Abstract
Olfactomedin-4 (OLFM-4) is an extracellular matrix protein that is highly expressed in human endometrium. We have examined the regulation and function of OLFM-4 in normal endometrium and in cases of endometriosis and endometrial cancer. OLFM-4 expression levels are highest in proliferative-phase endometrium, and 17β-estradiol up-regulates OLFM-4 mRNA in endometrial explant cultures. Using the luciferase reporter under control of the OLFM-4 promoter, it was shown that both 17β-estradiol and OH-tamoxifen induce luciferase activity, and epidermal growth factor receptor-1 is required for this estrogenic response. In turn, EGF activates the OLFM-4 promoter, and estrogen receptor-α is needed for the complete EGF response. The cellular functions of OLFM-4 were examined by its expression in OLFM-4-negative HEK-293 cells, which resulted in decreased vimentin expression and cell adherence as well as increased apoptosis resistance. In cases of endometriosis and endometrial cancer, OLFM-4 expression correlated with the presence of epidermal growth factor receptor-1 and estrogen receptor-α (or estrogen signaling). An increase of OLFM-4 mRNA was observed in the endometrium of endometriosis patients. No change in OLFM-4 expression levels were observed in patients with endometrial cancer relative with controls. In conclusion, cross-talk between estrogen and EGF signaling regulates OLFM-4 expression. The role of OLFM-4 in endometrial tissue remodeling before the secretory phase and during the predisposition and early events in endometriosis can be postulated but requires additional investigation.
In a recent gene expression profiling study we compared the expression of genes in late proliferative-phase endometrium to that of menstrual-phase endometrium in an attempt to identify genes that are potentially regulated by 17β-estradiol during endometrial growth and differentiation.1 One of the genes found to be up-regulated in late proliferative-phase endometrium but not reported in the previously published1 short list of up-regulated genes, was olfactomedin-4 (OLFM-4).
The OLFM-4 gene, also called human granulocyte colony stimulating clone-1 or GW112, is located on chromosome 13q21.1 and encodes for the 510-amino acid-long (55 kDa) extracellular matrix protein OLFM-4.2,3 National Center for Biotechnology Information databases4 revealed that OLFM-4 is expressed in human bone marrow, prostate, breast, pancreas, stomach, and colon.2,3,4 Moreover, this protein is overexpressed in gastrointestinal cancers,4,5,6,7,8,9,10 in inflamed colonic mucosa11 and in gastric biopsies from patients infected with Helicobacter pylori.12 Transcript levels of OLFM-4 are also higher in breast tumors compared to normal breast.4,13
Several authors have investigated the cellular functions of OLFM-4, but results do not always agree. Zhang et al4 overexpressed OLFM-4 cDNA in HEK-293 cells and described an antiapoptotic action of this protein. In another study on pancreatic cancer cells, down-regulation of OLFM-4 by small interfering RNA resulted in impaired S-phase transition and inhibition of proliferation, but no effect on apoptosis was observed.14 Liu et al3 used HEK-293 and NIH3T3 cells and showed that OLFM-4 directly binds to lectins and E-cadherin and leads to increased cell-cell interaction. The same authors in a separate study using myeloma cells, showed that OLFM-4 represses both motility and cell adherence.9 Therefore, this protein seems to play important roles in cell cycle control, cell adherence and motility, cell-cell interaction, and tissue architecture but probably its role may depend on the tissue type.
Because of the important role in multiple cellular functions and because of its strong association with estrogen exposure in human endometrium, we further investigated the regulation and the potential role of OLFM-4 in this tissue. We examined its expression throughout the menstrual cycle and studied its responsiveness to 17β-estradiol. In addition, we investigated the regulation of the OLFM-4 promoter using a luciferase reporter system. These experiments showed the OLFM-4 promoter is responsive to both 17β-estradiol and OH-tamoxifen and that this regulation depends on the cross-talk with the epidermal growth factor receptor (EGFR)-1. In OLFM-4-negative cells, its re-expression decreased vimentin level, cell adherence and increased resistance to apoptosis. OLFM-4 expression was also observed in clinical samples of endometriosis and endometrial cancer patients and correlated with the presence of estrogen receptor-α (ER-α) and EGFR1, thus confirming the observation obtained with the luciferase experiments. In addition, since OLFM-4 is overexpressed in the eutopic endometrium of endometriosis patients, our results suggest a possible role of OLFM-4 in the early events of disease onset.
Materials and Methods
Human Endometrial Tissue
Normal Pre- and Postmenopausal Endometrial Samples
Endometrial tissue was collected from healthy volunteers (n = 48) of 26–52 years of age with regular menstrual cycle. Twelve samples were collected during the menstrual phase (cycle day (CD) 1–5); 18 were collected during the proliferative phase, subdivided into early (CD 6–10, n = 9) and late (CD 11–14, n = 9); and 18 samples were collected in the secretory phase: early secretory phase (CD 15–19, n = 8), mid-secretory phase (CD 20–24, n = 8) and from the late secretory phase (CD 25–28, n = 2). The endometrial tissues were dated based on the start of the last menstrual period and confirmed by histological examination of the tissue by a pathologist as described previously.15 All women were documented not to be pregnant.
In addition, endometrium was obtained from 13 postmenopausal women. Pre- and postmenopausal tissue was collected from hysterectomy specimens or by pipelle biopsies during laparoscopy (Pipelle catheter; Unimar, Prodimed, Neuilly-Enthelle, France) in women who underwent surgery for benign indications other than endometriosis. After macroscopic inspection for abnormalities by a pathologist, tissue was transported to the laboratory. Portions of each sample were fixed in 10% buffered formalin for histological examinations, frozen in liquid nitrogen for DNA, RNA, and protein isolation, and frozen by immersion in isopentane (−80°C) for cryosectioning and microscopy. Tissue from the menstrual and proliferative phase endometrium was also used for the preparation of explant cultures (see below).
Endometriosis Tissue
An independent group of biopsies was used for the endometriosis study and consisted of eutopic and ectopic endometrial tissues from endometriosis patients and eutopic tissues from controls. Biopsies were collected at the University Hospital Gasthuisberg Leuven (Leuven, Belgium), and clinicopathological characteristics of these patients and controls have been described previously.16 In brief, controls consisted of 20 normal endometrial tissues from women with regular menstrual cycle undergoing surgery for benign indications and found free of endometriosis at laparoscopy/laparotomy. Samples of eutopic and ectopic endometrial tissue from endometriosis patients were collected from 14 women. Patients had moderate to severe disease, classified according to the revised system of the American Society for Reproductive Medicine.17 All samples were collected during the proliferative phase of the menstrual cycle as determined by the start of the last menstrual period and confirmed by histological examination by the pathologist.15
Endometrial Cancers and Controls
Endometrial carcinomas were primary and type I. Cancer samples (n = 9; age at diagnosis between 50 and 81; mean age: 64; additional clinical features are listed in Table 1) and postmenopausal endometrial control tissues with no malignancy (n = 14; age range: 50–80; mean age: 65) were collected from women undergoing hysterectomy at the Maastricht University Medical Centre (Maastricht, The Netherlands). Tissues were fixed in formalin for immunohistochemistry and immediately frozen in liquid nitrogen for RNA/DNA/protein isolation. A pathologist confirmed the presence of endometrial cancer in all specimens and the grade and stage of the disease (International Federation of Gynecology and Obstetrics criteria 2009).18
Table 1.
Clinical Features of Endometrial Cancer (according to the International Federation of Gynecology and Obstetrics Criteria 2009)18
| Specimen | Age at diagnosis | Stage | Grade | |
|---|---|---|---|---|
| Maastricht Medical Centre prospective collection | 1 | 50 | II | 3 |
| 2 | 58 | IA | 2 | |
| 3 | 61 | IB | 2 | |
| 4 | 62 | IB | 2 | |
| 5 | 62 | IB | 2 | |
| 6 | 67 | IB | 1 | |
| 7 | 70 | IA | 1 | |
| 8 | 76 | IVB | 1 | |
| 9 | 81 | IA | 1 | |
| PALGA (pathological anatomy national automated archive) collection | 1 | 54 | IA | 1 |
| 2 | 59 | IA | 2 | |
| 3 | 60 | III | 3 | |
| 4 | 62 | IB | 3 | |
| 5 | 68 | IA | 2 | |
| 6 | 68 | IA | 1 | |
| 7 | 68 | IA | 1 | |
| 8 | 69 | IB | 2 | |
| 9 | 71 | IA | 2 | |
| 10 | 78 | II | 3 | |
| 11 | 79 | IB | 2 | |
| 12 | 90 | IB | 2 | |
| 13 | PM∗ | IA | 2 | |
| 14 | PM∗ | IB | 1 | |
| 15 | PM∗ | IA | 3 | |
| 16 | PM∗ | IB | 1 | |
| 17 | PM∗ | IA | 2 | |
| 18 | PM∗ | IB | 3 | |
| 19 | PM∗ | IA | 1 | |
| 20 | PM∗ | IB | 1 | |
| 21 | PM∗ | IB | 1 |
The precise age at diagnosis was not recorded but was documented to be after menopause.
An additional group of endometrial cancers was used for immunohistochemistry analysis only and consisted of archival material selected from the Dutch national pathology database (PALGA), pathological anatomy national automated archive and described in an earlier study.19 For the present investigation, 21 type I endometrial cancer biopsies were used. Clinical features according to the International Federation of Gynecology and Obstetrics criteria 200918 are shown in Table 1. All specimens used in the present study originated from primary tumors. Nine patients developed recurrent disease.
All women prospectively enrolled in the present study did not receive steroid medications during the six months before tissue collection. All women agreed to participate in the study by signing an informed consent, according to a protocol approved by the local Medical Ethical Committee.
Steroid Hormones and Chemicals
OH-tamoxifen and 17β-estradiol were purchased from Sigma-Aldrich Chemie (Zwijndrecht, The Netherlands). ICI-164384 was a gift from Schering-Plough (Oss, The Netherlands). Human recombinant EGF was purchased from R&D Systems (Minneapolis, MN). The EGFR1 and erythroblastic leukemia viral oncogene homolog 2 (ERBB2) inhibitor GW2974 was purchased from Sigma-Aldrich Chemie. The inhibitors SB20190 (p38 mitogen-activated protein kinase (MAPK)), PD98059 (extracellular signal-regulated kinase 1), U-0126 (mitogen-activated protein kinase kinase (MEK)1/2), and Ly294002 (phosphatidylinositol 3-kinase (PI3K)) were purchased from Calbiochem (La Jolla, CA).
Explant Cultures
After collection, menstrual (n = 8) and late proliferative endometrium (n = 8) used for explant cultures was immediately transported to the laboratory in Dulbecco’s modified Eagle’s medium/Ham’s F12 medium (Life Technologies, Grand Island, NY) on ice. Explant cultures were prepared as described by Punyadeera et al.20
Cell Culture
Human endometrial cancer cell line ECC1, embryonic kidney cell line HEK-293, breast cancer cell line T47D, and cervical cancer cell line HeLa were purchased from the American Type Culture Collection (Manassas, VA). Cell maintenance, transfection, luciferase assay, and other in vitro procedures have been described previously.21,22
Plasmids
The expression plasmid for the ER-α was a gift from Prof. R. Schüle (Freiburg University, Germany) and has been described elsewhere.23 Expression plasmids for the EGFR1 and the EGFR-dominant negative variant (CD-533) were gifts from Dr. J. Theys (Maastricht University) and are described elsewhere.24,25
Luciferase Reporter Plasmids Driven by OLFM-4 Promoter
Nucleotide numbering refers to the study by Chin et al.26 The 1.3-kb fragment at the 5′ of OLFM-4 was cloned by PCR (TaqDNA polymerase; Fermentas, St. Leon-Rot, Germany) as follows: two PCR fragments corresponding to the nucleotides −1357/−606 (primers OLM-1/OLM-2; Table 2) and −816/+24 (primers OLM-3/OLM-4; Table 2) were amplified, cloned into vector pGEM-T-easy (Promega, Madison, WI), and reassembled after digestion with restriction enzyme BglII (−779). The complete 1.3-kb promoter or the 0.8-kb promoter proximal to the start site of transcription (fragment generated with primers OLM-3/OLM-4) was cloned in pGL3 basic vector (Promega). To delete the activator protein 1 (AP1) site, the 137-bp EcoRV/NcoI fragment (nucleotides −56/−192) from the 1.3-kb promoter vector (EcoRV lies 46 bp at the 5′ of the AP1 site) was substituted with the 47-bp EcoRV/NcoI fragment corresponding to nucleotides −56/−102, located at the 3′ of the AP1 site and therefore missing the AP1 site. This fragment was generated by PCR (primers OLM-5/OLM-6; Table 2), and the EcoRV site was included by primer extension at the 3′ of the fragment (primer OLM-5).
Table 2.
Primers Used for Cloning
| Name | Sequence | Cloned fragments |
|---|---|---|
| Cloning | ||
| OLM-1 | 5′-GACAAGCTTACTCTGTCACCCATGCTGCAGTG-3′ | −1357/−606 |
| OLM-2 | 5′-GAGAAAACACCCAAATAAACAAAG-3′ | |
| OLM-3 | 5′-TGCATCTTATTTCTCAGTTTTATG-3′ | −816/+24 |
| OLM-4 | 5′-TGTTTTTGGCGTCTTCCATCTTGTCCTCTTAGCTGGAGC-3′ | |
| OLM-5 | 5′-GGTGTTCTAGATATCGCAGTTCACAGTTCCCTGGC-3′ | AP1 site deletion |
| OLM-6 | 5′-GTGATTTCCCCATGGCCAGCAT-3′ | |
| OLM-7 | 5′-AAGAATTCCACCATGAGGCCCGGCCTCTCATTTCT-3′ | cDNA 5′ |
| OLM-8 | 5′-GATTTCTCGGCGAATGGCAAGG-3′ | |
| OLM-9 | 5′-GCTGGAGGTGGAGATAAGAAATATG-3′ | cDNA 3′ |
| OLM-10 | 5′-CTCCTCGAGACATTTACTGGGGCTTCTGCAAGACAG-3′ | |
| OLM-11 | 5′-CTAAACAGGATCCCTGGGGCTTCTGCAAGACAG-3′ | cDNA 3′ TAA-less |
| PY | 5′-CAGGGATCCATGGAATATATGCCAATGGAAATGGAATATATG CCAATGGAACATATATGA-3′ | PY-tag |
| Real-time PCR | ||
| EGFR1 | 5′-GGGCTCTGGAGGAAAAGAAA-3′ | EGFR1 expression |
| EGFR2 | 5′-AAATTCCCAAGGACCACCTCA-3′ |
OLM-4 Expression Plasmids
OLFM-4 cDNA was amplified by PCR (TaqDNA polymerase; Fermentas) as two fragments (nucleotides −12/+648 with primer pairs OLM-7/OLM-8 and nucleotides + 558/+ 1546 with OLM-9/OLM-10; numbering from accession NCBI NM_006418: +1 = translation start site; stop codon: 1531; primers are listed in Table 2). PCR fragments were cloned in pGEM-T-easy (Promega) and the complete cDNA was recomposed using HindIII restriction site (nucleotide + 597). Restriction sites EcoRI (−9) and XhoI (+1538) were included by primer extension at the 5′ and 3′ respectively of the cDNA, and used to clone the complete cDNA in pCDNA3.1 (Invitrogen, Breda, The Netherlands). Given that two EcoRI sites (−9 and +84) are present in the 5′ part of the cDNA, partial digestion was performed. To introduce the PY tag sequence, the BamHI restriction site was introduced at the 3′ of the OLFM-4 cDNA before the stop codon (nucleotide +1531) using primer OLM-11 in combination with primer OLM-9 (Table 2). The BamHI was used to clone the PY sequence generated by oligo PY (Table 2). All plasmids were checked by sequencing.
Total RNA Extraction and cDNA Synthesis
Total RNA was extracted from explant cultures and endometrial tissues using the SV total RNA isolation kit (Promega), according to the manufacturer’s protocol. RNA from the cell lines was isolated using the TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA) as recommended by the manufacturer. cDNA was synthesized using the Moloney murine leukemia virus reverse transcriptase (Invitrogen Life Technologies) as described earlier.21,22
Real-Time PCR
Primers and probes for OLFM-4 (GW112: Hs00197437-m1) were purchased from Applied Biosystems (Foster City, CA) as a predeveloped assay. EGFR1 was amplified using the Sybr Green ABGene system (ABGene, Epsom, UK), as recommended by the manufacturer. Primers EGFR1/EGFR2 (Table 2) purchased from MWG Biotech (Ebersberg, Germany) were used. Primers for TFF1, and for the house-keeping genes β-actin and cyclophilin, have been published previously.21 All real time-PCRs were performed using the BioRad MyIQ apparatus. Fold changes in the gene expression were assessed by comparing the ct values between the gene of interest and two housekeeping genes (β-actin and cyclophilin).
Additional Molecular Biology and Biochemistry
All molecular biology techniques were performed using standard protocols and have been described previously.21,22 For Western blots of OLFM-4, primary rabbit polyclonal antibody (1/500, IMG-5983; Imgenex, San Diego, CA) was used; EGFR1 was visualized with rabbit polyclonal antibody AB-6 (1/100; Oncogene, San Diego, CA); ERBB2 and ER-α were visualized with mouse monoclonal antibodies CB11 (1/250; GenWay Biotech, San Diego, CA) and rabbit polyclonal HC20 (1/1000; Santa Cruz Biotechnology, Heidelberg, Germany), respectively. Rabbit polyclonal antibodies against Akt, phospho-Akt (Ser473), phospho-p42/44 (Thr202/Tyr204) and phospho-p38 MAPK (Thr180/Tyr182) were purchased from Cell Signaling Technology (Danvers, MA) and used according to the manufacturer’s recommendations. As loading control, mouse monoclonal anti-human β-actin antibody AC-15 (Sigma-Aldrich Chemie) was used. For Western blotting using the kinase antibodies, cells were starved for 24 hours in medium without any fetal calf serum and supplemented with 0.1% bovine serum albumin before stimulation.
Proliferation, Apoptosis, and Adherence Assay
Cell viability was assessed with the 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide assay (Sigma-Aldrich Chemie) as described earlier.22 The cell cycle was analyzed using flow cytometry after propidium iodide staining as previously described22 with aid of the computer programs WinMDI 2.9 and Cylchred. Cell apoptosis was measured by flow cytometry after labeling the apoptotic cells with the M30 antibody (Roche Diagnostics Nederland, Almere, The Netherlands) as described previously.27
Cell adherence was assessed by plating equal numbers of cells in a 24-well plate and allowing cells to attach to the surface for different periods of time (as indicated). Wells were rinsed with PBS and the number of viable adhering cells was assessed with the 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide assay.
Immunohistochemistry, Fluorescence, and Confocal Laser Microscopy
Paraffin sections were stained using standard protocols. Sections were dewaxed, and endogenous peroxidases were blocked. Pepsin was used as antigen retrieval method for OLFM-4 and EGFR1, whereas Tris-EDTA buffer was used to retrieve the antigen for staining with ER-α. Antibodies IMG-5983 (1/40 rabbit polyclonal; Imgenex), 31G7 (1/20, monoclonal; Zymed/Invitrogen, Breda, The Netherlands), and D-5 (1/100, monoclonal; DakoCytomation, Glostrup, Denmark) were used to detect OLFM-4, EGFR1, and ER-α, respectively. Chemate Envision and 3,3-diaminobenzidine solution (DakoCytomation) were used to visualize antibody binding.
Immunofluorescence staining for microscopy and for confocal laser scanning microscopy was performed on cells plated on microscope coverglasses. Cells were fixed in 4% paraformaldehyde (10 minutes at room temperature) followed by permeabilization with PBS supplemented with 0.1% Tween 100.
OLFM-4 was stained with the IMG-5983 antibody (1:40) and vimentin with the M0725 antibody (1/500, monoclonal; DakoCytomation). Mouse-α-2PY was used to detect the PY tag. Goat-α-mouse IgG-FITC (1/100; DakoCytomation) and swine-anti-rabbit IgG-FITC (1/100; DakoCytomation) were used as secondary antibodies. For the co-staining protocols we combined the swine-anti-rabbit-FITC antibody (1/100; DakoCytomation) and goat-anti-mouse-Texas Red antibody (1/80; ITK Diagnostics, Uithoorn, The Netherlands).
Confocal laser scanning microscopy was performed using the Leica CTR 4000/CTC SPE and the Leica Application Suite/advance fluorescence software package (Leica Microsystems, Rijswijk, The Netherlands). The Image-J program was used to make three-dimensional images.
Accession Numbers
National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov, last accessed September 15, 2010) entries for human OLFM-4 gene name: OLFM4 (additional names GC1, OLM4, OlfD, GW112, human granulocyte colony stimulating clone-1; hOLfD, UNQ362, KIAA4294, bA209J19.1, and OLFM4) (GeneID: 10562).
Results
OLFM-4 Expression in the Endometrium During the Menstrual Cycle
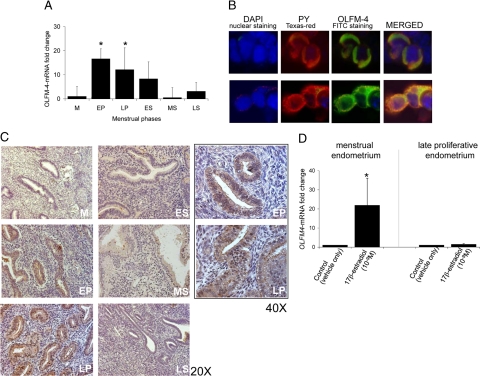
Expression of OLFM-4-mRNA is significantly increased in the early and late proliferative phase compared with the menstrual-phase endometrium and decreases after ovulation (Figure 1A). For the immunohistochemical staining of OLFM-4, the antibody clone IMG-5983 was used. Since this antibody was used for Western blot only, its specificity to detect the correct protein in whole cells was confirmed in the HEK-293 cells transfected with the OLFM-4 cDNA fused to a PY tag. Antibody IMG-5983 and the anti-PY antibody colocalized perfectly in costaining experiments (Figure 1B), and therefore, IMG-5983 was further used.
Figure 1.
OLFM-4 expression in the human endometrium. A: OLFM-4 mRNA expression in healthy cyclic endometrium. M, menstrual phase (n = 12); EP, early proliferative (n = 9); LP, late proliferative (n = 9); ES, early secretory (n = 8); MS, midsecretory (n = 8); and LS, late secretory (n = 2). mRNA levels were determined by quantitative real-time PCR. ∗P < 0.05 versus menstrual phase (Mann-Whitney U-test). B: Specificity of the IMG-5983 antibody for OLFM-4. We confirm perfect colocalization of OLFM-4 (polyclonal IMG-5983, Swine-anti-rabbit-FITC, green) and the PY tag (mouse-anti-2PY MMS-115R, goat-anti-mouse-Texas Red, red). Magnification ×40. C: OLFM-4 protein expression during the menstrual cycle determined by immunohistochemistry. M, menstrual phase; EP, early proliferative; LP, late proliferative; ES, early secretory; MS, midsecretory; and LS, late secretory. D: OLFM-4 mRNA expression in explant cultures of menstrual (n = 8) and late proliferative-phase endometrium (n = 8) exposed for 24 hours to vehicle (0.1% ethanol) or 17β-estradiol (1 nmol/L). mRNA levels were determined by quantitative real-time PCR. ∗P < 0.05 (Wilcoxon signed-rank test).
In the human endometrium, OLFM-4 protein is localized in the cytoplasm and the cell membrane of the glandular epithelium, although some immunoreactivity in the stroma cells is also observed (Figure 1C). Its expression is highest in the early and late proliferative phase (Figure 1C).
Using endometrial explant cultures, we confirmed the estrogen dependence of OLFM-4: its mRNA level is induced significantly by 17β-estradiol in explant cultures prepared from menstrual endometrium but not in explants prepared from late proliferative endometrium (Figure 1D; see Discussion).
Regulation of the OLFM-4 Gene Promoter
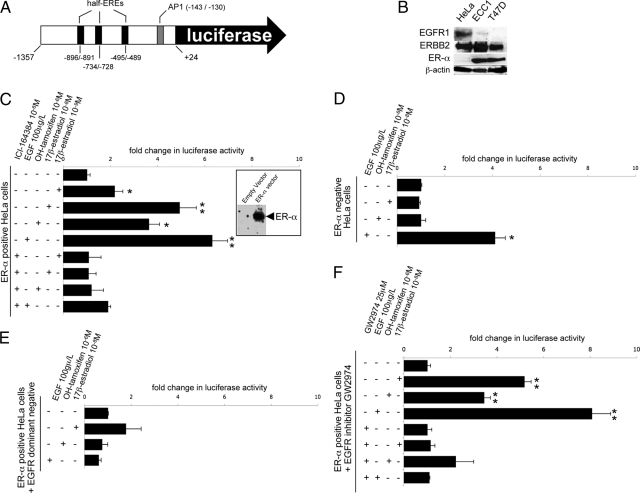
To further investigate the transcriptional regulation of OLFM-4, we cloned the 1.3-kb fragment upstream of the transcription start site in front of the luciferase gene (Figure 2A).26 The OLFM-4 promoter does not contain any canonical estrogen response element (ERE), but instead it has three half EREs (AGGTCA) and an AP1 site (TGACTCA). This construct was used for transfection of ER-α positive cell lines ECC1 (endometrial cancer) and T47D (breast cancer) as well as for transfection of two ER-α negative cell lines (endometrial cancer RL95.2 and cervical cancer HeLa) along with the ER-α expression plasmid (results not shown). The ER-α status for ECC1, T47D and HeLa cells, further used in this study, is shown in Figure 2B.
Figure 2.
OLFM-4 is regulated by estrogen and EGFR signaling. A: Scheme of the 1.3-kb OLFM-4 promoter that was cloned in front of the luciferase reporter. Nucleotide numbering as in the study of Chin et al.26 B: Western blot analysis for ER-α and EGFR1 and ERBB2 in HeLa, ECC1, and T47D cells. C–F: Luciferase response in HeLa cells transfected with the OLFM-4 promoter-luciferase along with: the expression plasmid for ER-α (the panel on the right shows ER-α expression after transfection with ER-α plasmid or with an empty vector) (C), and the ICI-164384 alone had no effect on the luciferase activity (not in figure); the empty vector (without the expression plasmid for ER-α) (D); the ER-α expression plasmid and a dominant-negative variant for of the EGFR (CD-533) (E); and the expression plasmid for ER-α in presence or absence of the EGFR inhibitor GW2974 (F). Bars indicate mean values (n = 3) ± SD. P values were calculated with the t t-test; ∗P <0.05; ∗∗P <0.01. All results were repeated in at least two independent experiments.
Surprisingly, the OLFM-4 promoter responds to estrogens in HeLa cells only after co-transfection with the OLFM-4-luciferase and the ER-α expression plasmid. Both 17β-estradiol and OH-tamoxifen induce luciferase activity and this response is antagonised by the anti-estrogen ICI-134384 (Figure 2C).
Given that an agonistic action of OH-tamoxifen is frequently associated with cross talks between ER-α and kinase pathways, and given that AP1 binding sites (activated by kinase cascades) are frequently functionally linked to half-EREs (both AP1 and half-EREs are present in the OLFM-4 3′; Figure 2A),26 we examined whether growth factor signaling is involved in the regulation of OLFM-4. EGF but not insulin-like growth factor (IGF) (results not shown) strongly enhances OLFM-4 promoter luciferase activity (Figure 2C). Interestingly, ICI-164384 impairs the response to EGF, besides the inhibition of the estrogen signaling.
The interaction between the ER-α and EGFR signaling pathways was further explored. ER-α is necessary for the estrogenic response of OLFM-4, because when ER-α is not cotransfected into the HeLa cells, the responses to 17β-estradiol and OH-tamoxifen are completely abolished (Figure 2D). Under these circumstances, also the response to EGF is partly impaired (∼50%; Figure 2D), indicating that ER-α is required for the full EGF response. This is further illustrated by the fact that the ER antagonist ICI-164384 abolishes the EGF-mediated response (Figure 2C).
Similarly, EGF signaling is needed for the estrogen response. In HeLa cells cotransfected with a dominant negative variant of EGFR (EGFR-CD-533), the activation of the OLFM-4 promoter by EGF, 17β-estradiol and OH-tamoxifen is impaired (Figure 2E). To confirm this observation, a general inhibitor of the membrane EGFR family (GW2974) was used. Pretreating HeLa cells with GW2974 inhibits the response to EGF as well as to 17β-estradiol and OH-tamoxifen (Figure 2F).
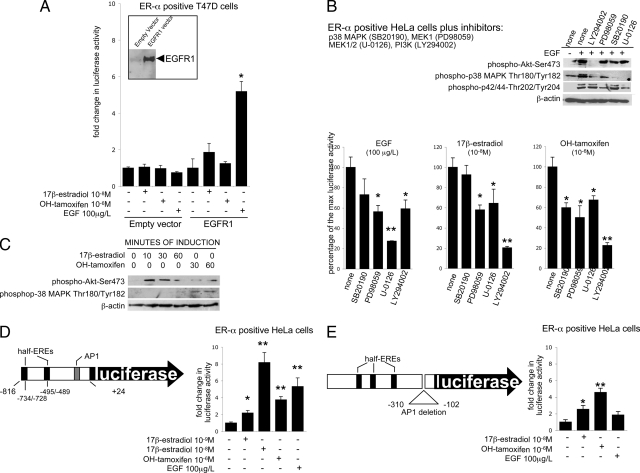
Several members of the EGFR family are known. Among them, EGFR1 and ERBB2 (HER2/NEU) are the most important ones. Western blot analysis (shown in Figure 2B) revealed that ERBB2 is expressed in HeLa (OLFM-4 promoter responsive to both estrogens and EGF) as well as in ECC1 and T47D cells (OLFM-4 promoter neither responsive to estrogens nor to EGF), whereas EGFR1 is expressed in HeLa cells only, but not in ECC1 and T47D cells. Therefore, we hypothesized that EGFR1 may be involved in the regulation of OLFM-4. Re-expression of EGFR1 by transient transfection in T47D cells is sufficient to recover the EGF response. However, EGFR1 re-expression recovers only partly (non-significantly) the estrogenic response of the OLFM-4 promoter (Figure 3A). Similar results were obtained with ECC1 cells (results not shown).
Figure 3.
Intracellular regulation of the OLFM-4 promoter. A: Luciferase response in T47D cells transfected with the OLFM-4 promoter-luciferase along with the expression plasmid for EGFR1 (the panel on the left shows the EGFR1 expression after transfection with EGFR1 plasmid or with an empty vector). B: Luciferase response of HeLa cells transfected with the OLFM-4 promoter-luciferase and the expression plasmid for ER-α. Cells were preincubated with the inhibitors for p38 MAPK (SB20190, 20 μmol/L), MEK1 (PD98059, 50 μmol/L), MEK1/2 (which is upstream both p38 MAPK and p42/44; U-0126, 10 μmol/L), and PI3K (Ly294002, 10 μmol/L). The Western blot on the top indicates that the inhibitory conditions used were efficient and specific: Ly294002 completely blocked EGF-induced phosphorylation of Akt (downstream of PI3K); PD98059 and U-0126 blocked phosphorylation of p42/44 MAPK; phosphorylation of p38 MAPK is partly inhibited by Ly294002, PD98059, and U-0126 as this kinase lies downstream of both PI3K/Akt and MEK1/2 but not by SB20190, which blocks phosphorylation downstream of p38 MAPK. C: Western blotting indicating the in HeLa cells transiently transfected with the expression plasmid for ER-α, stimulation with both 17β-estradiol and OH-tamoxifen activates the PI3K/Akt signaling, whereas OH-tamoxifen only activates p38 MAPK. D: Luciferase response of HeLa cells transfected with the expression plasmid for ER-α and the OLFM-4 promoter-luciferase, comprising only the 0.8-kb most proximal promoter part (shown on the left). E: Luciferase response of HeLa cells transfected with the expression plasmid for ER-α and the OLFM-4 promoter-luciferase where the AP1 site has been abolished. All inhibitors were also tested alone, and no effect on luciferase activity was observed. Bars indicate mean values (n = 3) ± SD. P values were calculated with the t-test; ∗P < 0.05; ∗∗P < 0.01. All results were repeated in at least two independent experiments.
We next investigated which pathways are involved in the EGFR-ER-α cross talk using specific intracellular signaling inhibitors (Figure 3, B and C). The signaling induced by EGF is efficiently blocked by the MEK1/2 inhibitor (U-0126), only partially blocked by inhibition of PI3K (LY294002) and not influenced by p38 MAPK inhibition (SB20190). The signaling induced by 17β-estradiol is most prominently blocked by pretreatment with the PI3K inhibitor LY294002 (Figure 3B) and partially inhibited by the MEK1/2 inhibitor (U-0126) and the MEK1 inhibitor (PD98059). The only difference between 17β-estradiol and OH-tamoxifen induced signaling is the fact that the p38 MAPK inhibitor SB20190 is able to repress the activity mediated by OH-tamoxifen but not the activity induced by 17β-estradiol. The Western blot in Figure 3B, top panel, shows that the used inhibitors were specific and efficient in our experimental conditions.
The activation by 17β-estradiol and OH-tamoxifen of PI3K/Akt was confirmed by Western blot using phospho-Akt-Ser473 antibody (Figure 3C). In addition, Western blotting using phospho-p38 MAPK (Thr180/Tyr182) antibody confirmed that p38 MAPK is phosphorylated by OH-tamoxifen but not by 17β-estradiol (Figure 3C).
Finally, we investigated the promoter sequences that are needed for estrogen and EGF responses. A truncated form of the 1.3-kb OLFM-4 promoter was generated, which contains the first 0.8-kb promoter fragment. This is enough to mediate the complete response of estrogen but only part of the EGF response (Figure 3D). The AP1 site is present in this 0.8-kb promoter (Figure 3D). Therefore, to examine whether this site is needed to mediate the estrogenic and the EGF response, the AP1 site was deleted from the 1.3-kb OLFM-4 promoter (Figure 3E). The responses to EGF and to 17β-estradiol are abolished after deletion of the AP1 binding sequence. The response to OH-tamoxifen, however, appears to be independent from the presence or absence of the AP1 binding site (Figure 3E).
OLFM-4 Decreases Cell Adherence and Vimentin Expression and Protects Against Apoptosis
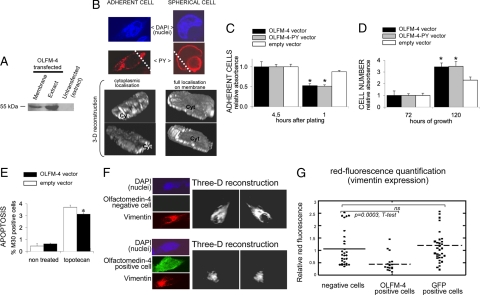
OLFM-4-negative HEK-293 cells were transiently transfected with the OLFM-4 expression plasmid to assess the function of this protein. After transfection, OLFM-4 is highly expressed both as native and as PY tagged (Figure 4, A and B) and is mainly present in the cytoplasm and the cell membrane. We observed that when OLFM-4 is expressed on the cell membrane (Figure 4B, right panels), cells assume a spherical shape and clearly detach from the plate surface, whereas in cells with a more adherent phenotype (Figure 4B, left panels) OLFM-4 is present throughout the cytoplasm (see the three-dimensional reconstruction in Figure 4B). In line with this observation, re-expression of OLFM-4 (native or PY fused) decreases the adherence to the plate surface compared with cells transfected with the empty vector (Figure 4C).
Figure 4.
OLFM-4 re-expression in HEK-293 cells infers apoptosis resistance and reduces cell adherence and vimentin expression. A–E: OLFM-4 was re-expressed in HEK-293 cells (OLFM-4 negative) by transient transfection with the OLFM-4 expression plasmid (or empty vector as control). A: Western blot with antibody IMG-5983 against the native OLFM-4. Equal amounts of protein were loaded on the “extract fractions.” B: Confocal laser scanning microscopy of HEK-293 cells transfected with OLFM-4 cDNA fused to PY tagging and detected with the anti-PY antibody (similar results were obtained with the native nontagged protein). On the left, a cell with predominant cytoplasmic localization of OLFM-4. On the right, membrane localization of OLFM-4. After three-dimensional reconstruction (ImageJ program), cells were (virtually) sectioned through the plane indicated on the top image (dashed line). The cell on the left is well attached to the plate surface, and staining for OLFM-4 is seen through the cytoplasmic section (Cyt). On the right, the OLFM-4 protein is fully localized on the membrane, which is accompanied by detachment (spherical shape) of the cell. We exclude that the rounded shape of the cell was associated to apoptosis because nuclear morphology looked normal, and apoptosis was diminished rather than augmented in cells overexpressing OLFM-4 (E). Magnification ×40. C: Adherence of cells to the culture plate is decreased after re-expression of OLFM-4 compared with cells transfected with the empty vector. ∗P < 0.05 versus empty vector, t-test. Transfected cells were seeded on a 24-well plate, and the number of attached cells was determined by 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide after different periods of time. Four and one-half hours after plating, all cells were seeded. D: The number of viable cells (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide assay) increased after OLFM-4 re-expression compared with cells transfected with the empty vector. ∗P < 0.05 versus empty vector, t-test. E: M30 staining/FACS analysis indicating that HEK-293 cells re-expressing OLFM-4 are more resistant to topotecan-induced apoptosis compared with cells transfected with the empty vector. ∗P < 0.05 versus empty vector, t-test. F: HEK-293 cells were transiently transfected with OLFM-4 cDNA and costained for OLFM-4 (FITC-green) and vimentin (Texas Red). Confocal laser scanning microscopy was used to visualize and quantify vimentin filament throughout the whole volume of cells that re-expressed OLFM-4 and cells that did not. The ImageJ program was used to make three-dimensional images. Magnification ×40. G: Quantification of the vimentin expression in cells transfected as described in F that were positive or negative for OLFM-4 expression. The same experiment was performed by transfecting HEK-293 cells with GFP expression plasmid, and no reduction in vimentin expression could be observed (GFP-positive cells).
Re-expressing OLFM-4 in HEK-293 cells also results in increased cell number compared with untransfected or empty vector transfected cells (Figure 4D). We did not find increased proliferative activity as determined by the DNA histograms generated with flow cytometry (results not shown); however, HEK-293 cells re-expressing OLFM-4 are slightly, but significantly, more resistant to topotecan induced apoptosis (Figure 4E).
Intermediate filaments, ie, vimentin, establish intimate interactions with extracellular matrix components (such as OLFM-4) and can influence a large number of cellular events including cell differentiation, adherence and apoptosis.28,29 Therefore, we examined whether vimentin expression and filament organization were affected by OLFM-4 expression. The level of vimentin in OLFM-4-positive and -negative cells was assessed by immunofluorescence (Figure 4, F and G). HEK-293 cells transiently transfected with the OLFM-4 cDNA were costained with the OLFM-4 antibody (green fluorescence) and with the vimentin antibody (red fluorescence). The efficiency of transient transfection never reaches 100%; therefore, in the same microscopic preparation, cells expressing and cells nonexpressing the transfected plasmid could be observed. Using confocal laser scanning microscopy, the red fluorescence (vimentin expression) was quantified through the complete volume of cells expressing OLFM-4 (green) or not (cells without green fluorescence) in the same transfection plate/reaction. Expression of OLFM-4 clearly results in reduced staining for vimentin (Figure 4, F and G). When we performed the same experiment in cells transfected with the green fluorescence protein expression vector, the vimentin staining in cells expressing and in cells nonexpressing green fluorescence protein did not differ (Figure 4G).
OLFM-4 Expression in Endometriosis and Endometrial Cancer
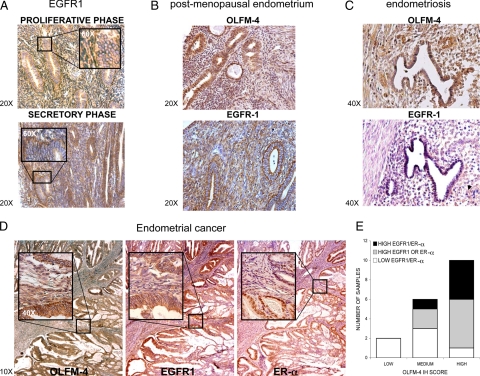
OLFM-4 and EGFR1 are expressed in pre- and postmenopausal endometrium (Figures 1 and 5, A and B), although significant individual variations are present. OLFM-4 is mostly localized in the glands. EGFR1 stains the membrane of the stroma cells and, less intensively, the epithelial cells, with the strongest immunoreactivity during the proliferative phase. OLFM-4 and EGFR1 are also expressed in endometriosis (Figure 5C) and endometrial cancer (Figure 5D). In endometriosis lesions, EGFR1 immunoreactivity is weak, and besides this, no additional difference in the staining intensities between normal endometrium and endometriosis lesions or endometrial cancer were observed.
Figure 5.
OLFM-4 and EGFR1 expression in endometriosis and endometrial cancer. A: EGFR1 expression in healthy premenopausal endometrium. Enlarged panels (×60) show the membrane expression of EGFR1. B: OLFM-4 and EGFR1 expression in postmenopausal endometrium. C: OLFM-4 and EGFR1 expression in endometriosis lesions. Little expression of EGFR1 is evident (area indicated with the arrowhead). D: OLFM-4, EGFR1, and ER-α expression in endometrial cancer. E: Correlation between OLFM-4, EGFR1, and ER-α expression in endometrial cancer (n = 18 biopsies from the pathological anatomy national automated archive (PALGA) collection; 3 of the 21 samples from this collection were not stained for OLFM-4, EGFR1, and ER-α because of lack of material). Protein expression was semiquantitatively assessed by calculating a staining index as described earlier.40 Afterward, samples were grouped in three categories for OLFM-4 expression (low, medium, high; x-axis) and in two for EGFR1 and ER-α (low and high). A combination of different expression levels of EGFR1 and ER-α are indicated with white, gray, and black. IH SCORE = staining index.
Interestingly, when comparing the immunohistochemical staining scores of OLFM-4 with that of EGFR1 and ER-α in endometrial cancer (Figure 5D), we observed that high OLFM-4 stainings correlate with high staining scores of EGFR1 and ER-α (Figure 5E). Quantifying immunohistochemical stainings is semiquantitative at best; therefore, we also assessed the mRNA levels of OLFM-4 and EGFR1 by real-time PCR. In addition, instead of assessing the level of ER-α, we examined the activation of the ER-α signaling by assessing the expression of the estrogen-responsive gene TFF1, as described previously.16 The levels of OLFM-4, EGFR1, and TFF1 were measured in endometriosis, endometrial cancer, and controls.
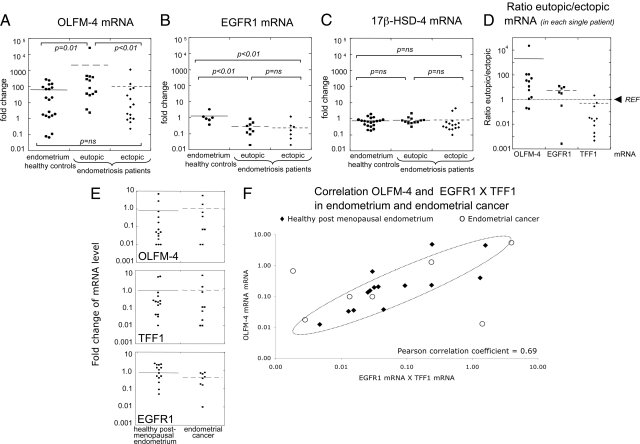
At the mRNA level, OLFM-4 expression is significantly higher in the eutopic endometrium of patients compared with the endometrium of controls but also compared with the endometriotic tissue (Figure 6A). EGFR1 mRNA levels are significantly lower in the eutopic and ectopic endometrium of patients when compared to the normal endometrium (Figure 6B), confirming the immonostaining. The expression level of the 17β-hydroxysteroid-dehydrogenase-4 (17β-HSD-4; Figure 6C) was used as a control of the endometriotic biopsies used for RNA isolation: this enzyme was previously shown to be similar in eutopic and ectopic endometrium and not to be expressed in nonendometrial tissue surrounding the lesions.16 Equal levels of 17β-HSD-4 in eutopic tissues (from both patients and controls) and ectopic material (Figure 6C) indicates the presence of comparable endometrial cells in all biopsies used for RNA isolation.
Figure 6.
mRNA levels of OLFM-4, EGFR1, and TFF1 in endometriosis and endometrial cancer. A: OLFM-4 mRNA levels in the endometrium of healthy controls and ectopic/eutopic endometrium of endometriosis patients (statistic significance was maintained after removal of the outsider sample with a fold change >104 in the “eutopic” group). B: EGFR1 mRNA levels in the endometrium of healthy controls and ectopic/eutopic endometrium of endometriosis patients. C: 17β-HSD4 mRNA in the endometrium of healthy controls and ectopic/eutopic endometrium of endometriosis patients. This enzyme is known to be expressed at comparable levels in the eutopic and ectopic endometrium,30 and it was used as a control for the presence of endometrial tissue in the ectopic lesions. D: Ratios between the levels of mRNA in the eutopic versus the corresponding ectopic endometrium (matched samples in each patient) for OLFM-4, EGFR1, and TFF1. The levels in the eutopic tissues were set to one (REF, indicated); therefore, a value higher than one indicates that in that patient the level of expression of the gene under investigation is higher inside the uterus than in the lesion; vice versa, a value lower than one indicates higher expression in the lesion compared to the endometrium at its normal location. E: OLFM-4, TFF1, and EGFR1 mRNA levels in the endometrium of postmenopausal healthy controls and endometrial cancer (none is significantly different between the two groups). F: Correlation between OLFM-4 mRNA levels and the product of EGFR1 and TFF1 mRNA levels in endometrium of healthy postmenopausal women (R2 value = 0.3) and endometrial cancer (R2 value = 0.8). All mRNA values were calculated by quantitative real-time PCR and were normalized using cyclophilin as a housekeeping gene. Results did not change when β-actin was used for normalization; data not shown). Nonparametric Mann-Whitney U statistical test was used and P values are indicated. Ns, nonsignificant.
We next examined the fold-change of the expression of TFF1, OLFM-4, and EGFR1 between the eutopic and ectopic endometrium in each single patient. In this way, we assessed if the expression of these genes differs in the endometrium located inside the uterus (eutopic) compared to the endometriotic lesions. As previously shown,16 TFF1 expression is lower in the eutopic compared to the ectopic endometrial tissue of each single patient (expression ratios lower than one; Figure 6D). However, OLFM-4 expression shows the opposite trend and is higher in the eutopic endometrial tissue compared with the levels in the ectopic endometrium in the same patient. The levels of EGFR1 mRNA are also slightly higher in the eutopic than in the ectopic lesions.
In contrast to the differences in expression levels observed in endometriotic tissue, mRNA levels of OLFM-4, EGFR1, and TFF1 are comparable in endometrial cancer and healthy postmenopausal endometrium (Figure 6E). In line with the correlation of high OLFM-4 protein level and high expression of EGFR1 and/or ER-α, the same relationship becomes evident in both healthy postmenopausal endometrium and endometrial cancer when the OLFM-4 mRNA levels are plotted against the product of the mRNA levels of EGFR1 and TFF1 (Figure 6F).
Discussion
OLFM-4 is an estrogen-responsive gene, highly up-regulated during the proliferative phase of the menstrual cycle. The response to 17β-estradiol was confirmed in vitro using explants cultures of human endometrial tissue (Figure 1D). The magnitude of the in vitro response was higher using menstrual-phase endometrial explants compared with late proliferative-phase explants. This phenomenon of differences in estrogen sensitivity in different preovulatory stages has been described previously.20 It can be attributed to the fact that late proliferative-phase endometrium has been exposed in vivo to 17β-estradiol for an extended period of time. This renders the endometrium refractory to further stimulation in vitro.
Despite its estrogenic response, the promoter region of the OLFM-4 gene does not contain any perfect EREs. Three half-palindromic EREs are present plus an AP-1 binding site. The presence of the AP1 site suggests that growth factor signaling may be involved in the full regulation of this gene. Indeed, OLFM-4 transcription is induced by EGF, 17β-estradiol, and OH-tamoxifen (Figures 2 and 3). From our experiments, we can infer that the signaling pathways activated by these three compounds are distinct but merge at various levels. In addition, both ER-α and EGFR signaling are required for full activation of the OLFM-4 promoter. The responses to EGF, 17β-estradiol, and OH-tamoxifen are impaired when EGFR is blocked by the dominant-negative receptor variant, or cells are pretreated with an EGFR inhibitor (GW2974). In turn, these same responses are impaired when ER-α is not expressed (only partly, in case of EGF) or when cells are exposed to the anti-estrogen ICI-164384 (Figure 2). In the HeLa clone used in these experiments, neither ER-α nor ER-β is endogenously expressed (as visualized by Western blot). Re-expression of ER-α clearly recovers the OLFM-4 promoter response, but the effects of introducing ER-β have not been investigated.
With respect to the EGF, its response is mediated by EGFR1, as indicated by the re-expression experiment in EGFR1-negative cells (ie, T47D cells; Figure 3A). The main signaling route of EGF-EGFR1 appears to be mediated via MEK1/2 and p42/44 MAPK and involves AP1 activation as well (Figure 3, B and E). Because the full EGF response also requires the presence of ER-α, the well-described tethering mechanism of ER-α and c-Fos/c-Jun at the AP1 region30 may control OLFM-4 promoter activation by EGF.
With respect to the estrogenic response, besides ER-α, EGFR is required, as mentioned earlier. However, re-expression of EGFR1 alone in T47D (Figure 3A) or in ECC1 cells (data not shown) is not enough to re-establish the full response of the OLFM-4 promoter to 17β-estradiol and OH-tamoxifen. Apparently, additional signaling paths are needed for the full estrogenic response. It is unlikely that other members of the EGFR family such as ERBB2, ERBB3, or ERBB4 may be involved. ERBB3 is expressed in T47D but not in HeLa cells (where we observed high OLFM-4 promoter activation); ERBB2 is expressed in both cells (Figure 3A), whereas ERBB4 is neither expressed in HeLa nor in T47D cells (expression data for ERBB3 and ERBB4 derived from GNF SymAtlas: http://biogps.gnf.org/, last accessed September 15, 2010). Preliminary results of our group have indicated that, in contrast to Akt Ser473, which becomes phosphorylated in an EGF/estrogen-dependent manner, Akt Thr308 is constitutively phosphorylated in HeLa cells but not in ECC1 or in T47D cells. PI3K-Akt signaling via Thr308 may be therefore needed for the estrogenic response of OLFM-4.
Subsequently, the estrogenic response requires PI3K-Akt/PKB and MEK1/2 activation (Figure 3, B and C). The further downstream events leading to the regulation of OLFM-4 transcription are not the same for 17β-estradiol and OH-tamoxifen. In case of 17β-estradiol-bound ER-α, the AP1 site in the promoter region is necessary, suggesting that, also in this case, a tethering mechanism of ER-α at the AP1 site may operate. However, induction of the OLFM-4 promoter by OH-tamoxifen-bound ER-α does not require the interaction with AP1. Instead, after activation of Akt/PKB and MEK1/2, it depends on p38 MAPK activation.
To corroborate our findings, we have assessed the presence of the half-EREs and the AP1 site in different species and observed that these sequences are conserved among rat, mouse, and human (results not shown).
The function of OLFM-4 in the endometrium is still elusive. In HEK-293 cells, OLFM-4 localizes in the cell membrane. Its overexpression decreases the adherence capability of cells and confers a slight resistance to apoptosis, confirming other reports.4,10 We have not investigated why in some cells OLFM-4 localizes on the cell membrane, whereas in other cells, the protein seems to remain trapped inside the cytoplasm (Figure 4B). Most probably, in this latter case, OLFM-4 is not correctly processed because of its overexpression after transient transfection.
The effects of OLFM-4 seem to be mediated by decreased vimentin expression (or filament re-organization). Similar results with respect to cell number/apoptosis and cell adherence were also obtained using ECC1 endometrial cancer cells (results not shown). Other authors have observed similar effects using colon cancer cells: OLFM-4 interfered with cytoskeleton filaments and caused cells to round up and detach.9
OLFM-4 is particularly associated with tissues with high regenerative potential (ie, bone marrow, the gastrointestinal tract, prostate, murine ovaries, rat testis,31 and endometrium). In addition, the fact that OLFM-4 affects processes such as cell adherence, cell-cell interaction, motility, and apoptosis, rather than proliferation, implies that this protein plays a determining role in differentiation of cells. Accordingly, Liu and coworkers9 have reported that OLFM-4 expression in colon carcinoma correlates with well-differentiated tumors, and we have shown that vimentin (a marker of cell de-differentiation during epithelial-mesenchymal transition)32 is down-regulated by OLFM-4 re-expression. These evidences and the fact that OLFM-4 expression is highest in proliferative endometrium indicate that OLFM-4 may promote the differentiation of endometrial epithelial cells at the late proliferative phase when the endometrium has finished the exponential growth and is preparing to respond to the luteal progesterone. Overall, our results are in line with previous investigations. Cross-talks between estrogens and EGF signaling are well documented,30,33 and they control cell migration and other remodeling and cell differentiation features of the endometrium.34
The synergy between ER-α and EGFR is corroborated by the fact that in the proliferative phase, when OLFM-4 expression is high, the levels of 17β-estradiol in the circulation are peaking and the expression of EGFR, in particular EGFR1,35 and its ligands (EGF and TGFα) are also significantly higher compared to the secretory phase.36,37
Investigating the human clinical samples gave support to our in vitro findings on the regulation of OLFM-4. OLFM-4 level is correlated to ER-α (or estrogen signaling measured as TFF1 expression) and EGFR1 expression in endometrial tumors and controls (mRNA and immunohistochemistry; Figures 6F and 5E, respectively). In endometriosis patients, the same trend is also observed. The ectopic locations have an increased estrogenic activity compared with the tissue inside the uterus as suggested by the lower TFF1 expression in the eutopic tissue compared with the corresponding ectopic endometrium in the same patient (16; see also Figure 6D). However, OLFM-4 mRNA levels are lower in the ectopic than in the eutopic endometrium of the same patient. This may be the result of the low level of EGFR1 expression in the lesions, which, despite their high estrogenic activity, have low EGF signaling and therefore low OLFM-4 transcription.
With respect to a possible role of OLFM-4 in endometrial pathologies, in endometrial cancer, the expression levels of OLFM-4 are comparable with those of healthy postmenopausal endometrium, indicating that this protein plays no role in the carcinogenesis of the endometrium. In contrast, the eutopic endometrium of women with endometriosis showed higher OLFM-4 expression compared with controls. Given that OLFM-4 reduces cell adherence, endometrial cells inside the uterus of patients may have increased ability to move into the peritoneum and to spread. At the same time, these cells may also have a survival advantage in the peritoneal cavity, because of the intrinsic apoptosis resistance conferred by OLFM-4. Interestingly, MEK1/2-p42/44 MAPK activation is elevated in endometrial stromal fibroblasts of women with endometriosis compared with women without disease,38 which supports the hypothesis that OLFM-4 has high expression in the eutopic endometrium of endometriosis patients and may predispose endometrial cells to spread, survive, and implant at ectopic locations. In addition, the fact that OLFM-4 expression is back to “healthy control” levels once the lesions are established (in the surgically excised ectopic lesions; Figure 6A) indicates that this protein may be involved in the initial steps of endometriosis onset only (spread, survival, infiltration), and it plays no additional role once the disease is established. In line with this, established disease is no longer characterized by tissue remodeling events, but rather by blood supply acquisition and exposure to estrogen/proliferative stimuli,16,39 events where OLFM-4 plays no role.
In conclusion, together with the recent finding that cell migration and other remodeling features of the endometrium are the result of an intimate interaction between estrogen signaling, PI3K, and extracellular signal-regulated kinase 1/2 signaling,34 the observations from this study suggest a role for OLFM-4 in these processes, under the tight regulation of 17β-estradiol and EGF. Unbalances in these regulations may favor the early onset of endometrial pathologies, like endometriosis.
Acknowledgments
We are grateful to Dr. Koen van de Vijver (Department of Pathology, Maastricht University and Medical Centre) for helping in the analysis of clinical samples; Dr. Bert Schutte and Mieke Hefling (Department of Molecular Cell Biology, Maastricht University and Medical Centre) for helping with the FACS analysis; Dr. Jos Broers and Frederiek Houben (Department Molecular Cell Biology, Maastricht University and Medical Centre) for helping with the confocal laser scanning microscope; Dr. Jan Theys (MAASTRO Lab, Maastricht University and Medical Centre) for providing the expression plasmids for EGFR1 (wild-type and dominant-negative variant); Prof. Ronald Schüle (Freiburg University, Germany) for providing the ER-α expression plasmid; and Dr. Griffin Rodgers and Dr. Wenli Liu (National Institutes of Health, Bethesda, MD) for useful discussion and advice for the functional studies.
Footnotes
Address reprint requests to Andrea Romano, Ph.D., Department of Obstetrics and Gynaecology, P Debyelaan 25, Maastricht University and Medical Centre, 6202 AZ, Maastricht, The Netherlands. E-mail: a.romano@og.unimaas.nl or andrearomano420@hotmail.com.
Supported by the Maastricht University and Medical Centre.
C.P. and B.D. and contributed equally to the study.
Current address of C.P.: Tissue Engineering and Microfluidics Laboratory, Australian Institute for Bioengineering and Nanotechnology, University of Queensland, Brisbane, Queensland, Australia; of P.G.: Women’s Health Department, Merck Sharp & Dohme BV, Oss, The Netherlands.
References
- Punyadeera C, Dassen H, Klomp J, Dunselman G, Kamps R, Dijcks F, Ederveen A, de Goeij A, Groothuis P. Oestrogen-modulated gene expression in the human endometrium. Cell Mol Life Sci. 2005;62:239–250. doi: 10.1007/s00018-004-4435-y. [DOI] [PubMed] [Google Scholar]
- Zhang J, Liu WL, Tang DC, Chen L, Wang M, Pack SD, Zhuang Z, Rodgers GP. Identification and characterization of a novel member of olfactomedin-related protein family, hGC-1, expressed during myeloid lineage development. Gene. 2002;283:83–93. doi: 10.1016/s0378-1119(01)00763-6. [DOI] [PubMed] [Google Scholar]
- Liu W, Chen L, Zhu J, Rodgers GP. The glycoprotein hGC-1 binds to cadherin and lectins. Exp Cell Res. 2006;312:1785–1797. doi: 10.1016/j.yexcr.2006.02.011. [DOI] [PubMed] [Google Scholar]
- Zhang X, Huang Q, Yang Z, Li Y, Li CY. GW112, a novel antiapoptotic protein that promotes tumor growth. Cancer Res. 2004;64:2474–2481. doi: 10.1158/0008-5472.can-03-3443. [DOI] [PubMed] [Google Scholar]
- Oue N, Aung PP, Mitani Y, Kuniyasu H, Nakayama H, Yasui W. Genes involved in invasion and metastasis of gastric cancer identified by array-based hybridization and serial analysis of gene expression. Oncology. 2005;69 Suppl 1:17–22. doi: 10.1159/000086627. [DOI] [PubMed] [Google Scholar]
- Oue N, Hamai Y, Mitani Y, Matsumura S, Oshimo Y, Aung PP, Kuraoka K, Nakayama H, Yasui W. Gene expression profile of gastric carcinoma: identification of genes and tags potentially involved in invasion, metastasis, and carcinogenesis by serial analysis of gene expression. Cancer Res. 2004;64:2397–2405. doi: 10.1158/0008-5472.can-03-3514. [DOI] [PubMed] [Google Scholar]
- Aung PP, Oue N, Mitani Y, Nakayama H, Yoshida K, Noguchi T, Bosserhoff AK, Yasui W. Systematic search for gastric cancer-specific genes based on SAGE data: melanoma inhibitory activity and matrix metalloproteinase-10 are novel prognostic factors in patients with gastric cancer. Oncogene. 2006;25:2546–2557. doi: 10.1038/sj.onc.1209279. [DOI] [PubMed] [Google Scholar]
- Yasui W, Oue N, Aung PP, Matsumura S, Shutoh M, Nakayama H. Molecular-pathological prognostic factors of gastric cancer: a review. Gastric Cancer. 2005;8:86–94. doi: 10.1007/s10120-005-0320-0. [DOI] [PubMed] [Google Scholar]
- Liu W, Liu Y, Zhu J, Wright E, Ding I, Rodgers GP. Reduced hGC-1 protein expression is associated with malignant progression of colon carcinoma. Clin Cancer Res. 2008;14:1041–1049. doi: 10.1158/1078-0432.CCR-07-4125. [DOI] [PubMed] [Google Scholar]
- Liu W, Zhu J, Cao L, Rodgers GP. Expression of hGC-1 is correlated with differentiation of gastric carcinoma. Histopathology. 2007;51:157–165. doi: 10.1111/j.1365-2559.2007.02763.x. [DOI] [PubMed] [Google Scholar]
- Shinozaki S, Nakamura T, Iimura M, Kato Y, Iizuka B, Kobayashi M, Hayashi N. Up-regulation of Reg 1α and GW112 in the epithelium of inflamed colonic mucosa. Gut. 2001;48:623–629. doi: 10.1136/gut.48.5.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannick EE, Schurr JR, Zapata A, Lentz JJ, Gastanaduy M, Cote RL, Delgado A, Correa P, Correa H. Gene expression in gastric biopsies from patients infected with Helicobacter pylori. Scand J Gastroenterol. 2004;39:1192–1200. doi: 10.1080/00365520410003588. [DOI] [PubMed] [Google Scholar]
- Koshida S, Kobayashi D, Moriai R, Tsuji N, Watanabe N. Specific overexpression of OLFM4(GW112/HGC-1) mRNA in colon, breast and lung cancer tissues detected using quantitative analysis. Cancer Sci. 2007;98:315–320. doi: 10.1111/j.1349-7006.2006.00383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi D, Koshida S, Moriai R, Tsuji N, Watanabe N. Olfactomedin 4 promotes S-phase transition in proliferation of pancreatic cancer cells. Cancer Sci. 2007;98:334–340. doi: 10.1111/j.1349-7006.2007.00397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes RW, Hertig AT, Rock J. Dating the endometrial biopsy. Am J Obstet Gynecol. 1975;122:262–263. doi: 10.1016/s0002-9378(16)33500-1. [DOI] [PubMed] [Google Scholar]
- Delvoux B, Groothuis P, D'Hooghe T, Kyama C, Dunselman G, Romano A. Increased production of 17β-estradiol in endometriosis lesions is the result of impaired metabolism. J Clin Endocrinol Metab. 2009;94:876–883. doi: 10.1210/jc.2008-2218. [DOI] [PubMed] [Google Scholar]
- Schenken RS, Guzick DS. Revised endometriosis classification: 1996. Fertil Steril. 1997;67:815–816. doi: 10.1016/s0015-0282(97)81390-8. [DOI] [PubMed] [Google Scholar]
- Pecorelli S. Revised FIGO staging for carcinoma of the vulva, cervix, and endometrium. Int J Gynaecol Obstet. 2009;105:103–104. doi: 10.1016/j.ijgo.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Pijnenborg JM, Romano A, Dam-de Veen GC, Dunselman GA, Fischer DC, Groothuis PG, Kieback DG. Aberrations in the progesterone receptor gene and the risk of recurrent endometrial carcinoma. J Pathol. 2005;205:597–605. doi: 10.1002/path.1738. [DOI] [PubMed] [Google Scholar]
- Punyadeera C, Dunselman G, Marbaix E, Kamps R, Galant C, Nap A, Goeij A, Ederveen A, Groothuis P. Triphasic pattern in the ex vivo response of human proliferative phase endometrium to oestrogens. J Steroid Biochem Mol Biol. 2004;92:175–185. doi: 10.1016/j.jsbmb.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Romano A, Adriaens M, Kuenen S, Delvoux B, Dunselman G, Evelo C, Groothuis P. Identification of novel ER-α target genes in breast cancer cells: gene- and cellselective co-regulator recruitment at target promoters determines the response to 17β-estradiol and tamoxifen. Mol Cell Endocrinol. 2010;314:90–100. doi: 10.1016/j.mce.2009.08.008. [DOI] [PubMed] [Google Scholar]
- Romano A, Delvoux B, Fischer DC, Groothuis P. The PROGINS polymorphism of the human progesterone receptor diminishes the response to progesterone. J Mol Endocrinol. 2007;38:331–350. doi: 10.1677/jme.1.02170. [DOI] [PubMed] [Google Scholar]
- Oehler MK, Greschik H, Fischer D-C, Tong X-W, Schuele S, Kieback DG. Somatic mutations affecting the function of the human estrogen receptor α (hER-α) in adenomyosis uteri. Mol Hum Reprod. 2004;10:853–860. doi: 10.1093/molehr/gah113. [DOI] [PubMed] [Google Scholar]
- Reardon DB, Contessa JN, Mikkelsen RB, Valerie K, Amir C, Dent P, Schmidt-Ullrich RK. Dominant negative EGFR-CD533 and inhibition of MAPK modify JNK1 activation and enhance radiation toxicity of human mammary carcinoma cells. Oncogene. 1999;18:4756–4766. doi: 10.1038/sj.onc.1202849. [DOI] [PubMed] [Google Scholar]
- Contessa JN, Abell A, Valerie K, Lin PS, Schmidt-Ullrich RK. ErbB receptor tyrosine kinase network inhibition radiosensitizes carcinoma cells. Int J Radiat Oncol Biol Phys. 2006;65:851–858. doi: 10.1016/j.ijrobp.2006.02.025. [DOI] [PubMed] [Google Scholar]
- Chin KL, Aerbajinai W, Zhu J, Drew L, Chen L, Liu W, Rodgers GP. The regulation of OLFM4 expression in myeloid precursor cells relies on NF-κB transcription factor. Br J Haematol. 2008;143:421–432. doi: 10.1111/j.1365-2141.2008.07368.x. [DOI] [PubMed] [Google Scholar]
- Schutte B, Henfling M, Kolgen W, Bouman M, Meex S, Leers MP, Nap M, Bjorklund V, Bjorklund P, Bjorklund B, Lane EB, Omary MB, Jornvall H, Ramaekers FC. Keratin 8/18 breakdown and reorganization during apoptosis. Exp Cell Res. 2004;297:11–26. doi: 10.1016/j.yexcr.2004.02.019. [DOI] [PubMed] [Google Scholar]
- Ivaska J, Pallari HM, Nevo J, Eriksson JE. Novel functions of vimentin in cell adhesion, migration, and signaling. Exp Cell Res. 2007;313:2050–2062. doi: 10.1016/j.yexcr.2007.03.040. [DOI] [PubMed] [Google Scholar]
- Bargagna-Mohan P, Hamza A, Kim YE, Khuan Abby Ho Y, Mor-Vaknin N, Wendschlag N, Liu J, Evans RM, Markovitz DM, Zhan CG, Kim KB, Mohan R. The tumor inhibitor and antiangiogenic agent withaferin A targets the intermediate filament protein vimentin. Chem Biol. 2007;14:623–634. doi: 10.1016/j.chembiol.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpino G, Wiechmann L, Osborne CK, Schiff R. Cross-talk between the estrogen receptor and the HER tyrosine kinase receptor family: molecular mechanism and clinical implications for endocrine therapy resistance. Endocr Rev. 2008;29:217–233. doi: 10.1210/er.2006-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schalburg KR, McCarthy SP, Rise ML, Hutson JC, Davidson WS, Koop BF. Expression of morphogenic genes in mature ovarian and testicular tissues: potential stem-cell niche markers and patterning factors. Mol Reprod Dev. 2006;73:142–152. doi: 10.1002/mrd.20359. [DOI] [PubMed] [Google Scholar]
- Demir AY, Groothuis PG, Nap AW, Punyadeera C, de Goeij AF, Evers JL, Dunselman GA. Menstrual effluent induces epithelial-mesenchymal transitions in mesothelial cells. Hum Reprod. 2004;19:21–29. doi: 10.1093/humrep/deh042. [DOI] [PubMed] [Google Scholar]
- Seval Y, Cakmak H, Kayisli UA, Arici A. Estrogen-mediated regulation of p38 mitogen-activated protein kinase in human endometrium. J Clin Endocrinol Metab. 2006;91:2349–2357. doi: 10.1210/jc.2005-2132. [DOI] [PubMed] [Google Scholar]
- Gentilini D, Busacca M, Di Francesco S, Vignali M, Vigano P, Di Blasio AM. PI3K/Akt and ERK1/2 signalling pathways are involved in endometrial cell migration induced by 17β-estradiol and growth factors. Mol Hum Reprod. 2007;13:317–322. doi: 10.1093/molehr/gam001. [DOI] [PubMed] [Google Scholar]
- Ejskjaer K, Sorensen BS, Poulsen SS, Mogensen O, Forman A, Nexo E. Expression of the epidermal growth factor system in human endometrium during the menstrual cycle. Mol Hum Reprod. 2005;11:543–551. doi: 10.1093/molehr/gah207. [DOI] [PubMed] [Google Scholar]
- Bonaccorsi G, Pansini F, Segala V, Bagni B, Bergamini CM, Mollica G. Modification of number and of affinity of endometrial EGF receptors during the menstrual cycle. Eur J Obstet Gynecol Reprod Biol. 1989;33:177–182. doi: 10.1016/0028-2243(89)90211-6. [DOI] [PubMed] [Google Scholar]
- Imai T, Kurachi H, Adachi K, Adachi H, Yoshimoto Y, Homma H, Tadokoro C, Takeda S, Yamaguchi M, Sakata M. Changes in epidermal growth factor receptor and the levels of its ligands during menstrual cycle in human endometrium. Biol Reprod. 1995;52:928–938. doi: 10.1095/biolreprod52.4.928. [DOI] [PubMed] [Google Scholar]
- Velarde MC, Aghajanova L, Nezhat CR, Giudice LC. Increased mitogen-activated protein kinase kinase/extracellularly regulated kinase activity in human endometrial stromal fibroblasts of women with endometriosis reduces 3′,5′-cyclic adenosine 5′-monophosphate inhibition of cyclin D1. Endocrinology. 2009;150:4701–4712. doi: 10.1210/en.2009-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nap AW, Groothuis PG, Demir AY, Evers JL, Dunselman GA. Pathogenesis of endometriosis. Best Pract Res Clin Obstet Gynaecol. 2004;18:233–244. doi: 10.1016/j.bpobgyn.2004.01.005. [DOI] [PubMed] [Google Scholar]