Abstract
Microsatellite instability (MSI) and genomic hypermethylation of methylated-in-tumor (MINT) loci are both strong prognostic indicators in a subgroup of patients with sporadic colorectal cancer (CRC). The present study was designed to determine whether the methylation of MINT loci during the progression of adenoma to CRC is related to MSI in CRC cases. Methylation index (MI) was measured by absolute quantitative assessment of methylated alleles at seven MINT loci in primary CRC with contiguous adenomatous and normal tissues of 79 patients. Results were then validated in primary CRC tissues from an independent group of 54 patients. Increased MI of both MINT loci 1 and 31 was significantly associated with MSI in CRC and was specific for adenoma. Total MI and the number of methylated loci were threefold (P = 0.02) and fivefold (P = 0.004) higher, respectively, in adenomas associated with microsatellite-stable CRC versus microsatellite-unstable CRC. MINT MI was found to be correlated with mismatch repair protein expression, MSI, BRAF (V600E) mutation status, mut-L homologue 1 methylation status, and disease-specific survival in the second independent validation group of patients. MI of specific MINT loci may be prognostic indicators of colorectal adenomas that will develop into sporadic microsatellite-unstable CRCs. Increased MINT locus methylation appears to precede MSI and may have utility in defining clinical pathology in the absence of features of malignant invasive tumors.
Epigenetic changes in epithelial cells, such as DNA methylation of CpG islands, have been related to the genesis and progression of some gastrointestinal cancers.1,2 Aberrations in DNA methylation are considered to be as important as genetic alterations in gastrointestinal tumor initiation and progression. In colorectal cancer (CRC), both hypomethylation and hypermethylation of promoter-region related CpG islands have been correlated with clinical and histopathological parameters.3,4 A number of tumor-related gene promoter regions are methylated in premalignant dysplastic lesions such as hyperplastic polyps,5 aberrant crypt foci,6,7 and adenomas.8,9,10 Furthermore, progressive genomic and epigenomic aberrations may be linked in CRC progression.11 For instance, it has been reported that widely increased methylation in sporadic CRCs overlaps with microsatellite instability (MSI).12,13,14,15 Studies to pinpoint the onset of MSI have been reported in hereditary nonpolyposis colorectal cancers that carry germline mutations (mt) in mismatch repair (MMR) genes,13,14,16,17 but as of yet, no study has examined this process in sporadic CRC.
Spontaneous regression of colorectal polyps is known to occur. Therefore, a critical issue in molecular analysis of colorectal precursor tumor lesions is whether a lesion with low or intermediate adenomatous dyplasia would develop into an invasive cancer. The DNA extracted from such lesions may not contain key premalignant aberrations. We recently described an on-slide sodium bisulfite modification (SBM) technique for gene methylation analysis in small (1 to 2 mm2) tissue areas isolated from a single section of paraffin-embedded archival tissue (PEAT).18 On-slide SBM allows comparison of gene methylation in the primary CRC, the contiguous adenoma lesion, and normal epithelium when these three tissue types are present on the same tissue section. Simultaneous assessment of methylation and MSI changes in CRC, adenoma, and normal tissues from the same patient using the on-slide SBM technique would provide an accurate analysis model to test development of these epigenetic and genetic events during CRC formation.
In a previous study, our group demonstrated the technical feasibility of using methylated-in-tumor (MINT) loci 1, 2, 12, and 31 to detect methylation differences between areas of the same colorectal tissue section.8 MINT loci are conservative human genomic sequences that adhere to the CpG island definition.19,20 They are found in noncoding genomic regions, and their gene regulatory purpose or other functional attributes are unknown. We have previously shown the clinical utility of methylation levels of specific MINT loci in rectal cancer20 and melanoma.4 Other groups have studied MINT locus methylation in colorectal20,21,22 and gastric cancers.20,21,22,23,24 Methylation of MINT loci has also been linked to MSI in CRC.25,26 However, whether this was present in the precursor adenoma is an important question that our analysis model may answer.
The objective was to determine whether MINT locus hypermethylation is associated with MMR during early stages of CRC development. Methylation at MINT loci was quantified by using the on-slide SBM technique combined with absolute quantitative assessment of methylated alleles (AQAMA). We further evaluated MSI status, KRAS mt at codons 12 and 13, BRAF V600E mt, and methylation status of the MutL homolog 1 MMR gene (MLH1) in primary CRC as well as the contiguous precursor lesion to rigorously assess whether these events occur at early stages of CRC development. Our hypothesis was that MINT locus methylation, MLH1 promoter region methylation, and MSI in adenoma tissue are higher in those adenomas adjacent to MSI-high (MSI-H) compared with microsatellite-stable (MSS) invasive CRC.
Materials and Methods
Patient Specimens
For the first phase of the study, we identified patients whose resected CRC specimen contained histopathologically confirmed areas of adenoma as well as invasive cancer in our cancer registry database. Excluded were any cases without available PEAT specimens. Consecutive patients were identified in reverse chronological order until an adequate sample size was reached. The final cohort of 115 patients who underwent surgical resection of CRC between 1996 and 2009 at the Saint John’s Health Center was selected for the first phase of the study.
A single H&E section was prepared and mounted; 7-μm sections were consecutively cut and mounted on silane-coated glass slides for DNA studies. Areas of adenoma (serrated and nonserrated), carcinoma, and normal tissue as well as the adenoma type were identified by a surgical pathologist (R.R.T.) with expertise in CRC.8,27
To further validate the findings of the first phase of the study, PEAT blocks of operative specimens were obtained from 54 patients who underwent surgical resection of CRC at the Leiden University Medical Center (LUMC) between 1990 and 2001. All specimens had been previously analyzed for MMR sufficiency at LUMC’s pathology department. From each PEAT block, a single section was cut for H&E staining, and 7-μm sections were consecutively cut on coated slides for on-slide SBM. Tumor areas were identified and marked by an expert CRC pathologist (H.M.). Study protocols for assessment of patient specimens were approved by the institutional review boards at the LUMC and at Saint John’s Health Center.
AQAMA and MLH1 Methylation Assessment
DNA from PEAT was modified in situ by sodium bisulfite according to our previously reported protocol.18 AQAMA of MINT loci 1, 2, 3, 12, 17, 25, and 31 was performed, and data were analyzed in a manner as previously described.8,28 MLH1 methylation status was analyzed by capillary-array-electrophoresis methylation-specific PCR.27,29,30 Primer sequences were selected based on previous literature and correlation with MLH1 protein expression determined.31,32,33,34,35
MSI, KRAS, and BRAF mt Analysis
For MSI assessment in the first phase of the study, a tissue section from each of the specimens was deparaffinized and stained with hematoxylin to identify adenoma and CRC cells for DNA isolation, as previously described.8 Normal epithelial cells were harvested from a separate tissue block of the same specimen (ie, from noninvolved resection margins). As biomarkers for MSI assessment, we used three quasi-monomorphic mononucleotide repeats (BAT25, BAT26, and BAT40) and two microsatellite, dinucleotide repeats (D2S123 and D5S346) found in the revised Bethesda guidelines to interrogate the specimens.36 Forward primers were dye-labeled for automated high-throughput multiplex detection by capillary array electrophoresis37 (CEQ 8000XL; Beckman Coulter, Fullerton, CA). Forward primer sequences for BAT25, BAT26, BAT40, D2S123, and D5S346 were as follows: 5′-CCTCGCCTCCAAGAATGTAA-3′, 5′-GCAGTCAGAGCCCTTAACCTT-3′, 5′-AAGATTAACTTCCTACACCACAACC-3′, 5′-TGGCCAGAGAA ATTAGACACA-3′, and 5′-TTCAGGGAATTGAGAGTTACAGG-3′, respectively; corresponding reverse primer sequences were as follows: 5′-TGCTTTTGGTTACCACACTTCA-3′, 5′-CCATTTAAAGCTAGTTATCTAATCCA-3′, 5′-GTAGAGCAAGACCACCTTGT-3′, 5′-TCT GACTTGGATACCATCTATCTATCT-3′ and 5′-TCACTCTAGTGATAAATCGGGAAA-3′. Differences in PCR product fragment length among different tissue categories were visualized by the CEQ software (Beckman Coulter). PCR products from the five amplified microsatellite regions in adenoma and cancer were compared with the reference normal epithelium. KRAS (codons 12 and 13) and BRAF (V600E) mt were assessed by a peptide nucleic acid clamp-based quantitative real-time PCR assay as previously described.38,39 These assays were performed in triplicate and carried out at least twice to confirm accuracy. A gene was considered mutated when the results were uniformly positive in the triplicates under the optimal conditions. Respective normal and positive PEATs as well as cell line controls were included in each assay.
MMR and mt Status Analysis
The diagnostic techniques used by the LUMC’s pathology department for MSI status and MMR protein expression assessment are described previously.40 Briefly, MSI status was assessed by using MSI Analysis System (Promega Corp., Madison, WI; five mononucleotide and two pentanucleotide repeats).41 Immunostaining of MMR proteins was performed with anti-MLH1 (clone G168-728; 1:50; BD Biosciences, Franklin Lakes, NJ) and anti-PMS2 (clone A16-4; 1:50; BD Biosciences). Immunohistochemistry (IHC) staining was performed on PEAT sections (4 μm in thickness) from tissue microarray (TMA). IHC staining patterns of these MMR proteins were evaluated by using normal epithelial, stromal, or inflammatory cells, or the centers of lymphoid follicles as internal controls, as previously published.42 TMAs contained three cores punched from each primary tumor. Individual cores of the TMA were scored as either positive (showing nuclear staining in at least some tumor cells) or negative. Cases were considered positive if at least one TMA tissue core showed nuclear staining; if not, cases were considered negative. Cases in which both tumor and internal control stained negative were not included in the study. Cases were scored by two independent reviewers (H.M. and N.F.C.C. de M.); in case of a discrepancy, both reviewers reassessed the slides for consensus. KRAS43 and BRAF44 mt were detected by means of sequencing as previously described.
Biostatistical Analyses
Significance of changes in methylation index (MI) at individual MINT loci and total MI was evaluated with nonparametric tests for related and independent sample sets (Wilcoxon’s rank-sum test; Mann-Whitney’s U-test). Fisher’s exact test (two-tailed) was used to assess significance of intergroup differences in the prevalence of MLH1 methylation, BRAF, or KRAS mt.
Spearman’s rank correlation coefficient between quantitative MINT MI and MLH1 MI was assessed as a nonparametric measure of correlation. Correlations with clinical parameters were tested with Pearson’s χ2 test and, for ordinal variables, with Mann-Whitney’s U-test or Kruskal-Wallis’ test. Postoperative distant recurrence probability and disease-free and overall survival were estimated with Kaplan-Meier plots, and significance was assessed with the log-rank test. Cox regression models considered the following variables that were entered in a stepwise manner: age, nodal stage, MSI status, and tumor differentiation. P < 0.05 (two-sided) was considered significant. SPSS (SPSS Inc., Chicago, IL) statistical software version 16.0.1 was used for all analyses.
Results
MINT Locus Methylation and MSI during CRC Development
In the first phase of the study, we assessed operative specimens from 115 patients who underwent open resection of CRC and whose specimen, according to the pathology report, contained adenoma as well as cancer (see Table 1 for patient characteristics). Fifty of these specimens included normal tissue. Twenty-seven of the 115 patients were excluded from the study because the adenomas contained high-grade dysplasia and/or had carcinoma without evidence of submucosal invasion. Of the remaining 88 specimens that contained low- or medium-grade adenomatous dysplasia, 79 still had invasive carcinoma tissue after reviewing the cut sections. MSI status was analyzed in normal and cancer tissue of the 79 patients by using five established genomic markers.36 Specimens from nine patients (11%) showed instability in ≥4 biomarkers; these patients comprised the MSI-H group. Specimens from 65 patients (83%) did not show a shift in any of the markers, and specimens from five (6%) patients had a single dinucleotide repeat affected; these 70 patients with ≤1 aberrant MSI marker comprised the MSS group. The median age of MSI-H patients was 81.4 years (range, 69 to 91). Figures 1 and 2 show representative PCR product analyses of AQAMA and capillary array electrophoresis.
Table 1.
Patient Characteristics
| Characteristics | Total patient group (n = 115) | Patients with cancer tissues (n = 79) | Microsatellite unstable (n = 9) | Microsatellite stable (n = 70) | P |
|---|---|---|---|---|---|
| Sex, n (%) | |||||
| Male | 45 (39) | 35 (44) | 3 (33) | 33 (47) | 0.44 |
| Female | 70 (61) | 44 (56) | 6 (66) | 37 (53) | |
| Age, year | |||||
| Mean (SE) | 76.1 (1.2) | 75.0 (1.0) | 81.4 (7.3) | 76.1 (10.6) | 0.15 |
| Tumor location, n (%) | |||||
| Cecum | 24 (21) | 16 (20) | 3 (33) | 14 (20) | 0.03 |
| Colon ascendens | 15 (13) | 9 (11) | 4 (44) | 5 (7) | |
| Hepatic flexure | 6 (5) | 4 (5) | 0 | 4 (6) | |
| Colon transversum | 17 (15) | 12 (13) | 2 (22) | 9 (13) | |
| Colon descendens | 4 (4) | 3 (4) | 0 | 3 (4) | |
| Sigmoid colon | 13 (11) | 9 (11) | 0 | 8 (11) | |
| Rectosigmoid | 12 (10) | 9 (10) | 0 | 9 (13) | |
| Rectum | 24 (21) | 17 (22) | 0 | 18 (26) | |
| Adenoma type, n (%) | |||||
| Serrated | 9 (8) | 5 (6) | 2 (22) | 3 (4) | 0.10* |
| Classic | 106 (92) | 74 (94) | 7 (88) | 67 (96) | |
| Villoglandular | 7 (6) | 4 (5) | 0 | 4 (6) | |
| Tubular | 11 (10) | 8 (10) | 2 (22) | 6 (9) | |
| Tubulovillous | 15 (13) | 10 (13) | 1 (11) | 9 (13) | |
| Villous | 73 (63) | 52 (66) | 4 (44) | 48 (68) |
Fisher’s exact test evaluating MSI status in serrated versus classic adenoma in patients with adenoma as well as carcinoma available for analysis (n = 72).
Figure 1.
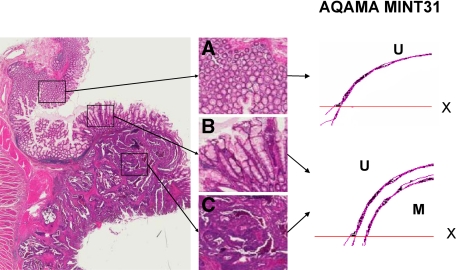
Representative molecular analyses in normal (A), adenoma (B), and cancer (C) tissues. AQAMA (MINT31) real-time PCR results showing results in triplicate of exponential amplification of methylated (M) and unmethylated (U) dye probe signals. X; PCR threshold cut-off. The vertical axis represents signal intensity, horizontal axis represents PCR cycle number.
Figure 2.
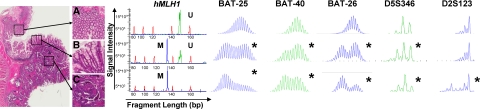
Representative molecular analyses in normal (A), adenoma (B), and cancer (C) tissues. Capillary array electrophoresis results of amplified dye-labeled PCR products of five MSI biomarkers and MLH1 methylation-specific (M, in blue) and unmethylated-specific sequence (U, in green) primer sets. Red peaks represent the DNA ladder signal. The horizontal axis represents size of the PCR product in bp. The vertical axis represents arbitrary units of signal intensity of detected amplicons. Asterisk indicates MSI positive.
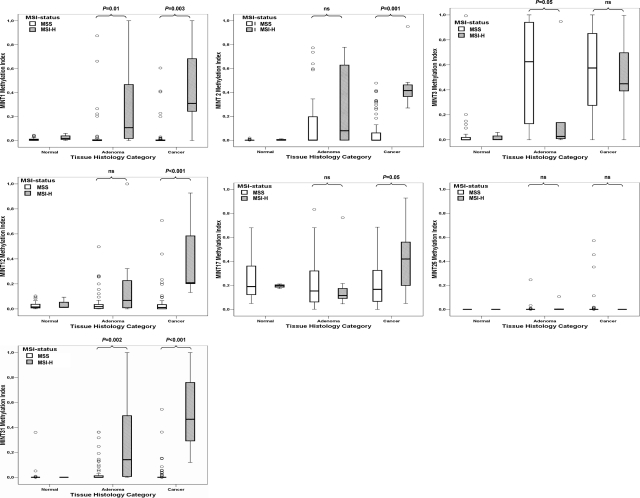
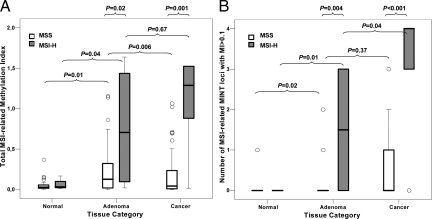
The MI of normal, adenoma, and cancer tissue was determined for each MINT locus of patients in MSI-H and MSS groups (Figure 3). Methylation levels of MINT loci 1, 2, 3, 12, and 31 were significantly higher in CRC than in normal epithelium. Methylation levels of MINT loci 1, 2, 12, 17, and 31 were significantly higher in MSI-H CRC compared with MSS CRC. MINT17 MI was MSI-related in CRC but was also present in normal tissue. Methylation levels of MINT loci 1 and 31 were significantly higher in adenomas contiguous to MSI-H CRC than in adenomas contiguous to MSS CRC. The average number of tumor-specific, MSI-related MINT loci showing MI >0.1 was 1.5 (SD ± 1.4) in adenomas from specimens containing MSI-H CRC, whereas on average only 0.3 (SD ± 0.6) MINT loci had MI >0.1 in adenomas from specimens containing MSS CRC (P = 0.004; Figure 4B). The total MI of MINT loci 1, 2, 12, and 31 was 4.5 times higher in adenomatous tissue from specimens containing MSI-H CRC (P = 0.02; Figure 4A). MSI status was significantly correlated to right-sided tumor location and not to serrated adenoma type (Table 1). MI was significantly higher in serrated versus nonserrated adenomas only at MINT1 and MINT31 (P = 0.002 and P = 0.02, respectively). There were no significant differences in MINT methylation level of cancer tissues contiguous to serrated versus classic adenoma. Methylation levels were relatively low in the two serrated/MSI-H adenomas compared with the nonserrated/MSI-H adenomas.
Figure 3.
Box plots summarizing MI values for each MINT locus in specimens containing normal, adenoma, and cancer tissues from patients in MSS and MSI-H groups.
Figure 4.
Box plots of MI data for specimens containing normal, adenoma, and cancer tissues from patients in MSS and MSI-H groups. A: y axis represents total MI of four MSI-related MINT loci (MINT1, 2, 12, and 31). B: y axis represents the number of MSI-related MINT loci with MI greater than 0.1.
From this we conclude that increased methylation of MINT loci 1, 2, 12, 17, and 31 in adenoma correlates with MSI-H sporadic CRC, and increased methylation of MINT loci 1, 2, 12, and 31 is specific for MSI-H CRC. Increased methylation of MINT 1 and 31 was adenoma-/CRC-/MSI-specific, and these loci were differentially methylated in serrated versus nonserrated adenomas. The adenomas associated with MSI-H CRC with high MINT loci methylation did not show any distinct histopathology features. Results indicate that total MI and number of affected MSI-specific MINT loci are indicators of colorectal adenomas that will become microsatellite-unstable CRC.
MLH1 Methylation Status and MSI during CRC Development
We subsequently determined whether the correlation between MINT methylation levels and MSI in colorectal adenomas could be explained by methylation of the MLH1 MMR gene. The relation between MLH1 methylation status and MSI was assessed in a case-control approach. Normal, adenomatous, and cancer tissues were acquired from all nine patients in the MSI-H group and from 13 randomly selected patients with sufficient available tissue in the MSS control group. This case-control approach was opted for because after multiple sectioning of the PEAT blocks, some cases lost either their cancer or their adenoma tissue. There were no significant differences between cases and controls with respect to age, sex, and polyp histopathology type. MLH1 was methylated in all cancers from the MSI-H group compared with two cancers from the MSS group (P < 0.001). MLH1 was methylated in 6 of 9 MSI-H adenomas compared with 2 of 13 adenomas paired to MSS cancers (P = 0.02). Only three of six (50%) MSI-H adenomas with MLH1 methylation showed to be MSI-H. In combination, MI of MINT1, 2, 12, and 31 with collateral methylation of MLH1 constitute a specific biomarker panel of adenomatous colorectal tissue that shows MSI or will develop into a sporadic MSI-H CRC. The results further indicate that MINT locus methylation along with MLH1 methylation precedes MSI and appears before histopathology signs of CRC development.
BRAF and KRAS mt during CRC Development
Studies in CRC have shown correlations between increased methylation of important tumor-related genes and mt of BRAF (V600E) and KRAS (codons 12 and 13), often in combination with MSI-H.15,19,45,46 The significance is not clearly understood; however, it has been suggested that these events synergistically induce a high-risk phenotype that results in a clinically distinct subtype of CRC. We determined BRAF mt and KRAS mt in the same group analyzed for MLH1 methylation status analysis (9 MSI-H cases and 13 MSS cases); the results are summarized in Table 2. BRAF mt status was significantly correlated to MSI and increased MINT MI. KRAS mt was more frequent in MSS adenomas and carcinomas, although this did not reach statistical significance. These results were as expected based on previous reports. This assessment demonstrated that genetic and epigenetic events synergize in the earliest phase of CRC development.
Table 2.
Correlation of KRAS mt, BRAF mt, Tissue Histopathology, Microsatellite Instability, and MINT Methylation Index
| Mutation status | Carcinoma (n = 22)
|
Adenoma (n = 21)
|
Carcinoma (n = 22)
|
Adenoma (n = 21)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BRAF mt+ | BRAF mt− | P | BRAF mt+ | BRAF mt− | P | KRAS mt+ | KRAS mt− | P | KRAS mt+ | KRAS mt− | P | |
| Case-control group, n (%) | 4 (18) | 18 (82) | 5 (24) | 16 (76) | 10 (45) | 12 (55) | 8 (38) | 13 (62) | ||||
| MSS group (N = 13), n (%) | 0 | 13 (100) | 0.02 | 0 | 13 (100) | 0.003 | 8 (62) | 5 (38) | 0.10 | 7 (54%) | 6 (46) | 0.09 |
| MSI-H group (N = 9), n (%) | 4 (44) | 5 (56) | 5 (62)† | 3 (38)† | 2 (22) | 7 (78) | 1 (13)† | 7 (87)† | ||||
| Total MINT* MI, mean (SEM) | 1.9 (0.6) | 0.5 (0.2) | 0.009 | 1.4 (0.6) | 0.3 (0.1) | 0.03 | 0.6 (0.4) | 0.9 (0.3) | 0.39 | 0.2 (0.1) | 0.8 (0.3) | 0.16 |
| MINTs* with MI >0.1, mean (SEM) | 3.8 (0.3) | 1.2 (0.4) | 0.02 | 2.6 (0.9) | 0.8 (0.2) | 0.09 | 1.0 (0.5) | 2.3 (0.6) | 0.09 | 0.8 (0.3) | 1.6 (0.5) | 0.34 |
MINT1, 2, 12, and 31.
n = 8 for MSI-H adenoma cases.
Quantitative MINT Locus Methylation and Mismatch Repair
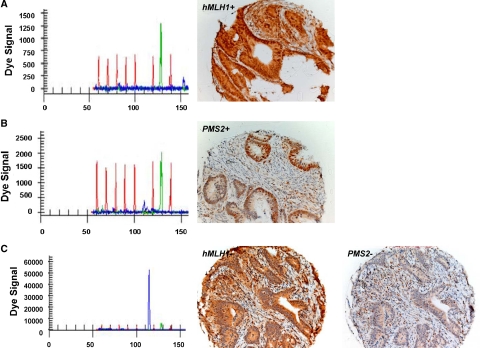
Methylation levels of five different MINT loci were related to methylation of MLH1, BRAF mt status, and to MSI in CRC specimens from 79 patients in the first phase of the study. These findings were corroborated by using an independent cohort of 54 patients with CRC whose MLH1 and PMS2 expression as well as MSI status were known. We first corroborated whether increased MINT MI linearly correlates to MLH1 MI, and subsequently whether it affects MMR at the protein level. MIs at the MSI-associated MINT loci (1, 2, 12, 17, and 31) and MLH1 MI were measured in 54 CRC specimens that were previously analyzed for MMR sufficiency. Two outcome parameters were analyzed: total MI (defined by the sum of MIs of MINT loci 1, 2, 12, 17, and 31) and number of methylated MINT loci. The number of methylated MINT loci in a specimen was determined as the number of MINT loci (1, 2, 12, 17, and 31) that exceeded the MSI-H related MI cut-off established during the first phase of the study (average MI ± 1 SD in MSS CRC specimens). Correlation analysis showed that MLH1 MI was closely associated with total MI (ρ = 0.43; P = 0.002) and with the number of methylated MINT loci (ρ = 0.400; P < 0.001). Table 3 shows the association between MLH1 protein expression and MI. Epigenomic down-regulation of MLH1 was demonstrated by the lower expression of its co-protein PMS2 in all cases.47 Examples of IHC and capillary-array-electrophoresis methylation-specific PCR results are given in Figure 5. Subsequently, all MMR protein-deficient cases showed MSI by PCR analysis. Again we analyzed two outcome parameters: number of affected MINT loci and total MI. These results demonstrated a linear correlation between quantitative MINT methylation, MLH1 down-regulation, and subsequent MMR deficiency, and showed the importance of the number of involved loci as well as total MI.
Table 3.
Relation of Methylated Loci and MI to MLH1 Methylation Status and Protein Expression
| No. of MINT loci methylated | No. of patients |
MLH1
|
MLH1
|
||
|---|---|---|---|---|---|
| Average methylation index (SEM) | P* | IHC Nonexpressing† (%) | P* | ||
| 0 | 32 | 0.14 (0.22) | 0.007* | 5 (16) | 0.002* |
| 1 | 7 | 0.08 (0.10) | 2 (29) | ||
| 2 | 4 | 0.01 (0.01) | 1 (25) | ||
| 3 | 5 | 0.72 (0.34) | 1 (20) | ||
| 4 | 2 | 0.42 (0.25) | 2 (100) | ||
| 5 | 4 | 0.43 (0.37) | 4 (100) | ||
Kruskal-Wallis test for correlation between methylated MINT loci and MLH1 MI or MLH1 protein expression.
Confirmed by underexpression of PMS2.
Figure 5.
Representative IHC staining of MLH1 and PMS2 proteins. A and B: MLH1-expressing CRC and PMS2-expressing CRC, respectively, with their corresponding capillary-array-electrophoresis methylation-specific PCR results for MLH1 showing unmethylated signals only. C: CRC showing methylation of the MLH1 promoter region with corresponding absence of nuclear staining of MLH1, which is confirmed by underexpression of PMS2.
BRAF mt and KRAS mt were also assessed in this patient group. BRAF mt cases (n = 11, 20%) were significantly associated with MSI-H (P = 0.003) and underexpression of MLH1 (P = 0.003). KRAS mt tumors (n = 14, 26%) did not show any significant associations with mismatch repair parameters. BRAF mt tumors had significantly increased methylation at MINT1, 2, 12, 17, and 31 (P < 0.001, P < 0.001, P = 0.001, P < 0.001, and P = 0.003, respectively). None of the BRAF mt tumors had KRAS mt. Methylation levels did not differ significantly between KRAS mt tumors and KRAS wild-type tumors.
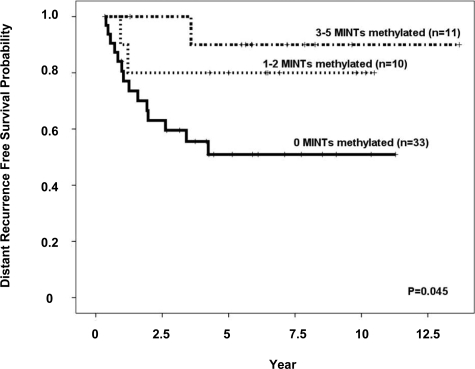
Clinical outcome in our validation group was analyzed with respect to the number of MINT loci that showed MSI-related methylation. The result (Figure 6) confirmed previous reports of a direct correlation between methylation levels and recurrence-free survival. Multivariate analysis showed that the number of methylated, MSI-related MINT loci was an independent predictor of distant recurrence-free survival (hazard ratio: 0.20; 95% confidence interval: 0.04 to 0.96; P = 0.02), disease-free survival (hazard ratio: 0.33; 95% confidence interval: 0.12 to 0.87; P = 0.04), and overall survival (hazard ratio: 0.38; 95% confidence interval: 0.14 to 1.00; P = 0.05), along with nodal stage and patient age.
Figure 6.
Kaplan-Meier curves for distant recurrence probability in patients stratified by 0, 1 to 2, or 3 to 5 MINT loci with MSI-related methylation.
Discussion
Our study adds important findings on genomic and epigenomic events at an early stage of CRC development. Several groups have identified epigenetic silencing of MLH1 as the cause for MSI in nonhereditary CRC.13,14,15,48 By using a unique assay system, we demonstrated a reproducible correlation between MSI, BRAF mt, MLH1, and MINT locus methylation between CRC and its associated precursor adenoma. This concurrent analysis of contiguous invasive and noninvasive tissue is preferable to analysis of adenoma tissue from “premalignant” polyps collected during colonoscopy, because it is known that not all polyp-associated adenomas will progress to CRC. Genetic and/or epigenetic aberrations in the adenoma tissue with contiguous cancer that we analyzed may have progressed compared with adenoma tissue from polyps without invasive component. The histopathological stage, however, was premalignant, and the main point of our study is that MINT markers may identify histopathological negative lesions that are likely to become MSI-H CRCs.
In this study methylation levels were measured for seven genomic MINT loci related to cancer and located on different chromosomes. The MINT biomarkers were originally detected in a study by Toyota et al.49 These investigators used a genome-wide screening method to show differential methylation in DNA from a colon cancer cell line (Caco2) compared with normal epithelium. We used MINT loci as biomarkers in this study, as we have previously shown that quantitative assessment of MINT biomarkers can distinguish between early CRC and benign disease.8 It is unclear whether MINT loci are representative of genomic hypermethylation. However, they have been well associated with the CpG island methylator phenotype (CIMP) in CRC. There is currently no consensus on CIMP biomarkers such as the Bethesda guidelines for MSI assays. Clinically relevant correlations between methylation markers, MSI, and BRAF mt/KRAS mt in CRC have been reported by using different techniques and combinations of three,50 eight,51 or five52,53 different methylation biomarkers. Our results show new application of MINT biomarkers to identify precursor lesions of MSI-H CRC disease and further demonstrate significant associations with CIMP-features (BRAF mt, MHL1 methylation, MSI, and disease survival).
In adenomas associated with MSI-H CRCs, there was significantly more hypermethylation of MLH1. This suggests that CRC precursor lesions with a higher MI at MINT loci may be more likely to have methylation of the MLH1 MMR gene. This has not been reported to date. The relationship between degree of methylation and MSI was further validated at the MMR protein level in an independent patient group. MINT MI linearly correlated with MLH1 MI, and the underexpression of MLH1 protein and MSI were demonstrated in the more methylated CRCs. This suggests that sporadic MSI is due to deregulation of a CRC’s MMR system, which may be a direct result of MLH1 epigenetic gene silencing in a tumor with increased MINT methylation. The level of DNA methylation appears to be an underlying mechanism of sporadic MSI, and the accumulation of DNA methylation affects the MMR system early during CRC development. We further corroborated the validity of the quantitative MINT methylation AQAMA assay by reproducing its previously reported prognostic value in colorectal and gastric cancer.51,53,54
Recent studies on DNA hypermethylation in CRC include BRAF mt and KRAS mt analyses. BRAF mt is more frequent in sporadic CRCs showing MSI-H (approximately 50%), CIMP and MLH1 methylation,55,56 and is possibly associated with shorter survival.51 Specific single-nucleotide mt of KRAS have predictive value in stage III/IV CRC, likely related to systemic treatment response.57,58,59 However, as of yet, no study has demonstrated a functional relation between MMR impairment and reduced cell signaling properties of KRAS mt or BRAF mt containing CRCs. In both the first-phase analysis and the validation patient groups, we could not show significant correlations between KRAS mt status, MSI, and MINT methylation. Because colon cancer has a >40% to 60% frequency of KRAS mt, it is likely that there is no strong relation to MSI. Two recent, large studies could not either identify a correlation between KRAS and MSI in colon cancer.60,61 Our results do confirm the association between BRAF mt and MINT locus hypermethylation, and add that these events occur early in the development of CRC in nonserrated adenomas with low or intermediate dysplasia.
This study demonstrated that AQAMA of MINT loci may have clinicopathologic utility for early identification of patients at risk for developing MSI-H phenotype sporadic cancers. A potentially useful approach would be to examine MINT locus methylation in polyps collected during routine colonoscopies; changes in MINT methylation over time might correlate with a high risk for CRC development. Patients whose adenomas have high MINT locus methylation may benefit from scheduling more frequent screening colonoscopies. Our results further show that MINT loci methylation levels can discriminate normal from adenomatous tissue. MINT biomarkers could be explored to be used as a fecal DNA screening tool to identify patients who should undergo colonoscopy. MSI-H precursor lesions may be identified by MINT biomarkers 1, 2, 12, and 31, and MSS lesions may be identified by MINT3 methylation. Furthermore, it is known that MSI-H cancers respond differentially to common chemotherapeutics. For instance, irinotecan is suggested to be more effective than 5-Fluro-uracil in these tumors. In the development of new targeted drugs, preventive treatment regimens may be available for high-risk patients with adenomas that show high MINT locus methylation. In addition, there is increasing evidence for MSI-induced generation of novel tumor-specific carboxy-terminal frameshift peptides in MSI-H cancers.62 Polyps with high MINT locus methylation may become a stratification factor in trials of frameshift peptide-based vaccines.
This is the first major study to demonstrate a correlation between MSI and MINT hypermethylation in CRC precursor lesions, supporting that MMR may be dampened by progressive epigenetic events. The degree of genomic hypermethylation in CRC precursors may be a potential cause of sporadic MSI CRC. Specific MINT locus methylation may have utility for early identification of colorectal polyps with high likelihood of developing into sporadic MSI-H cancers.
Acknowledgments
We thank Linhda Nguyen for technical assistance and Kana Rivera for editorial assistance.
Footnotes
Address reprint requests to Dr. Dave S.B. Hoon, Department of Molecular Oncology, John Wayne Cancer Institute, 2200 Santa Monica Blvd, Santa Monica, CA 90404. E-mail: hoon@jwci.org.
Supported in part by the Leslie and Susan Gonda (Goldschmied) Foundation (Los Angeles, CA) and Ruth and Martin H. Weil Fund (Los Angeles, CA).
N.N. and A.B. contributed equally to this work.
None of the authors disclosed any relevant financial relationships.
References
- Haydon AM, Jass JR. Emerging pathways in colorectal-cancer development. Lancet Oncol. 2002;3:83–88. doi: 10.1016/s1470-2045(02)00649-6. [DOI] [PubMed] [Google Scholar]
- Herman JG. Epigenetic changes in cancer and preneoplasia. Cold Spring Harb Symp Quant Biol. 2005;70:329–333. doi: 10.1101/sqb.2005.70.036. [DOI] [PubMed] [Google Scholar]
- Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Tanemura A, Terando AM, Sim MS, van Hoesel AQ, de Maat MF, Morton DL, Hoon DS. CpG island methylator phenotype predicts progression of malignant melanoma. Clin Cancer Res. 2009;15:1801–1807. doi: 10.1158/1078-0432.CCR-08-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynter CV, Walsh MD, Higuchi T, Leggett BA, Young J, Jass JR. Methylation patterns define two types of hyperplastic polyp associated with colorectal cancer. Gut. 2004;53:573–580. doi: 10.1136/gut.2003.030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AO, Broaddus RR, Houlihan PS, Issa JP, Hamilton SR, Rashid A. CpG island methylation in aberrant crypt foci of the colorectum. Am J Pathol. 2002;160:1823–1830. doi: 10.1016/S0002-9440(10)61128-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Chen WD, Pretlow TP. CpG island methylation in aberrant crypt foci and cancers from the same patients. Int J Cancer. 2005;115:747–751. doi: 10.1002/ijc.20936. [DOI] [PubMed] [Google Scholar]
- de Maat MF, Umetani N, Sunami E, Turner RR, Hoon DS. Assessment of methylation events during colorectal tumor progression by absolute quantitative analysis of methylated alleles. Mol Cancer Res. 2007;5:461–471. doi: 10.1158/1541-7786.MCR-06-0358. [DOI] [PubMed] [Google Scholar]
- Jass JR. Serrated adenoma of the colorectum and the DNA-methylator phenotype. Nat Clin Pract Oncol. 2005;2:398–405. doi: 10.1038/ncponc0248. [DOI] [PubMed] [Google Scholar]
- Wynter CV, Kambara T, Walsh MD, Leggett BA, Young J, Jass JR. DNA methylation patterns in adenomas from FAP, multiple adenoma and sporadic colorectal carcinoma patients. Int J Cancer. 2006;118:907–915. doi: 10.1002/ijc.21363. [DOI] [PubMed] [Google Scholar]
- Baylin SB, Ohm JE. Epigenetic gene silencing in cancer: a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- Ahuja N, Mohan AL, Li Q, Stolker JM, Herman JG, Hamilton SR, Baylin SB, Issa JP. Association between CpG island methylation and microsatellite instability in colorectal cancer. Cancer Res. 1997;57:3370–3374. [PubMed] [Google Scholar]
- Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW, Kane MF, Kolodner RD, Vogelstein B, Kunkel TA, Baylin SB. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veigl ML, Kasturi L, Olechnowicz J, Ma AH, Lutterbaugh JD, Periyasamy S, Li GM, Drummond J, Modrich PL, Sedwick WD, Markowitz SD. Biallelic inactivation of hMLH1 by epigenetic gene silencing, a novel mechanism causing human MSI cancers. Proc Natl Acad Sci USA. 1998;95:8698–8702. doi: 10.1073/pnas.95.15.8698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, Koh H, Simms L, Barker M, Leggett B, Levine J, Kim M, French AJ, Thibodeau SN, Jass J, Haile R, Laird PW. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- Shibata D. When does MMR loss occur during HNPCC progression? Cancer Biomark. 2006;2:29–35. doi: 10.3233/cbm-2006-21-204. [DOI] [PubMed] [Google Scholar]
- Cunningham JM, Christensen ER, Tester DJ, Kim CY, Roche PC, Burgart LJ, Thibodeau SN. Hypermethylation of the hMLH1 promoter in colon cancer with microsatellite instability. Cancer Res. 1998;58:3455–3460. [PubMed] [Google Scholar]
- Umetani N, de Maat MF, Sunami E, Hiramatsu S, Martinez S, Hoon DS. Methylation of p16 and Ras association domain family protein 1a during colorectal malignant transformation. Mol Cancer Res. 2006;4:303–309. doi: 10.1158/1541-7786.MCR-05-0199. [DOI] [PubMed] [Google Scholar]
- Toyota M, Ohe-Toyota M, Ahuja N, Issa JP. Distinct genetic profiles in colorectal tumors with or without the CpG island methylator phenotype. Proc Natl Acad Sci USA. 2000;97:710–715. doi: 10.1073/pnas.97.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Maat MF, van de Velde CJ, van der Werff MP, Putter H, Umetani N, Klein-Kranenbarg EM, Turner RR, van Krieken JH, Bilchik A, Tollenaar RA, Hoon DS. Quantitative analysis of methylation of genomic loci in early-stage rectal cancer predicts distant recurrence. J Clin Oncol. 2008;26:2327–2335. doi: 10.1200/JCO.2007.14.0723. [DOI] [PubMed] [Google Scholar]
- Park SJ, Rashid A, Lee JH, Kim SG, Hamilton SR, Wu TT. Frequent CpG island methylation in serrated adenomas of the colorectum. Am J Pathol. 2003;162:815–822. doi: 10.1016/S0002-9440(10)63878-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham SC, Park SJ, Cruz-Correa M, Houlihan PS, Half EE, Lynch PM, Wu TT. Frequent CpG island methylation in sporadic and syndromic gastric fundic gland polyps. Am J Clin Pathol. 2004;122:740–746. doi: 10.1309/4QUN-J4F2-7QK7-RR0G. [DOI] [PubMed] [Google Scholar]
- Toyota M, Ahuja N, Suzuki H, Itoh F, Ohe-Toyota M, Imai K, Baylin SB, Issa JP. Aberrant methylation in gastric cancer associated with the CpG island methylator phenotype. Cancer Res. 1999;59:5438–5442. [PubMed] [Google Scholar]
- Whitehall VL, Wynter CV, Walsh MD, Simms LA, Purdie D, Pandeya N, Young J, Meltzer SJ, Leggett BA, Jass JR. Morphological and molecular heterogeneity within nonmicrosatellite instability-high colorectal cancer. Cancer Res. 2002;62:6011–6014. [PubMed] [Google Scholar]
- Samowitz WS, Albertsen H, Herrick J, Levin TR, Sweeney C, Murtaugh MA, Wolff RK, Slattery ML. Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology. 2005;129:837–845. doi: 10.1053/j.gastro.2005.06.020. [DOI] [PubMed] [Google Scholar]
- de Maat MF, van de Velde CJ, Umetani N, de Heer P, Putter H, van Hoesel AQ, Meijer GA, van Grieken NC, Kuppen PJ, Bilchik AJ, Tollenaar RA, Hoon DS. Epigenetic silencing of cyclooxygenase-2 affects clinical outcome in gastric cancer. J Clin Oncol. 2007;25:4887–4894. doi: 10.1200/JCO.2006.09.8921. [DOI] [PubMed] [Google Scholar]
- Umetani N, de Maat MF, Mori T, Takeuchi H, Hoon DS. Synthesis of universal unmethylated control DNA by nested whole genome amplification with phi29 DNA polymerase. Biochem Biophys Res Commun. 2005;329:219–223. doi: 10.1016/j.bbrc.2005.01.088. [DOI] [PubMed] [Google Scholar]
- Hoon DS, Spugnardi M, Kuo C, Huang SK, Morton DL, Taback B. Profiling epigenetic inactivation of tumor suppressor genes in tumors and plasma from cutaneous melanoma patients. Oncogene. 2004;23:4014–4022. doi: 10.1038/sj.onc.1207505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Kim J, Yamano T, Takeuchi H, Huang S, Umetani N, Koyanagi K, Hoon DS. Epigenetic up-regulation of C-C chemokine receptor 7 and C-X-C chemokine receptor 4 expression in melanoma cells. Cancer Res. 2005;65:1800–1807. doi: 10.1158/0008-5472.CAN-04-3531. [DOI] [PubMed] [Google Scholar]
- Arnold CN, Goel A, Boland CR. Role of hMLH1 promoter hypermethylation in drug resistance to 5-fluorouracil in colorectal cancer cell lines. Int J Cancer. 2003;106:66–73. doi: 10.1002/ijc.11176. [DOI] [PubMed] [Google Scholar]
- Arnold CN, Goel A, Compton C, Marcus V, Niedzwiecki D, Dowell JM, Wasserman L, Inoue T, Mayer RJ, Bertagnolli MM, Boland CR. Evaluation of microsatellite instability, hMLH1 expression and hMLH1 promoter hypermethylation in defining the MSI phenotype of colorectal cancer. Cancer Biol Ther. 2004;3:73–78. doi: 10.4161/cbt.3.1.590. [DOI] [PubMed] [Google Scholar]
- Ashktorab H, Smoot DT, Carethers JM, Rahmanian M, Kittles R, Vosganian G, Doura M, Nidhiry E, Naab T, Momen B, Shakhani S, Giardiello FM. High incidence of microsatellite instability in colorectal cancer from African Americans. Clin Cancer Res. 2003;9:1112–1117. [PubMed] [Google Scholar]
- Ashktorab H, Smoot DT, Farzanmehr H, Fidelia-Lambert M, Momen B, Hylind L, Iacosozio-Dononue C, Carethers JM, Goel A, Boland CR, Giardiello FM. Clinicopathological features and microsatellite instability (MSI) in colorectal cancers from African Americans. Int J Cancer. 2005;116:914–919. doi: 10.1002/ijc.21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brim H, Mokarram P, Naghibalhossaini F, Saberi-Firoozi M, Al-Mandhari M, Al-Mawaly K, Al-Mjeni R, Al-Sayegh A, Raeburn S, Lee E, Giardiello F, Smoot DT, Vilkin A, Boland CR, Goel A, Hafezi M, Nouraie M, Ashktorab H. Impact of BRAF. MLH1 on the incidence of microsatellite instability high colorectal cancer in populations based study. Mol Cancer. 2008;7:68–79. doi: 10.1186/1476-4598-7-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Ruschoff J, Fishel R, Lindor NM, Burgart LJ, Hamelin R, Hamilton SR, Hiatt RA, Jass J, Lindblom A, Lynch HT, Peltomaki P, Ramsey SD, Rodriguez-Bigas MA, Vasen HF, Hawk ET, Barrett JC, Freedman AN, Srivastava S. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto A, Takeuchi H, Taback B, Hsueh EC, Elashoff D, Morton DL, Hoon DS. Allelic imbalance of 12q22–23 associated with APAF-1 locus correlates with poor disease outcome in cutaneous melanoma. Cancer Res. 2004;64:2245–2250. doi: 10.1158/0008-5472.can-03-2932. [DOI] [PubMed] [Google Scholar]
- Shinozaki M, O'Day SJ, Kitago M, Amersi F, Kuo C, Kim J, Wang HJ, Hoon DS. Utility of circulating B-RAF DNA mutation in serum for monitoring melanoma patients receiving biochemotherapy. Clin Cancer Res. 2007;13:2068–2074. doi: 10.1158/1078-0432.CCR-06-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Reber HA, Dry SM, Elashoff D, Chen SL, Umetani N, Kitago M, Hines OJ, Kazanjian KK, Hiramatsu S, Bilchik AJ, Yong S, Shoup M, Hoon DS. Unfavourable prognosis associated with K-ras gene mutation in pancreatic cancer surgical margins. Gut. 2006;55:1598–1605. doi: 10.1136/gut.2005.083063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong AE, van Puijenbroek M, Hendriks Y, Tops C, Wijnen J, Ausems MG, Meijers-Heijboer H, Wagner A, van Os TA, Brocker-Vriends AH, Vasen HF, Morreau H. Microsatellite instability, immunohistochemistry, and additional PMS2 staining in suspected hereditary nonpolyposis colorectal cancer. Clin Cancer Res. 2004;10:972–980. doi: 10.1158/1078-0432.ccr-0956-3. [DOI] [PubMed] [Google Scholar]
- Murphy KM, Zhang S, Geiger T, Hafez MJ, Bacher J, Berg KD, Eshleman JR. Comparison of the microsatellite instability analysis system and the Bethesda panel for the determination of microsatellite instability in colorectal cancers. J Mol Diagn. 2006;8:305–311. doi: 10.2353/jmoldx.2006.050092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks Y, Franken P, Dierssen JW, De Leeuw W, Wijnen J, Dreef E, Tops C, Breuning M, Brocker-Vriends A, Vasen H, Fodde R, Morreau H. Conventional and tissue microarray immunohistochemical expression analysis of mismatch repair in hereditary colorectal tumors. Am J Pathol. 2003;162:469–477. doi: 10.1016/S0002-9440(10)63841-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eijk R, van Puijenbroek M, Chhatta AR, Gupta N, Vossen RH, Lips EH, Cleton-Jansen AM, Morreau H, van Wezel T. Sensitive and specific KRAS somatic mutation analysis on whole-genome amplified DNA from archival tissues. J Mol Diagn. 12:27–34. doi: 10.2353/jmoldx.2010.090028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-de-Abajo A, de la Hoya M, van Puijenbroek M, Tosar A, Lopez-Asenjo JA, Diaz-Rubio E, Morreau H, Caldes T. Molecular analysis of colorectal cancer tumors from patients with mismatch repair proficient hereditary nonpolyposis colorectal cancer suggests novel carcinogenic pathways. Clin Cancer Res. 2007;13:5729–5735. doi: 10.1158/1078-0432.CCR-06-2996. [DOI] [PubMed] [Google Scholar]
- Goel A, Nagasaka T, Arnold CN, Inoue T, Hamilton C, Niedzwiecki D, Compton C, Mayer RJ, Goldberg R, Bertagnolli MM, Boland CR. The CpG island methylator phenotype and chromosomal instability are inversely correlated in sporadic colorectal cancer. Gastroenterology. 2007;132:127–138. doi: 10.1053/j.gastro.2006.09.018. [DOI] [PubMed] [Google Scholar]
- Nagasaka T, Koi M, Kloor M, Gebert J, Vilkin A, Nishida N, Shin SK, Sasamoto H, Tanaka N, Matsubara N, Boland CR, Goel A. Mutations in both KRAS and BRAF may contribute to the methylator phenotype in colon cancer. Gastroenterology. 2008;134:1950–1960. doi: 10.1053/j.gastro.2008.02.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S, Lindor NM, Burgart LJ, Smalley R, Leontovich O, French AJ, Goldberg RM, Sargent DJ, Jass JR, Hopper JL, Jenkins MA, Young J, Barker MA, Walsh MD, Ruszkiewicz AR, Thibodeau SN. Isolated loss of PMS2 expression in colorectal cancers: frequency, patient age, and familial aggregation. Clin Cancer Res. 2005;11:6466–6471. doi: 10.1158/1078-0432.CCR-05-0661. [DOI] [PubMed] [Google Scholar]
- Hawkins N, Norrie M, Cheong K, Mokany E, Ku SL, Meagher A, O'Connor T, Ward R. CpG island methylation in sporadic colorectal cancers and its relationship to microsatellite instability. Gastroenterology. 2002;122:1376–1387. doi: 10.1053/gast.2002.32997. [DOI] [PubMed] [Google Scholar]
- Toyota M, Ho C, Ahuja N, Jair KW, Li Q, Ohe-Toyota M, Baylin SB, Issa JP. Identification of differentially methylated sequences in colorectal cancer by methylated CpG island amplification. Cancer Res. 1999;59:2307–2312. [PubMed] [Google Scholar]
- Yagi K, Akagi K, Hayashi H, Nagae G, Tsuji S, Isagawa T, Midorikawa Y, Nishimura Y, Sakamoto H, Seto Y, Aburatani H, Kaneda A. Three DNA methylation epigenotypes in human colorectal cancer. Clin Cancer Res. 16:21–33. doi: 10.1158/1078-0432.CCR-09-2006. [DOI] [PubMed] [Google Scholar]
- Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Meyerhardt JA, Loda M, Giovannucci EL, Fuchs CS. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S, Cantor M, Kawasaki T, Brahmandam M, Kirkner GJ, Weisenberger DJ, Campan M, Laird PW, Loda M, Fuchs CS. CpG island methylator phenotype (CIMP) of colorectal cancer is best characterised by quantitative DNA methylation analysis and prospective cohort studies. Gut. 2006;55:1000–1006. doi: 10.1136/gut.2005.082933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barault L, Charon-Barra C, Jooste V, de la Vega MF, Martin L, Roignot P, Rat P, Bouvier AM, Laurent-Puig P, Faivre J, Chapusot C, Piard F. Hypermethylator phenotype in sporadic colon cancer: study on a population-based series of 582 cases. Cancer Res. 2008;68:8541–8546. doi: 10.1158/0008-5472.CAN-08-1171. [DOI] [PubMed] [Google Scholar]
- An C, Choi IS, Yao JC, Worah S, Xie K, Mansfield PF, Ajani JA, Rashid A, Hamilton SR, Wu TT. Prognostic significance of CpG island methylator phenotype and microsatellite instability in gastric carcinoma. Clin Cancer Res. 2005;11:656–663. [PubMed] [Google Scholar]
- Koinuma K, Shitoh K, Miyakura Y, Furukawa T, Yamashita Y, Ota J, Ohki R, Choi YL, Wada T, Konishi F, Nagai H, Mano H. Mutations of BRAF are associated with extensive hMLH1 promoter methylation in sporadic colorectal carcinomas. Int J Cancer. 2004;108:237–242. doi: 10.1002/ijc.11523. [DOI] [PubMed] [Google Scholar]
- Shen L, Toyota M, Kondo Y, Lin E, Zhang L, Guo Y, Hernandez NS, Chen X, Ahmed S, Konishi K, Hamilton SR, Issa JP. Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proc Natl Acad Sci USA. 2007;104:18654–18659. doi: 10.1073/pnas.0704652104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreyev HJ, Norman AR, Cunningham D, Oates J, Dix BR, Iacopetta BJ, Young J, Walsh T, Ward R, Hawkins N, Beranek M, Jandik P, Benamouzig R, Jullian E, Laurent-Puig P, Olschwang S, Muller O, Hoffmann I, Rabes HM, Zietz C, Troungos C, Valavanis C, Yuen ST, Ho JW, Croke CT, O'Donoghue DP, Giaretti W, Rapallo A, Russo A, Bazan V, Tanaka M, Omura K, Azuma T, Ohkusa T, Fujimori T, Ono Y, Pauly M, Faber C, Glaesener R, de Goeij AF, Arends JW, Andersen SN, Lovig T, Breivik J, Gaudernack G, Clausen OP, De Angelis PD, Meling GI, Rognum TO, Smith R, Goh HS, Font A, Rosell R, Sun XF, Zhang H, Benhattar J, Losi L, Lee JQ, Wang ST, Clarke PA, Bell S, Quirke P, Bubb VJ, Piris J, Cruickshank NR, Morton D, Fox JC, Al-Mulla F, Lees N, Hall CN, Snary D, Wilkinson K, Dillon D, Costa J, Pricolo VE, Finkelstein SD, Thebo JS, Senagore AJ, Halter SA, Wadler S, Malik S, Krtolica K, Urosevic N. Kirsten ras mutations in patients with colorectal cancer: the “RASCAL II” study. Br J Cancer. 2001;85:692–696. doi: 10.1054/bjoc.2001.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnanasampanthan G, Elsaleh H, McCaul K, Iacopetta B. Ki-ras mutation type and the survival benefit from adjuvant chemotherapy in Dukes’ C colorectal cancer. J Pathol. 2001;195:543–548. doi: 10.1002/path.990. [DOI] [PubMed] [Google Scholar]
- Tol J, Koopman M, Cats A, Rodenburg CJ, Creemers GJ, Schrama JG, Erdkamp FL, Vos AH, van Groeningen CJ, Sinnige HA, Richel DJ, Voest EE, Dijkstra JR, Vink-Borger ME, Antonini NF, Mol L, van Krieken JH, Dalesio O, Punt CJ. Chemotherapy, bevacizumab, and cetuximab in metastatic colorectal cancer. N Engl J Med. 2009;360:563–572. doi: 10.1056/NEJMoa0808268. [DOI] [PubMed] [Google Scholar]
- Roth AD, Tejpar S, Delorenzi M, Yan P, Fiocca R, Klingbiel D, Dietrich D, Biesmans B, Bodoky G, Barone C, Aranda E, Nordlinger B, Cisar L, Labianca R, Cunningham D, Van Cutsem E, Bosman F. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60–00 trial. J Clin Oncol. 28:466–474. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- Ogino S, Meyerhardt JA, Irahara N, Niedzwiecki D, Hollis D, Saltz LB, Mayer RJ, Schaefer P, Whittom R, Hantel A, Benson AB, 3rd, Goldberg RM, Bertagnolli MM, Fuchs CS. KRAS mutation in stage III colon cancer and clinical outcome following intergroup trial CALGB 89803. Clin Cancer Res. 2009;15:7322–7329. doi: 10.1158/1078-0432.CCR-09-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwitalle Y, Kloor M, Eiermann S, Linnebacher M, Kienle P, Knaebel HP, Tariverdian M, Benner A, von Knebel Doeberitz M. Immune response against frameshift-induced neopeptides in HNPCC patients and healthy HNPCC mutation carriers. Gastroenterology. 2008;134:988–997. doi: 10.1053/j.gastro.2008.01.015. [DOI] [PubMed] [Google Scholar]