Abstract
Microglia, the primary immune effector cells in the brain, continually monitor the tissue parenchyma for pathological alterations and become activated in Alzheimer’s disease. Loss of signaling between neurons and microglia via deletion of the microglial receptor, CX3CR1, worsens phenotypes in various models of neurodegenerative diseases. In contrast, CX3CR1 deficiency ameliorates pathology in murine stroke models. To examine the role of CX3CR1 in Alzheimer’s disease–related β-amyloid pathology, we generated APPPS1 and R1.40 transgenic mouse models of Alzheimer’s disease deficient for CX3CR1. Surprisingly, CX3CR1 deficiency resulted in a gene dose-dependent reduction in β-amyloid deposition in both the APPPS1 and R1.40 mouse models of AD. Immunohistochemical analysis revealed reduced staining for CD68, a marker of microglial activation. Furthermore, quantitative immunohistochemical analysis revealed reduced numbers of microglia surrounding β-amyloid deposits in the CX3CR1-deficient APPPS1 animals. The reduced β-amyloid pathology correlated with reduced levels of TNFα and CCL2 mRNAs, but elevated IL1β mRNA levels, suggesting an altered neuroinflammatory milieu. Finally, to account for these seemingly disparate results, both in vitro and in vivo studies provided evidence that CX3CL1/CX3CR1 signaling alters the phagocytic capacity of microglia, including the uptake of Aβ fibrils. Taken together, these results demonstrate that loss of neuron-microglial fractalkine signaling leads to reduced β-amyloid deposition in mouse models of AD that is potentially mediated by altered activation and phagocytic capability of CX3CR1-deficient microglia.
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder characterized by extracellular deposits of β-amyloid (Aβ) peptides in senile plaques. Accumulating evidence suggests that neuroinflammatory processes may contribute to the pathophysiology of AD. Microglia, the resident inflammatory cells of the brain, are found in a highly activated state in the AD brain, including morphological alterations, proliferation, increased expression of cell surface receptors, and secretion of inflammatory cytokines and chemokines.1,2 However, recent in vivo imaging studies demonstrated that activated microglia migrate to newly formed Aβ deposits in mouse models of AD and may restrict their growth by removing Aβ fibrils.3,4,5 Therefore, depending on the local conditions, microglia may exhibit beneficial or detrimental activation programs.6 Indeed, various genetically modified mouse models have convincingly demonstrated the role for various inflammatory pathways in altering microglial activation and Aβ deposition, including chemokines/cytokines,7,8,9 cyclooxygenase,10 and complement.11 While these studies suggest a correlative link between neuroinflammation and AD pathophysiology, the molecular mechanisms underlying beneficial or deleterious microglial activities remain incompletely understood.
The chemokine fractalkine (CX3CL1) and its cognate receptor, CX3CR1, play an important role in neuroinflammation via paracrine signaling between neurons and microglia.12,13,14 While CX3CR1 is expressed by many leukocyte cell types in the periphery,15 in the brain CX3CR1 is exclusively expressed by microglia13 whereas CX3CL1 is highly expressed within neurons.14 CX3CL1 can signal to CX3CR1 in a membrane bound form, or as a secreted ligand after constitutive or inducible cleavage by the ADAM10 and ADAM17 metalloproteases, respectively.16 Genetic variants in CX3CR1, which result in defective CX3CL1-CX3CR1 interactions, have been associated with age-related macular degeneration, a neuroinflammatory disorder.17,18 Finally, disruption of CX3CL1-CX3CR1 signaling promotes neurodegeneration in mouse models of Parkinson’s disease, amyotrophic lateral sclerosis, as well as in neuroinflammation induced by systemic administration of lipopolysaccharide.13 Conversely, lack of CX3CR1 is protective in murine stroke models.19,20
To examine the role of CX3CR1 signaling in amyloid pathology associated with AD, we mated Cx3cr1 knockout mice to two different mouse models of AD that exhibit either rapid deposition of primarily Aβ42 (the APPPS1 model)21 or gradual deposition of primarily Aβ40 (the R1.40 model).22,23,24 Surprisingly, CX3CR1 deficiency resulted in a gene dose-dependent reduction in Aβ deposition as well as the numbers of microglia surrounding the Aβ deposits in both mouse models of AD. CX3CR1-deficient mice with amyloid pathology exhibited altered microglial activation with reduced immunohistochemical staining for the microglial activation marker CD68 and altered expression of cytokines and chemokines. Finally, we demonstrate that CX3CL1-CX3CR1 signaling reduces microglial phagocytic capabilities and that blocking CX3CL1-CX3CR1 signaling increases uptake of Aβ, suggesting that CX3CR1 deficiency leads to enhanced Aβ clearance.
Materials and Methods
Mice
The APPPS1-21 (APPPS1) mouse line coexpresses human amyloid precursor protein harboring the K670M/N671L familial AD mutation and presenilin 1 harboring the L166P familial AD mutation, under the control of the neuron-specific Thy1 promoter.21 APPPS1 mice (in an isogenic C57BL/6J background; kindly provided by Mathias Jucker) were mated to Cx3cr1−/− animals15; congenic on the C57BL/6J background) and subsequently intercrossed to generate APPPS1;Cx3cr1+/− and APPPS1;Cx3cr1−/− genotypes on a defined C57BL/6J genetic background. Aged-matched APPPS1;Cx3cr1+/+ animals were used as controls.
The R1.40 transgene is a full genomic copy of human APP (carried on a 650-kb yeast artificial chromosome clone) carrying the K670M/N671L familial AD mutation associated with early-onset familial AD. Creation of the R1.40 transgenic mouse strain and subsequent backcrossing to inbred strains has been described previously.22,23,25 Homozygous R1.40 animals, maintained on the C57BL/6J genetic background, were mated to Cx3cr1−/− animals (also maintained on the C57BL/6J genetic background) and subsequently intercrossed to generate animals homozygous for the R1.40 transgene and either homozygous or heterozygous for the Cx3cr1 knockout allele and controls. Animals were housed at the Cleveland Clinic Biological Resources Unit, a facility fully accredited by the Association of Assessment and Accreditation of Laboratory Animal Care. All procedures were approved by the Institutional Animal Care and Use Committee of the Cleveland Clinic.
Western Blot Analysis
Mice were sacrificed by cervical dislocation, and their brains were removed, sagittally bisected, and snap frozen. Tissues were subsequently homogenized in 10 volumes of Tris-buffered saline (50 mmol/L Tris; pH 7.4, 150 mmol/L NaCl, 1 mmol/L EDTA, 0.1% Triton-X100) or 20 volumes of T-PER reagent (Thermo Scientific, Rockford, IL) with a protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO). Total brain homogenates were subsequently sonicated to shear the DNA and centrifuged to remove nuclei and cellular debris. Total protein concentration was determined using the Pierce BCA Protein Assay Kit (Thermo Scientific). Brain protein (25–30 μg) was run on a Novex NuPage, 4 to 12% Bis-Tris gel (Invitrogen) and transferred to a PVDF membrane. The resulting Western blots were blocked with 5% milk and subsequently incubated with CT15 antibody (rabbit polyclonal; 1:5000, kindly provided by Edward H. Koo) raised against the C terminus of APP26 or 6E10 antibody (mouse monoclonal; 1:2000; Signet Labs) raised against amino acids 1-16 of human Aβ.
After overnight incubation at 4°C with primary antibodies, blots were washed with PBS and detected using anti-mouse or anti-rabbit secondary antibodies conjugated to horseradish peroxidase (1:20,000; Jackson ImmunoResearch Laboratories, West Grove, PA) diluted in 5% milk with 0.1% Tween 20. To confirm equal protein loading, anti–α-tubulin antibody (mouse monoclonal; 1:5000; Neomarkers, Fremont, CA) or anti–glyceraldehyde 3-phosphate dehydrogenase (GAPDH; mouse monoclonal; 1:20,000; Millipore, Billerica, MA) was used. For quantification, blots were incubated with secondary antibody mixtures (goat anti-mouse IRDye 700 and goat anti-rabbit IRDye 800; 1:20,000; Rockland Immunochemicals, Gilbertsville, PA) for one hour at RT. Infrared signals were measured and quantified in an Odyssey infrared imaging system (Li-Cor Biosciences, Lincoln, NE). The integrated intensity ratio for APP/α-tubulin or C-terminal fragment (CTF) β/α-tubulin was calculated, and the mean ± SEM for eight independent experiments were used for statistical analysis.
Aβ ELISA
For determination of Aβ42 levels, a commercially available ELISA kit was used (Invitrogen). Mouse hemi-brains were extracted with 5 mol/L guanidine HCl and incubated at room temperature for four hours. Homogenates were centrifuged to remove insoluble debris, supernatants were diluted 1:5000 in standard diluent buffer, and ELISA was carried out according to manufacture instructions. The plate was read using a microtiter plate reader. Optical density values were fitted against the standard curve and pmol/g wet brain calculated. The results were tabulated as mean ± SEM and compared using an unpaired t-test.
Histology
Mice were deeply anesthetized with Avertin and transcardially perfused with ice-cold 0.1 mol/L sodium phosphate buffer, followed by 4% paraformaldehyde in PBS as described previously.27 The brain was dissected, immediately removed from the cranium, and transferred to fresh 4% paraformaldehyde in PBS at 4°C overnight. The brains were subsequently cryoprotected by sinking in 30% sucrose/PBS at 4°C overnight. After bisecting across the midline, the brains were embedded in Optimal Cutting Temperature (OCT) Compound and 30 μm free-floating cryostat sections cut and stored in PBS at 4°C until use.
Thioflavine S Staining
Sections were pretreated in 10 mmol/L sodium citrate buffer (pH 6.0, 0.05% Triton-X100) for 15 minutes at 95°C to quench eGFP expression in CX3CR1-deficient animals, which reduced background fluorescence to that observed in non-transgenic controls (modified from Ref. 28; data not shown). Sections were mounted onto SuperPlus glass slides and air-dried and immersed in 1% Thioflavine S (in dH2O) for five minutes. Sections were subsequently hydrated in 70% ethanol and washed twice in PBS. The slides were coverslipped with 1:1 PBS:glycerol, and edges were sealed with nail polish.
Low-power images of cortex and hippocampus were compiled to form a series of overlapping images that were used to obtain an integrated image using Photoshop. The number of Thioflavine S–positive plaques in the cortex (below the pial surface and above the corpus callosum) and the hippocampus (excluding the subiculum and the fimbriae) was quantified. For all experiments, four separate sections, 360 μm apart, were quantified from each mouse, and all sections were taken from the same brain regions. All data were expressed as mean density or total count of Thioflavine S–positive plaques ± SEM and statistically significant alterations between the threes genotypes determined by one-way analysis of variance followed by Newman-Keuls post hoc test (GraphPad Prism, La Jolla, CA).
Immunohistochemistry
For diaminobenzidine immunohistochemistry, sections were first rinsed in PBS containing 0.1% Triton-X100 (PBST), pretreated with 10 mmol/L sodium citrate buffer (pH 6.0, 0.05% Triton-X100) for 15 minutes at 95°C for epitope retrieval. Sections were subsequently cooled for 30 minutes at room temperature, immersed in 0.3% hydrogen peroxide in PBS for 30 minutes to remove endogenous peroxidase activity, and blocked in 5% normal goat serum in PBS containing 0.3% Triton X-100 for one hour at room temperature. After incubation overnight at 4°C in blocking buffer containing either rat monoclonal anti-CD45 (diluted 1:500; Serotec, Oxford, UK), rat monoclonal anti-CD68 (diluted 1:500; Serotec, Oxford, UK), mouse monoclonal anti-human Aβ (clone 6E10; diluted 1:500; Signet Labs), rabbit polyclonal anti-glial fibrillary acidic protein (GFAP; diluted 1:500; Sigma-Aldrich), mouse monoclonal anti-phosphorylated tau (clone AT8; diluted 1:500; Thermo Scientific), or anti-APP (clone CT15; diluted 1:500) antibodies, slides were washed three times in PBST and incubated for one hour at room temperature in blocking buffer containing biotintylated anti-rat, anti-mouse, or anti-rabbit IgG antibody (diluted 1:200; Vector, Burlingame, CA). After three additional PBST washes, sections were incubated in Vectastain ABC Elite reagent (Vector) for one hour at room temperature and developed using diaminobenzidine with nickel chloride enhancement, according to the manufacturer’s instructions. Sections stained for CD45 or CD68 were additionally counterstained with Congo Red to visualize dense core Aβ deposits. Finally, free-floating sections were mounted onto SuperPlus glass slides and coverslipped with Permount.
For double fluorescent immunohistochemistry, sections were rinsed in PBST, pretreated with sodium citrate buffer to quench microglial eGFP, cooled for 30 minutes at room temperature, blocked for one hour at room temperature, and incubated overnight at 4°C in blocking buffer containing anti-Iba1 rabbit polyclonal antibody (diluted 1:1000; Wako, Richmond, VA) and the anti-Aβ mouse monoclonal antibody 4G8 (diluted 1:500; Signet Labs). After three PBST washes, sections were incubated for one hour at room temperature in blocking buffer containing secondary antibodies conjugated to fluorescent Alexa dyes (diluted 1:1000; Invitrogen). The sections were finally washed three times in PBST, incubated with TO-PRO-3 (diluted 1:1000 in PBS; Invitrogen) for 10 minutes at room temperature, mounted onto SuperPlus glass slides, and coverslipped with hard-set Vectashield mounting media (Vector). Controls included sections in which the fluorescent dyes were swapped as well as no primary antibody (data not shown).
Quantification of Microglia
Twenty- to thirty-μm thick Z-stacks were collected at ×40 magnification with 1 μm between each slice using a Leica SP5 confocal microscope, and images were postprocessed using ImageJ. To minimize any potential experimental bias, high-resolution confocal Z-stacks that spanned the entire thickness of the section were obtained and quantified via an unambiguous parameter (ie, number of cells rather than percent area or optical density). In addition, all sections were immunostained and imaged together using the same parameters. To determine the number of microglia surrounding each Aβ deposit, Iba1-positive cells whose nuclei or primary processes directly overlapped 4G8 immunoreactivity in a given plane of focus were counted. Only those microglia whose nuclei were present in the Z-stack were included in the analysis. The size of the Aβ deposits were calculated from arbitrarily thresholded maximum projections of 4G8 immunoreactivity and subsequently used to determine the number of microglia surrounding Aβ deposits of different sizes. Three random fields within the cortex were analyzed from three independent nonadjacent sections per mouse, with four animals analyzed per genotype. Data from 35 plaques of each size category were included for statistical analyses using one-way analysis of variance followed by Newman-Keuls post hoc test (GraphPad Prism) and presented as mean ± SEM.
RNA Extraction and Reverse Transcription-PCR
Hemi-brains of APPPS1;Cx3cr1+/+, APPPS1;Cx3cr1+/−, APPPS1;Cx3cr1−/−, and nontransgenic C57BL/6J animals were snap frozen and stored at −80°C until RNA extractions were performed. RNA was extracted using the Tri Reagent as described by the manufacturer (Invitrogen). Total RNA (50 ng/uL) was converted to cDNA using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems Inc. [ABI], Foster City, CA) and amplified using specific TaqMan probes on the ABI 7300 Real-Time PCR System. TaqMan probes used in the current studies include: chemokine (C-C motif) ligand 2 (CCL2; Mm00441241_ml), CX3CL1 (Mm01243036_ m1), interferon γ (IFNγ; Mm00801778_m1), interleukin 1 β (IL1β; Mm00434228_ml), IL4 (Mm00445259_m1), IL10 (Mm00439615_g1), IL6 (Mm00446191_m1), and tumor necrosis factor α (TNFα; Mm00443258_m1). The relative amount of each transcript was determined via normalization across all samples to the endogenous control, GAPDH (part #4308313) as recommended by ABI. In addition, before initiation of the studies, cDNA was serially diluted and amplified using the respective TaqMan probes to ensure that the amount of cDNA used was in the linear range. RNA samples from each individual animal were run in triplicate.
To quantify the relative expression levels of the different transcripts for each genotype, we calculated the difference (ΔCt) between the cycle threshold of the genes of interest (GOI) and the housekeeping gene Gapdh. From this data, the −ΔΔCt (−[ΔCtGOI −ΔCtB6]) was computed and converted to a value relative to the entire brain using the formula 2−ΔΔCt. The results were tabulated as mean ± SEM and compared between genotypes via one-way analysis of variance followed by Newman-Keuls post hoc test.
Cell Culture
Primary microglia were cultured from postnatal day 0–3 Cx3cr1+/+ and Cx3cr1−/− mice as described previously.29 Briefly, dissected cerebral cortices were sequentially rinsed with PBS containing 1 g/L glucose, minced, and digested with 0.05% trypsin-EDTA for 20 minutes at 37°C. Cells were triturated in DMEM/F12 containing 20% heat-inactivated fetal bovine serum (HI-FBS) and 1% penicillin-streptomycin and plated in cell culture flasks. Media was changed the next day to DMEM/F12 with 10% HI-FBS.
After 14 days of incubation, microglia were isolated as described previously30 by removal of astrocytes by incubation in 0.25% trypsin/HBSS diluted 1:4 in serum-free DMEM for 45 minutes at 37°C. 0.25% trypsin/PBS was then added to remove adherent microglia, and cells were replated in DMEM containing 2% HI-FBS at a density of 250,000 per well in 6-well plates.
Phagocytosis Assay
The phagocytosis assay was modified from previous studies.31,32,33 After overnight incubation in serum-free DMEM in the absence or presence of 100 nmol/L CX3CL1 (chemokine domain; R&D Systems, Minneapolis, MN), Nile Red fluorescent microspheres (Invitrogen) washed in PBS containing 1 mg/ml BSA were added to the cells for 90 minutes. Cells were then extensively washed with PBS and fixed in 2% paraformaldehyde. The numbers of phagocytosed microspheres per cell were counted from random fields totaling 60–100 cells on an inverted microscope. The results were tabulated as mean ± SEM and compared between genotypes using an unpaired t-test. Concentration of CX3CL1 used in the study has been shown to promote microglial survival34 and cell adhesion in vitro.35
To confirm uptake, cells plated on coverslips were immunostained with antibody against Iba1. After permeabilization with 0.1% PBST for 10 minutes at room temperature, cells were blocked in 5% normal goat serum in PBS containing 0.3% Triton X-100 for one hour at room temperature. Cells were then incubated with rabbit polyclonal Iba1 antibody (Wako) diluted 1:1000 in blocking buffer for 1 hour at room temperature, washed with PBS, incubated with Alexa 648 conjugated anti-rabbit IgG secondary antibody (Invitrogen) for one hour at room temperature, washed with PBS, and coverslipped with Vectashield mounting media (Vector). Single plane images were taken using a Leica SP5 confocal microscope.
Preparation of Fibrillar Aβ
Unlabeled Aβ1-42 (American Peptide Company, Sunnyvale, CA) or HiLyte 555-conjugated Aβ1-42 (AnaSpec, Fremont, CA) was diluted 1 mg/ml in ddH2O and fibrillarized for 24 hours at 37°C as previously described.29 Control Aβ42-1 peptide (Invitrogen), composed of same amino acids as Aβ1-42 but in reverse sequence, was prepared in the same manner.
Stereotaxic Intracerebral Aβ Microinjection
Five- to seven-month-old Cx3cr1+/+ and Cx3cr1−/− were deeply anesthetized with ketamine/xylazine cocktail (100/10 mg/kg) and immobilized in a Kopf stereotaxic apparatus. A 0.5-mm burr hole was drilled in the skull, and 2 μl of fibrillar Aβ1-42 containing solution was injected at coordinates anteroposterior: −1.0 mm, lateral: 2.0 mm, and dorsoventral: 1.0 mm with respect to bregma in the right cortex using a 10-μl Hamilton syringe. Some animals were injected with 2 μl of Aβ42-1-containing solution at mirroring coordinates on the left cortex, providing an internal control. Mice were sacrificed 48 hours after injection and prepared for histology as described above. Coronal sections containing the needle track were immunostained for Iba1 (and for Aβ using the 6E10 antibody in animals in which unlabeled Aβ1-42 or Aβ42-1 was injected) and visualized using appropriate Alexa dye-conjugated secondary antibodies. Iba1-positive microglia within 50 μm of the needle track were counted and examined for the presence of intracellular Aβ in confocal Z-stacks spanning 20–30 μm in depth to calculate percent phagocytic microglia. Four Aβ1-42 injected brains and three Aβ42-1 injected brains were analyzed per genotype. The results were tabulated as mean ± SEM and compared using an unpaired t-test. Unconjugated and HiLyte 555-conjugated Aβ1-42 injections triggered identical microglial reactions (data not shown), and thus the results were combined into a single group. Furthermore, Aβ42-1-injected Cx3cr1+/+ and Cx3cr1−/− controls showed no significant differences in microglial counts (data not shown). Finally, control sections incubated with fluorescent anti-mouse IgG secondary antibodies reveled no specific labeling (data not shown), thereby ruling out nonspecific mouse IgG binding at the injection site.
Results
CX3CR1 Deficiency Results in Reduced Aβ Deposition in APPPS1 Mice
APPPS1 mouse line coexpresses APP K670M/N671L and PS1 L166P mutations under the control of the neuron-specific Thy1 promoter and exhibits early and robust Aβ deposition beginning at 6–8 weeks of age in the cortex and 3–4 months of age in the hippocampus.21 As reported previously, nearly all plaques are fibrillar. There is also marked microglial activation around the Aβ deposits from the earliest time-points.
Cx3cr1 knockout animals were bred to the rapid-onset APPPS1 mouse model of AD to generate the following genotypes: APPPS1;Cx3cr1+/+, APPPS1;Cx3cr1+/−, and APPPS1;Cx3cr1−/−. Brain sections from four-month-old mice were stained with Thioflavine S, a fluorescent dye specific for fibrillar Aβ. Consistent with published reports, APPPS1;Cx3cr1+/+ controls exhibited numerous fibrillar Aβ deposits in the cortex and hippocampus (Figure 1A). Surprisingly, however, APPPS1;Cx3cr1+/− (Figure 1B) and APPPS1;Cx3cr1−/− (Figure 1C) animals demonstrated a gene dose-dependent reduction in Thioflavine S–positive Aβ deposits in both the cortex as well as the hippocampus. In a quantitative analysis of fibrillar Aβ burden, cortical Thioflavine S positive Aβ deposition was reduced by 35% in APPPS1;Cx3cr1+/− and by 57% in APPPS1;Cx3cr1−/− mice when compared to APPPS1;Cx3cr1+/+ controls (P < 0.001; Figure 1D). The APPPS1;Cx3cr1−/− mice demonstrated significantly reduced cortical deposition when compared to APPPS1;Cx3cr1+/− mice, indicating a gene dose-dependent effect (P < 0.05). Aβ deposition was also reduced in the hippocampus by 70% and 85% in the APPPS1;Cx3cr1+/− and APPPS1;Cx3cr1−/− animals (P < 0.001), when compared to control mice (Figure 1E). To determine whether the Aβ deposits recognized by Thioflavine S were representative of the total Aβ within the brain, Western blots of Cx3cr1+/+, APPPS1;Cx3cr1+/+, and APPPS1;Cx3cr1−/− brain extracts were probed with the human-specific anti-Aβ monoclonal antibody 6E10 (Figure 1F). Confirming results obtained with Thioflavine S, APPPS1;Cx3cr1−/− brains exhibited drastically reduced levels of biochemically extracted Aβ when compared to APPPS1;Cx3cr1+/+ brains. In support of the Thioflavine S straining, ELISA analysis revealed reduced Aβ42 levels in APPPS1;Cx3cr1−/− brains (1375 ± 803 pmol/g wet brain; Figure 1G) when compared to APPPS1;Cx3cr1+/+ brains (4608 ± 861 pmol/g wet brain; Figure 1G). Finally, 6E10 immunohistochemistry also confirmed a gene dose-dependent reduction Aβ deposits in the cortex (see Supplemental Figure S1 at http://ajp.amjpathol.org) and hippocampus (data not shown). Taken together, these data indicate that CX3CR1 deficiency results in a gene dose-dependent reduction in Aβ deposition in the APPPS1 mouse model of AD.
Figure 1.
Gene dose-dependent reduction in fibrillar Aβ deposition in the APPPS1 mouse model of AD with CX3CR1 deficiency. Brain sections (30 μm) from APPPS1;Cx3cr1+/+ (A, n = 7), APPPS1;Cx3cr1+/− (B, n = 6), and APPPS1;Cx3cr1−/− (C, n = 7) mice at 4 months of age were stained with Thioflavine S. A series of low-power images (Scale bar = 1 mm) were used to reconstruct the cortex and hippocampus from four sections from each animal. As expected based on the published literature,21 APPPS1;Cx3cr1+/+ animals exhibit abundant fibrillar Aβ deposition throughout the cortex with reduced Aβ deposition in the hippocampus (A). By contrast, age-matched APPPS1;Cx3cr1+/− (B) and APPPS1;Cx3cr1−/− (C) animals exhibited a gene dose-dependent reduction in fibrillar Aβ deposition throughout the cortex and hippocampus. Quantification of Thioflavine S staining in the cortex (D) and hippocampus (E) across all animals revealed a statistically significant decrease in APPPS1 mice with either one or two copies of Cx3cr1 loss of function alleles when compared with age-matched APPPS1;Cx3cr1+/+ controls (*P < 0.001). Notably, there was also a significant difference in Aβ deposition in the cortex between APPPS1;Cx3cr1+/− and APPPS1;Cx3cr1−/− genotypes (P < 0.05). To confirm results obtained using Thioflavine S staining, Western blots of brain extracts from Cx3cr1+/+, APPPS1;Cx3cr1+/+, and APPPS1;Cx3cr1−/− animals were probed with antibodies to human Aβ and GAPDH as a loading control. Consistent with Thioflavine S staining, APPPS1;Cx3cr1−/− brains exhibited lower steady-state Aβ levels when compared with age-matched APPPS1;Cx3cr1+/+ controls (F). Furthermore, ELISAs performed on APPPS1;Cx3cr1+/+ (n = 6) and APPPS1;Cx3cr1−/− brain homogenates (n = 5) revealed reduced Aβ42 levels in CX3CR1-deficient animals (G; *P < 0.05).
CX3CR1 Deficiency Results in Reduced Aβ Deposition in R1.40 Mice
To address the effect of CX3CR1 deficiency in another AD mouse model, we bred Cx3cr1 knockout animals to the R1.40 mouse model of AD. The R1.40 transgene is a full genomic copy of the human APP gene expressing the K670M/N671L mutation carried on a yeast artificial chromosome. The R1.40 transgenic mouse line develops gradual AD-like Aβ pathology, exhibiting both diffuse and fibrillar Aβ deposition beginning at 14–16 months of age in the cortex and 18–20 months of age in the hippocampus.36 Notably, fibrillar Aβ deposits in R1.40 mice are associated with reactive microglia.
Brain sections from R1.40;Cx3cr1+/+, R1.40;Cx3cr1+/−, and R1.40;Cx3cr1−/− animals at 20–24 months of age were stained with Thioflavine S to visualize fibrillar Aβ. R1.40;Cx3cr1+/+ controls exhibited abundant Thioflavine S positive fibrillar Aβ deposits in the cortex (see Supplemental Figure S2 at http://ajp.amjpathol.org). Similar to the results in CX3CR1-deficient APPPS1 mice, R1.40;Cx3cr1+/− and R1.40;Cx3cr1−/− animals also demonstrated a gene dose-dependent reduction in Thioflavine S–positive Aβ deposits in the cortex. Quantification of the total number of Thioflavine S–positive Aβ deposits in the cortex revealed that R1.40;Cx3cr1+/− and R1.40;Cx3cr1−/− exhibited markedly fewer cortical Thioflavine S–positive plaques when compared to site-matched brain regions in R1.40;Cx3cr1+/+ mice (P < 0.05). Taken together, these data indicate that CX3CR1 deficiency results in a gene dose-dependent reduction in Aβ deposition in mouse models of AD, regardless of the kinetics of deposition.
CX3CR1 Deficiency Reduces Aβ-Associated Dystrophic Neurites in APPPS1 and R1.40 Mice
To assess neuronal integrity in the current studies, several additional experiments were performed. First, TUNEL and cleaved caspase 3 staining of brain sections from CX3CR1-deficient APPPS1 and R1.40 animals did not reveal any gross alterations in neurodegeneration (data not shown), consistent with previous studies demonstrating no overt neurodegeneration in either the APPPS1 or R1.40 animals.21,36 Second, additional staining with antibodies for dystrophic neurites, including APP (antibody CT15) and tau phosphorylation (antibody AT8) revealed a reduction in staining in both the APPPS1;Cx3cr1−/− (See Supplemental Figure S3 at http://ajp.amjpathol.org) and R1.40;Cx3cr1−/− animals (data not shown) that likely reflects the reduction in Aβ deposition observed in these mice.
CX3CR1 Deficiency Does Not Alter APP Levels or APP Processing
One potential explanation for the alterations in Aβ deposition observed in the APPPS1;Cx3cr1+/− and APPPS1;Cx3cr1−/− animals was that either APP transgene expression or APP processing was altered. To examine this possibility, brain extracts from two-month-old APPPS1;Cx3cr1+/+, APPPS1;Cx3cr1+/−, and APPPS1;Cx3cr1−/− animals were analyzed by Western blot for the levels of holo-APP and APP CTFs. No significant differences in the steady state levels of holo-APP were observed in the samples from three different genotypes (Figure 2, A [top] and B). The steady-state levels of APP CTFs, including CTFβ, the direct precursor of Aβ, were unaltered (Figure 2, A [middle] and C). Similar results were also obtained in the R1.40 mouse model of AD with CX3CR1 deficiency (see Supplemental Figure S4 at http://ajp.amjpathol.org). In addition, there were no alterations in the steady-state levels of brain Aβ in pre-depositing APPPS1 and R1.40 mice (data not shown). Taken together, these results indicate that the differences in Aβ deposition observed in this study are not due to altered expression or processing of APP by neurons and instead suggest involvement of brain microglia, where CX3CR1 is exclusively expressed.
Figure 2.
Lack of effect of CX3CR1 genotype on APP processing. A: Western blots of brain extracts from the APPPS1;Cx3cr1+/+ (Lanes 1–3), APPPS1;Cx3cr1+/− (Lanes 4–6). and APPPS1;Cx3cr1−/− (Lanes 7–9) animals were probed with an antibody to the C terminus of APP (top two panels) and subsequently stripped and reprobed with an antibody against α-tubulin as a loading control. Shown on the right is the approximate size in kDa. B: Relative levels of holo-APP were quantified (n = 8) from each genotype by normalizing the intensity values of APP to α-tubulin. C: Relative values of CTFβ were quantified similarly averaging the values of APP CTFβ to α-tubulin. No significant differences were observed in the relative levels of either holo-APP or CTFβ between genotypes.
CX3CR1 Deficiency Leads to an Altered Neuroinflammatory Milieu
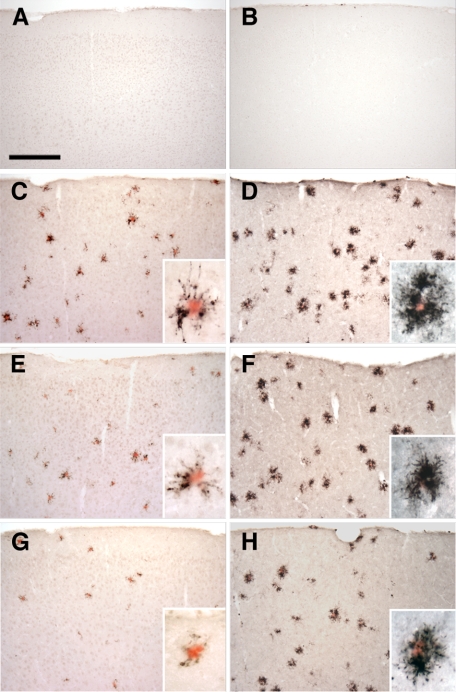
One potential explanation for the alterations in Aβ deposition observed in the APPPS1;Cx3cr1+/− and APPPS1;Cx3cr1−/− animals was that loss of CX3CL1-CX3CR1 signaling leads to altered microglial activation. To examine this possibility, brain sections from four-month-old wild-type C57BL/6J mice (Figure 3, A and B), APPPS1;Cx3cr1+/+ (Figure 3, C and D), APPPS1;Cx3cr1+/− (Figure 3, E and F), and APPPS1;Cx3cr1−/− (Figure 3, G and H) were immunostained with either an antibody to CD68 (Figure 3, A, C, E, and G), a marker for macrophages (including microglia-derived macrophages), or CD45 (Figure 3, B, D, F, and H), a marker for cells of the hematopoietic lineage which is moderately up-regulated in activated microglia. Sections were subsequently counterstained with the amyloid dye Congo Red to visualize dense-core Aβ deposits. Wild-type non-transgenic sections demonstrated no Congo Red–positive deposits and sparse CD68 (Figure 3A) and CD45 immunoreactivity (Figure 3B). CD68- and CD45-postive microglia are closely associated with Congo Red–positive deposits in all APPPS1 genotypes (Figure 3, insets). CD68 immunoreactivity was qualitatively reduced in APPPS1;Cx3cr1+/− (Figure 3E) and APPPS1;Cx3cr1−/− animals (Figure 3G) compared to APPPS1;Cx3cr1+/+ controls (Figure 3C). By contrast, CD45 immunoreactivity between the APPPS1;Cx3cr1+/+ (Figure 3D), APPPS1;Cx3cr1+/− (Figure 3F) and APPPS1;Cx3cr1−/− (Figure 3H) genotypes appeared relatively unchanged. These data suggest that CX3CR1 deficiency alters the microglial activation program in reaction to dense-core Aβ deposits.
Figure 3.
Altered expression of microglial markers in the APPPS1 mouse model of AD with CX3CR1 deficiency. Brain sections (30 μm) from wild-type C57BL/6J (A and B), APPPS1;Cx3cr1+/+ (C and D), APPPS1;Cx3cr1+/− (E and F), and APPPS1;Cx3cr1−/− (G and H) mice at 4 months of age were immunostained with antibodies to either CD68, a marker for phagocytic microglia/macrophages (A, C, E, and G), or CD45, a marker for cells of hematopoietic lineage which is up-regulated in activated microglia (B, D, F, and H). Sections were also counterstained with Congo Red, a dye specific for dense-core Aβ deposits. Wild-type sections contained no Congo Red–positive deposits and rare CD68 (A) and CD45 (B) immunoreactivity. Consistent with the published literature, APPPS1;Cx3cr1+/+ animals show abundant CD68 (C) and CD45 immunoreactivity (D), especially in the immediate vicinity of Congo Red–positive Aβ deposits (insets). While age-matched APPPS1;Cx3cr1+/− and APPPS1;Cx3cr1−/− animals exhibit gene dose-dependent reduction in CD68 immunoreactivity compared to APPPS1;Cx3cr1+/+ controls (E and G, respectively), CD45 immunoreactivity is relatively unchanged in APPPS1;Cx3cr1+/− and APPPS1;Cx3cr1−/− animals compared with APPPS1;Cx3cr1+/+ controls (F and H, respectively). As expected, higher magnification revealed that CD68- and CD45-positive microglia are mostly clustered around Congo Red–positive Aβ deposits (insets). Scale bar = 250 μm.
To determine whether microglial CX3CR1 signaling mediates non–cell-autonomous effects in other inflammatory cells in the brain, sections from four-month-old wild-type, APPPS1;Cx3cr1+/+, and APPPS1;Cx3cr1−/− animals were stained with an antibody to GFAP, an astrocytic marker and Congo Red (see Supplemental Figure S5 at http://ajp.amjpathol.org). As expected, wild-type controls exhibited no Congo Red–positive Aβ deposits and GFAP immunoreactivity was limited to the meninges and perivascular regions. By contrast, APPPS1; Cx3cr1+/+ mice exhibited abundant Congo Red–positive Aβ deposits that were ringed by numerous GFAP-positive astrocytes. In addition to a substantial reduction in the amount of Aβ deposition in the APPPS1;Cx3cr1−/− animals (Figure 1), there was also significantly reduced GFAP immunoreactivity. Taken together, these results indicate that, in addition to alterations in microglial activation, there is also reduced astrocytosis in CX3CR1-deficient APPPS1 mice, although both of these alterations could be due to the reduced Aβ deposition observed.
CX3CR1 Deficiency Leads to Reductions in Plaque-Associated Microglia
Our immunohistochemical studies suggested that CX3CR1 deficiency leads to reductions in CD68-positive microglia. This finding could be due to the reduced Aβ deposition observed in CX3CR1-deficient mice. To address this possibility, we quantified the number of microglia on a per-plaque basis. Brain sections from four-month-old APPPS1;Cx3cr1+/+ (Figure 4, A–C), APPPS1;Cx3cr1+/− (Figure 4, D–F), and APPPS1;Cx3cr1−/− mice (Figure 4, G–I) were coimmunostained with monoclonal anti-Aβ antibody, 4G8 (red; Figure 4, A, D, and G) and an antibody against the pan-microglial marker Iba1 (green; Figure 4, B, E, and H). Sections were subsequently counterstained with a TO-PRO-3 nuclear dye (blue; Figure 4C, F, and I) and imaged via confocal microscopy to obtain Z–stacks. We observed a gene dose-dependent reduction in the number of plaque-associated microglia in the cortex of APPPS1;Cx3cr1+/− (Figure 4, D–F) and APPPS1;Cx3cr1−/− (Figure 4, G–I) animals when compared to APPPS1;Cx3cr1+/+ controls (Figure 4, A–C).
Figure 4.
Reduced plaque-associated microglia in the APPPS1 mouse model of AD with CX3CR1 deficiency. Brain sections (30 μm) from APPPS1;Cx3cr1+/+ (A–C), APPPS1;Cx3cr1+/− (D–F), and APPPS1;Cx3cr1−/− (G–I) mice at 4 months of age were immunostained with monoclonal Aβ antibody 4G8 (red; A, D, and G), with an antibody against the pan-microglial marker Iba1 (green; B, E, and H), and counterstained with the nuclear TO-PRO-3 dye (blue, C, F, and I). Confocal microscopy was used to obtain maximum projections reconstructed from Z-stacks spanning 20–30 μm in depth. As expected, APPPS1;Cx3cr1+/+ controls exhibit extensive accumulation of Iba1-positive microglia around senile plaques (A–C). However, age-matched APPPS1;Cx3cr1+/− (D–F) and APPPS1;Cx3cr1−/− animals (G–I) exhibit gene dose-dependent reduction in the number of Iba1-positive microglia surrounding the senile plaques. The number of Iba1-positive microglia associated with senile plaques in the three genotypes was quantified in three nonadjacent sections from each of the four animals per genotype. APPPS1 mice with either one or two copies of Cx3cr1 loss-of-function alleles exhibited a statistically significant reduction in microglia surrounding both large (>1000 μm2; J), medium (>500 μm2, <1000 μm2; K), and small (<500 μm2; L) Aβ deposits when compared to age-matched APPPS1;Cx3cr1+/+ controls (*P < 0.05; **P < 0.001, respectively). The APPPS1;Cx3cr1+/− and APPPS1;Cx3cr1−/− genotypes exhibited a statistically significant difference in microglial accumulation around medium Aβ deposits (P < 0.05), suggesting a gene dose-dependent effect. n = 35 for all analyses. Scale bar = 25 μm.
To evaluate the possibility that CX3CR1 signaling to microglia might mediate a differential response to larger plaques, Aβ deposits were grouped based on area and numbers of Iba1-positive microglia whose nuclei or primary processes directly overlapped Aβ deposits in a given tissue section were quantified. Large plaques (>1000 μm2) in APPPS1;Cx3cr1+/+ controls exhibited an average of ∼13 plaque associated microglia, whereas those in APPPS1;Cx3cr1+/− and APPPS1;Cx3cr1−/− animals exhibited only ∼11 and ∼9 plaque-associated microglia, respectively (Figure 4J). For medium plaques (>500 μm2 and <1000 μm2), APPPS1;Cx3cr1+/− and APPPS1;Cx3cr1−/− mice demonstrated an average of ∼7 and ∼5 plaque associated microglia, respectively, which was significantly lower than ∼9 plaque associated microglia in the medium plaques of APPPS1;Cx3cr1+/+ controls (Figure 4K). In addition, for medium plaques, APPPS1;Cx3cr1−/− mice exhibited a significant decrease in plaque associated microglia when compared to APPPS1;Cx3cr1+/− mice, indicating a gene dose-dependent effect. For small plaques (<500 μm2), differences were not statistically significant among the genotypes (Figure 4L). In summary, these results indicate that CX3CR1 deficiency reduces the number of microglia around Aβ deposits in a gene dose-dependent manner in the APPPS1 mouse model of AD.
CX3CR1 Deficiency Leads to Altered Expression of Inflammatory Cytokines/Chemokines
To gain insight into the potential mechanisms underlying the concomitant reduction in Aβ deposition and plaque-associated microglia, the expression levels of various cytokines and chemokines was examined by quantitative RT-PCR. Total brain RNA from four-month-old B6 controls, APPPS1;Cx3cr1+/+, APPPS1;Cx3cr1+/−, and APPPS1;Cx3cr1−/− mice was reverse transcribed and assayed in triplicate for expression of the CCL2, CX3CL1, IFNγ, IL1β, IL4, IL6, IL10, and TNFα mRNAs using TaqMan probes. Expression of both TNFα and CCL2 mRNAs were increased in APPPS1;Cx3cr1+/+ mice at four months of age relative to B6 control mice (Figure 5, A and C, respectively). Notably, age-matched APPPS1;Cx3cr1+/− and APPPS1;Cx3cr1−/− mice demonstrated reduced expression levels of both TNFα (P < 0.001) and CCL2 mRNAs (P < 0.01) relative to APPPS1;Cx3cr1+/+ mice (Figure 5, A and C, respectively). By contrast, expression of IL1β mRNA was up-regulated in APPPS1;Cx3cr1+/− and APPPS1; Cx3cr1−/− animals (P < 0.05 and P < 0.01, respectively; Figure 5B) as previously reported in other neuroinflammatory models.13 Levels of mRNAs for CX3CL1, IFNγ, IL4, IL6, and IL10 did not exhibit significant differences among the genotypes relative to B6 control mice (data not shown). These results suggest that there are alterations in the expression of the chemokines/cytokines CCL2, IL1β, and TNFα in the brains of the APPPS1 mouse model of AD with CX3CR1 deficiency.
Figure 5.
Altered expression of inflammatory markers in CX3CR1-deficient APPPS1 mouse model of AD. Relative mRNA expression of TNFα (A), IL1β (B), and MCP-1 (C) in APPPS1;Cx3cr1+/+ (n = 7), APPPS1;Cx3cr1+/− (n = 8), and APPPS1;Cx3cr1−/− (n = 9) mice at 4 months. The mRNA levels for each cytokine were normalized to the mRNA levels of GAPDH and expressed relative to that of nontransgenic C57BL/6J mice. Results from APPPS1;Cx3cr1+/− and APPPS1;Cx3cr1−/− animals were compared with results from age-matched APPPS1;Cx3cr1+/+ controls (*P < 0.05, **P < 0.01, ***P < 0.001).
CX3CR1 Signaling Inhibits Microglial Phagocytosis
Our observations that reduced amyloid deposition in CX3CR1-deficient mice, along with less microglia per plaque, were somewhat counterintuitive. One potential explanation is that CX3CL1-CX3CR1 signaling could alter the phagocytic capacity of microglia. To examine this possibility, primary microglia from Cx3cr1+/+ (Figure 6, A and B) and Cx3cr1−/− animals (Figure 6, C and D) were assessed for their ability to phagocytose fluorescent microspheres. On average, Cx3cr1+/+ microglia phagocytosed ∼4 microspheres per cell (Figure 6, A and E) whereas CX3CL1-treated Cx3cr1+/+ microglia phagocytosed only ∼2 microspheres per cell (Figure 6, B and E; P < 0.05). Cx3cr1−/− microglia demonstrated equivalent phagocytic responses either with (Figure 6, C and E) or without fractalkine treatment (Figure 6, D and E), indicating that CX3CL1-mediated inhibition of microglial phagocytic capacity required CX3CR1 signaling. These results suggest that alterations in CX3CL1-CX3CR1 signaling can lead to altered phagocytic capabilities of microglia and could account for the apparent discrepancy between the reduced numbers of microglia per plaque and reduced Aβ deposition observed in CX3CR1-deficient animals.
Figure 6.
CX3CL1-CX3CR1 signaling inhibits microglial phagocytosis. Cx3cr1+/+ (A and B) and Cx3cr1−/− (C and D) primary microglia were incubated in the absence (A and C) or presence (B and D) of CX3CL1 (100 nmol/L) overnight, and fluorescent microspheres were added for 90 minutes. The cells were fixed and stained with antibody against Iba1 and visualized with Alexa 648-conjugated secondary antibody (pseudo-colored green; A–D). Single-plane confocal images are shown. CX3CL1-treated Cx3cr1+/+ microglia exhibited reduced numbers of phagocytosed microspheres per cell when compared with untreated Cx3cr1+/+ and CX3CL1-treated Cx3cr1−/− microglia (E; *P < 0.05). n = 3 for each group. Scale bar = 15 μm.
CX3CR1 Deficiency Enhances Aβ Phagocytosis
Our in vitro data suggested that CX3CL1-CX3CR1 signaling inhibits microglial phagocytic capacity. To examine whether loss of CX3CL1-CX3CR1 signaling could lead to enhanced uptake of Aβ in vivo and thus account for findings in the CX3CR1-deficient AD mouse models, fibrillar Aβ1-42 was stereotactically injected into the cortices of Cx3cr1+/+ (Figure 7A) and Cx3cr1−/− animals (Figure 7B). On average, 46% of Cx3cr1+/+ microglia within 50 μm of the needle track contained fibrillar Aβ, whereas 66% of Cx3cr1−/− microglia contained Aβ (Figure 7D; P < 0.01). Notably, however, both Cx3cr1+/+ and Cx3cr1−/− microglia accumulated in similar numbers near the fibrillar Aβ1-42 injection sites (Figure 7E; P < 0.05), suggesting that there was no significant alteration in the ability of microglia to migrate to the Aβ injection site. Stereotaxic injections with a control Aβ42-1 peptide triggered significantly reduced microglial reaction in both the Cx3cr1+/+ and Cx3cr1−/− brains (Figure 7, C and E; P < 0.05), indicating that the microglial responses were specific to fibrillar Aβ1-42. In addition, the Cx3cr1−/− microglia in the Aβ1-42 injected animals exhibited a different morphology, with a more rounded appearance, than either Cx3cr1+/+ Aβ1-42 injected animals or Aβ42-1 injected controls (compare Figure 7B to 7A and 7C), which is consistent with an altered phagocytic capacity of these cells. These data demonstrate that blocking CX3CL1-CX3CR1 signaling leads to enhanced uptake of fibrillar Aβ within microglia and could thus explain the reductions in Aβ deposition observed in CX3CR1-deficient APPPS1 and R1.40 animals.
Figure 7.
Enhanced Aβ phagocytosis in CX3CR1-deficient mice. Five- to 7-month-old Cx3cr1+/+ (n = 4; A) and Cx3cr1−/− (n = 4; B) mice were injected with fibrillar Aβ1-42 (red; HiLyte 555-conjugated) or with control Aβ42-1 peptide (n = 6; C). Sections (30 μm) containing the needle track were immunostained with an antibody against Iba1 and visualized with Alexa 648-conjugated secondary antibody (pseudo-colored green; A–C). From confocal Z-stacks spanning 20–30 μm in depth, we obtained maximum projections (A–C) and slices in the x- and y-plane showing Aβ internalization (A and B). The number of microglia (E) and percent phagocytic microglia (D) within 50 μm of the needle track were quantified. Cx3cr1−/− microglia exhibited enhanced Aβ phagocytosis when compared with control Cx3cr1+/+ microglia (*P < 0.01; D), whereas the number of microglia around the injection site did not significantly differ between genotypes (E). Furthermore, as expected, control Aβ42-1 peptide injection triggered significantly reduced microglial reaction compared to fibrillar Aβ1-42 injection (C and E; *P < 0.05).
Discussion
The findings presented here offer insights into the role of microglial CX3CR1 signaling in the development of amyloid pathology in the APPPS1 and R1.40 mouse models. Previous studies showed that in the CNS, CX3CL1 is expressed by neurons and signals to CX3CR1 on microglia.13,14 Furthermore, recent reports implicated impaired CX3CL1-CX3CR1 signaling in neurodegeneration. First, in various models of neurodegeneration, including the 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine model of Parkinson’s disease; a transgenic mouse model of amyotrophic lateral sclerosis; and induction of neuroinflammation via systemic lipopolysaccharide administration, CX3CR1 deficiency resulted in increased neuronal cell death and worsening of phenotypes via mechanisms cell-autonomous to microglia.13 Second, Cx3cr1 knockout mice exhibited age-dependent subretinal microglial accumulation and retinal degeneration.37,38 Third, the V249I and T280M polymorphisms in CX3CR1 are associated with altered risk for age-related macular degeneration in humans.17,18 Taken together these results support a role of CX3CL1-CX3CR1 signaling to microglia in modulating neurodegenerative disease phenotypes.
The present studies tested the hypothesis that CX3CR1 deficiency would alter AD phenotypes in transgenic mouse models of the disease. Unexpectedly, CX3CR1 deficiency in two different mouse models of AD leads to an amelioration of the amyloid pathology in both the rapid onset APPPS1 (Figure 1) as well as the gradual onset R1.40 (see Supplemental Figure S2 at http://ajp.amjpathol.org) mouse models of AD. To examine the effects of CX3CR1 deficiency on neurons, TUNEL and cleaved caspase 3 staining of brain sections from CX3CR1-deficient APPPS1 and R1.40 animals revealed no evidence for any gross neurodegeneration (data not shown) while staining with antibodies for dystrophic neurites (CT15 and AT8) revealed a reduction in staining in both the APPPS1;Cx3cr1−/− (see Supplemental Figure S3 at http://ajp.amjpathol.org) and R1.40;Cx3cr1−/− animals (data not shown) that appears to reflect the reduction in Aβ deposition observed in these mice.
Significantly, we also observed a gene dose-dependent effect of CX3CR1 deficiency, with heterozygous Cx3cr1 mice exhibiting an intermediate phenotype. This suggests that alterations in CX3CR1 signaling could influence human AD pathogenesis, a hypothesis that could be tested via genetic association studies with CX3CR1 variants implicated in age-related macular degeneration or via therapeutic strategies aimed at reducing CX3CR1 signaling. The finding of a heterozygous phenotype also has implications for previous studies in the field, as heterozygous Cx3cr1GFP/+ mice have been used to study microglial dynamics in AD mouse models.3,5
Multiple mechanisms could account for the reduction in Aβ deposition observed in the CX3CR1-deficient mouse models of AD. To determine whether CX3CR1 deficiency altered APP production or processing, we showed that the levels of steady-state APP and APP CTFs remained unchanged in CX3CR1 deficient APPPS1 and R1.40 mice (Figure 2 and see Supplemental Figure S4 at http://ajp.amjpathol.org). Given that CX3CR1 is exclusively expressed by microglia within the brain and the effects of CX3CR1 deficiency are autonomous to microglia,13 we postulated that the effect of CX3CR1 deficiency on Aβ deposition was mediated via alterations within microglia.
Analysis of microglial markers demonstrated a gene dose-dependent reduction in both the numbers of Iba1-positive microglia surrounding individual Aβ deposits (Figure 4) as well as staining for the microglial activation marker CD68 (Figure 3) in CX3CR1-deficient APPPS1 mice. Thus, our results showed a reduction in the amount of Aβ deposition and alterations in the activation of microglia surrounding the Aβ deposits. These observations were unexpected, as it seemed more likely that reduced amyloid burden might be mediated by augmented microglial activation.
In addition to alterations in microglial activation, we also observed a substantial reduction in astrocytosis in CX3CR1-deficient APPPS1 animals (see Supplemental Figure S5 at http://ajp.amjpathol.org), suggesting that there is an altered neuroinflammatory milieu within these animals. However, this was not unexpected given that there is reduced Aβ deposition in these animals. In addition, because CX3CR1 is exclusively expressed by microglia in the murine CNS,13,15 the reduced astrogliosis is by definition a secondary response to primary alterations in microglia. Thus, while we cannot formally exclude the role of astrocytes in the reduced Aβ deposition observed in CX3CR1-deficient animals, the current studies point to enhanced Aβ phagocytosis by Cx3cr1−/− microglia as the more proximal and significant contributor.
Interestingly, we also found selective alterations in the levels of brain mRNAs for specific cytokines/chemokines depending on the genotype of the animal (Figure 5). Similar to previous findings in AD brains and mouse models,39,40,41,42 the levels of CCL2 and TNFα mRNAs were elevated in the APPPS1;Cx3cr1+/+ mouse brain. Notably, CX3CR1 deficiency reduced the levels of CCL2 and TNFα mRNA to that observed in the non-transgenic controls, while brain levels of IL1β mRNA were increased.
In addition, in vitro phagocytosis assay revealed that CX3CL1-CX3CR1 signaling regulates the phagocytic capacity of microglia (Figure 6). CX3CL1 treatment of primary microglia led to a 50% reduction in the number of phagocytosed microspheres in wild-type Cx3cr1+/+ primary microglia. Importantly, the phagocytic capacity of Cx3cr1−/− primary microglia did not change with CX3CL1 treatment, indicating that the reduced microglial phagocytosis observed is mediated by CX3CR1 signaling. We did not observe an enhancement of phagocytosis in Cx3cr1−/− compared to untreated Cx3cr1+/+ microglia.
Finally, we confirmed that microglial Aβ phagocytosis was enhanced in CX3CR1-deficient APPPS1 animals (Figure 7). Sixty-six percent of Cx3cr1−/− microglia around fibrillar Aβ1-42 injection sites were phagocytic, whereas only 46% of Cx3cr1+/+ microglia were phagocytic. However, Cx3cr1+/+ and Cx3cr1−/− microglia accumulated in similar numbers around the injection sites, suggesting that microglial migration was not impaired by CX3CR1 deficiency.
In the AD brain, microglia accumulate at the site of newly formed Aβ deposits via migration or local proliferation and may help restrict plaque growth.4,5 We hypothesize that the drastic reduction in Aβ deposition in CX3CR1 deficient R1.40 and APPPS1 animals is due to alterations in the ability of CX3CR1 deficient microglia to i) remove Aβ, and/or ii) accumulate around Aβ deposits via proliferation and/or migration. The postulated sequence of events is that Aβ accumulation is associated with neuronal stress and CX3CL1 proteolytic release,43 leading to reduced phagocytosis by Cx3cr1+/+ but not Cx3cr1−/− microglia.
Microglial Removal of Aβ
Recent in vivo imaging studies using mouse models of AD have shown that microglia may help restrict the growth of senile plaques.4,5 However, considerable evidence suggests that in vivo Aβ removal is critically dependent on microglial activation status.33,44,45,46,47 Microglia can exist in multiple activation states that are determined by the local stimuli for activation and reflected in production of specific cytokines and chemokines.48 Within the AD brain, there is a chronic activation of microglia associated with inflammatory cytokines including TNFα that can substantially block of ability of the microglia to remove47,49,50 or degrade Aβ.51,52 Recent studies demonstrated that overexpression of IL1β, an inflammatory cytokine leads to reduced Aβ pathology in mouse models of AD.9,53
One partial explanation for our results is that CX3CR1 deficiency alters the microglial activation status and enhances Aβ clearance. In support of this possibility, TNFα mRNA levels were selectively down-regulated in CX3CR1 deficient APPPS1 animals, and TNFα has been shown to directly reduce microglial expression of Aβ-degrading enzymes such as insulin-degrading enzyme and neprilysin.51,52 Furthermore, IL1β mRNA levels were significantly increased in a gene dose-dependent manner in the CX3CR1-deficient APPPS1 animals, and previous studies demonstrated that IL1β overexpression enhances Aβ clearance.9,53 Finally, mRNA levels of MCP-1, which promotes microglia-mediated Aβ oligomerization,54 was reduced in CX3CR1-deficient APPPS1 animals and thus could lead to reduced Aβ oligomerization/deposition. These observations suggest that CX3CR1 deficiency leads to a cytokine environment that harnesses the beneficial effects of microglial activation in response to Aβ. Notably, increased IL1β levels were also observed in Cx3cr1 knockout mice in response to lipopolysaccharide injections, where it led to enhanced neurotoxicity.13 Indeed, the effect of CX3CR1 deficiency appears to depend on the nature of the CNS insult as recent studies demonstrated that Cx3cr1 knockout mice exhibit smaller infarcts after transient middle cerebral artery occlusion.19,20
Another relevant observation is that CX3CR1 signaling directly blunts microglial phagocytosis and might prevent effective Aβ clearance. In support of this hypothesis, our in vitro and in vivo data demonstrate that CX3CL1-CX3CR1 signaling inhibits microglial phagocytosis, including fibrillar Aβ. Notably, CX3CL1 is found in high concentrations in both normal and diseased CNS,55 and our results suggest that CX3CL1-CX3CR1 signaling establishes a permissive environment for Aβ deposition in the AD brains and transgenic mouse models by down-regulating the microglial phagocytic response toward Aβ. Enhanced phagocytosis in the absence of CX3CR1 signaling could also help to explain the heterogeneity of phenotypes in CX3CR1-deficient CNS disease models. Depending on the nature of the CNS insult, increased microglial phagocytosis may ameliorate46 or worsen pathology.56
Finally, a recent study provided evidence that gancyclovir-mediated depletion of microglia expressing thymidine kinase had no demonstrable effect on Aβ pathology in two mouse models of AD.57 While these studies suggest that relatively short term depletion of microglia had only negligible effect on Aβ deposition, numerous other studies have suggested that altering the proliferation,58 signaling,8,9,11 and migration7,59 of microglia directly impact development of Aβ pathology. Taken together, these data suggest multiple redundant pathways that regulate Aβ clearance and deposition in different disease states and experimental conditions.
Microglial Proliferation/Migration
In mouse models of AD, the numbers of plaque-associated microglia increase over time and restrict the growth in plaque volume.4,5 However, in other situations, excess microglial accumulation appears to exacerbate Aβ pathology.7 In CX3CR1-deficient animals, there was a gene dose–dependent reduction in number of microglia associated with Aβ deposition. We postulate that our observations could be explained by the following: i) enhanced microglial clearance of Aβ, so that the retention of cells per plaque was reduced; ii) alterations in microglial proliferation/cell death; and/or iii) alterations in migration of microglia/macrophages to Aβ deposits.
Microglia surrounding Aβ deposits undergo extensive turnover in a mouse model of AD, exhibiting both apoptosis and proliferation.60 Interestingly, CX3CR1 signaling inhibits microglial apoptosis via Fas-mediated mechanisms in vitro.34 Future studies will be needed to determine whether the reduction in microglial number associated with Aβ deposition in CX3CR1-deficient animals is due to alterations in microglial cell death or proliferation. However, our preliminary analysis suggests that microglial proliferation and cell death were unaltered in APPPS1 mice deficient for CX3CR1 as assessed via bromodeoxyuridine incorporation and cleaved caspase 3 labeling, respectively (data not shown).
Another possibility is altered migration of either microglia or peripheral monocytes/macrophages to Aβ deposits in the CX3CR1-deficient animals. Various genetically modified mouse models have convincingly demonstrated the role for other chemokines/cytokine pathways in altering the accumulation of mononuclear phagocytes (monocytes, macrophages, and microglia) and Aβ pathology, including CCR259 and IL1β.9 However, in the present studies, despite the reduction in the number of microglia around the Aβ deposits in CX3CR1-deficient APPPS1 animals, there was a significant reduction in Aβ deposition, consistent with an enhanced capacity of microglia to remove Aβ. Accordingly, the simplest interpretation of our data are that fewer Cx3cr1−/− microglia were associated with each plaque due to their enhanced phagocytic capacity, leading to the associated findings of reduced Aβ deposition and fewer microglia per plaque.
A recent study (published while the current study was under review) suggested that CX3CR1 deficiency can prevent neuronal loss in the 3xTg mouse model of AD.61 A 1.8% loss of neurons per month observed in the 3xTg mouse model by two-photon microscopy was prevented in the CX3XR1 deficient 3xTg animals. However, this study examined 3xTg animals at 4–6 months, an age at which there is no extracellular deposition of Aβ but instead abundant intracellular Aβ immunoreactivity. At the single age examined, the levels of Aβ remain relatively unchanged in these animals, although their assessment of insoluble Aβ appears to show a non-significant reduction in the knockout mice.
Fuhrmann et al argue that these studies show that the lack of CX3CR1 does not “interfere with the Aβ-phagocytosing activity of microglia.” However, our studies demonstrate that CX3CR1 deficiency reduces extracellular Aβ deposition in two different mouse models of AD via a mechanism that likely involves enhanced phagocytic clearance of extracellular Aβ fibrils. Thus, the apparent discrepancy in the results of the two studies may be reflective of the cellular location of the Aβ aggregates and is supported by our findings that the steady-state levels of Aβ in predepositing CX3CR1-deficient APPPS1 and R1.40 animals remained unchanged (data not shown).
Furthermore, we did not observe any obvious alterations in neurodegeneration in the APPPS1;Cx3cr1−/− and R1.40;Cx3cr1−/− animals, consistent with previous studies demonstrating no overt neurodegeneration in either the APPPS1 or R1.40 animals.21,36 Notably, similar types of studies also did not detect robust neuronal loss in aged 3xTg animals,62 and it remains to be determined how the neuronal loss observed by Fuhrmann et al via two photon microscopy relates to the amyloid toxicity observed in the 3xTg, APPPS1 and R1.40 mouse models. Notably, the reduction in Aβ deposition in the CX3CR1-deficient APPPS1 and R1.40 mice also led to a reduction in the formation of dystrophic neurites, which perhaps could, in turn, impact local neurodegeneration. Clearly, additional studies are warranted to further define the role of CX3CL1/CX3CR1 signaling in AD pathogenesis
Acknowledgments
We thank Mathias Jucker (University of Tuebingen, Germany) for the APPPS1 mice; Gary Landreth (Case Medical School, Cleveland) and Mathias Jucker for helpful comments about the manuscript; Nicole Maphis, Cathleen Chang and Grahame Kidd for technical support; and Edward Koo (University of California San Diego, San Diego, CA) for providing the CT15 antibody.
Footnotes
Address reprint requests to Bruce T. Lamb, Ph.D., Lerner Research Institute, The Cleveland Clinic, Department of Neurosciences, 9500 Euclid Avenue, NC30, Cleveland, OH 44195-0001. E-mail: lambb@ccf.org.
Supported by a grant from the American Health Assistance Foundation (07-997, to B.T.L.), National Institutes of Health grant AG023012 (B.T.L.), an investigator-initiated research grant from the Alzheimer’s Association (B.T.L.), a grant from the Charles A. Dana Foundation (R.M.R.), a Career Transition Award from the National Multiple Sclerosis Society (TA-3021, to A.E.C.), National Research Service Award Fellowship F30NS068003 (S.L.), and an anonymous foundation.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
Current address of N.H.V.: Department of Cellular Neurology, Hertie-Institute for Clinical Brain Research, University of Tübingen, Tübingen, Germany; of A.E.C.: Department of Biology and STCEID, University of Texas San Antonio, San Antonio, Texas.
References
- Wyss-Coray T, Mucke L. Inflammation in neurodegenerative disease–a double-edged sword. Neuron. 2002;35:419–432. doi: 10.1016/s0896-6273(02)00794-8. [DOI] [PubMed] [Google Scholar]
- Akiyama H, Barger S, Barnum S, Bradt B, Bauer J, Cole GM, Cooper NR, Eikelenboom P, Emmerling M, Fiebich BL, Finch CE, Frautschy S, Griffin WS, Hampel H, Hull M, Landreth G, Lue L, Mrak R, Mackenzie IR, McGeer PL, O'Banion MK, Pachter J, Pasinetti G, Plata-Salaman C, Rogers J, Rydel R, Shen Y, Streit W, Strohmeyer R, Tooyoma I, Van Muiswinkel FL, Veerhuis R, Walker D, Webster S, Wegrzyniak B, Wenk G, Wyss-Coray T. Inflammation and Alzheimer’s disease. Neurobiol Aging. 2000;21:383–421. doi: 10.1016/s0197-4580(00)00124-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenigsknecht-Talboo J, Meyer-Luehmann M, Parsadanian M, Garcia-Alloza M, Finn MB, Hyman BT, Bacskai BJ, Holtzman DM. Rapid microglial response around amyloid pathology after systemic anti-Abeta antibody administration in PDAPP mice. J Neurosci. 2008;28:14156–14164. doi: 10.1523/JNEUROSCI.4147-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolmont T, Haiss F, Eicke D, Radde R, Mathis CA, Klunk WE, Kohsaka S, Jucker M, Calhoun ME. Dynamics of the microglial/amyloid interaction indicate a role in plaque maintenance. J Neurosci. 2008;28:4283–4292. doi: 10.1523/JNEUROSCI.4814-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Luehmann M, Spires-Jones TL, Prada C, Garcia-Alloza M, de Calignon A, Rozkalne A, Koenigsknecht-Talboo J, Holtzman DM, Bacskai BJ, Hyman BT. Rapid appearance and local toxicity of amyloid-beta plaques in a mouse model of Alzheimer’s disease. Nature. 2008;451:720–724. doi: 10.1038/nature06616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan D, Gordon MN, Tan J, Wilcock D, Rojiani AM. Dynamic complexity of the microglial activation response in transgenic models of amyloid deposition: implications for Alzheimer therapeutics. J Neuropathol Exp Neurol. 2005;64:743–753. doi: 10.1097/01.jnen.0000178444.33972.e0. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Horiba M, Buescher JL, Huang D, Gendelman HE, Ransohoff RM, Ikezu T. Overexpression of monocyte chemotactic protein-1/CCL2 in beta-amyloid precursor protein transgenic mice show accelerated diffuse beta-amyloid deposition. Am J Pathol. 2005;166:1475–1485. doi: 10.1016/s0002-9440(10)62364-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Town T, Laouar Y, Pittenger C, Mori T, Szekely CA, Tan J, Duman RS, Flavell RA. Blocking TGF-beta-Smad2/3 innate immune signaling mitigates Alzheimer-like pathology. Nat Med. 2008;14:681–687. doi: 10.1038/nm1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaftel SS, Kyrkanides S, Olschowka JA, Miller JN, Johnson RE, O'Banion MK. Sustained hippocampal IL-1 beta overexpression mediates chronic neuroinflammation and ameliorates Alzheimer plaque pathology. J Clin Invest. 2007;117:1595–1604. doi: 10.1172/JCI31450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Z, Ho L, Yemul S, Zhao Z, Qing W, Pompl P, Kelley K, Dang A, Teplow D, Pasinetti GM. Cyclooxygenase-2 promotes amyloid plaque deposition in a mouse model of Alzheimer’s disease neuropathology. Gene Expr. 2002;10:271–278. doi: 10.3727/000000002783992352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier M, Peng Y, Jiang L, Seabrook TJ, Carroll MC, Lemere CA. Complement C3 deficiency leads to accelerated amyloid beta plaque deposition and neurodegeneration and modulation of the microglia/macrophage phenotype in amyloid precursor protein transgenic mice. J Neurosci. 2008;28:6333–6341. doi: 10.1523/JNEUROSCI.0829-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limatola C, Lauro C, Catalano M, Ciotti MT, Bertollini C, Di Angelantonio S, Ragozzino D, Eusebi F. Chemokine CX3CL1 protects rat hippocampal neurons against glutamate-mediated excitotoxicity. J Neuroimmunol. 2005;166:19–28. doi: 10.1016/j.jneuroim.2005.03.023. [DOI] [PubMed] [Google Scholar]
- Cardona AE, Pioro EP, Sasse ME, Kostenko V, Cardona SM, Dijkstra IM, Huang D, Kidd G, Dombrowski S, Dutta R, Lee JC, Cook DN, Jung S, Lira SA, Littman DR, Ransohoff RM. Control of microglial neurotoxicity by the fractalkine receptor. Nat Neurosci. 2006;9:917–924. doi: 10.1038/nn1715. [DOI] [PubMed] [Google Scholar]
- Harrison JK, Jiang Y, Chen S, Xia Y, Maciejewski D, McNamara RK, Streit WJ, Salafranca MN, Adhikari S, Thompson DA, Botti P, Bacon KB, Feng L. Role for neuronally derived fractalkine in mediating interactions between neurons and CX3CR1-expressing microglia, Proc Natl Acad Sci USA. 1998;95:10896–10901. doi: 10.1073/pnas.95.18.10896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S, Aliberti J, Graemmel P, Sunshine MJ, Kreutzberg GW, Sher A, Littman DR. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol. 2000;20:4106–4114. doi: 10.1128/mcb.20.11.4106-4114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazan JF, Bacon KB, Hardiman G, Wang W, Soo K, Rossi D, Greaves DR, Zlotnik A, Schall TJ. A new class of membrane-bound chemokine with a CX3C motif. Nature. 1997;385:640–644. doi: 10.1038/385640a0. [DOI] [PubMed] [Google Scholar]
- Tuo J, Smith BC, Bojanowski CM, Meleth AD, Gery I, Csaky KG, Chew EY, Chan CC. The involvement of sequence variation and expression of CX3CR1 in the pathogenesis of age-related macular degeneration. FASEB J. 2004;18:1297–1299. doi: 10.1096/fj.04-1862fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CC, Tuo J, Bojanowski CM, Csaky KG, Green WR. Detection of CX3CR1 single nucleotide polymorphism and expression on archived eyes with age-related macular degeneration. Histol Histopathol. 2005;20:857–863. doi: 10.14670/hh-20.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denes A, Ferenczi S, Halasz J, Kornyei Z, Kovacs KJ. Role of CX3CR1 (fractalkine receptor) in brain damage and inflammation induced by focal cerebral ischemia in mouse. J Cereb Blood Flow Metab. 2008;28:1707–1721. doi: 10.1038/jcbfm.2008.64. [DOI] [PubMed] [Google Scholar]
- Soriano SG, Amaravadi LS, Wang YF, Zhou H, Yu GX, Tonra JR, Fairchild-Huntress V, Fang Q, Dunmore JH, Huszar D, Pan Y. Mice deficient in fractalkine are less susceptible to cerebral ischemia-reperfusion injury. J Neuroimmunol. 2002;125:59–65. doi: 10.1016/s0165-5728(02)00033-4. [DOI] [PubMed] [Google Scholar]
- Radde R, Bolmont T, Kaeser SA, Coomaraswamy J, Lindau D, Stoltze L, Calhoun ME, Jaggi F, Wolburg H, Gengler S, Haass C, Ghetti B, Czech C, Holscher C, Mathews PM, Jucker M. Abeta42-driven cerebral amyloidosis in transgenic mice reveals early and robust pathology. EMBO Rep. 2006;7:940–946. doi: 10.1038/sj.embor.7400784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb BT, Sisodia SS, Lawler AM, Slunt HH, Kitt CA, Kearns WG, Pearson PL, Price DL, Gearhart JD. Introduction and expression of the 400 kilobase amyloid precursor protein gene in transgenic mice [corrected]. Nat Genet. 1993;5:22–30. doi: 10.1038/ng0993-22. [DOI] [PubMed] [Google Scholar]
- Lamb BT, Call LM, Slunt HH, Bardel KA, Lawler AM, Eckman CB, Younkin SG, Holtz G, Wagner SL, Price DL, Sisodia SS, Gearhart JD. Altered metabolism of familial Alzheimer’s disease-linked amyloid precursor protein variants in yeast artificial chromosome transgenic mice. Hum Mol Genet. 1997;6:1535–1541. doi: 10.1093/hmg/6.9.1535. [DOI] [PubMed] [Google Scholar]
- Lehman EJH, Kulnane LS, Lamb BT. Alterations in β-amyloid production and deposition in brain regions of two transgenic models. Neurobiol Aging. 2003;24:645–653. doi: 10.1016/s0197-4580(02)00153-7. [DOI] [PubMed] [Google Scholar]
- Lehman EJ, Kulnane LS, Gao Y, Petriello MC, Pimpis KM, Younkin L, Dolios G, Wang R, Younkin SG, Lamb BT. Genetic background regulates β-amyloid precursor protein processing and β-amyloid deposition in the mouse. Hum Mol Genet. 2003;12:2949–2956. doi: 10.1093/hmg/ddg322. [DOI] [PubMed] [Google Scholar]
- Martin LJ, Sisodia SS, Koo EH, Cork LC, Dellovade TL, Weidemann A, Beyreuther K, Masters C, Price DL. Amyloid precursor protein in aged nonhuman primates. Proc Natl Acad Sci USA. 1991;88:1461–1465. doi: 10.1073/pnas.88.4.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Varvel NH, Lamb BT, Herrup K. Ectopic cell cycle events link human Alzheimer’s disease and amyloid precursor protein transgenic mouse models. J Neurosci. 2006;26:775–784. doi: 10.1523/JNEUROSCI.3707-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andorfer C, Acker CM, Kress Y, Hof PR, Duff K, Davies P. Cell-cycle reentry and cell death in transgenic mice expressing nonmutant human tau isoforms. J Neurosci. 2005;25:5446–5454. doi: 10.1523/JNEUROSCI.4637-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed-Geaghan EG, Savage JC, Hise AG, Landreth GE. CD14 and toll-like receptors 2 and 4 are required for fibrillar A{beta}-stimulated microglial activation. J Neurosci. 2009;29:11982–11992. doi: 10.1523/JNEUROSCI.3158-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saura J, Tusell JM, Serratosa J. High-yield isolation of murine microglia by mild trypsinization. Glia. 2003;44:183–189. doi: 10.1002/glia.10274. [DOI] [PubMed] [Google Scholar]
- Koenigsknecht J, Landreth G. Microglial phagocytosis of fibrillar beta-amyloid through a beta1 integrin-dependent mechanism. J Neurosci. 2004;24:9838–9846. doi: 10.1523/JNEUROSCI.2557-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DC, Kim SH, Jeong MW, Baek NI, Kim KT. Effect of rottlerin, a PKC-delta inhibitor, on TLR-4-dependent activation of murine microglia. Biochem Biophys Res Commun. 2005;337:110–115. doi: 10.1016/j.bbrc.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Koenigsknecht-Talboo J, Landreth GE. Microglial phagocytosis induced by fibrillar beta-amyloid and IgGs are differentially regulated by proinflammatory cytokines. J Neurosci. 2005;25:8240–8249. doi: 10.1523/JNEUROSCI.1808-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehme SA, Lio FM, Maciejewski-Lenoir D, Bacon KB, Conlon PJ. The chemokine fractalkine inhibits Fas-mediated cell death of brain microglia. J Immunol. 2000;165:397–403. doi: 10.4049/jimmunol.165.1.397. [DOI] [PubMed] [Google Scholar]
- Haskell CA, Hancock WW, Salant DJ, Gao W, Csizmadia V, Peters W, Faia K, Fituri O, Rottman JB, Charo IF. Targeted deletion of CX(3)CR1 reveals a role for fractalkine in cardiac allograft rejection. J Clin Invest. 2001;108:679–688. doi: 10.1172/JCI12976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulnane LS, Lamb BT. Neuropathological characterization of mutant amyloid precursor protein yeast artificial chromosome transgenic mice. Neurobiol Dis. 2001;8:982–992. doi: 10.1006/nbdi.2001.0446. [DOI] [PubMed] [Google Scholar]
- Combadiere C, Feumi C, Raoul W, Keller N, Rodero M, Pezard A, Lavalette S, Houssier M, Jonet L, Picard E, Debre P, Sirinyan M, Deterre P, Ferroukhi T, Cohen SY, Chauvaud D, Jeanny JC, Chemtob S, Behar-Cohen F, Sennlaub F. CX3CR1-dependent subretinal microglia cell accumulation is associated with cardinal features of age-related macular degeneration. J Clin Invest. 2007;117:2920–2928. doi: 10.1172/JCI31692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raoul W, Keller N, Rodero M, Behar-Cohen F, Sennlaub F, Combadiere C. Role of the chemokine receptor CX3CR1 in the mobilization of phagocytic retinal microglial cells. J Neuroimmunol. 2008;198:56–61. doi: 10.1016/j.jneuroim.2008.04.014. [DOI] [PubMed] [Google Scholar]
- Ishizuka K, Kimura T, Igata-yi R, Katsuragi S, Takamatsu J, Miyakawa T. Identification of monocyte chemoattractant protein-1 in senile plaques and reactive microglia of Alzheimer’s disease. Psychiatry Clin Neurosci. 1997;51:135–138. doi: 10.1111/j.1440-1819.1997.tb02375.x. [DOI] [PubMed] [Google Scholar]
- Simard AR, Soulet D, Gowing G, Julien JP, Rivest S. Bone marrow-derived microglia play a critical role in restricting senile plaque formation in Alzheimer’s disease. Neuron. 2006;49:489–502. doi: 10.1016/j.neuron.2006.01.022. [DOI] [PubMed] [Google Scholar]
- Heneka MT, Sastre M, Dumitrescu-Ozimek L, Dewachter I, Walter J, Klockgether T, Van Leuven F. Focal glial activation coincides with increased BACE1 activation and precedes amyloid plaque deposition in APP[V717I] transgenic mice. J Neuroinflammation. 2005;2:22. doi: 10.1186/1742-2094-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janelsins MC, Mastrangelo MA, Oddo S, LaFerla FM, Federoff HJ, Bowers WJ. Early correlation of microglial activation with enhanced tumor necrosis factor-alpha and monocyte chemoattractant protein-1 expression specifically within the entorhinal cortex of triple transgenic Alzheimer’s disease mice. J Neuroinflammation. 2005;2:23. doi: 10.1186/1742-2094-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman GA, Moores K, Harrison D, Campbell CA, Stewart BR, Strijbos PJ. Fractalkine cleavage from neuronal membranes represents an acute event in the inflammatory response to excitotoxic brain damage. J Neurosci. 2000;20:RC87. doi: 10.1523/JNEUROSCI.20-15-j0004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend KP, Town T, Mori T, Lue LF, Shytle D, Sanberg PR, Morgan D, Fernandez F, Flavell RA, Tan J. CD40 signaling regulates innate and adaptive activation of microglia in response to amyloid beta-peptide. Eur J Immunol. 2005;35:901–910. doi: 10.1002/eji.200425585. [DOI] [PubMed] [Google Scholar]
- Zelcer N, Khanlou N, Clare R, Jiang Q, Reed-Geaghan EG, Landreth GE, Vinters HV, Tontonoz P. Attenuation of neuroinflammation and Alzheimer’s disease pathology by liver x receptors. Proc Natl Acad Sci USA. 2007;104:10601–10606. doi: 10.1073/pnas.0701096104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissonneault V, Filali M, Lessard M, Relton J, Wong G, Rivest S. Powerful beneficial effects of macrophage colony-stimulating factor on beta-amyloid deposition and cognitive impairment in Alzheimer’s disease. Brain. 2009;132:1078–1092. doi: 10.1093/brain/awn331. [DOI] [PubMed] [Google Scholar]
- Shie FS, Montine KS, Breyer RM, Montine TJ. Microglial EP2 is critical to neurotoxicity from activated cerebral innate immunity. Glia. 2005;52:70–77. doi: 10.1002/glia.20220. [DOI] [PubMed] [Google Scholar]
- Ransohoff RM, Perry VH. Microglial physiology: unique stimuli, specialized responses. Annu Rev Immunol. 2009;27:119–145. doi: 10.1146/annurev.immunol.021908.132528. [DOI] [PubMed] [Google Scholar]
- Jimenez S, Baglietto-Vargas D, Caballero C, Moreno-Gonzalez I, Torres M, Sanchez-Varo R, Ruano D, Vizuete M, Gutierrez A, Vitorica J. Inflammatory response in the hippocampus of PS1M146L/APP751SL mouse model of Alzheimer’s disease: age-dependent switch in the microglial phenotype from alternative to classic. J Neurosci. 2008;28:11650–11661. doi: 10.1523/JNEUROSCI.3024-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsonego A, Zota V, Karni A, Krieger JI, Bar-Or A, Bitan G, Budson AE, Sperling R, Selkoe DJ, Weiner HL. Increased T cell reactivity to amyloid beta protein in older humans and patients with Alzheimer disease. J Clin Invest. 2003;112:415–422. doi: 10.1172/JCI18104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Kiyota T, Walsh SM, Liu J, Kipnis J, Ikezu T. Cytokine-mediated inhibition of fibrillar amyloid-beta peptide degradation by human mononuclear phagocytes. J Immunol. 2008;181:3877–3886. doi: 10.4049/jimmunol.181.6.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu E, Kawahara K, Kajizono M, Sawada M, Nakayama H. IL-4-induced selective clearance of oligomeric beta-amyloid peptide(1-42) by rat primary type 2 microglia. J Immunol. 2008;181:6503–6513. doi: 10.4049/jimmunol.181.9.6503. [DOI] [PubMed] [Google Scholar]
- Quinn J, Montine T, Morrow J, Woodward WR, Kulhanek D, Eckenstein F. Inflammation and cerebral amyloidosis are disconnected in an animal model of Alzheimer’s disease. J Neuroimmunol. 2003;137:32–41. doi: 10.1016/s0165-5728(03)00037-7. [DOI] [PubMed] [Google Scholar]
- Kiyota T, Yamamoto M, Xiong H, Lambert MP, Klein WL, Gendelman HE, Ransohoff RM, Ikezu T. CCL2 accelerates microglia-mediated Abeta oligomer formation and progression of neurocognitive dysfunction. PLoS One. 2009;4:e6197. doi: 10.1371/journal.pone.0006197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajetto A, Bonavia R, Barbero S, Florio T, Schettini G. Chemokines and their receptors in the central nervous system. Front Neuroendocrinol. 2001;22:147–184. doi: 10.1006/frne.2001.0214. [DOI] [PubMed] [Google Scholar]
- Gowing G, Lalancette-Hebert M, Audet JN, Dequen F, Julien JP. Macrophage colony stimulating factor (M-CSF) exacerbates ALS disease in a mouse model through altered responses of microglia expressing mutant superoxide dismutase. Exp Neurol. 2009;220:267–275. doi: 10.1016/j.expneurol.2009.08.021. [DOI] [PubMed] [Google Scholar]
- Grathwohl SA, Kalin RE, Bolmont T, Prokop S, Winkelmann G, Kaeser SA, Odenthal J, Radde R, Eldh T, Gandy S, Aguzzi A, Staufenbiel M, Mathews PM, Wolburg H, Heppner FL, Jucker M. Formation and maintenance of Alzheimer’s disease beta-amyloid plaques in the absence of microglia. Nat Neurosci. 2009;12:1361–1363. doi: 10.1038/nn.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manczak M, Mao P, Nakamura K, Bebbington C, Park B, Reddy PH. Neutralization of granulocyte macrophage colony-stimulating factor decreases amyloid beta 1–42 and suppresses microglial activity in a transgenic mouse model of Alzheimer’s disease. Hum Mol Genet. 2009;18:3876–3893. doi: 10.1093/hmg/ddp331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Khoury J, Toft M, Hickman SE, Means TK, Terada K, Geula C, Luster AD. Ccr2 deficiency impairs microglial accumulation and accelerates progression of Alzheimer-like disease. Nat Med. 2007;13:432–438. doi: 10.1038/nm1555. [DOI] [PubMed] [Google Scholar]
- Bondolfi L, Calhoun M, Ermini F, Kuhn HG, Wiederhold KH, Walker L, Staufenbiel M, Jucker M. Amyloid-associated neuron loss and gliogenesis in the neocortex of amyloid precursor protein transgenic mice. J Neurosci. 2002;22:515–522. doi: 10.1523/JNEUROSCI.22-02-00515.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann M, Bittner T, Jung CK, Burgold S, Page RM, Mitteregger G, Haass C, LaFerla FM, Kretzschmar H, Herms J. Microglial Cx3cr1 knockout prevents neuron loss in a mouse model of Alzheimer’s disease. Nat Neurosci. 13:411–413. doi: 10.1038/nn.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes JP, Blurton-Jones M, Yamasaki TR, Agostinho P, LaFerla FM. Activation of cell cycle proteins in transgenic mice in response to neuronal loss but not amyloid-beta and tau pathology. J Alzheimers Dis. 2009;16:541–549. doi: 10.3233/JAD-2009-0993. [DOI] [PubMed] [Google Scholar]