in natural hosts. Trends Immunol. 2008;29:419–28. doi: 10.1016/j.it.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Janot-Sardet C, Assouline B, Cheynier R, Morre M, Beq S. A validated assay to measure soluble IL-7 receptor shows minimal impact of IL-7 treatment. J Immunol Methods. 2010;353:115–23. doi: 10.1016/j.jim.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 46.Madakamutil LT, Christen U, Lena CJ, et al. CD8αα-mediated survival and differentiation of CD8 memory T cell precursors. Science. 2004;304:590–3. doi: 10.1126/science.1092316. [DOI] [PubMed] [Google Scholar]
- 47.Konno A, Okada K, Mizuno K, et al. CD8αα memory effector T cells descend directly from clonally expanded CD8α+βhigh TCRαβ T cells in vivo. Blood. 2002;100:4090–7. doi: 10.1182/blood-2002-04-1136. [DOI] [PubMed] [Google Scholar]
- 48.Boulassel MR, Mercier F, Gilmore N, Routy JP. Immunophenotypic patterns of CD8+ T cell subsets expressing CD8αα and IL-7Rα in viremic, aviremic and slow progressor HIV-1-infected subjects. Clinical Immunol (Orlando, Fla) 2007;124:149–57. doi: 10.1016/j.clim.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 49.Chandele A, Kaech SM. Cutting edge: memory CD8 T cell maturation occurs independently of CD8αα. J Immunol. 2005;175:5619–23. doi: 10.4049/jimmunol.175.9.5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhong W, Reinherz EL. CD8αα homodimer expression and role in CD8 T cell memory generation during influenza virus A infection in mice. Eur J Immunol. 2005;35:3103–10. doi: 10.1002/eji.200535162. [DOI] [PubMed] [Google Scholar]
- 51.Leishman AJ, Naidenko OV, Attinger A, et al. T cell responses modulated through interaction between CD8αα and the nonclassical MHC class I molecule, TL. Science (New York, NY) 2001;294:1936–9. doi: 10.1126/science.1063564. [DOI] [PubMed] [Google Scholar]
- 52.Magalhaes I, Vudattu NK, Jager E, Maeurer MJ. Tumor antigen-specific T-cells are Present in the CD8αα+ T-cell effector-memory pool. J Immunother. 2008;31:840–8. doi: 10.1097/CJI.0b013e31818883a1. [DOI] [PubMed] [Google Scholar]
- 53.Magalhaes I, Sizemore DR, Ahmed RK, et al. rBCG induces strong antigen-specific T cell responses in rhesus macaques in a prime-boost setting with an adenovirus 35 tuberculosis vaccine vector. PLoS ONE. 2008;3:e3790. doi: 10.1371/journal.pone.0003790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheroutre H, Lambolez F. Doubting the TCR coreceptor function of CD8αα. Immunity. 2008;28:149–59. doi: 10.1016/j.immuni.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 55.Bosselut R, Kubo S, Guinter T, Kopacz JL, Altman JD, Feigenbaum L, Singer A. Role of CD8β domains in CD8 coreceptor function: importance for MHC I binding, signaling, and positive selection of CD8+ T cells in the thymus. Immunity. 2000;12:409–18. doi: 10.1016/s1074-7613(00)80193-4. [DOI] [PubMed] [Google Scholar]
- 56.Cebecauer M, Guillaume P, Hozak P, Mark S, Everett H, Schneider P, Luescher IF. Soluble MHC–peptide complexes induce rapid death of CD8+ CTL. J Immunol. 2005;174:6809–19. doi: 10.4049/jimmunol.174.11.6809. [DOI] [PubMed] [Google Scholar]
- 57.Sala P, Tonutti E, Feruglio C, Florian F, Colombatti A. Persistent expansions of CD4+ CD8+ peripheral blood T cells. Blood. 1993;82:1546–52. [PubMed] [Google Scholar]
- 58.Nascimbeni M, Shin EC, Chiriboga L, Kleiner DE, Rehermann B. Peripheral CD4+ CD8+ T cells are differentiated effector memory cells with antiviral functions. Blood. 2004;104:478–86. doi: 10.1182/blood-2003-12-4395. [DOI] [PubMed] [Google Scholar]
- 59.Macchia I, Gauduin MC, Kaur A, Johnson RP. Expression of CD8α identifies a distinct subset of effector memory CD4+ T lymphocytes. Immunology. 2006;119:232–42. doi: 10.1111/j.1365-2567.2006.02428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Addison EG, North J, Bakhsh I, et al. Ligation of CD8α on human natural killer cells prevents activation-induced apoptosis and enhances cytolytic activity. Immunology. 2005;116:354–61. doi: 10.1111/j.1365-2567.2005.02235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 62.Kondo T, Takata H, Matsuki F, Takiguchi M. Cutting edge: phenotypic characterization and differentiation of human CD8+ T cells producing IL-17. J Immunol. 2009;182:1794–8. doi: 10.4049/jimmunol.0801347. [DOI] [PubMed] [Google Scholar]
- 63.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+ CD25+ regulatory T cells. Nat Immunol. 2003;4:330–6. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 64.Cosmi L, Liotta F, Lazzeri E, et al. Human CD8+ CD25+ share phenotypic and functional features with CD4+ CD25+ regulatory thymocytes. Blood. 2003;102:4107–14. doi: 10.1182/blood-2003-04-1320. [DOI] [PubMed] [Google Scholar]
- 65.Jarvis LB, Matyszak MK, Duggleby RC, Goodall JC, Hall FC, Gaston JS. Autoreactive human peripheral blood CD8+ T cells with a regulatory phenotype and function. Eur J Immunol. 2005;35:2896–908. doi: 10.1002/eji.200526162. [DOI] [PubMed] [Google Scholar]
- 66.Nigam P, Velu V, Kannanganat S, Chennareddi L, Kwa S, Siddiqui M, Amara RR. Expansion of FOXP3+ CD8 T cells with suppressive potential in colorectal mucosa following a pathogenic simian immunodeficiency virus infection correlates with diminished antiviral T cell response and viral control. J Immunol. 2010;184:1690–701. doi: 10.4049/jimmunol.0902955. [DOI] [PubMed] [Google Scholar]
- 67.Hohlfeld R, Wekerle H. Autoimmune concepts of multiple sclerosis as a basis for selective immunotherapy: from pipe dreams to (therapeutic) pipelines. Proc Natl Acad Sci U S A. 2004;101(Suppl. 2):14599–606. doi: 10.1073/pnas.0404874101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dieckmann D, Plottner H, Berchtold S, Berger T, Schuler G. Ex vivo isolation and characterization of CD4+ CD25+ T cells with regulatory properties from human blood. J Exp Med. 2001;193:1303–10. doi: 10.1084/jem.193.11.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Seddiki N, Santner-Nanan B, Martinson J, et al. Expression of interleukin (IL)-2 and IL-7 receptors discriminates between human regulatory and activated T cells. J Exp Med. 2006;203:1693–700. doi: 10.1084/jem.20060468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Michel L, Berthelot L, Pettre S, et al. Patients with relapsing-remitting multiple sclerosis have normal Treg function when cells expressing IL-7 receptor α-chain are excluded from the analysis. J Clin Invest. 2008;118:3411–9. doi: 10.1172/JCI35365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vudattu NK, Magalhaes I, Schmidt M, Seyfert-Margolis V, Maeurer MJ. Reduced numbers of IL-7 receptor (CD127) expressing immune cells and IL-7-signaling defects in peripheral blood from patients with breast cancer. Int J Cancer. 2007;121:1512–9. doi: 10.1002/ijc.22854. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


