Abstract
The rpoH regulatory region of different members of the enteric bacteria family was sequenced or downloaded from GenBank and compared. In addition, the transcriptional start sites of rpoH of Yersinia frederiksenii and Proteus mirabilis, two distant members of this family, were determined. Sequences similar to the σ70 promoters P1, P4 and P5, to the σE promoter P3 and to boxes DnaA1, DnaA2, cAMP receptor protein (CRP) boxes CRP1, CRP2 and box CytR present in Escherichia coli K12, were identified in sequences of closely related bacteria such as: E.coli, Shigella flexneri, Salmonella enterica serovar Typhimurium, Citrobacter freundii, Enterobacter cloacae and Klebsiella pneumoniae. In more distant bacteria, Y.frederiksenii and P.mirabilis, the rpoH regulatory region has a distal P1-like σ70 promoter and two proximal promoters: a heat-induced σE-like promoter and a σ70 promoter. Sequences similar to the regulatory boxes were not identified in these bacteria. This study suggests that the general pattern of transcription of the rpoH gene in enteric bacteria includes a distal σ70 promoter, >200 nt upstream of the initiation codon, and two proximal promoters: a heat-induced σE-like promoter and a σ70 promoter. A second proximal σ70 promoter under catabolite-regulation is probably present only in bacteria closely related to E.coli.
INTRODUCTION
Cells of almost any organism respond to a sudden up-shift of temperature and to several other stress conditions by a transient increase in the cellular concentration of a set of proteins, the heat-shock proteins (HSPs). In Escherichia coli K12, approximately 40 genes that encode the HSPs define the heat-shock stimulon (1,2). Most of these genes, including the main chaperone and protease genes, are under the positive control of σ32, encoded by rpoH; while approximately 10 genes, including rpoH, rpoE, fkpA and degP, have a promoter for σE, encoded by rpoE (1–3).
During the heat-shock response there is an increase in σ32. This increase is primarily due to an enhanced translation of the rpoH mRNA and stabilization of the protein (1,2); however, transcription of rpoH is subject to complex regulation. This gene has four promoters: P1, P3, P4 and P5. Promoters P1, P4 and P5 are σ70 promoters, while P3 is controlled by σE. Promoters P1 and P4 are responsible for rpoH transcription under most growth conditions, while promoter P3 is strongly induced by heat and promoter P5 is a weak catabolite-sensitive promoter (1,2,4). Promoter P1 overlaps the terminator of the cell division operon ftsYEX (5). In addition, the regulatory region of the rpoH gene, ∼250 bp, contains two cAMP receptor protein (CRP) boxes, two DnaA boxes and one CytR box (4,6,7). It has been proposed that the binding of CRP to box CRP1 increases transcription from P5 and decreases that from P4 (4), while the binding of the CRP–CytR complex to the CRP1–CytR–CRP2 sites reduces transcription from P3, P4 and P5 (7). Finally, the binding of DnaA to the DnaA boxes represses transcription from P3 and P4 (6). The in vivo physiological significance of these boxes has not been completely evaluated.
The presence of rpoH genes and σ32-like factors has been determined in several species of the α and γ subdivisions of the Proteobacteria (8–14). In addition to a σ32 factor, a highly conserved controlling inverted repeat of chaperone expression (CIRCE), has also been detected in front of genes groE and dnaK of the α Proteobacteria Agrobacterium tumefaciens, Bradyrhizobium japonicum and Caulobacter crescentus (15,16). In bacteria of other divisions, instead of rpoH homologs and σ32-dependent promoters, the sequence CIRCE has been detected in the regulatory region of the main heat-shock genes, dnaK and groE (15).
Although in several bacteria the structure and function of the σ32 protein are similar to those of its E.coli counterpart, the architecture, copy number and regulation of the rpoH gene are different. In C.crescentus and Pseudomonas aeruginosa this gene is transcribed from two promoters, regulated by σ70 and σ32, and σ70 and σE, respectively (17,18). Bradyrhizobium japonicum has three copies of the rpoH gene regulated by different mechanisms (11). Finally, the predicted sequence that generates an mRNA secondary structure involved in translational control of rpoH, and a σE-putative promoter were found in the γ but not in the α subdivision of the Proteobacteria (19).
In this study, the rpoH regulatory region of several members of the enteric bacteria was sequenced, analyzed and compared with sequences and regulatory elements previously determined for E.coli and other enteric bacteria. In addition, the transcriptional start sites of rpoH of Yersinia frederiksenii and Proteus mirabilis, two distant members of this family, were determined.
MATERIALS AND METHODS
Bacterial strains
The bacteria used in this study as DNA source were: E.coli, Salmonella enterica serovar Typhimurium, Shigella flexneri, Klebsiella pneumoniae, Citrobacter freundii, Y.frederiksenii and Erwinia amylovora. With the exception of E.amylovora, all strains used were isolated at hospitals in Mexico City, Mexico (14).
Polymerase chain reaction (PCR) amplification and cloning of the regulatory region of rpoH homologs from enteric bacteria
The regulatory region of rpoH homologs was PCR amplified using different primers complementary to ftsX (forward: F1, 5′-TGATTGGTGCGACAGATG; F2, 5′-AGAAATTCTGGTGCTGCG; F3, 5′-CCTGCTATTGCTGCTGGTAT; F4, 5′-TTTACGCCACTTTACGCC) and rpoH (reverse: R1, 5′-CAGTCATTCAAATCCTCTCA; R2, 5′-GTTACCTTCCTGAATCAAATCC; R3, 5′-CGAGCAATATGAACAACAAACC) coding regions of E.coli K12. The PCR protocol included an initial step of 1 min of denaturation at 95°C, followed by 30 cycles of 30 s of denaturation at 95°C, 30 s of annealing at 50–58°C and 1 min of polymerase extension. A final extension was performed for 5 min at 72°C. Escherichia coli DNA was amplified with primers F1/R2 and primer annealing was at 58°C. Primers F3/R1 were used to amplify DNAs from S.flexneri, Y.frederiksenii, S.enterica and C.freundii and primer annealing was performed at 58, 55, 55 and 53°C, respectively. Klebsiella pneumoniae and E.amylovora were amplified using primers F4/R3 and F2/R3, respectively, and 50°C as annealing temperature. The amplified DNA fragments were electrophoresed in 1% agarose gels in 1× TBE buffer (20) using a 100 bp ladder as size marker and purified by Qiaquick PCR purification kit (Qiagen). The purified DNAs were cloned in the pMOSBlue T-vector (Amersham Life Science). Plasmids carrying the rpoH amplified regulatory region were selected for DNA sequencing.
DNA sequence determination
DNA was sequenced using the Thermo sequenase cycle sequencing kit (Amersham Life Science) and M13 primers. Internal primers were used to complete the sequences.
Primer extension analysis
Synthetic oligonucleotides complementary to nucleotides –15 to –39 of the rpoH regulatory sequence (5′-TGCATGCATGTTTAGTGCAAATTTC) of Y.frederiksenii and to +15 to +37 of the rpoH coding sequence (5′-GCGGAACCAATGCTAAGGATTGC) of P.mirabilis were 5′-end labeled with T4 polynucleotide kinase and [γ-32P]ATP. Total RNA was extracted by the acid-hot phenol method from Y.frederiksenii and P.mirabilis cells harvested before and 5 and 10 min after a heat shock from 30 to 43°C and from 30 to 47°C, respectively. The labeled primers were hybridized to 40 µg of total RNA and primer extension reactions were performed using AMV reverse transcriptase (Promega). DNA sequencing reactions carried out with the same primers were used to identify the transcriptional start sites.
Analysis of the nucleotide sequences
The nucleotide sequences were first aligned using the Clustal algorithm of PC/GENE release 6.85 (IntelliGenetics) and were then manually corrected. The relationships among these aligned sequences were estimated by two methods: maximum-parsimony (MP) and neighbor-joining (NJ) (21). The MP method was applied using the branch and bound model (TBR option) of program PAUP 3.1.1 (22), and the NJ method was applied as implemented by the MEGA program (23). The genetic distances among rpoH regulatory sequences were calculated by using the two-parameter model (24). Robustness of MP and NJ trees was determined by analyzing 100 bootstrap replicates using the PHYLIP (25) and MEGA programs, respectively.
Putative CRP and DnaA regulatory sites as well as other sites were initially identified by the method described by Thieffry et al. (26). Sequences in which no CRP and DnaA sites were identified, were further analyzed by eye searching for sequences that matched any of the three consensus sequences proposed by Messer and Weigel (27). The putative CytR sites were identified by eye, taking advantage of the consensus sequence defined by Pedersen and Valentin-Hansen (28).
Nucleotide sequence accession numbers
Erwinia amylovora (AF127098), C.freundii (AF127099), K.pneumoniae (AF127100), S.enterica serovar Typhimurium (AF127101), S.flexneri (AF127102), Y.frederiksenii (AF127103) and E.coli (AF127104).
RESULTS AND DISCUSSION
DNA nucleotide sequence comparison of the regulatory region of the rpoH homologs
A total of 10 regulatory region sequences of rpoH homologs from different Enterobacteria were analyzed. The seven sequences obtained in this work were from E.coli, S.enterica serovar Typhimurium, S.flexneri, K.pneumoniae, C.freundii, Y.frederiksenii and E.amylovora. Three sequences were downloaded from GenBank: reference strain E.coli K12 strain MG1655 (AE000422), Enterobacter cloacae (D50829) and P.mirabilis (D50830). The reported partial sequence, 122 nt 5′ to the first rpoH codon of C.freundii (X14960), was also downloaded for comparison with the C.freundii sequence of 276 nt obtained in this work. Both sequences were identical in the overlapping region.
DNA sequences of bacteria closely related to E.coli K12, that is E.coli, S.flexneri, S.enterica, K.pneumoniae, E.cloacae and C.freundii, showed a high degree of nucleotide identity (80–98.8%) compared with that of the reference strain. A lower degree of identity was observed for more distant bacteria, Y.frederiksenii (65.3%), P.mirabilis (55.5%) and E.amylovora (53.0%). The G+C content was similar for all sequences (42–46%), with the exception of P.mirabilis in which the G+C content was much lower, 31.3%. These percentages are lower than the G+C content of the rpoH coding region and the genome of these bacteria (29).
To analyze the regulatory region of gene rpoH homologs of different enteric bacteria, the E.coli K12 regulatory region was subdivided into distal and proximal regions. The distal region comprises promoter P1 and the putative transcriptional terminator of operon ftsYEX; while the proximal region comprises promoters P3, P4 and P5, and boxes DnaA1, DnaA2, CRP1, CRP2 and CytR.
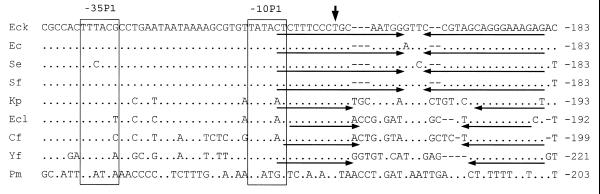
The alignment of the sequences of the rpoH distal regulatory region of the studied bacteria, is shown in Figure 1. Erwinia amylovora was excluded from this analysis since its sequence differs greatly from the reference strain. In all aligned sequences, a putative promoter P1 was identified. In P.mirabilis, the –35 and –10 sites differ from those of E.coli K12 in three bases each. In E.coli, S.enterica and S.flexneri, promoter P1 is located at exactly the same position as the E.coli P1 promoter. In K.pneumoniae, E.cloacae, C.freundii, Y.frederiksenii and P.mirabilis, this promoter is located upstream to that of E.coli K12, in relation to the beginning of the gene. The distance separating the –35 and –10 sites was the same in all the strains. The presence of at least one TAA termination codon located between the putative –35 and –10 sites was detected in all the strains. In addition to these termination codons, inverted repeats representing a putative transcriptional terminator were identified 3′ to the –10 sites. The sequence and position of this terminator are identical in E.coli K12, E.coli, S.enterica and S.flexneri. The length of the inverted repeats is lower and the distance separating both inverted repeats is larger in K.pneumoniae, E.cloacae, C.freundii and Y.frederiksenii (Fig. 1). No similar inverted repeat sequence was identified in the P.mirabilis and E.amylovora distal region. However, it is possible that the E.amylovora and P.mirabilis rpoH regulatory distal sequences obtained were not complete, since the putative transcriptional terminator of operon ftsYEX located upstream of rpoH, was not identified in these sequences.
Figure 1.
Sequence comparison of the distal rpoH regulatory region of different enteric bacteria. Distal rpoH regulatory regions were compared with that of E.coli K12. Sites similar to the –35 and –10 sites of the E.coli K12 promoter P1 are boxed. Vertical arrow, transcription start-site for promoter P1; opposite arrows under the sequences, inverted repeats of the transcriptional terminator of ftsX. Nucleotide positions relative to the +1 of each sequence are indicated on the right. EcK, E.coli K12; Ec, E.coli; Se, S.enterica serovar Typhimurium; Sf, S.flexneri; Kp, K.pneumoniae; Ecl, E.cloacae; Cf, C.freundii; Yf, Y.frederiksenii; Pm, P.mirabilis. Identical residues are represented by a dot and substituted residues are indicated by the relevant letter. Dashed lines, introduced gaps to maximize the alignment.
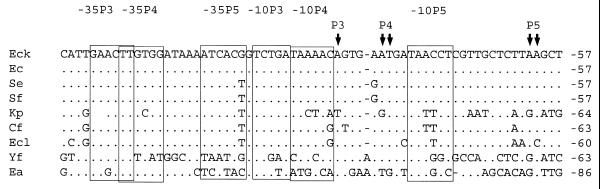
The sequences of the proximal regulatory region of gene rpoH aligned for their analysis were those of E.coli K12, E.coli, S.enterica, S.flexneri, K.pneumoniae, C.freundii, E.cloacae, Y.frederiksenii and E.amylovora (Fig. 2). The sequence of P.mirabilis corresponding to this region differs greatly from those of the other bacteria, and was not included in this alignment.
Figure 2.
Sequence comparison of the proximal regulatory region of rpoH homologs of different enteric bacteria. The –10 and –35 sites similar to the E.coli K12 promoters P3, P4 and P5 are boxed. Ea, E.amylovora. Vertical arrows, transcription start sites. For key, see Figure 1 legend.
In all aligned sequences, a putative σE promoter (P3) was identified (Fig. 2). A similar promoter is also present in the P.mirabilis proximal region; however, it is located at a different position (–10 and –35 sites at –121 and –142, respectively). The presence of this promoter was previously reported by Nakahigashi et al. (9).
The –10 and –35 sites of promoter P4 of E.coli, S.enterica and S.flexneri were identical to that of E.coli K12. In general, the –35P4 sites of the other bacteria presented a lower number of nucleotide changes than those presented by the –10P4 sites. For example, the –35P4 sites of C.freundii, E.cloacae and E.amylovora were identical to that of E.coli K12, while the –10P4 sites differed in 1, 1 and 5 nt, respectively. This conservation may be related to the CRP sites present in these sequences overlapping the –35P4 site.
Sequences similar to the –10 region of promoter P5 of E.coli K12 were found in almost all analyzed bacteria (Fig. 2). A putative –35P5 region should be located at 21 nt 5′ to the –10P5 site of E.coli K12. Similar –35P5 sites were identified in other bacteria, with the exception of Y.frederiksenii and E.amylovora. Sequences similar to promoters P4 and P5 were not identified in the proximal regulatory region of P.mirabilis.
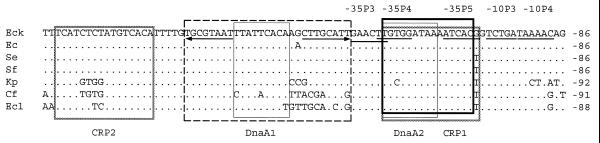
Sequences similar to the DnaA1, DnA2, CRP1, CRP2 and CytR boxes of E.coli K12 were identified at almost the same position in the E.coli, S.enterica, S.flexneri, K.pneumoniae, C.freundii and E.cloacae sequences (Fig. 3). The CytR box overlaps DnaA1, and CRP1 overlaps DnaA2. On the other hand, DnaA2 overlaps the –35P4 site, while CRP1 overlaps the –34P4 and –35P5 sites (4,6,7). In P.mirabilis, one putative DnaA2 box (GTTGATAAA) was identified downstream of the σE promoter. The location of this box suggests that binding of protein DnaA could repress transcription from this promoter, as it does in E.coli K12 (6). Sequences similar to the DnaA, CRP and CytR boxes were not identified in the corresponding region of Y.frederiksenii and E.amylovora. In the sequences in which putative DnaA and CRP boxes were present, an AraC binding site was also identified. The fact that only one putative AraC box was identified and that it overlaps boxes DnaA2 and CRP1 (Fig. 3), suggests that this box could be non-functional.
Figure 3.
Predicted binding sites of regulatory proteins at the regulatory region of the rpoH homologs. CytR, DnaA, AraC and CRP binding sites are boxed with discontinuous, thin, thick and gray lines, respectively. Opposite arrows underline the inverted repeats present in box CytR. The –10 and –35 sites for E.coli K12 promoters P3, P4 and P5 are underlined. For key, see Figure 1 legend.
Phylogenetic relationships among rpoH regulatory regions
The alignment of the regulatory region sequence data of gene rpoH homologs consisted of 307 nucleotide sites for each of the 10 species of the enteric bacteria analyzed. Of these sites, 274 were variable and when sites with gaps were excluded to reduce systematic errors, 166 and 179 sites were variable and informative in the MP and NJ analysis, respectively.
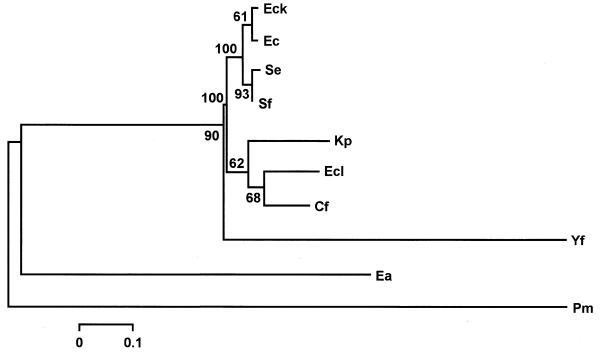
The trees obtained with the MP and NJ analysis were further analyzed by carrying out bootstrap searches with 100 replicates. In both cases, a single tree was obtained. The tree obtained by NJ analysis is shown in Figure 4. The topology of this tree shows three main groups. The first group includes the rpoH regulatory sequences of E.coli K12, E.coli, S.enterica and S.flexneri. The second group includes three species, K.pneumoniae, E.cloacae and C.freundii. Finally, the sequences of E.amylovora and P.mirabilis form a basal group while Y.frederiksenii appears as a sister species of this group. The topology of the tree (Fig. 4) shows that the sequences of the first group exhibited slower evolution at nucleotide level; therefore, this group has shorter branch lengths than the second group. The MP tree obtained after a moderated bootstrap analysis (data not shown), also supports all these relationships among the rpoH regulatory sequences of the enteric bacteria studied.
Figure 4.
Neighbor-joining tree of the regulatory region of rpoH homologs of different enteric bacteria. The number above each fork represents the percentage of 100 bootstrap replicates that supported that branch. For bacterial nomenclature see legends to Figures 1 and 2.
The relationships inferred from the MP and NJ analysis of the regulatory region of rpoH homologs suggests that the regions of E.coli K12, E.coli, S.enterica and S.flexneri conform a closely related group (Fig. 4). Klebsiella pneumoniae, E.cloacae and C.freundii constitute another group that shows a higher nucleotide substitution rate than that of the previous group. Finally, the sequences of Y.frederiksenii, E.amylovora and P.mirabilis were found to be distant to these two groups. Similar relationships among enteric bacteria have been found comparing the conserved 2.1–2.4 coding region of σ32 (9). The relationships among the sequences of the regulatory region of gene rpoH reported in this work are similar to the phylogenetic relationships among enteric bacteria (30). Given that the topology of the tree with regulatory regions reproduces the evolutionary distances among these bacteria, we assume that the evolution of these regulatory regions did not involve major shuffling events in these regions.
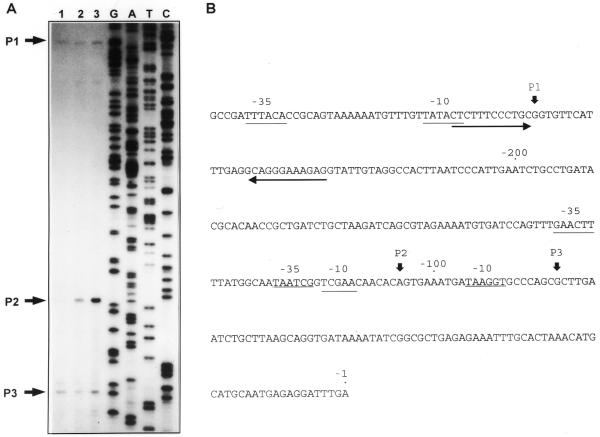
Mapping of rpoH transcription start sites in Y.frederiksenii and P.mirabilis
To determine the presence of functional promoters in the regulatory region of the rpoH genes of Y.frederiksenii and P.mirabilis the 5′-end of the transcript was identified by primer extension analysis. In Y.frederiksenii, three transcription start sites were detected at positions –253 (P1), –105 (P2) and –82 (P3) (Fig. 5A). Transcription from P2 increased ∼10-fold after a 10 min heat shock from 30 to 43°C, while that from P1 and P3 showed only a slight increase (Fig. 5A). The proposed –35 and –10 sites for the putative σ70 promoters P1 and P3, as well as the –35 and –10 sites of the putative σE promoter P2 (Fig. 5B), correspond to those predicted by the sequence comparison analysis shown in Figure 1. Promoters P1, P2 and P3 seem to be homologous to promoters P1, P3 and P5 of E.coli (Figs 1 and 2). However, in Y.frederiksenii the transcripts from the putative P5-like promoter P3 are present in growth conditions in which those of the catabolite-sensitive E.coli P5 promoter are not detected (1,31). This result and the lack of sequences homologous to the CRP box in the rpoH regulatory region of Y.frederiksenii suggest that, in this bacterium, P3 is not under CRP regulation.
Figure 5.
Identification of the transcription start sites in the rpoH gene in Y.frederiksenii. (A) Major products from primer extension experiments are indicated by numbers and black arrows (P1, P2 and P3). Lane 1, cells grown at 30°C; lane 2, cells grown at 30°C and exposed to 43°C for 5 min; lane 3, cells grown at 30°C and exposed to 43°C for 10 min. (B) Sequence of the regulatory region of the Y.frederiksenii rpoH gene. Vertical arrows, transcription start sites. Potential –10 and –35 sequences of the promoters are underlined. Opposite arrows underline the transcriptional terminator of ftsX.
Three transcription start sites were also detected in P.mirabilis. These sites are localized at positions –242 (P1), –121 (P2) and –115 (P3) (Fig. 6A). At 30°C, the predominant transcripts were those corresponding to P1 and P3. A 10 min heat shock from 30–47°C induced a 2-fold and a 10-fold increase in the amount of the P1 and P2 transcripts, respectively. In contrast, under the same experimental conditions, there was a clear decrease in the level of transcription from P3 (Fig. 6A). The proposed –35 site for promoter P1 (Fig. 6B) is the same as that predicted by sequence comparison (Fig. 1). However, in order to optimize the distance between the transcription start site and its –10 box, a different sequence (AATAAT), 2 nt upstream of that shown in Figure 1, is proposed (Fig. 6B). The σE-like –35 (GATATG) and –10 (GCTGG) sites are proposed for P2 instead of the sequences predicted by Nakahigashi et al. (9) and by our sequence analysis. The –10 sequence previously predicted is localized at only 1 nt upstream of the transcriptional start site, while the –10 box proposed after the primer extension analysis is 5 nt upstream of that site. Finally, –10 and –35 sites are proposed for promoter P3 (Fig. 6B), a promoter that was not identified by sequence comparison analysis. Promoters P1, P2 and P3 seem to be homologous to the E.coli promoters P1, P3 and P4. In P.mirabilis, as in E.coli, promoters P2 and P3 and promoters P3 and P4, respectively, are overlapped (Figs 2 and 6B). In both strains a heat shock induces an increase in the level of transcription from the σE promoter and a decrease in that from the overlapped σ70 promoter. Primer extension analysis performed with total RNA from P.mirabilis cells grown in minimal medium with glucose or glycerol did not detect a catabolite-sensitive promoter distal to P3 (data not shown). These results and the lack of CRP-like boxes in the rpoH regulatory region of P.mirabilis suggest that this bacterium does not have an E.coli P5-like promoter.
Figure 6.
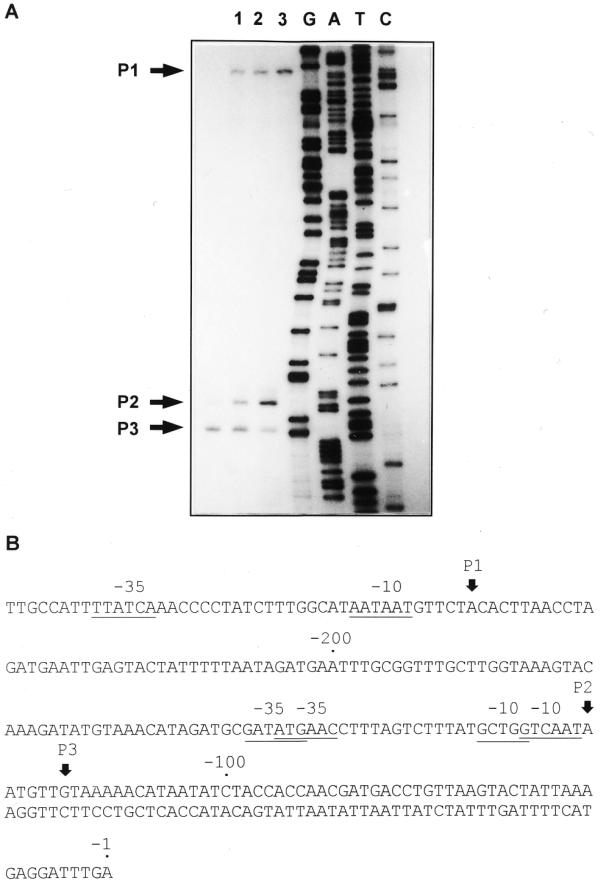
Identification of the transcription start sites in the rpoH gene in P.mirabilis. (A) Major products from primer extension experiments are indicated by numbers and black arrows (P1, P2 and P3). Lane 1, cells grown at 30°C; lane 2, cells grown at 30°C and exposed to 47°C for 5 min; lane 3, cells grown at 30°C and exposed to 47°C for 10 min. (B) Sequence of the regulatory region of the P.mirabilis rpoH gene. For key see Figure 5 legend.
Analysis of the regulatory region sequences of the enteric bacteria rpoH genes, of the phylogenetic study of these sequences and the mapping of the rpoH transcription start sites in Y.frederiksenii and P.mirabilis, suggest that in most enteric bacteria, rpoH should be transcribed from one σE-like promoter and from at least two σ70-like promoters. A common organization of the enteric bacteria regulatory region shows a distal σ70-like promoter up to 200 nt upstream of the initiation codon, a proximal σE-like heat-induced promoter and a second σ70-like promoter localized downstream from this promoter. The two proximal promoters seem to be overlapped in most of the members of the enteric bacteria family, including the more distant member of the family, P.mirabilis. However, in Y.frederiksenii, another distant member, these proximal promoters do not overlap. A third σ70 promoter localized downstream of these two promoters and regulated by CRP, is probably present only on those members of the enteric family closely related to E.coli, i.e. Shigella, Salmonella, Klebsiella, Enterobacter and Citrobacter.
The pattern of regulation of the rpoH gene probably varies slightly among members of the two groups closely related to E.coli (Fig. 4). In these bacteria, transcription from the rpoH promoters P3, P4 and P5, appears to be modulated by various regulatory proteins, DnaA, CytR and CRP. This is probably true mainly for Shigella and Salmonella, since the CRP2 and CytR boxes of the other bacteria of these groups show a sequence with less consensus. The pattern of rpoH regulation could show higher differences for bacteria of the third group, Y.frederiksenii, E.amylovora and P.mirabilis. It is important to mention that E.amylovora is a plant bacterium and that P.mirabilis has a lower G+C content compared with the other enteric bacteria.
To better elucidate the mechanisms governing the transcription of gene rpoH in E.coli K12 and other enteric bacteria, a direct experimental approach is necessary. This approach must be focused mainly on the regulatory mechanisms that couple rpoH transcription of different promoters, to the carbon source, cellular response to several stress conditions and probably DNA replication.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to M. Paéz for preparing bacterial media, A. M. Huerta for running the programs to search for binding sites, J. Membrillo-Hernández for advice and support in primer extension experiments and I. Pérez-Montfort for advice on manuscript style. This study was partially supported by grants 3086P-N9607 and 27980-N from CONACyT, México.
DDBJ/EMBL/GenBank accession nos AF127098–AF127104
References
- 1.Gross C.A. (1996) Function and regulation of the heat shock proteins. In Neidhardt,F.C., Curtiss,R.,III, Ingraham,J.L., Lin,E.C.C., Low,K.B., Magasanik,B., Reznikoff,W.S., Riley,M., Schaechter,M. and Umbarger,H.E. (eds), Escherichia coli and Salmonella. Cellular and Molecular Biology, 2nd Edn. ASM Press, Washington, DC, Vol. 1, pp. 1382–1399.
- 2.Yura T. and Nakahigashi,K. (1999) Regulation of the heat-shock response. Curr. Opin. Microbiol., 2, 153–158. [DOI] [PubMed] [Google Scholar]
- 3.Rouvière P.E., De Las Peñas,A., Mecsas,J., Lu,C.Z., Rudd,K.E. and Gross,C.A. (1995) rpoE, the gene encoding the second heat-shock sigma factor, σE, in Escherichia coli. EMBO J., 14, 1032–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagai H., Yano,R., Erickson,J.W. and Yura,T. (1990) Trancriptional regulation of the heat shock regulatory gene rpoH in Escherichia coli: involvement of a novel catabolite-sensitive promoter. J. Bacteriol ., 172, 2710–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gill D.R., Hatfull,G.F. and Salmond,G.P.C. (1986) A new cell division operon in Escherichia coli. Mol. Gen. Genet., 205, 134–145. [DOI] [PubMed] [Google Scholar]
- 6.Wang Q. and Kaguni,J.M. (1989) DnaA protein regulates transcription of the rpoH gene of Escherichia coli. J. Biol. Chem., 264, 7338–7344. [PubMed] [Google Scholar]
- 7.Kallipolitis B.H. and Valentin-Hansen,P. (1998) Transcription of rpoH, encoding the Escherichia coli heat-shock regulator σ32, is negatively controlled by the cAMP-CRP/CytR nucleoprotein complex. Mol. Microbiol., 29, 1091–1099. [DOI] [PubMed] [Google Scholar]
- 8.Fleischmann R.D., Adams,M.D., White,O., Clayton,R.A., Kirkness,E.F., Kerlavage,A.R., Bult,C.J., Tomb,J.-F., Dougherty,B.A., Merrick,J.M. et al. (1995) Whole-genome random sequencing and assembly of the Haemophilus influenzae Rd. Science, 269, 496–512. [DOI] [PubMed] [Google Scholar]
- 9.Nakahigashi K., Yanagi,H. and Yura,T. (1995) Isolation and sequence analysis of rpoH genes σ32 homologs from gram negative bacteria: conserved mRNA and protein segments for heat shock regulation. Nucleic Acids Res., 23, 4383–4390. [PMC free article] [PubMed] [Google Scholar]
- 10.Wu J. and Newton,A. (1996) Isolation, identification, and transcription specificity of the heat shock sigma factor sigma 32 from Caulobacter crescentus. J. Bacteriol ., 178, 2094–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Narberhaus F., Krummenacher,P., Fischer,H.M. and Hennecke,H. (1997) Three disparately regulated genes for σ32-like transcription factors in Bradyrhizobium japonicum. Mol. Microbiol., 21, 93–104. [DOI] [PubMed] [Google Scholar]
- 12.Sahu G.K., Chowdhury,R. and Das,J. (1997) The rpoH gene encoding σ32 homolog of Vibrio cholerae. Gene, 189, 203–207. [DOI] [PubMed] [Google Scholar]
- 13.Karls R.K., Brooks,J., Rossmeissl,P., Luedke,J. and Donohue,T.J. (1998) Metabolic roles of a Rhodobacter sphaeroides member of the σ32 family. J. Bacteriol ., 180, 10–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramírez-Santos J. and Gómez-Eichelmann,M.C. (1998) Identification of σ32-like factors and ftsX-rpoH gene arrangements in enteric bacteria. Can. J. Microbiol., 44, 565–568. [PubMed] [Google Scholar]
- 15.Segal G. and Ron,E.Z. (1996) Regulation and organization of the groE and dnaK operons in eubacteria. FEMS Microbiol. Lett., 138, 1–10. [DOI] [PubMed] [Google Scholar]
- 16.Baldini R.L., Avedissian,M. and Lopes,S. (1998) The CIRCE element and its putative repressor control cell cycle expression of the Caulobacter crescentus groESL operon. J. Bacteriol ., 180, 1632–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naczynski Z.N., Mueller,C. and Kropinski,A.M. (1995) Cloning the gene for the heat shock response positive regulator (sigma 32 homolog) from Pseudomonas aeruginosa. Can. J. Microbiol., 41, 75–87. [DOI] [PubMed] [Google Scholar]
- 18.Wu J. and Newton,A. (1997) The Caulobacter heat shock sigma factor gene rpoH is positively autoregulated from a σ32-dependent promoter. J. Bacteriol ., 179, 514–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakahigashi K., Yanagi,H. and Yura,T. (1998) Regulatory conservation and divergence of σ32 homologs from gram-negative bacteria: Serratia marcescens, Proteus mirabilis, Pseudomonas aeruginosa, and Agrobacterium tumefaciens. J. Bacteriol ., 180, 2402–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Saitou N. and Nei,M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol., 4, 406–425. [DOI] [PubMed] [Google Scholar]
- 22.Swofford D. (1993) PAUP: Phylogenetic Analysis Using Parsimony, version 3.1.1. Illinois Natural History Survey, Champaign, IL.
- 23.Kumar S., Tamura,K. and Nei,M. (1994) MEGA: molecular evolutionary genetics analysis software for microcomputers. Comput. Appl. Biosci., 10, 189–191. [DOI] [PubMed] [Google Scholar]
- 24.Kimura M. (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol., 16, 111–120. [DOI] [PubMed] [Google Scholar]
- 25.Felsestein J. (1993) PHYLIP: Phylogenetic Inference Package, version 3.5c. University of Washington, Seattle, WA.
- 26.Thieffry D., Salgado,H., Huerta,A.M. and Collado-Vides,J. (1998) Prediction of transcriptional regulatory sites in the complete genome sequence of Escherichia coli K12. Bioinformatics, 14, 391–400. [DOI] [PubMed] [Google Scholar]
- 27.Messer W. and Weigel,C. (1997) DnaA initiator – also a transcription factor. Mol. Microbiol., 24, 1–6. [DOI] [PubMed] [Google Scholar]
- 28.Pedersen H. and Valentin-Hansen,P. (1997) Protein-induced fit: the CRP activator protein changes sequence-specific DNA recognition by the CytR repressor, a highly flexible LacI member. EMBO J., 16, 2108–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brenner D.J. (1984) Family I. Enterobacteriaceae. In Krieg,N.R. and Holt,J.G. (eds), Bergey’s Manual of Systematic Bacteriology. The Williams & Wilkins Co., Baltimore, MD, Vol. 1, pp. 408–516.
- 30.Ochman H. and Wilson,A.C. (1987) Evolutionary history of the enteric bacteria. In Neidhardt,F.C., Ingraham,J.L., Low,K.B., Magasanik,B., Schaechter,M. and Umbarger,H.E. (eds), Escherichia coli and Salmonella typhimurium. Cellular and Molecular Biology. ASM Press, Washington, DC, Vol. 1, pp. 1649–1654.
- 31.Erickson J.W., Vaughn,V., Walter,W.A., Neidhardt,F.C. and Gross,C.A. (1987) Regulation of the promoters and transcripts of rpoH, the Escherichia coli heat shock regulatory gene. Genes Dev., 1, 419–432. [DOI] [PubMed] [Google Scholar]