Abstract
Reelin is a secreted, signaling protein associated with neuronal cell positioning and migration. Recently, reelin was found to be epigenetically silenced in gastric and pancreatic cancers in which down-regulation was associated with increased migratory ability and reduced survival. Here we analyzed reelin expression by immunohistochemistry in 17 normal breast tissue samples from reduction mammoplasties and in two independent tissue microarrays of 136 and more than 2000 breast cancer biopsy samples, respectively. Results were analyzed with regard to clinical parameters, including BRE (Bloom, Richardson, Elston) grade, nodal status, estrogen receptor and HER2 status, and overall survival. Reelin was expressed in the luminal epithelium and myoepithelium of the normal human breast but not in cancerous breasts. Loss of reelin protein expression correlated significantly with decreased survival (P = 0.01) and positive lymph node status (P < 0.001). By measuring reelin expression and promoter methylation status in 39 primary breast tumors, as well as in breast cancer-derived cell lines before and after decitabine treatment, we established that reelin expression levels correlated inversely with promoter methylation status, whereas demethylation increased reelin mRNA expression in vitro. Reelin overexpression in MDA-MB231 cells, as well as incubation with recombinant reelin, suppressed cell migration, invadopodia formation, and invasiveness in vitro. We conclude that reelin may play an important role in controlling invasiveness and metastatic potential of breast cancer cells and that its expression is controlled by promoter methylation.
Reelin is a secreted signaling protein and an important regulator of neuronal cell positioning during embryonic brain development (for review, see refs. 1, 2). It was first described as a regulator of neuronal cell migration of the neocortex during the development of the early cortical plate.3 In mice mutated in reelin (reeler) or in members of its downstream signaling pathway (reeler-like mice) cortical cell layering is severely disturbed.3,4,5,6,7,8
In the brain reelin acts mainly through binding to the low-density lipoprotein receptors VLDLR and ApoER2.9 On reelin binding the receptors cluster and signal down to the adaptor protein Dab1, which becomes phosphorylated10 in an Src family kinase-dependent process.11,12 pDab1 can activate the phosphatidylinositol 3-kinase/protein kinase B pathway, leading to glycogen synthase kinase-3β inhibition and actin remodeling.13,14 In another process, pDab1 can interact with the Lis1 complex, leading to changes in microtubular formation and nucleokinesis.15
The reelin promoter contains large CpG islands, and its expression in neurons is strongly controlled by promoter methylation and acetylation.16 Demethylation of the promoter or treatment with histone deacetylase inhibitors increases reelin mRNA expression in neuronal NT2 cells in vitro.16 In addition, hypermethylation of the reelin promoter can be found in brain regions of patients with schizophrenia.16
The physiological function of reelin has so far only been described in the brain. Although it has been shown to be present in other organs of the adult mouse including spinal cord, liver, kidney, testis, and ovary17 as well as in serum,18 no biological function has so far been attributed to reelin outside the brain. However, recent publications established a role for the reelin signaling pathway in different cancers. Whereas strong reelin expression was associated with high-grade prostate cancer,19 transcriptional silencing of reelin in gastric and pancreatic cancers through epigenetic control mechanisms was associated with a poor outcome.20,21 Knockdown of reelin, of its receptors VLDLR and ApoER2, or of Dab1 in a pancreatic cancer cell line increased cell migration and colony formation, whereas demethylation and treatment with trichostatin A (TSA) increased reelin expression.20
In the light of these somewhat contradictory results in three different cancers, we aimed to establish whether reelin was expressed in the human breast and whether its expression played a role in breast cancer. Here we describe, for the first time, that reelin is expressed in the normal breast epithelium and that reelin is down-regulated in breast cancer. Overexpression of reelin in a breast cancer cell line strongly reduces invadopodia formation and migration, as well as invasiveness in a collagen I matrix, whereas loss of reelin expression in breast cancers is significantly associated with positive lymph node status and decreased overall survival. We show that regulation of reelin expression is strongly linked to promoter methylation in breast cancer cell lines and in primary breast cancers.
Materials and Methods
Cell Lines–Growth and Treatment
Cell lines MDA-MB 231 (HTB-26), MDA-MB361 (HTB-27), MDA-MB435S (HTB-129D), MDA-MB 436 (HTB-130), MDA-MB453 (HTB-131), MDA-MB468 (HTB-132), MCF-7 (HTB-22), BT-20 (HTB-19), BT-474 (HTB-20), BT-549 (HTB-122), and ZR-75-30 (CRL-1504) were purchased from the American Type Culture Collection (LGC Standards, Teddington, UK) and grown according to the conditions set out by the American Type Culture Collection.
Reelin-overexpressing HEK293 cells were a generous gift from Drs. Foerster and Frotscher (Freiburg, Germany) and have been described previously.22 These were grown in Dulbecco’s modified Eagle’s medium (DMEM)/10% fetal calf serum in the presence of 1 mg/ml G418 (Invitrogen, Paisley, UK).
Cells were treated with 1 μmol/L 5-aza-2′-deoxycytidine (Decitabine) and 300 nmol/L TSA (Sigma-Aldrich, Dorset, UK) alone or in combination. Cells were treated with 5-aza-2′-deoxycytidine for 7 days with fresh inhibitor added every day. No obvious signs of cell death were observed during this period. TSA was added before the last day of 5-aza-2′-deoxycytidine treatment. Doxorubicin (200 ng/ml, Sigma-Aldrich) was used to treat cells for 24 hours.
Production of Stable Cell Lines
MDA-MB231 cells were transfected with a reelin expression vector (pCrl; generous gift from T. Curran, The Children’s Hospital of Philadelphia, Philadelphia, PA). This vector expresses full-length reelin under control of a cytomegalovirus promoter in a pcDNA3 backbone (neomycin-resistant) and was first cloned by D’Arcangelo et al.23 Cells were transfected at 80% confluence with 2 μg of unlinearized plasmid DNA, using Lipofectamine 2000 (Invitrogen) for 24 hours. Stable transfectants were selected by growing cells in 500 μg/ml G418. After serial dilution in a 96-well plate, 10 clones were cultured, and reelin protein levels were analyzed by Western blotting (data not shown). Finally, one clone expressing the full-length reelin and one clone expressing a truncated fragment of reelin were selected for further investigation. The procedure was repeated using linearized pCrl and two further clones were selected that showed either full-length or truncated reelin expression.
Quantitative RT-PCR
RNA was isolated using a Mini-RNeasy Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Total RNA (500 ng) was used for cDNA synthesis with Roche Transcriptor reverse transcriptase, and 1 μl of cDNA was used per PCR reaction. The PCR was performed using a Roche Light-Cycler with probe 72 (Roche Probe Library, Roche Diagnostics Ltd., Burgess Hill, UK) and the following primers: forward 5′-TGCTGGAATACACTAAGGATGC-3′ and reverse 5′-GAAGGCACTGGGTCTGTACG-3′. All reactions were run in triplicate, and the results were normalized to an internal control (β-actin, forward 5′-ATTGGCAATGAGCGGTTC-3′ and reverse 5′-GGATGCCACAGGACTCCAT-3′; probe 11).
Methylation-Specific PCR
Genomic DNA was isolated using a DNeasy Blood and Tissue Kit (Qiagen) according to the manufacturer’s instructions, and 200 ng of genomic DNA was modified by sodium bisulfate using a Zymo EZ DNA Methylation Kit (Zymo Research, Orange, CA). Approximately 20 ng of modified DNA was used in each PCR reaction. Primers used were as follows: unmethylated forward primer 5′-GGGTGTTTTTTTAGGTTTGGTTG-3′; unmethylated reverse primer 5′-CAACTCCCAAAATTACTTTAAACCAC-3′; methylated forward primer 5′-CGTTTTTTTAGGTTTGGTCGA-3′; and methylated reverse primer 5′-GACTCCCAAAATTACTTTAAACCG-3′.
Cycling conditions were as follows: an initial 8 cycles of 95°C for 2 minutes, 60°C for 30 seconds, and 72°C for 30 seconds followed by 32 cycles of 95°C for 30 seconds, 60°C for 30 seconds, and 72°C for 30 seconds, followed by a final extension at 72°C for 5 minutes. Products were separated on 2% agarose gels and visualized by ethidium bromide staining. Unmethylated control DNA was extracted from normal peripheral blood; chemically modified methylated control DNA (CpGenome Universal Methylated DNA) was purchased from Chemicon Europe (Cambridge Bioscience, Cambridge, UK).
Bisulfite Sequencing of Cell Lines
DNA (500 ng) was modified using the Zymo EZ DNA Methylation Kit (Zymo Research) according to the manufacturer’s instructions. Primers for bisulfite sequencing were designed using the MethPrimer protocol.24 Modified DNA (25 ng) was amplified using Thermo-Start Master Mix (Abgene Ltd., Epsom, UK) containing 500 nmol/L of each primer (forward 5′-GTAATATGTAGGGAAATGAGTATTT-3′ and reverse 5′-CCCAAAAAAAACAAAACACACTAA-3′) using an amplification program consisting of an initial incubation for 15 minutes at 95°C, followed by 40 cycles of 30 seconds at 95°C, 30 seconds annealing at 50°C, and 30 seconds extension at 72°C and a final 5-minute incubation at 72°C. Products were eluted from agarose gels using Zymoclean Gel DNA Recovery Kits (Zymo Research) and cloned into TA vectors (Invitrogen) according to the manufacturer’s instructions. DNA from at least 12 individual clones was isolated using TempliPhi amplification (GE Healthcare UK Limited, Bucks, UK) and analyzed by Sanger sequencing.
Pyrosequencing Analysis
After bisulfite treatment and PCR, the degree of methylation at each CpG position in a sequence was determined from the ratio of T and C. PCR primers (forward 5′-BIOT-GGGTTTTAAGAAGGTGTG-3′ and reverse 5′-CTCCCAAAATTACTTTAAAC-3′) were designed to amplify a 158-bp fragment across the CpG islands and optimized for software dedicated to methylation analysis.
PCR conditions were as follows: one initial 95°C step for 8 minutes followed by 40 cycles of 95°C for 30 seconds, 53°C for 30 seconds, 72°C for 40 seconds, and a final extension for 7 minutes at 72°C. The PCR products were separated on a 2% agarose gel and visualized using a transilluminator. The PCR products were finally analyzed by pyrosequencing using a Biotage Sample Prep kit (Biotage, Uppsala, Sweden) and the specific sequencing primer (PCR F). After the pyrosequencing run, analysis of the methylation percentage of each CpG site was performed using PyroMark ID software (Biotage). Placental DNA was used as a negative control of methylation (0% average methylation) and a commercial methylated DNA (Millipore, Watford, UK) was used as a positive control (98% average methylation).
Immunocytochemistry
Reelin-specific antibody clone 142 (Millipore) was used for all immunocytochemical assays (dilution 1:800), using the IMPRESS kit (Vector Laboratories UK, Peterborough, UK) and antigen retrieval with EDTA/Tris buffer (pH 8). In brief, after dewaxing in xylene, tissues were treated with 0.3% H2O2 before antigen retrieval and overnight antibody treatment; 2.5% horse serum was used as a blocking agent. Tissue sections were counterstained with Hematoxylin Z (Cellpath Ltd., Newtown, UK). Because no blocking peptides were available and the epitope of antibody 142 is not known, antibody specificity was tested by a 30-minute preincubation with 10 μl of reelin protein solution derived from stably transfected HEK293 cells.22 Reelin-expressing cells and green fluorescent protein-transfected HEK293 control cells were grown to near confluence, washed twice with PBS, and incubated for 48 hours in serum-free DMEM. Ten milliliters of supernatant was concentrated ∼50 times using Amicon Ultra-15 filter (Millipore) devices with a 30-kDa cutoff and washed once with 10 ml of PBS to obtain 200 μl of concentrated solution.
Breast Tissues and Arrays
The material used in the pilot study was taken from a cohort of patients who were enrolled into the Ludwig Trial V Ludwig Breast Cancer Study Group.25 The cancers used were representative of the trial cohort. From 1981 to 1985, the International Breast Cancer Study Group (formerly the Ludwig Breast Cancer Study Group) conducted a randomized clinical study to assess the effect of early commencement of adjuvant cyclophosphamide, methotrexate, and 5-fluorouracil chemotherapy in patients with lymph node-negative and lymph node-positive breast carcinoma.
The larger set of tissue arrays consisted of 2197 cases of primary breast cancer collected from 1985 to 1999 at the University Hospital Basel, Institute of Clinical Pathology and Institute of Pathology and Triemli Hospital Zürich26 and have been used previously in our laboratory.27 The median age of these patients was 62 years (range, 26 to 101 years). The mean follow-up time was 68 months (range, 1 to 176 months). The tissue microarrays were reviewed by two pathologists to define histological grade according to the Elston and Ellis BRE scoring system28 and histological tumor type. In addition, pathological stage, tumor diameter, and nodal status were known from the primary pathology reports. HER2 status by fluorescence in situ hybridization analysis and estrogen receptor (ER) status by immunohistochemistry were analyzed in Al-Kuraya et al29 and Ruiz et al.26
Reelin staining of tumor cells was scored for intensity (0, no; 1, weak; 2, moderate; and 3, strong staining) and percentage of positive cells (1, 0 to 4%; 2, 5 to 19%; 3, 20 to 39%; 4, 40 to 59%; 5, 60 to 79%; 6, 80 to 100%). These values were multiplied to obtain an H-score (0 to 18) and grouped into no (H-score = 0), low (H-score = 1–6), and high reelin staining (H-score > 6).
Fluorescent-Gelatin Degradation Assay
Coverslips were precleaned with 20% nitric acid for 20 minutes followed by an extensive wash with water. Ethanol-sterilized coverslips were then coated with 50 μg/ml poly-l-lysine (Sigma-Aldrich) for 20 minutes at room temperature, followed by fixation with 0.5% glutaraldehyde for 15 minutes. The coverslips were washed three times with PBS, inverted on a 50-μl drop of Oregon Green 488 gelatin matrix (Invitrogen), and incubated for 20 minutes at room temperature. After washing with PBS, the residual reactive groups in the gelatin matrix were quenched with 5 mg/ml sodium borohydride for 15 minutes followed by extensive washing with PBS. The coverslips were then ethanol-sterilized for 5 minutes followed by quenching in DMEM for 1 hour.
To assess the ability of cells to form invadopodia and gelatin degradation, 5 × 104 cells were seeded on the coverslips coated with 488 gelatin and incubated at 37°C for 5 hours. Foci of degraded matrix were visible as dark areas that lacked fluorescence. The total pixels of the degradation area corresponding to the cells (n = 60 for each cell line; in triplicate) were analyzed and quantified by the Gelatin Degradation plug-in of ImageJ.
Confocal Fluorescence and Live Cell Microscopy
Cells on coverslips were fixed with 4% paraformaldehyde for 10 minutes followed by permeabilization with 0.1% Triton X-100 for 10 minutes. Actin filaments were visualized with rhodamine phalloidin (Molecular Probes, Eugene, OR). Confocal images were collected using an Olympus laser scanning microscope (FluoView FV1000) with ×63/1.4 NA oil objectives. Images were processed with Volocity.
Scratch Wound Migration Assay
Monolayers of confluent cells in a six-well plate were wounded by scraping with a P200 plastic pipette tip and rinsed three times with growth medium to remove floating cells. Images were periodically collected every 10 minutes over the next 36 hours with a ×10 objective on an inverted phase microscope (Nikon, Tokyo, Japan) equipped with a chamber to maintain the atmosphere of 5% CO2 and 37°C. Cell motility was evaluated by the time the cells took to close the same distance of wound.
Transwell Migration Assay
MDA-MB231 cells (1 × 105) were plated in serum-free medium in the presence or absence of reelin. Reelin protein was harvested as described above; 20 μl of concentrated protein or control supernatant was used per well, and cells were incubated for 20 hours at 5% CO2 and 37°C. Cells were stained with calcein AM (Invitrogen) and visualized with an inverted fluorescent microscope (Olympus XI51). Cells that had not migrated through the pores were removed from the top chamber before image capture at ×100 magnification. Cells from three randomly selected areas were counted from each well.
Inverted Invasion Assay
Inverted invasion assays were performed as described previously.30 In brief, collagen I (PureCol, concentrate to 11 mg/ml and mix 3:1 with 10× DMEM, 2 mmol/L glutamine, and 170 mmol/L NaHCO3) was allowed to polymerize in transwell inserts (Costar, Cambridge, MA) for 1 hour at 37°C. Inserts were then inverted, and 4 × 104 cells were seeded directly onto the opposite face of the filters. After a 4-hour incubation at 37°C, the inserts were reversed. Transwell inserts were finally placed in serum-free DMEM, and DMEM supplemented with 10% fetal calf serum, 25 ng/ml epidermal growth factor (MDA-MB-231), or 10% FCS, 25 ng/ml epidermal growth factor plus 100 μg/ml G418 (reelin stable clones) was placed on top of the matrix, providing a chemotactic gradient. At 120 hours after seeding, invading cells were stained with calcein AM and visualized by Leica confocal microscopy. Serial optical sections were captured at 15-μm intervals.
Statistics
Statistical analyses were performed using JMP (SAS Institute Inc., Cary, NC). Association of reelin expression with clinicopathologic parameters and HER2 and ER status were evaluated using a χ2 test. The Kaplan-Meier method was used to visualize association of reelin expression with cancer-specific survival, and the log-rank test was used to test the significance between stratified groups. Cox proportional multivariate analysis was used to test the statistical independence and significance of reelin expression with other potential prognostic factors. All statistical tests were two-sided, and P < 0.05 was considered as significant.
Results
Reelin Protein Is Expressed in the Breast
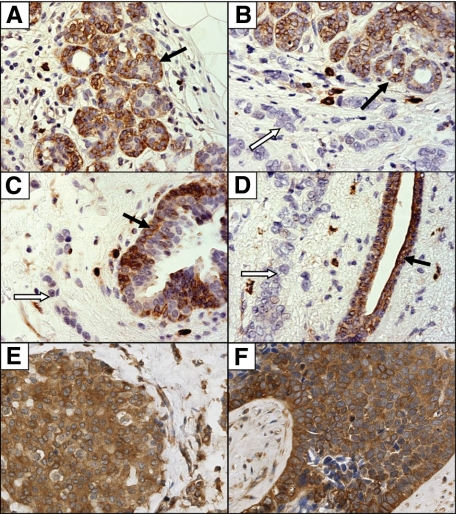
To see whether reelin was expressed in the breast epithelium we stained 17 samples of morphologically normal human breast tissue from reduction mammoplasties for reelin protein using immunocytochemistry. All sections showed a distinct immunoreactivity in the cytoplasm of the myoepithelium. This could be found in the major ducts, terminal ducts, and lobuloalveolar units. The luminal epithelia showed varying degrees of reelin-positive staining (Figure 1, A–F). This staining was blocked on preincubation with reelin protein derived from reelin-overexpressing HEK293 cells, but not when the antibody was preincubated with a control medium from green fluorescent protein-expressing HEK293 cells (Supplemental Figure S1, see http://ajp.amjpathol.org). Other immunoreactive cells were endothelial cells, mast cells, myofibroblasts in reactive stroma, and smooth muscle cells in vessel walls and peripheral nerves.
Figure 1.
Immunohistochemical staining for reelin protein in normal and malignant breast tissue. Breast sections were stained using antibody clone 142 and horseradish peroxidase-conjugated secondary antibody/diaminobenzidine. Black arrows indicate reelin-positive normal alveolar (A and B) and ductal tissue (C and D). White arrows highlight the adjacent reelin-negative cancer tissue (B–D). E and F: Reelin-positive malignant breast tissue. Original magnification, ×400.
To study a possible change in reelin expression in breast cancer, we performed a pilot study using a small, well characterized group of 39 breast cancers of varying grades and compared reelin-staining intensity between the tumor tissue and adjacent normal epithelium. Of these, 27 showed complete loss of staining for the protein in the tumor tissue (Figure 1, B–D), whereas 3 showed reduced expression relative to the normal breast epithelium. Nine samples were positive for reelin in the normal as well as the tumor tissue (Figure 1, E and F).
Loss of Reelin Protein Expression Is Associated with Decreased Survival
To establish whether down-regulation of reelin expression in breast cancers had any clinical relevance with regard to outcome, two tissue array studies were performed. First, a pilot study was performed on 136 breast cancers from patients who were randomized to the International Breast Cancer Study Group Trial V. These were representative of the trial cohort. Staining intensity for reelin immunoreactivity was scored on a scale of 0 (negative) to 3 (strong). In this subgroup, 52.2% of patients were scored 0; 18.4% were scored 1; 12.5% were scored 2; and 16.9% were scored 3. Although a small number of patients were examined, there was a clear trend toward a poorer outcome in patients with loss of reelin staining, without reaching statistical significance. Another trend showed an association between the loss of strong reelin immunoreactivity and positive lymph node status (data not shown).
To be able to gain statistically significant data, we analyzed reelin protein expression in a larger set of patients, using a tissue microarray of 2200 breast cancer cases. Reelin expression levels in cancer cells could be assessed for 2060 cases by immunocytochemistry using the H-score method. When these data were compared with other clinical parameters, loss of reelin staining correlated significantly with positive ER (P < 0.001) and lymph node status (P < 0.001), but no correlation was found with BRE grade, tumor size, or HER2 status (Table 1). Loss of expression was significantly correlated with lobular carcinoma (P < 10–4) and cribriform carcinoma (P = 0.031) compared with invasive ductal carcinoma of no special type. The trend for decreased survival of patients with loss of reelin staining, indicated in the pilot study, was confirmed and reached statistical significance when cancers with no reelin staining were compared with those with low (H-score 1 to 6) or high (H-score > 6) levels of reelin (P = 0.01) (Figure 2) or when strong reelin staining (H-score > 6) was compared with negative and low reelin-positive staining (H-score 0 to 6) combined (P = 0.003; data not shown). Multivariate analysis using Cox regression showed that reelin staining was not an independent prognostic indicator in the presence of BRE grade, nodal status, ER status, and HER2 status (Table 2).
Table 1.
Comparison of Reelin Staining with Other Clinical Parameters and Cancer Histology
| Parameters | On array | Analyzable | Reelin immunohistochemical result
|
P | ||
|---|---|---|---|---|---|---|
| Negative % (score 0) | Weak % (score 1–6) | Strong % (score > 6) | ||||
| All | 2197 | 2060 | 58.7 | 27.5 | 13.8 | |
| Histology | ||||||
| Ductal carcinoma | 1531 | 1454 | 57.8 | 28.1 | 14.2 | |
| Lobular carcinoma | 311 | 283 | 68.6 | 24.7 | 6.7 | 0.0002* |
| Mucinous carcinoma | 69 | 55 | 40.0 | 29.1 | 30.9 | |
| Medullary carcinoma | 57 | 55 | 45.5 | 30.9 | 23.6 | |
| Tubular carcinoma | 56 | 48 | 43.8 | 35.4 | 20.8 | |
| Cribriform carcinoma | 64 | 59 | 74.6 | 16.9 | 8.5 | 0.0305* |
| Papillary carcinoma | 30 | 30 | 63.3 | 13.3 | 23.3 | |
| Others | 79 | 76 | 59.2 | 31.6 | 9.2 | |
| pT stage | ||||||
| 1 | 804 | 377 | 59.2 | 35.5 | 18.3 | <0.0001 |
| 2 | 1015 | 589 | 44.8 | 19.4 | 11.8 | |
| 3 | 124 | 81 | 25.9 | 14.8 | 10.5 | |
| 4 | 242 | 158 | 34.8 | 13.9 | 9.4 | |
| pN stage | ||||||
| 0 | 936 | 865 | 53.8 | 28.3 | 17.9 | <0.0001 |
| 1 | 783 | 741 | 62.5 | 27.4 | 10.1 | |
| 2 | 121 | 115 | 64.3 | 31.3 | 4.3 | |
| BRE | ||||||
| 1 | 539 | 488 | 60.0 | 26.6 | 13.3 | 0.1520 |
| 2 | 839 | 790 | 62.8 | 25.1 | 12.2 | |
| 3 | 646 | 625 | 56.0 | 29.6 | 14.4 | |
| ER | ||||||
| ER+ | 1545 | 1349 | 61.9 | 26.0 | 12.1 | 0.0004 |
| ER− | 474 | 441 | 51.7 | 31.7 | 16.6 | |
| HER2 | ||||||
| Amplification | 293 | 268 | 53.9 | 31.4 | 14.6 | 0.7698 |
| Normal | 1551 | 1213 | 55.7 | 29.2 | 15.0 | |
Reelin staining was scored as either negative (H-score = 0), weak (H-score 1 to 6), or high (H-score > 6) as described in the Materials and Methods. These results were compared with those for other clinical parameters, including cancer histology, pT stage (tumor spread), pN stage (lymph node status), BRE grade, ER status, and HER2 status (fluorescence in situ hybridization). P < 0.05 was considered to be statistically significant.
Compared with invasive ductal carcinoma of no special type.
Figure 2.
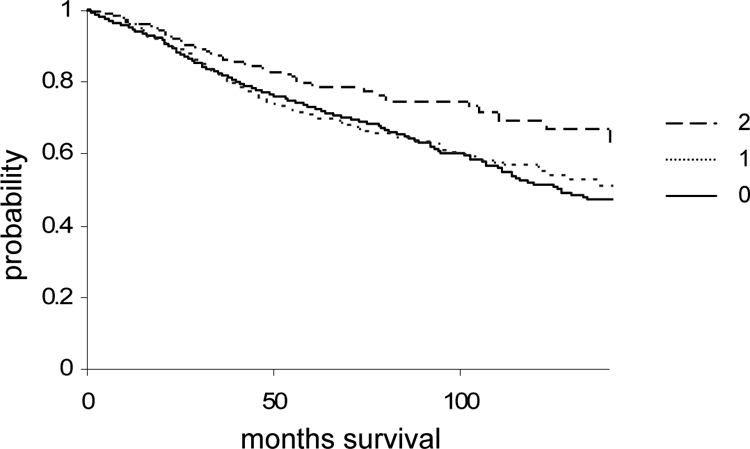
Kaplan-Meier curve for breast cancers with high, low, and no reelin staining. Overall survival of patients with breast cancer over a period of more than 10 years for cancers that showed negative (0, H-score = 0), low (1, H-score 1 to 6), and high (2, H-score > 6) is shown. Reelin tumor = r staining. Patients with loss of reelin staining had a significantly (P = 0.01) lower chance of survival.
Table 2.
Multivariate Analysis of Reelin Breast Cancer Prognosis Array
| Source | No. parameters | df | Likelihood ratio χ2 | P |
|---|---|---|---|---|
| pT | 1 | 1 | 15.04 | 0.0001 |
| pN | 1 | 1 | 94.47 | 0.0000 |
| BRE grade | 1 | 1 | 44.78 | 0.0000 |
| ER | 1 | 1 | 4.469 | 0.0345 |
| HER2, FISH | 1 | 1 | 5.47 | 0.0194 |
| Reelin | 2 | 2 | 0.20 | 0.9045 |
Cox regression analysis to measure whether reelin staining was an independent prognostic indicator in the presence of pT status (tumor spread), pN status (lymph node status), BRE grade, ER status, or HER2 status.
FISH, fluorescence in situ hybridization.
A small number of cancers showed strong stromal staining for reelin. However, there was no correlation of the stromal staining with the staining of the malignant tissue, survival, or other clinical parameters (data not shown).
Promoter Methylation Suppresses Reelin mRNA Expression in Breast Cancer-Derived Cell Lines
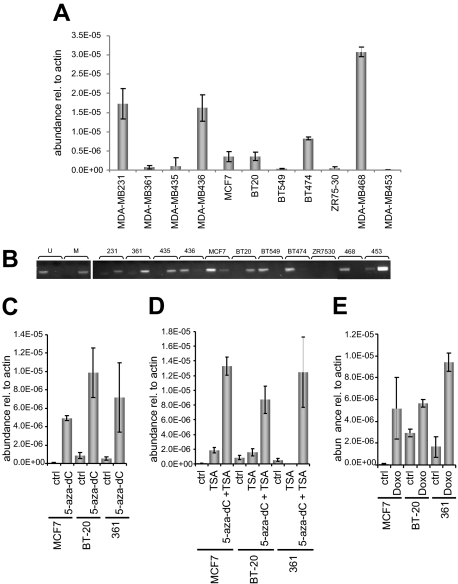
Because reelin expression is controlled in gastric and pancreatic cancer cells by promoter methylation we tested whether the same was true in breast cancer cells. Eleven breast cancer-derived cell lines were initially tested for reelin mRNA expression, using quantitative RT-PCR, and expression was compared with their promoter methylation status (Figure 3, A and B). The results showed that cell lines expressing high reelin mRNA levels had either unmethylated (MDA-MB468, MDA-MB 436, and BT474) or partly methylated (MDA-MB 231) reelin CpG islands, whereas those with low or no expression showed hypermethylation of the reelin promoter (MDA-MB361, MDA-MB453, MCF-7, BT20, and MDA-MB435), with the exception of BT549 cells, which showed no reelin expression but also had no detectable CpG methylation (Figure 3B). The methylation status was confirmed in a selection of cell lines using bisulfite sequencing (Supplemental Figure S2, see http://ajp.amjpathol.org). This result indicated that reelin mRNA expression could at least in part be controlled by promoter methylation in breast cancer cells.
Figure 3.
Reelin mRNA expression in breast cancer-derived cell lines and promoter methylation. A: Reelin mRNA levels in 11 breast cancer-derived cell lines. Expression levels were normalized to β-actin. B: Methylation-specific PCR using the same cell lines. Unmethylated control DNA extracted from normal peripheral blood (U) and chemically modified methylated control DNA (M) were used as controls. C: Quantitative RT-PCR of reelin mRNA after treatment of cells with decitabine (5-aza-dC) for 7 days. D: Reelin mRNA levels after treatment with TSA or with TSA and decitabine. E: Reelin mRNA levels after treatment with doxorubicin (Doxo) for 24 hours. All experiments were run in triplicate. ctrl, control.
Promoter Demethylation Can Lead to Reexpression of Reelin mRNA
To test this hypothesis further, MCF7, BT20, and MDA-MB361 cells were treated with the demethylating agent decitabine, and reelin mRNA levels were measured by quantitative RT-PCR. All cell lines showed significantly increased levels of reelin mRNA after 7 days of treatment (Figure 3C). Because deacetylation in pancreatic cancer cells also increased reelin mRNA expression, reelin mRNA levels were measured after treatment with TSA alone or in combination with decitabine in the same breast cancer cell lines (Figure 3D). The results showed that TSA alone had no detectable effect on reelin mRNA expression, whereas TSA plus decitabine treatment led to increased expression in all three cell lines. These results indicate that promoter methylation but not histone acetylation plays a role in controlling reelin mRNA expression in breast cancer-derived cell lines.
It has previously been shown that treatment of the neuronal precursor cell line NT2 with doxorubicin led to a 100-fold increase in reelin mRNA expression after 12 hours, presumably via DNA methyltransferase 1 inhibition.31 To test whether the same could be found in breast epithelial cells, MCF7, BT20, and MDA-MB361 cells were treated with doxorubicin for 24 hours. Reelin mRNA levels increased in all three cell lines (Figure 3E). Because the cell lines BT20 and MDA-MB361 express mutant p53, the results show that the effect of doxorubicin treatment was p53-independent. These results further strengthen the hypothesis that promoter methylation controls reelin expression in the breast epithelium.
Promoter Methylation Correlates with Reduced Reelin Protein Expression in Breast Cancer Tissue
To test whether loss of reelin expression in breast cancers is associated with promoter methylation in vivo, the reelin promoter methylation status was assessed in the previously characterized group of 39 invasive breast carcinomas using methylation-specific PCR. Of these, 25 showed promoter methylation, of which 22 were negative for the protein. The other 3 had either reduced expression relative to the normal breast epithelium or were weakly positive. Of the 14 cases that did not show methylation 9 were positive for reelin and 5 were negative (Table 3). These results reveal a very close association between methylation in the reelin CpG island and down-regulation of reelin expression, and therefore imply that reelin promoter methylation is a major controlling element for reelin down-regulation in human breast cancer in vivo. Methylation in the CpG island of the reelin gene was confirmed in 12 samples using pyrosequencing technology, which allows simultaneous quantification of the degree of methylation at several CpGs in close proximity with high quantitative resolution. This analysis showed again a close relationship between negative reelin staining and promoter methylation (Supplemental Figure S3, see http://ajp.amjpathol.org).
Table 3.
Reelin Promoter Methylation and Protein Expression Status in Primary Breast Cancers
| Promoter status | Reelin staining
|
|
|---|---|---|
| Positive | Negative | |
| Methylated | 3* | 22 |
| Unmethylated | 9 | 5 |
Reelin expression in breast tumors was measured by immunohistochemical analysis and compared with that in adjacent normal tissue. Promoter methylation was measured by methylation-specific PCR.
Weak staining.
Reelin Reduces the Migratory Behavior of MDA-MB231 Cells
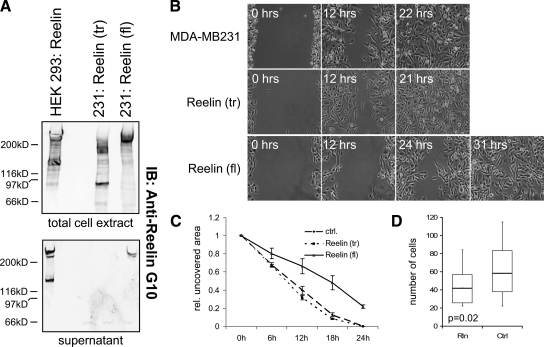
During cortical brain development, reelin controls cell positioning and migration of neuronal progenitors in the outer cortex. To test whether reelin could also control the migration of breast epithelial cells, we tested the migratory behavior of MDA-MB 231 cells that had been stably transfected with full-length reelin compared with cells expressing a C-terminally truncated reelin (as established by RT-PCR; data not shown) (Figure 4A),9,22 as well as the parental cell line, using a “wound-healing” assay. Reelin expression as well as secretion was assessed by Western blot (Figure 4A)9,22 using total cell extract and cell supernatant, respectively. Only those cells expressing full-length reelin were able to secrete reelin, confirming that the C terminus was required for secretion as described previously by de Bergeyck et al.32 This protein did not show the processing of the reelin found in the supernatant of stably transfected HEK293 cells but was similar to the reelin found in rat adrenal gland or liver extracts in previous studies.18 Although the parental cell line and the cells expressing the truncated version of reelin showed a fast closure of the gap (21 to 22 hours), similar closure was only detected after 31 hours when full-length reelin was overexpressed (Figure 4, B and C). Similar results were obtained using a transwell migration assay, in which MDA-MB231 cells migrated in the presence or absence of reelin protein (Figure 4D), showing that reelin overexpression reduces cell migratory behavior in this cell system. Time-lapse analysis of the stably transfected MDA-MB231 cells further showed that cells expressing full-length reelin had reduced lamellipodia and pseudopodia formation compared with cells expressing the truncated form of reelin, as well as a more flattened morphology (data not shown).
Figure 4.
Migratory ability of MDA-MB231 cells overexpressing reelin. MDA-MB231 cells were stably transfected with a truncated (tr) form of reelin or full-length (fl) reelin and grown from a single colony. A: Western blot for reelin protein using total cell extract and cell supernatants. Stably transfected HEK293 cells22 were used as a positive control, showing the typical reelin banding pattern as described previously.9 B: Wound healing assay using untransfected or stably transfected MDA-MB231 cell lines. Cells were photographed at the indicated time points after scratching. IB, immunoblot. C: The graph shows the average relative (rel.) uncovered scratch area in each microscopic field with SD for each cell line from three independent experiments over a period of 24 hours. D: MDA-MB231 cells were plated in transwells in medium without serum in the presence of reelin (Rln) protein or control (Ctrl) supernatant and were allowed to migrate toward epidermal growth factor-containing medium. The box and whisker plot shows the results obtained from six replicates.
Reelin Suppresses Invadopodia Formation and Invasiveness into Collagen I
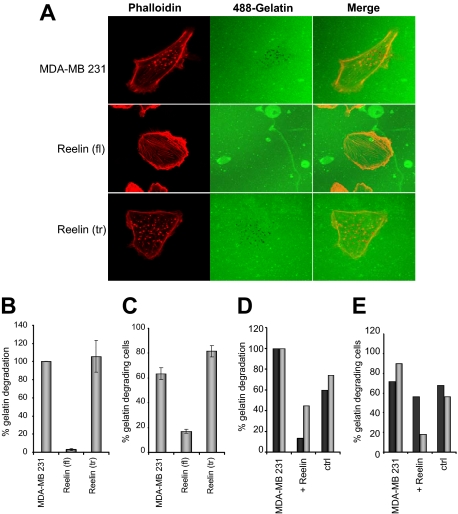
Reelin expression was significantly (P < 0.001) associated with reduced tumor spread (Table 1, pT stage). It is therefore possible that reelin negatively affects tumor invasiveness. Invadopodia are invasion-associated protrusions that have been suggested to facilitate cancer cell-induced degradation of the extracellular matrix and subsequent invasion and can therefore be used as a measure for the invasive potential of a cell. Invadopodia can be visualized when cells are grown on a fluorescent Oregon Green 488 gelatin matrix as areas of gelatin degradation (dark spots) with punctate actin staining. The invadopodia formation ability of reelin-overexpressing MDA-MB231 cells was therefore compared with that of the highly invasive parental cell line and with that of MDA-MB231 cells expressing the C-terminally truncated form of reelin. Cells were grown on fluorescent gelatin for 5 hours, and invadopodia formation was assessed by counting the total number of invadopodia and of invadopodia-forming cells. Cells that overexpressed full-length reelin showed a strongly reduced number of invadopodia and a rounded appearance (Figure 5, A, middle row), whereas those cells expressing the truncated reelin (bottom row) showed a level of invadopodia formation similar to that of the parental cell line (Figure 5A, top row; B and C). In addition, full-length reelin-expressing MDA-MB231 cells showed increased actin stress fiber formation (phalloidin staining) instead of the punctate actin pattern associated with invadopodia, which could be seen in the other two cell lines. This result was reproducible using independently selected reelin-expressing MDA-MB231 stable transfectants (data not shown). A similar, although weaker, effect was observed when the parental MDA-MB231 cell line was incubated with reelin collected from reelin-expressing HEK293 cells. In two separate experiments (gray and black bars) a stronger reduction in gelatin degradation and in the number of gelatin-degrading cells was observed in the presence of reelin-containing medium than with control medium from green fluorescent protein-overexpressing HEK293 cells (Figure 5, D and E).
Figure 5.
Invadopodia formation in MDA-MB231 clones. Parental MDA-MB231 cells and those overexpressing truncated (tr) or full-length reelin (fl) were grown on fluorescent 488 gelatin. A: Phalloidin staining (red) was used to visualize actin fibers, whereas gelatin degradation was detected as dark spots. Original magnification, ×640. B: Bar graph showing the percentage of gelatin degradation compared with that in parental MDA-MB231 cells. C: Percentage of gelatin-degrading cells. D: Percentage of gelatin degradation from two (gray bars and black bars, respectively) independent experiments of untreated MDA-MB231 cells and those treated with either reelin-containing supernatant or control supernatant. E: Percentage of gelatin-degrading cells from two (gray bars and black bars, respectively) independent experiments of untreated MDA-MB231 cells and those treated with either reelin-containing supernatant or control supernatant. For all experiments, n = 60; error bars represent the SD. ctrl, control.
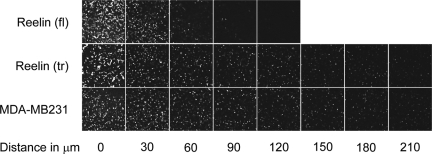
To test whether this decreased ability to form invadopodia would indeed lead to a reduced invasiveness, an invasiveness assay was performed using a collagen I matrix (Figure 6). Cells were plated at the bottom of a transwell plate and allowed to migrate against gravity through the transwell and invade the overlaying collagen matrix in the top chamber. Cells were subsequently stained with calcein, and the distance that the cells had invaded the collagen was measured, using confocal microscopy. In this assay, the parental cell line and those transfected with the truncated form of reelin behaved similarly, whereas overexpression of full-length reelin led to a strongly reduced invasion (>200 and 90 μm, respectively). These results show that reelin can control breast cancer cell invasiveness in vitro.
Figure 6.
Invasiveness of reelin overexpressing MDA-MB231 cells into a collagen I matrix. Invasiveness of parental MDA-MB231 cells and those stably transfected with either full-length (fl) or truncated (tr) reelin was measured by the distance of invasion, using confocal microscopy.
Discussion
Until recently, a biological function for reelin had only been found in the developing and adult brain, although its expression has been described in many other organs.17,18 The idea that reelin could indeed function outside the brain was supported by recent studies,19,20,21 which linked the reelin signaling pathway to prostate, gastric, and pancreatic cancer, respectively. These studies showed that local reelin expression is an important prognostic factor in these systems, albeit in opposite directions. Whereas reelin down-regulation was found in pancreatic cancer and gastric cancer, in prostate cancer reelin expression was associated with high-grade disease. Using two independent cohorts of breast cancer patients, we have now shown for the first time that reelin protein is expressed in the human breast epithelium and that loss of expression is associated with poor prognosis in breast cancer. Although one study did not reach statistical significance because of the low number of patients, our study of more than 2000 tissue cores clearly showed a statistically significant link between loss of strong reelin immunoreactivity and poor survival. Loss of reelin expression was also associated with positive lymph node status, suggesting that it may play a role in suppressing metastasis formation. This finding is in agreement with the findings by Sato et al20 and Dohi et al21 that epigenetic silencing of reelin is associated with pancreatic and gastric cancer. Sato et al suggested that loss of reelin expression is accompanied by an increase in migratory and invasive ability, as shown in an in vitro invasion assay. We have now been able to confirm these results in breast cancer. In the developing cerebral cortex, it is thought that one of the functions of reelin may be to provide a stop signal in a paracrine fashion for migrating neurons to inhibit these cells from entering the marginal zone.33,34,35 Reeler mice and those with a defect in the receptor VLDLR both showed increased invasion of the marginal zone by migrating neurons. It is conceivable that in the breast epithelium reelin could act in an autocrine fashion to suppress cell migration/invasion. However, it is not clear whether reelin signals in the breast through the same pathways as those that have been described in the brain. Indeed, Dulabon et al34 showed that reelin can inhibit cortical neuron migration through modulation of integrin-dependent cell adhesion, in particular integrins α3β1, which have also been associated with breast cancer metastasis formation.36 It is therefore feasible that reelin may reduce cell migration through modulation of integrin-matrix interaction. This hypothesis is currently under investigation.
Our studies further indicate that reelin may be a metastasis-suppressing factor. Unfortunately, reeler mice that show no reelin expression are runted, have a short life span, with many dying after a few weeks, and can therefore not be used as a tumor model. Thus, reexpression of reelin in a metastatic breast cancer cell line that shows low levels of reelin in combination with xenograft studies in nude mice may be useful to address this question.
In gastric21 and pancreatic cancer,20 as well as in the brains of patients with schizophrenia, reelin is epigenetically silenced.37,38 We have now shown that epigenetic silencing of reelin expression can also be found in breast cancers. In a selection of 39 breast cancers and in 12 breast cancer-derived cell lines, down-regulation of reelin mRNA and protein expression was clearly associated with aberrant methylation in the CpG island of the reelin gene. Furthermore, decitabine treatment of MDA-MB361, BT20, and MCF7 cells led to a significant increase in reelin mRNA levels. Further, as shown previously for the neuronal precursor cell line NT2,31 doxorubicin treatment also increased reelin mRNA levels significantly in three breast cancer cell lines in a p53-independent manner. Our data therefore strongly support the idea that epigenetic factors including promoter methylation control reelin expression in the breast epithelium.
It is of interest to note that reelin staining did not correlate with BRE grade or mitotic figures (data not shown); hence, there was no significant correlation with proliferation, suggesting that reelin does not affect proliferation of the breast epithelium. Indeed, treatment of the breast cancer-derived epithelial cell line MCF7 with a reelin-containing supernatant did not have any effect on proliferation in a 5-bromo-2′-deoxyuridine-labeling assay (unpublished data). Instead, reelin significantly suppressed invadopodia formation and invasiveness using in vitro assays. The mechanisms of this suppression are so far unknown. Recent studies have shown that Dab1 can induce filopodia formation through N-Wasp in a CDC42-independent mechanism, but that phosphorylation of Dab1 by the Src-family kinase Fyn reduces filopodia formation and targets Dab1 for proteasomal degradation.39 It might be predicted that overexpression of reelin could therefore lead to phosphorylation of Dab1 in our system. We have investigated this possibility, and despite finding that Dab1 and the putative receptors VLDLR and ApoER2 are expressed in MDA-MB231 cells, we have not been able to demonstrate increased Dab1 phosphorylation (data not shown), indicating that an alternative pathway(s) may be involved.
In conclusion, we have established that loss of reelin expression correlates with worse clinical outcome and increased lymph node metastasis formation in breast cancer and that reelin suppresses invadopodia formation. By identifying the way in which reelin may potentially suppress metastasis formation we might be able to open up a new potential therapeutic route, and it will be interesting to learn whether loss of reelin, through epigenetic silencing or other means, is also associated with other highly metastatic cancers, as has been shown for pancreatic and gastric cancer.
Acknowledgments
We thank Karen Price for preparing the results from the pilot study, based on data from the International Breast Cancer Study Group Trial V. We also thank Drs. Yves Jossin and Andre Goffinet for helpful discussions and technical support and Dr. Camille Huser for critical reading of the manscript. Further, we thank Drs. Eckart Foerster and Michael Frotscher for the reelin overexpressing HEK cells.
Footnotes
Address reprint requests to Dr. Torsten Stein, Institute of Cancer Sciences, College of Medical, Veterinary and Life Sciences, University of Glasgow, Glasgow, G11 6NT, UK. E-mail: torsten.stein@glasgow.ac.uk.
Supported by Breakthrough Breast Cancer (project grant to T.S. and B.A.G.) and by the International Breast Cancer Study Group for production of the Trial V tissue array (B.A.G.). L.M.M. is funded by a Medical Research Council UK Senior Fellowship (G117/569).
None of the authors disclosed any relevant financial relationships.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
Current address of P.R.S.: Cancer Genomics Group, Medical Oncology Centre, Barts and the London School of Medicine, Charterhouse Square, London, UK; of T.C.: Department of Medical Oncology, Charing Cross Hospital, London, UK.
References
- Tissir F, Goffinet AM. Reelin and brain development. Nat Rev Neurosci. 2003;4:496–505. doi: 10.1038/nrn1113. [DOI] [PubMed] [Google Scholar]
- Jossin Y. Neuronal migration and the role of reelin during early development of the cerebral cortex. Mol Neurobiol. 2004;30:225–251. doi: 10.1385/MN:30:3:225. [DOI] [PubMed] [Google Scholar]
- D'Arcangelo G, Miao GG, Chen SC, Soares HD, Morgan JI, Curran T. A protein related to extracellular matrix proteins deleted in the mouse mutant reeler. Nature. 1995;374:719–723. doi: 10.1038/374719a0. [DOI] [PubMed] [Google Scholar]
- Goldowitz D, Cushing RC, Laywell E, D'Arcangelo G, Sheldon M, Sweet HO, Davisson M, Steindler D, Curran T. Cerebellar disorganization characteristic of reeler in scrambler mutant mice despite presence of reelin. J Neurosci. 1997;17:8767–8777. doi: 10.1523/JNEUROSCI.17-22-08767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BW, Hawkes R, Soriano P, Cooper JA. Neuronal position in the developing brain is regulated by mouse disabled-1. Nature. 1997;389:733–737. doi: 10.1038/39607. [DOI] [PubMed] [Google Scholar]
- Sheldon M, Rice DS, D'Arcangelo G, Yoneshima H, Nakajima K, Mikoshiba K, Howell BW, Cooper JA, Goldowitz D, Curran T. Scrambler and yotari disrupt the disabled gene and produce a reeler-like phenotype in mice. Nature. 1997;389:730–733. doi: 10.1038/39601. [DOI] [PubMed] [Google Scholar]
- Trommsdorff M, Gotthardt M, Hiesberger T, Shelton J, Stockinger W, Nimpf J, Hammer RE, Richardson JA, Herz J. Reeler/Disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell. 1999;97:689–701. doi: 10.1016/s0092-8674(00)80782-5. [DOI] [PubMed] [Google Scholar]
- Falconer DS. Two new mutations, trembler and reeler, with neurological actions in the house mouse. J Genet. 1951;50:192–201. doi: 10.1007/BF02996215. [DOI] [PubMed] [Google Scholar]
- D'Arcangelo G, Homayouni R, Keshvara L, Rice DS, Sheldon M, Curran T. Reelin is a ligand for lipoprotein receptors. Neuron. 1999;24:471–479. doi: 10.1016/s0896-6273(00)80860-0. [DOI] [PubMed] [Google Scholar]
- Hiesberger T, Trommsdorff M, Howell BW, Goffinet A, Mumby MC, Cooper JA, Herz J. Direct binding of Reelin to VLDL receptor and ApoE receptor 2 induces tyrosine phosphorylation of disabled-1 and modulates tau phosphorylation. Neuron. 1999;24:481–489. doi: 10.1016/s0896-6273(00)80861-2. [DOI] [PubMed] [Google Scholar]
- Arnaud L, Ballif BA, Forster E, Cooper JA. Fyn tyrosine kinase is a critical regulator of disabled-1 during brain development. Curr Biol. 2003;13:9–17. doi: 10.1016/s0960-9822(02)01397-0. [DOI] [PubMed] [Google Scholar]
- Keshvara L, Benhayon D, Magdaleno S, Curran T. Identification of reelin-induced sites of tyrosyl phosphorylation on disabled 1. J Biol Chem. 2001;276:16008–16014. doi: 10.1074/jbc.M101422200. [DOI] [PubMed] [Google Scholar]
- Ballif BA, Arnaud L, Cooper JA. Tyrosine phosphorylation of Disabled-1 is essential for Reelin-stimulated activation of Akt and Src family kinases. Brain Res Mol Brain Res. 2003;117:152–159. doi: 10.1016/s0169-328x(03)00295-x. [DOI] [PubMed] [Google Scholar]
- Beffert U, Morfini G, Bock HH, Reyna H, Brady ST, Herz J. Reelin-mediated signaling locally regulates protein kinase B/Akt and glycogen synthase kinase 3β. J Biol Chem. 2002;277:49958–49964. doi: 10.1074/jbc.M209205200. [DOI] [PubMed] [Google Scholar]
- Assadi AH, Zhang G, Beffert U, McNeil RS, Renfro AL, Niu S, Quattrocchi CC, Antalffy BA, Sheldon M, Armstrong DD, Wynshaw-Boris A, Herz J, D'Arcangelo G, Clark GD. Interaction of reelin signaling and Lis1 in brain development. Nat Genet. 2003;35:270–276. doi: 10.1038/ng1257. [DOI] [PubMed] [Google Scholar]
- Chen Y, Sharma RP, Costa RH, Costa E, Grayson DR. On the epigenetic regulation of the human reelin promoter. Nucleic Acids Res. 2002;30:2930–2939. doi: 10.1093/nar/gkf401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y, Terashima T. Expression of reelin, the gene responsible for the reeler mutation, in embryonic development and adulthood in the mouse. Dev Dyn. 1997;210:157–172. doi: 10.1002/(SICI)1097-0177(199710)210:2<157::AID-AJA8>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Smalheiser NR, Costa E, Guidotti A, Impagnatiello F, Auta J, Lacor P, Kriho V, Pappas GD. Expression of reelin in adult mammalian blood, liver, pituitary pars intermedia, and adrenal chromaffin cells. Proc Natl Acad Sci USA: 2000;97:1281–1286. doi: 10.1073/pnas.97.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone G, Vincenzi B, Zagami M, Santini D, Panteri R, Flammia G, Verzi A, Lepanto D, Morini S, Russo A, Bazan V, Tomasino RM, Morello V, Tonini G, Rabitti C. Reelin expression in human prostate cancer: a marker of tumor aggressiveness based on correlation with grade. Mod Pathol. 2007;20:344–351. doi: 10.1038/modpathol.3800743. [DOI] [PubMed] [Google Scholar]
- Sato N, Fukushima N, Chang R, Matsubayashi H, Goggins M. Differential and epigenetic gene expression profiling identifies frequent disruption of the RELN pathway in pancreatic cancers. Gastroenterology. 2006;130:548–565. doi: 10.1053/j.gastro.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Dohi O, Takada H, Wakabayashi N, Yasui K, Sakakura C, Mitsufuji S, Naito Y, Taniwaki M, Yoshikawa T. Epigenetic silencing of RELN in gastric cancer. Int J Oncol. 36:85–92. [PubMed] [Google Scholar]
- Forster E, Tielsch A, Saum B, Weiss KH, Johanssen C, Graus-Porta D, Muller U, Frotscher M. Reelin, Disabled 1, and β1 integrins are required for the formation of the radial glial scaffold in the hippocampus. Proc Natl Acad Sci USA: 2002;99:13178–13183. doi: 10.1073/pnas.202035899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Arcangelo G, Nakajima K, Miyata T, Ogawa M, Mikoshiba K, Curran T. Reelin is a secreted glycoprotein recognized by the CR-50 monoclonal antibody. J Neurosci. 1997;17:23–31. doi: 10.1523/JNEUROSCI.17-01-00023.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- Ludwig Breast Cancer Study Group Combination adjuvant chemotherapy for node-positive breast cancer. Inadequacy of a single perioperative cycle. N Engl J Med. 1988;319:677–683. doi: 10.1056/NEJM198809153191104. [DOI] [PubMed] [Google Scholar]
- Ruiz C, Seibt S, Al Kuraya K, Siraj AK, Mirlacher M, Schraml P, Maurer R, Spichtin H, Torhorst J, Popovska S, Simon R, Sauter G. Tissue microarrays for comparing molecular features with proliferation activity in breast cancer. Int J Cancer. 2006;118:2190–2194. doi: 10.1002/ijc.21581. [DOI] [PubMed] [Google Scholar]
- Stein T, Price KN, Morris JS, Heath VJ, Ferrier RK, Bell AK, Pringle MA, Villadsen R, Petersen OW, Sauter G, Bryson G, Mallon EA, Gusterson BA. Annexin A8 is up-regulated during mouse mammary gland involution and predicts poor survival in breast cancer. Clin Cancer Res. 2005;11:6872–6879. doi: 10.1158/1078-0432.CCR-05-0547. [DOI] [PubMed] [Google Scholar]
- Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19:403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- Al-Kuraya K, Schraml P, Sheikh S, Amr S, Torhorst J, Tapia C, Novotny H, Spichtin H, Maurer R, Mirlacher M, Simon R, Sauter G. Predominance of high-grade pathway in breast cancer development of Middle East women. Mod Pathol. 2005;18:891–897. doi: 10.1038/modpathol.3800408. [DOI] [PubMed] [Google Scholar]
- Hennigan RF, Hawker KL, Ozanne BW. Fos-transformation activates genes associated with invasion. Oncogene. 1994;9:3591–3600. [PubMed] [Google Scholar]
- Kundakovic M, Chen Y, Costa E, Grayson DR. DNA methyltransferase inhibitors coordinately induce expression of the human reelin and glutamic acid decarboxylase 67 genes. Mol Pharmacol. 2007;71:644–653. doi: 10.1124/mol.106.030635. [DOI] [PubMed] [Google Scholar]
- de Bergeyck V, Nakajima K, Lambert de Rouvroit C, Naerhuyzen B, Goffinet AM, Miyata T, Ogawa M, Mikoshiba K. A truncated Reelin protein is produced but not secreted in the ‘Orleans’ reeler mutation (Reln[rl-Orl]). Brain Res Mol Brain Res. 1997;50:85–90. doi: 10.1016/s0169-328x(97)00166-6. [DOI] [PubMed] [Google Scholar]
- Hack I, Hellwig S, Junghans D, Brunne B, Bock HH, Zhao S, Frotscher M. Divergent roles of ApoER2 and Vldlr in the migration of cortical neurons. Development. 2007;134:3883–3891. doi: 10.1242/dev.005447. [DOI] [PubMed] [Google Scholar]
- Dulabon L, Olson EC, Taglienti MG, Eisenhuth S, McGrath B, Walsh CA, Kreidberg JA, Anton ES. Reelin binds α3β1 integrin and inhibits neuronal migration. Neuron. 2000;27:33–44. doi: 10.1016/s0896-6273(00)00007-6. [DOI] [PubMed] [Google Scholar]
- Frotscher M. Cajal-Retzius cells. Reelin, and the formation of layers. Curr Opin Neurobiol. 1998;8:570–575. doi: 10.1016/s0959-4388(98)80082-2. [DOI] [PubMed] [Google Scholar]
- Morini M, Mottolese M, Ferrari N, Ghiorzo F, Buglioni S, Mortarini R, Noonan DM, Natali PG, Albini A. The α3β1 integrin is associated with mammary carcinoma cell metastasis, invasion, and gelatinase B (MMP-9) activity. Int J Cancer. 2000;87:336–342. [PubMed] [Google Scholar]
- Grayson DR, Chen Y, Costa E, Dong E, Guidotti A, Kundakovic M, Sharma RP. The human reelin gene: transcription factors (+), repressors (−) and the methylation switch (+/−) in schizophrenia. Pharmacol Ther. 2006;111:272–286. doi: 10.1016/j.pharmthera.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Grayson DR, Jia X, Chen Y, Sharma RP, Mitchell CP, Guidotti A, Costa E. Reelin promoter hypermethylation in schizophrenia. Proc Natl Acad Sci USA: 2005;102:9341–9346. doi: 10.1073/pnas.0503736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu S, Tezuka T, Morimura T, Hattori M, Mikoshiba K, Yamamoto T, Takenawa T. Regulation of actin cytoskeleton by mDab1 through N-WASP and ubiquitination of mDab1. Biochem J: 2004;384:1–8. doi: 10.1042/BJ20041103. [DOI] [PMC free article] [PubMed] [Google Scholar]