Abstract
In this brief communication, we describe the Allele Name Translation Tool (ANTT) and Update NomenCLature (UNCL), free programs developed to facilitate the translation of HLA allele names recorded using the December 2002 version of the HLA allele nomenclature (e.g., A*01010101) to those recorded using the colon-delimited version of the HLA allele nomenclature (e.g., A*01:01:01:01) adopted in April of 2010. In addition, the ANTT and UNCL translate specific HLA allele-name changes (e.g., DPB1*0502 is translated to DPB1*104:01), as well as changes to the locus-prefix for HLA-C (i.e., Cw* is translated to C*). The ANTT and UNCL will also translate allele names that have been truncated to two, four or six digits, as well as ambiguous allele-strings. The ANTT is a locally installed and run application, while UNCL is a web-based tool that requires only an internet connection and a modern browser. The ANTT accepts a variety of HLA data-presentation and allele-name formats. In addition, the ANTT can translate using user-defined conversion settings (e.g., the names of alleles that encode identical peptide binding domains can be translated to a common ‘P-code’), and can serve as a preliminary data-sanity tool. The ANTT is available for download, and UNCL for use, at www.igdawg.org/software.
Over the last four decades, World Health Organization (WHO) Nomenclature Committee for factors of the human leukocyte antigen (HLA) system has developed a rigorous hierarchical nomenclature for the representation of the complex polymorphisms that comprise HLA alleles (1-7). During that period, technological advances have required a succession of nomenclature versions, each of which incorporates more information about HLA polymorphism into an allele name. However, in the absence of an automated system for converting between different versions of the nomenclature, HLA data consistent with one version may not be updated upon the advent of a new version, because even simple allele name translations [e.g., a change from 01011 to 010101 (7)] become time consuming when changes must be made to large datasets. In addition, non-obvious nomenclature changes [e.g., changing B*1522 to B*3543 (7)] may not be adopted in a timely fashion. Furthermore, transcription errors may be introduced when allele names are manually updated to conform to new versions of the nomenclature, and commonly used spreadsheet applications can routinely introduce a variety of errors (e.g., 01010101 might be presented as 1010101 or 1,010,101), as these were not designed with HLA in mind.
From April of 2010, HLA allele nomenclature will incorporate colons to explicitly define the domains that specify polymorphisms among allele families, protein sequences, synonymous coding nucleotide sequences, and noncoding nucleotide sequences; at the same time, the names of many alleles will change in nonobvious ways (e.g., DPB1*0502 will change to DPB1*104:01), and the locus prefix for HLA-C alleles will change from Cw* to C* (8). Many of the above-mentioned challenges to the timely application of new nomenclature versions to extant data will likely pertain to these changes, and while these are logical and necessary improvements to the current version of the nomenclature, their inherent complexity may present obstacles to the continuity of HLA data-analysis. An automated system for consistently translating allele names to the new nomenclature versions can help ensure that extant HLA data are updated easily and consistently across datasets, reducing the likelihood of analytical and reporting errors related to allele-name variation within and between datasets.
Here, we describe the Allele Name Translation Tool (ANTT) and Update NomenCLature (UNCL), software tools designed to translate allele names recorded using the 2002 Nomenclature for Factors of the HLA System (7) to allele names recorded under the above described April 2010 version of the nomenclature (8) in an automated fashion. Using either UNCL or the ANTT, the HLA allele names recorded since 2002 can be translated to their April 2010 versions quickly, and with minimal effort on the part of the researcher, for entire datasets. Both applications are capable of quickly translating between versions of the nomenclature, and have successfully translated datasets including millions of allele names.
The ANTT is a Microsoft .NET framework (9) software application that can run under Windows, Apple OS X and Linux operating systems to automatically translate the allele names in entire data files. The .NET framework is native in all Windows 7 systems. A setup application distributed with the ANTT will allow ANTT users to install the .NET framework on Windows XP and Vista systems if it has not already been installed. Apple OS X and Linux operating system users must install the Mono open source .NET development framework in order to use the ANTT.
The ANTT has been designed with flexibility and general utility in mind; in addition to translating HLA allele names between the 2002 and April 2010 nomenclature versions, it can be used to translate between any pair of user-defined allele-naming conventions, so that alleles recorded using pre-2002 nomenclatures, or allele names that have been changed, can be translated to the April 2010 version of the nomenclature without an intermediate naming step (e.g., A*01011 can be translated to A*01:01:01, or B*1522 can be translated to B*35:43). Similarly, alleles that share the same protein sequence for the peptide binding domain can be translated to the same ‘P-code’ (e.g., A*02:01 and A*02:09 could both be translated to A*02:01P), and alleles that share identical nucleotide sequences for the exons that encode the peptide binding domain can be translated to the same ‘G-code’ (e.g., A*02:01:01:01 and A*02:01:01:03 could both be translated to A*02:01:01G). National Marrow Donor Program (NMDP) allele-codes can also be translated using the ANTT (e.g., the A*02CAVZ code can be translated to the P-code A*02:01P). Finally, the ANTT can also “back-translate” allele names from one version of the nomenclature to an older version (e.g., from the April 2010 version to the 2002 version).
The ANTT accepts allele name data that are arranged in columns and stored in a tab-delimited text file as input, and generates a tab-delimited text file containing the translated alleles in the same order and organization as output. Columns containing non-allelic data may be included in the input files and will be included in the same order and organization in the output file (c.f. Table 1). The first row of each column must contain a field name that identifies either the locus of the alleles to be translated or the non-allelic content of the column, as defined by the user in an associated configuration file (described below). So long as the column headers are defined in the configuration file, data from multiple sources may be combined in a single input file. Similarly, allele data with and without locus prefixes (e.g., A*01010101 and 01010101) can be included in the input file, and will be translated accordingly. This flexibility in the format and organization of the input data permits the ANTT to translate genotype data, as well as allele-count and allele-frequency data, with minimal requirements for reformatting.
Table 1.
Examples of ANTT input and output files
| ANTT input file | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | hla-a | hla-a | hla-a | acount | hla-b | bfreq | id | drb1 | drb1 | hla-c | hla-c | ccount | cfreq |
| s001 a | 0101 b | 0201 b | A*301102 | 20 | 070205 | 0.25 | 2230 | 0802 b | 1602 b | Cw*020102 d | 080301 | 223 | 0.19 |
| s001 a | 0109 | 0207 | A*2310 | 15 | 1509 | 0.1 | 2231 | 0402 | 1401 b | Cw*033801 | 1801 | 13 | 0.004 |
| s002 a | 2602/ | 0211 | A*24020102L | 2 | 150108 | 0.4 | HK35 | 1102 b | 1304 | 06020102 | Cw*1507 | 19 | 0.11 |
| 2604 c | |||||||||||||
| s002 a | 2607 b | 0209 | A*680301 | 14 | 9558 | 0.15 | Cw*030203 | 020206 | 45 | 0.001 | |||
| s003 a | 1107 | 2302 | A*9211 | 23 | 1803 | 0.1 | Cw*04010101/ | 12030101/ | 319 | 0.032 | |||
| Cw*04010103 | Cw*12030102 c | ||||||||||||
| s003 a | 1107 | 2301 b | A*110201 | 9 | |||||||||
| s004 | **** e | 2402 b | A*7404 | 8 | |||||||||
| A*1104 | 7 | ||||||||||||
| ANTT output file | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | hla-a | hla-a | hla-a | acount | hla-b | bfreq | id | drb1 | drb1 | hla-c | hla-c | ccount | cfreq |
| s001 a | 01:01 b | 02:01 b | A*30:11:02 | 20 | 07:02:05 | 0.25 | 2230 | 08:02 b | 16:02 b | Cw*020102 d | 08:03:01 | 223 | 0.19 |
| s001 a | 01:09 | 02:07 | A*23:10 | 15 | 15:09 | 0.1 | 2231 | 04:02 | 14:01 b | C*03:38:01 | 18:01 | 13 | 0.004 |
| s002 a | 26:02/ | 02:11 | A*24:02:01:02L | 2 | 15:01:08 | 0.4 | HK35 | 11:02 b | 13:04 | 06:02:01:02 | C*15:07 | 19 | 0.11 |
| 26:04 c | |||||||||||||
| s002 a | 26:07 b | 02:09 | A*68:03:01 | 14 | 15:158 | 0.15 | C*03:02:03 | 02:02:06 | 45 | 0.001 | |||
| s003 a | 11:07 | 23:02 | A*02:111 | 23 | 18:03 | 0.1 | C*04:01:01:01/ | 12:03:01:01/ | 319 | 0.032 | |||
| C*04:01:01:03 c | C*12:03:01:02 c | ||||||||||||
| s003 a | 11:07 | 23:01 b | A*11:02:01 | 9 | |||||||||
| s004 | **** e | 24:02 b | A*74:04 | 8 | |||||||||
| A*11:04 | 7 | ||||||||||||
ANTT, Allele Name Translation Tool; HLA, human leukocyte antigen.
Ambiguous genotypes, represented by multiple rows of genotypes for a given sample, can be translated by the ANTT.
Truncated alleles are translated by the ANTT.
Ambiguous alleles, represented here by strings of alleles separated by a '/', can be translated by the ANTT.
Invalid alleles (e.g., Cw*020102) are not translated, but are reported in the output file.
Missing data, as defined in the configuration file, are reported in the output file.
In addition, the ANTT will translate the individual alleles included in ambiguous allele strings; multiple genotype entries for ambiguous genotype sets will be translated as well. Allele names that have been truncated to a smaller number of polymorphic domains than recognized in the current nomenclature version will be translated to a name that corresponds to the same truncated level of organization under the new nomenclature version (e.g., A*0101 will be translated to A*01:01, and A*01:01 will be translated back to A*0101). However, the ANTT will not translate truncated allele names that violate the expectations of the 2002 or 2010 versions of the nomenclature (e.g., A*01010 and A*01:01:0 will not be translated). Instead, an error message will be included in a log file, and the untranslated allele will be included in the output file. Similarly, allele names that are not recognized for a given locus will not be translated, but will be noted in the error log, and included in the output file. In this capacity, the ANTT can be used for preliminary data “sanity” checks, identifying invalid allele names prior to analysis.
The ANTT uses a text-formatted configuration file to identify the column-header field names that pertain to allelic and nonallelic data, the character or characters used to identify missing or untyped allele data, the character or characters used to separate the alleles in an ambiguous string, and the location and names of the translation files that identify the appropriate translation between successive nomenclature versions or user-defined naming conventions for each locus.
The translation files accepted by the ANTT must contain two columns of data in a tab-delimited text format. The left column contains allele names recorded using the nomenclature version of the data in the input file. The right column contains the corresponding allele name recorded using the nomenclature version to which allele names will be translated. Both columns must have headers. The translation files distributed with the ANTT are derived from the table of current and new allele correspondences available from the hla.alleles.org web site (http://hla.alleles.org/data/txt/Nomenclature_2009.txt). As these correspondences are updated, a new set of translation tables can be generated from downloaded copies of the Nomenclature_2009.txt file, or a user-defined file of multi-locus allele name correspondences, using the File Manager utility distributed with the ANTT.
UNCL is a less flexible but more generally accessible version of the ANTT. It is a platform independent, web-based tool that utilizes R, the open source language and environment for statistical computing (10). UNCL uses the table of old and new allele correspondences available from the hla.alleles.org web site to automatically translate columns of allele names from the 2002 version of the nomenclature to the April 2010 version for entire data files. Where the ANTT is designed for flexibility, UNCL is designed for utility and accessibility; it requires only a modern web-browser (e.g., Internet Explorer 8, Firefox 3, or Safari 4) and internet-access to function, and does not need to be installed on a user’s system. UNCL functions to translate allele data arranged in columns; there is no hard limit on the number of columns permitted, and allelic data from multiple loci can be included in the same file, along with non-allelic data. UNCL generates columns of translated-allele names in the same order and organization as the original data, along with unmodified columns of nonallelic data, and presents them for review in the user’s web browser as well as for download as a text file.
As with the ANTT, UNCL requires that data be stored in tab-delimited text files. Each column must include a header describing its contents; for allelic data, these headers must correspond to the locus prefix of the alleles in that column. Complete locus prefixes (e.g. ‘DRB1’, ‘dpa1,’ ‘A’, ‘b’, or ‘Cw’) are required; incomplete locus prefixes (e.g., ‘drb’, ‘dq’, or ‘C’) are not recognized. A list of accepted locus prefix column headers is available on the UNCL web page (www.igdawg.org/sofware), along with detailed instructions for input data formatting. UNCL will translate allele names that have been truncated to two, four, or six digits (as appropriate), and will translate slash-delimited strings of allele names. However, the uploaded text file cannot include empty cells; missing data can be represented by any character or string of characters that does not correspond to a valid allele name. Missing data, non-allelic data and allelic data that do not conform to the 2002 version of the nomenclature will be returned unchanged in the translated output file.
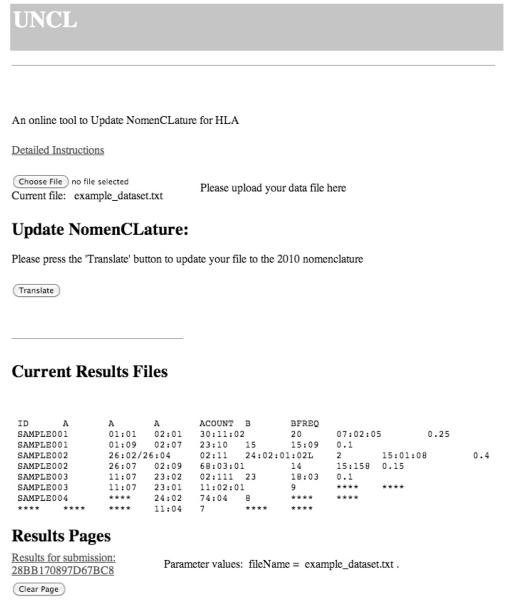
Data files are easily uploaded for translation on the UNCL web page. Uploaded allele-data files persist on the UNCL server for the duration of the translation session only, and a user can clear their uploaded data from the server at any time during the session (Figure 1).
Figure 1.
Screenshot of the Update NomenCLature web page showing a completed nomenclature translation.
UNCL provides a quick and easy method for updating HLA allele data files to the April 2010 version of the nomenclature with very little requirement for the user with regard to platform, proprietary software or expertise. Future versions of UNCL will expand on its current functions, for example, allowing users to perform custom translations, as with the ANTT, by uploading custom translation tables.
The ANTT and UNCL have been developed in keeping with the goals of the immunogenomic data analysis working group (IDAWG), an international collaboration of investigators interested in issues of immunogenomics data management and analysis, and are available for free public download and use from www.igdawg.org/software. The IDAWG aims to foster consistency in the management and analysis of immunogenomic data, as well as to increase accessibility of analytical tools, and it is our hope that the tools presented here will contribute to that goal.
Acknowledgements
The work presented here was carried out with the support of NIH grant U01AI067068. We thank Glenys Thomson, Erik Sowa, Henry A. Erlich, Katrina R. Eaton, Jorge Oksenberg and Pierre-Antoine Gourraud, Pawinee Kupatawintu, Steven G. E. Marsh, James Robinson and Hazael Maldonado Torres, Mike Varney and the Australian Red Cross Blood Service, Rachel Yedlin, Martin Maiers, and Thomas Smith for testing of the ANTT and UNCL and for their helpful comments.
Literature Cited
- 1.WHO Nomenclature Committee Nomenclature for factors of the HL-A system. Bull World Health Organ. 1968;39:483–6. [PMC free article] [PubMed] [Google Scholar]
- 2.WHO Nomenclature Committee Nomenclature for factors of the HLA system. Bull World Health Organ. 1975;52:261–5. [PMC free article] [PubMed] [Google Scholar]
- 3.WHO Nomenclature Committee Nomenclature for factors of the HLA system--1977. Tissue Antigens. 1978;11:81–6. doi: 10.1111/j.1399-0039.1978.tb01231.x. [DOI] [PubMed] [Google Scholar]
- 4.WHO Nomenclature Committee Nomenclature for factors of the HLA system, 1987. Tissue Antigens. 1988;32:177–87. doi: 10.1111/j.1399-0039.1988.tb01655.x. [DOI] [PubMed] [Google Scholar]
- 5.Bodmer JG, Marsh SG, Albert ED, et al. Nomenclature for factors of the HLA system, 1990. Tissue Antigens. 1991;37:97–104. doi: 10.1111/j.1399-0039.1991.tb01853.x. [DOI] [PubMed] [Google Scholar]
- 6.Bodmer JG, Marsh SG, Albert ED, et al. Nomenclature for factors of the HLA system, 1995. Tissue Antigens. 1995;46:1–18. doi: 10.1111/j.1399-0039.1995.tb02470.x. [DOI] [PubMed] [Google Scholar]
- 7.Marsh SG, Albert ED, Bodmer WF, et al. Nomenclature for factors of the HLA system, 2002. Tissue Antigens. 2002;60:407–64. doi: 10.1034/j.1399-0039.2002.600509.x. [DOI] [PubMed] [Google Scholar]
- 8.Marsh SG, Albert ED, Bodmer WF, et al. Nomenclature for factors of the HLA system, 2010. Tissue Antigens. 2010;75:291–455. doi: 10.1111/j.1399-0039.2010.01466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balena F. Programming Microsoft Visual Basic.NET. Microsoft Press; Redmond, WA USA: 2002. [Google Scholar]
- 10.R Development Core Team. R . A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2008. [Google Scholar]