Abstract
The cell of origin and pathogenesis of the majority of adult soft tissue sarcomas (STS) remains poorly understood. Because mutations in both the P53 and RB tumor suppressor genes are frequent in STS in humans, we inactivated these genes by Cre-loxP–mediated recombination in mice with floxed p53 and Rb. Ninety-three percent of mice developed spindle cell/pleomorphic sarcomas after a single subcutaneous injection of adenovirus carrying Cre-recombinase. Similar to human STS, these sarcomas overexpress Cxcr4, which contributes to their invasive properties. Using irradiation chimeras generated by transplanting bone marrow cells from mice carrying either the Rosa26StoploxPLacZ or the Z/EG reporter, as well as the floxed p53 and Rb genes, into irradiated p53loxP/loxPRbloxP/loxP mice, it was determined that sarcomas do not originate from bone marrow–derived cells, such as macrophages, but arise from the local resident cells. At the same time, dermal mesenchymal stem cells isolated by strict plastic adherence and low levels of Sca-1 expression (Sca-1low, CD31negCD45neg) have shown enhanced potential for malignant transformation according to soft agar, invasion, and tumorigenicity assays, after the conditional inactivation of both p53 and Rb. Sarcomas formed after transplantation of these cells have features typical for undifferentiated high-grade pleomorphic sarcomas. Taken together, our studies indicate that local Sca-1low dermal mesenchymal stem/progenitor cells are preferential targets for malignant transformation associated with deficiencies in both p53 and Rb.
Soft tissues sarcomas (STS) are malignant neoplasms arising from nonepithelial and extraskeletal tissues, such as connective tissue, muscles, vessels, and peripheral nerves. While some sarcomas resemble a specific differentiated tissue (eg, muscle, fat, etc), many are poorly differentiated and their cell of origin remains a debatable topic. Not surprisingly, diagnosis of STS is a difficult task, and few reliable prognostic criteria, mainly based on sarcoma grade and stage, have been identified.1,2,3,4
During recent years, a concept has been developed that many STS, particularly those without significant presentation of cell lineage-specific markers, may arise from mesenchymal stem cells (MSC). The majority of those studies have been based on MSC derived from the bone marrow.5,6,7,8 The possibility that STS may develop from bone marrow MSC has been raised by a report that, according to cell lineage tracing experiments in irradiation chimeras, STS arise from bone marrow–derived cells.9
Genetically, STS can be divided into two groups. One group has a consistent set of recurrent genetic alterations, such as translocations (eg, PAX3-FOXO1A in alveolar rhabdomyosarcoma) and specific mutations (eg, KIT activating mutation in gastrointestinal stromal tumors). Another group, which comprises the majority of STS, is characterized by complex genomic profiles and frequent presence of mutations in tumor suppressor genes P53 and Retinoblastoma 1 (RB).1,2,3,4 Further supporting importance of these genes for sarcomagenesis, patients with Li-Fraumeni syndrome, which is associated with germline P53 mutations, as well as patients with germline RB mutation have a higher frequency of STS.2,4 In agreement with known cooperation between P53 and RB pathways, mutations in both genes are frequent in STS.10 It has been recently reported that conditional Cre-loxP–mediated inactivation of p53 by expression of Osterix-Cre transgene in committed osteoblast progenitors results in formation of osteosarcomas, and loss of Rb potentiates osteosarcomagenesis.11,12 Similar cooperation between p53 and Rb inactivation in acceleration of sarcomagenesis was also observed after expression of Prx1-Cre transgene in mesenchymal cells of mouse embryonic limbs.13 In addition to predominant osteosarcoma formation, development of poorly differentiated STS was also reported in that model. However, given that the majority of human STS affect adults,4 interpretation of this model has been somewhat complicated due to expression of Prx1-Cre transgene in the early mesenchymal tissues.
By using conditional activation of K-ras and inactivation of p53 by intramuscular injection of adenovirus expressing Cre recombinase (AdCMVCre) into adult mice, Kirsch et al14 have established a mouse model of adult high-grade sarcomas with myofibroblastic differentiation. We have used a similar approach for initiation of STS associated with p53 and Rb deficiency by subcutaneous AdCMVCre administration and demonstrate that the majority of these neoplasms are undifferentiated high-grade pleomorphic sarcomas (UPS), also known as malignant fibrous histiocytomas (MFH). Notably, similar to their human counterparts, mouse sarcomas overexpress Cxcr4, and its knockdown results in reduction of invasive properties of sarcoma cells. Based on bone marrow reconstitution experiments we have determined that STS have local as opposed to bone marrow origin. Finally, by using enhanced purification of dermal MSC we have demonstrated that these cells have superior transformation potential and form UPS after p53 and Rb inactivation.
Materials and Methods
Experimental Animals
Mice with floxed copies of p53 and Rb genes were prepared as described previously.15,16 FVB/N mice were used for controls. Reporter mice Z/EG (Tg(ACTB-Bgeo/GFP)21Lbe/J;17), Rosa26StoploxPLacZ (B6;129-Gt(ROSA)26Sortm1Sor/J;18,19), B5/EGFP (FVB.Cg-Tg (ACTB-EGPF)B5Nagy/J20) and Rosa26LacZ (B6; 129S-Gt(ROSA)26Sor/J;21) were obtained from the Jackson Laboratory (Bar Harbor, ME). All experiments were conducted under identical conditions, following recommendations of the Institutional Laboratory Animal Use and Care Committee of Cornell University.
Genotyping
p53loxP/loxP and RbloxP/loxP mice were identified by PCR genotyping essentially as previously described.22 Mice carrying LacZ were detected with PCR primers LACZ5′ (5′-GCGTTGGCAATTTAACCGCCAGTCA-3′) and LACZ3′ (5′-TCAGCACCGCATCAGCAAGTGTATC-3′) yielding 240-bp DNA fragment. Mice carrying EGFP were identified with PCR primers ZEGneo1 (5′-AGAGGCTATTCGGCTATGACTG-3′) and ZEGneo2 (5′-TTCGTCCAGATCATCCTGATC-3′) yielding 430-bp DNA fragment.
Adenovirus Administration
Recombinant adenoviruses AdCMVLacZ, AdCMVCre, AdCMVeGFP, and AdCMVCre-EGFP are modifications of the adenovirus-5 genome, from which the ela and elb regions required for viral replication have been deleted and replaced with Escherichia coli-derived LacZ, Cre, or enhanced green fluorescent protein (EGFP) adjacent to the CMV immediate early regulatory sequence23 (Gene Transfer Vector Core; University of Iowa, Iowa City, IA). After deep anesthesia with intraperitoneal (IP) Avertin (2.5% vol/vol in 0.85% NaCl; 0.02 ml/g body weight), mice were shaved and adenovirus (5 × 108 pfu/μl in saline) was injected into the subcutaneous (SC) tissue of the dorsal area with 21-gauge needle under the control of a dissection microscope. The needle was slowly withdrawn to avoid any accidental leak of adenovirus. According to experiments with AdCMVEGFP, AdCMVCre, and C-dots,24 as well as evaluation of the early stages of sarcomagenesis (Choi and Nikitin, unpublished), labeled cells were located in the reticular layer of the dermis and in the adjacent loose connective tissue of the hypodermis. Given the continuum of both areas, cells of both regions were used in all experiments and indicated as “dermal” cells.
Generation of Irradiation Chimeras
Six- to 7-week-old wild-type FVB/N or p53loxP/loxPRbloxP/loxP mice were irradiated at 11 Gy, 2 Gy/min) by using PRIMUS Linear Accelerator (SIEMENS, Malvern, PA) or a sealed cesium137 source irradiator Mark 1-68 (JL Shepherd and Associates, San Fernando, CA). According to the preliminary test experiments, irradiation in both devices resulted in comparable depletion of bone marrow cells. Within 4 hours after irradiation, mice were rescued by tail vein injection of 106 bone marrow cells derived from the Z/EG or the Rosa26StoploxPLacZ p53loxP/loxPRBloxP/loxP mice as described previously.25,26 To generate positive control mice, bone marrow cells from B5/EGFP or Rosa26LacZ reporter mice were administrated into irradiated mice. Fourteen days after bone marrow reconstitution, AdCMVCre was administrated subcutaneously into chimeras to induce a sarcoma as described above. In addition to tumor collection, blood for PCR genotyping was collected from the orbital sinus after anesthesia.
To evaluate a proportion of donor bone marrow cells in chimeras, bone marrow cells were collected from the femur and the dermis of chimeras rescued by bone marrow cells from B5/EGFP reporter mice. Bone marrow cells were stained with PerCP-Cy5.5 anti-mouse Gr-1 (#15-5931-82, eBioscience, Inc., San Diego, CA) for 30 minutes on ice and then analyzed by a BD LSRII analyzer (BD Biosciences, Franklin Lakes, NJ). All collected data were analyzed by FlowJo software (Tree Star, Inc. Ashland, OR). The skin tissues were fixed in 4% paraformaldehyde overnight and embedded in paraffin.
Pathological Evaluation
For long-term survival experiments mice were monitored daily for 600 days, until becoming moribund or until tumor reached 1 cm in diameter. For short-term experiments, mice were killed at 1, 3, 7, and 14 days postadministration of adenovirus. Mice were anesthetized with Avertin and subjected to cardiac perfusion at 90 mm Hg with phosphate-buffered 4% paraformaldehyde. Tumor, skin, lung, liver, and spleen, as well as organs with pathological changes, were examined during necropsy and placed into 4% paraformaldehyde for postfixation overnight. For short-term experiments with EGFP adenoviruses, the entire dorsal skin from the shoulder to hip was collected and visualized using fluorescence microscopy. Representative specimens were further characterized by microscopic analysis of paraffin sections, cut 4 μm thick, and stained with hematoxylin and eosin. All neoplasms were classified according to current histological classification of mouse27 and human4 STS.
Immunohistochemical Analyses
For avidin-biotin-peroxidase-based (ABC) detection, deparaffinized and rehydrated sections were subjected to antigen retrieval by either boiling in 10 mmol/L sodium citrate buffer (pH 6.0, 10 minutes) for antibodies to desmin, procollagen type I, EGFP, and CXCR4 or trypsin pretreatment for antibodies to CD31 and F4/80. The antibodies to F4/80 (1:200; #MCP497G, Serotec, Raleigh, NC), procollagen type I (Y-18; 1:50; #sc-8787, Santa Cruz Biotechnology, Santa Cruz, CA), CD31 (PECAM-1; 1:100; #551262, BD Biosciences), smooth muscle actin (1:300; #E2464, Spring bioscience, Fremont, CA), desmin (1:200; #YMPS31, Accurate Chemical & Scientific Corporation, Wesbury, NY), CXCR4 antibody (1:200; #sc-9046, Santa Cruz Biotechnology, Inc., Santa Cruz, CA), β-galactosidase (1:500; #ab616-1, Abcam, Cambridge, MA), or EGFP (1:500; #NB600-303, NOVUS biologicals, Littleton, CO) were incubated for 1 hour at room temperature (RT) with the exception of CD31 which was used overnight at 4°C. The antibody to α-sarcomeric actin (1:1500; #A2172, Sigma, St. Louis, MO) was incubated using MOM Kit (Vector, Burlingame, CA) for 20 minutes at RT. After treatment with 0.3% H2O2 in methanol, sections were incubated with biotinylated secondary antibodies and ABC complex and visualized using 3,3′-diaminobenzidine tetrahydrochloride (DAB) and counterstaining with hematoxylin essentially as described previously.28 As a control, primary antibodies were replaced with non-immune sera of the same species.
For immunostaining of human STS tissue arrays (#BC24011, US Biomax, Inc., Rockville, MD), slides were incubated at 60°C for 2 hours according to the manufacturer’s instructions and then processed for staining with antibody to CXCR4 as described above. The semiquantitative assessment of staining level was performed as follows: −, no detectable staining; +, <30% of cells are positive, ++ 30 to 60% of cells are positive, +++, >60% of cells are positive.
For double immunofluorescence analysis the deparaffinized sections of 4% paraformaldehyde-fixed tissue were incubated with rabbit antibodies to EGFP (1:300) for 1 hour at 37°C or rabbit antibodies to CXCR4 (1:100) or rat antibodies to F4/80 (1:100) for 1 hour, RT, followed by Alexa Fluor 488 conjugated donkey anti-rabbit or anti-rat secondary antibody (Invitrogen, Carlsbad, CA) for 30 minutes, RT. Subsequently, goat antibodies to procollagen type I (1:50), Iba-1 (1:200; abcam) or rabbit antibodies to CXCR4 (1:100) were applied for 1 hour, RT, followed by Alexa Fluor 594 conjugated donkey anti-goat or anti-rabbit secondary antibody (Invitrogen) was incubated with sections.
For immunostaining on frozen sections, tissues were frozen in liquid nitrogen with Tissue-Tek (SAKURA, McGaw Park, IL). Seven-micrometer-thick tissue sections were placed onto Superfrost plus-coated slides (Fisher Scientific, Pittsburgh, PA) and fixed in cold acetone (−20°C) for 10 minutes. Sections were incubated with primary antibody to F4/80 (1:100), Iba-1 (1:200), CD31 (1:100), smooth muscle actin (1:200), and procollagen type I (1:50) at RT for 1 hour or to CD31 (1:100) at 4°C overnight followed by secondary antibodies conjugated with Alexa Fluor 594. Counterstaining was performed by incubation for 1 minute with DAPI (1 mg/ml of an aqueous solution diluted 1:100 in TBS). Slides were mounted with GEL/MOUNT (Biomeda corp, Foster City, CA) and viewed with a Zeiss LSM 510-META confocal microscope.
Cell Culture
Tissue cultures were maintained using Dulbecco’s modification of Eagle’s medium (DMEM; Cellgro, Mediatech, Inc., Manassas, VA), containing 10% heat-inactivated fetal bovine serum (FBS; Sigma), 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mmol/L l-glutamine, and 1 mmol/L sodium pyruvate (Cellgro, Mediatech, Inc.). Primary fibroblast culture was prepared from tail dermis of adult p53loxP/loxPRbloxP/loxP mice. The collected tissue was washed three times in 70% EtOH and three times in PBS, minced finely, and disaggregated with 0.05% trypsin/0.53 mmol/L EDTA (Cellgro, Mediatech, Inc.) for 1 hour at 37°C. The trypsin was removed by centrifugation, and the cells were resuspended in medium. The cells were plated in two 6-cm-diameter tissue culture dishes.
For adenovirus infection adherent or suspension cells were plated at a concentration of 105 cells in 3 ml of complete growth medium per well in six-well plates. When cells reached 70% confluence, they were washed twice with PBS and AdCMVCre or AdCMVEGFP was added at a concentration of 5 × 108 pfu/ml in 1 ml of serum free growth medium. After incubation for 2 hours at 37°C with 5% CO2, the virus containing medium was removed and the cells were washed two times with PBS. The cells were given at least 3 days to recover in complete growth medium before use in further assays.
Cell lines STN1 and STN2 were established from angiosarcoma and UPS, respectively, developed in p53loxP/loxPRbloxP/loxP mice after SC AdCMVCre injection. FBN1 cell line was established 40 passages after Cre-loxP-mediated deletion of p53 and Rb in plastic adherent dermal cells. Cultures were maintained at 37°C and 5% CO2, and medium replaced every 3–4 days.
Western Blotting
Cells were lysed in RIPA (25 mmol/L Tris, pH 8.2, 50 mmol/L NaCl, 0.1% SDS, 0.5% Nonidet P-40, and 0.5% deoxycholate) buffer with protease inhibitors as described previously.29 Proteins were resolved on SDS-PAGE gels and transferred to nitrocellulose via semidry transfer (BioRad, Hercules, CA). Immunoblots were blocked with 5% milk in TBST for 1 hour and washed three times with fresh TBST. CXCR4 antibody (1:200; #sc-9046, Santa Cruz Biotechnology, Inc.) was incubated with immunoblots overnight at 4°C followed by HRP-conjugated anti-rabbit antibody (1 hour at RT) and detection with SuperSignal West Pico Chemiluminescent Substrate (Pierce Biotechnology, Inc., Rockland, IL).
Cxcr4-siRNA Transfection
One day before transfection, cells were plated with 2 ml of growth medium without antibiotics in a six-well plate. Cxcr4-siRNA (5 nmol; Accell SMARTpool siRNA, #E-060184-00-0005, Thermo Scientific, Waltham, MA) or control-siRNA oligomers (Accell nontargeting pool, #D-001910-10) was diluted in siRNA buffer (Thermo Scientific) to make a 100 μmol/L siRNA solution. 7.5 μl of the 100 μmol/L siRNA was combined with 750 μl Accell delivery media (Thermo Scientific), and 100 μl of the appropriate delivery mix was added to each well containing cells with ∼50% of confluence. Cells were incubated at 37°C in a 5% CO2 incubator for 72 hours until using for an invasion assay.
Isolation of Mesenchymal Stem Cells
Plastic adherent dermal (PAD) cells were prepared from 6- to 7-week-old p53loxP/loxPRbloxP/loxP male transgenic mice according to a protocol by Wilson et al30 with modifications. Briefly, euthanized mice were shaved and treated with Nair (Church & Dwight Co., Inc., Princeton, NJ) before skin removal. Harvested skin was cut into 1-cm2 pieces and incubated for 1 hour at 37°C in enzyme mix A, consisting of 5 U/ml dispase (Sigma) and 3% FBS in PBS. After this, the epidermis was discarded by mechanical dissociation and the remaining tissue was incubated overnight at 4°C in enzyme mix A. Tissue was then incubated for 2 hours at 37°C in enzyme mix B, consisting of 1.0 U/ml dispase, 0.13 U/ml collagenase (Sigma), 700 U/ml hyaluronidase (Sigma), and 20 U/ml DNase I (Sigma) in 1×PBS. Both enzyme mix A and B were combined and forced through a 70-μm strainer for a single cell suspension. Cells were subsequently plated in complete medium at 5 × 106 cells per 10-cm plate. PAD cells in passage 1 were detached and suspended in antibody staining buffer, consisting of 0.1% bovine serum albumin (BSA; Sigma) in PBS. Negative selection was carried out using magnetic Dynalbeads (Invitrogen) with CD31 (Platelet Endothelial Cell Adhesion Molecule-1, PECAM-1) and CD45 (Leukocyte Common Antigen, Ly-5) biotinylated antibodies (#553371 and #553077, respectively, BD Biosciences). Cells were incubated at 4°C for 30 minutes at a concentration 1 μg of antibodies per 106 cells, according to the manufacturer’s instructions. Supernatant containing CD31−/CD45− cells was used for Fluorescence-Activated Cell Sorting (FACS) after incubation with fluorescein-conjugated antibodies to Ly-6A/E (#553335, Sca-1; BD Biosciences) at a concentration of 1 μg of antibodies per 106 cells at 4°C, in the dark, for 30 minutes. Appropriate isotype controls were used for accurate gating parameters. FACS was conducted using a FACSAria cell sorter (BD Biosciences).
Differentiation Assay
Sorted cells in passage three were replated onto six-well plates at a concentration of 5 × 105 cells per well in 3 ml of a complete growth medium. When cells were confluent, medium was removed and replaced with appropriate differentiation or control medium. Control medium consisted of DMEM with 1% FBS to reduce nonspecific differentiation. Cells were cultured from 6 to 21 days, depending on the assay type, and had medium replaced every 2 days. Adipocyte differentiation medium contains 5 μg/ml insulin (Sigma), 1 μmol/L dexamethasone (Sigma), 0.5 mmol/L IBMX (Sigma), and 10% FBS in DMEM:F12 (Cellgro, Mediatech, Inc.). The cells were maintained for 7 days, then fixed with 4% paraformaldehyde, stained with 0.21% (wt/vol) Oil Red O (Sigma) in 60% isopropanol for 10 minutes, and washed with tap water. Chondrocytes were maintained in medium consisting of 200 μmol/L L-ascorbic acid 2-phosphate, 100 nmol/L dexamethasone, 10 ng/ml TGF- β1 (R&D Systems, Minneapolis, MN), 40 μg/ml L-proline (Sigma), 1:100 ITS + Premix (BD Biosciences), and 10% FBS in DMEM for 14 days. Then, cells were fixed in 2.5% glutaraldehyde, 25 mmol/L sodium acetate, and 0.4 mol/L MgCl2 containing 0.05% (wt/vol) Alcian Blue (Sigma) for 48 hours and washed in steps with 3% acetic acid, 3% acetic acid/25% ethanol, and 3% acetic acid/50% ethanol. For the osteocyte differentiation assay, cells were cultured in 300 μmol/L L-ascorbic acid 2-phosphate (Sigma), 10 nmol/L dexamethasone, 10 mmol/L β-glycerophosphate (Sigma), and 10% FBS in DMEM:F12 for 21 days. Then, cells were fixed in 70% ice-cold ethanol for 1 hour, stained with 2% (wt/vol) Alizarin Red (Sigma) pH 4.2, for 10 minutes, and rinsed with 1×PBS. Ten random images of each well were taken at ×20 magnification for quantification.
Soft Agar Colony Formation, Invasion, and Tumorigenicity Assays
All assays were performed as previously described.31,32,33 Briefly, for soft agar colony formation, PAD cells in passage 3 were suspended in an agar mix at 5 × 105 cells per 6-cm plate and incubated at 37°C and 5% CO2. Analysis was conducted by comparing the number of colonies present in each group after 3 weeks of growth in soft agar. For uniformity, a colony was considered to be a group of four or more cells. For invasion assay, PAD cells in passage 3 or tumor cells were placed into Matrigel Invasion Chambers (BD Biosciences, Franklin Lakes, NJ) at concentration 2.5 × 104 cells per well. For PAD cells, complete medium was added in each chamber. For tumor cells treated with either Cxcr4-siRNA or scrambled siRNA, 400 ng of Sdf-1 (R&D Systems, Inc.) in serum-free medium was added in lower chamber. The plates were then incubated for 22 hours at 37°C and 5% CO2, and invasion ability was determined by comparing cell number on the bottom of control and invasion wells, respectively, to the original plated density. For tumorigenicity assay, 106 PAD cells in passage 3 were injected subcutaneously into 7-week-old male SCID/NCr mice (Animal production program, NCI-Frederick, Frederick, MD), and monitoring of tumor growth and tumor collection were performed as described under Pathological Evaluation.
Statistical Analyses
All statistical analyses were performed with the programs InStat 3.05 and Prism 4.03 (GraphPad, Inc. San Diego, CA). Survival fractions were calculated using the Kaplan–Meier method. Survival curves were compared by log rank Mantel–Haenszel tests. Means were compared by estimation of the two-tailed P value.
Results
Conditional Inactivation of p53 and Rb in Dermal Connective Tissue Cells Results in Formation of Soft Tissue Tumors
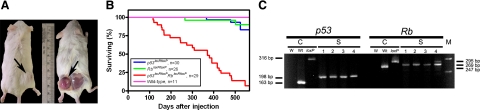
To test whether inactivation of p53 and/or Rb induces soft tissue sarcomas, AdCMVCre was subcutaneously injected into the right lower dorsal quadrant of wild-type or p53loxP/loxP and/or RbloxP/loxP adult male mice. By 600 days of age, three of 30 p53loxP/loxP mice (10%) and two of 26 RbloxP/loxP mice (8%) developed subcutaneous tumors (Figure 1A and Table 1). At the same time, tumors were formed in 27 of 29 (93%) p53loxP/loxPRbloxP/loxP mice starting at 116 days postinjection. After initial detection of a small, hard mass on the site of injection, tumors developed rapidly and aggressively over a period of 1 to 2 weeks and frequently showed ulceration, necrosis, hemorrhage, and decreased survival. No tumors were found in the place of AdCMVLacZ injection, the left lower dorsal quadrant of the same mice. Similarly, no tumors were found in wild-type mice injected with AdCMVCre. PCR genotyping of primary cells, as well as established cell lines from tumors of p53loxP/loxPRbloxP/loxP mice, showed deletion of both p53 and Rb (Figure 1C).
Figure 1.
Induction of soft tissue sarcomas by conditional inactivation of p53 and/or Rb. A: Sarcomas are detected at the site of AdCMVCre (right lower dorsal quadrant, arrow) but not AdCMVLacZ injection (left lower dorsal quadrant) of p53loxp/loxPRbloxP/loxP mice. In the right mouse, a sarcoma mass was flipped over together with attached skin during dissection. B: Survival of wild-type, p53loxP/loxP, RbloxP/loxP, and p53loxP/loxPRbloxP/loxP male mice after a single subcutaneous injection of AdCMVCre. Mice were sacrificed when sarcoma diameter reached 10 mm in diameter. All remaining mice were euthanized on day 600. Median survival for p53loxP/loxPRbloxP/loxP mice was 370 days. Compared to wild-type mice, P values for survival curves of p53loxP/loxPRbloxP/loxP, p53loxP/loxP, and RbloxP/loxP mice were <0.0001, 0.4096, and 0.4616, respectively. n, number of mice in the experimental group. C: Loss of the p53 and Rb genes in soft tissue tumors according to PCR analysis. Lanes 1 and 2: STN1 and STN2 cell lines, respectively. Lanes 3 and 4: Cells from primary tumors negatively sorted for F4/80 and CD11b and analyzed one passage (lane 3) and directly after sorting (lane 4). The 295-, 269-, and 247-bp fragments are diagnostic for floxed, excised, and wild-type alleles of the Rb gene, respectively. The 585-, 467-, and 432-bp fragments are diagnostic for floxed, excised, and wild-type alleles of the p53 gene, respectively. W, blank control, Wt, wild-type DNA, loxP, DNA with floxed gene, M, DNA marker.
Table 1.
Soft Tissue Sarcomas Induced by a Single Subcutaneous Injection of AdCMVCre into Mice Carrying floxed p53 and/or Rb
| Genotype | p53loxP/loxP | RbloxP/loxP | p53loxP/loxPRbloxP/loxP | Wild-type |
|---|---|---|---|---|
| Mice with neoplasms, total, %* | 9.1 (3/33)† | 7.7 (2/31) | 93 (27/29) | 0 (0/19) |
| UPS, %‡ | 33.3 (1/3) | 0 (0/2) | 70 (19/27) | 0 (0/0) |
| Leiomyosarcoma, % | 33.3 (1/3) | 0 (0/2) | 11 (3/27) | 0 (0/0) |
| Rhabdomyosarcoma, % | 33.3 (1/3) | 0 (0/2) | 7 (2/27) | 0 (0/0) |
| Angiosarcoma, % | 0 (0/3) | 50 (1/2) | 4 (1/27) | 0 (0/0) |
| Histocytic sarcoma, % | 0 (0/3) | 0 (0/2) | 4 (1/27) | 0 (0/0) |
| Fibrosarcoma, % | 0 (0/3) | 0 (0/2) | 4 (1/27) | 0 (0/0) |
| Dermatofibroma, % | 0 (0/3) | 50 (1/2) | 0 (0/27) | 0 (0/0) |
Numbers in parentheses indicate number of mice with neoplasm of total number of mice.
Fisher’s exact test. P is 0.2907, 0.5192, and <0.0001 for p53loxP/loxP, RbloxP/loxP, and p53loxP/loxPRbloxP/loxP mice, respectively, compared with wild-type mice.
Numbers in parentheses indicate number of particular neoplasm of total number of neoplasms.
Characterization of Soft Tissue Sarcomas
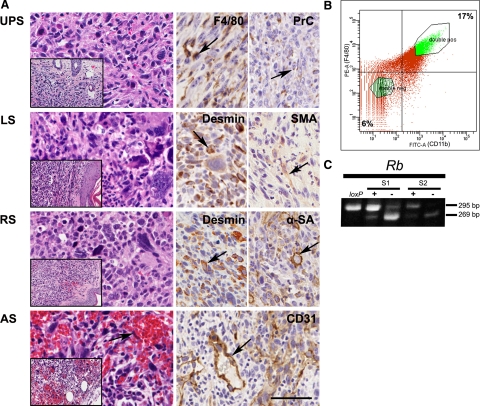
The majority of tumors were high-grade pleomorphic sarcomas and contained highly variable areas consisting of plump spindle cells intermixed with more polygonal or round, frequently giant cells with hyperchromatic irregular nuclei (Figure 2A). Multiple mitotic figures and some apoptotic cells were observed and the stroma consisted of delicate collagen fibrils encircling individual cells. Invasion of surrounding tissues was commonly observed and pulmonary metastasis were detected in 1 of 3 (33%) and 7 of 29 (24%) sarcomas of p53loxP/loxP and p53loxP/loxPRbloxP/loxP mice, respectively. Few neoplasms had distinct histological features consistent with angiosarcoma, histiocytic sarcoma, fibrosarcoma, and dermatofibroma (Table 1). Angiosarcoma and histiocytic sarcoma, respectively expressed markers of endothelial (CD31) and histiocytic (F4/80) differentiation.
Figure 2.
Characterization of soft tissue sarcomas. A: Pathology of soft tissue sarcomas induced by p53 and/or Rb inactivation. Expression (arrows, brown color) of F4/80 and procollagen type I (PrC), desmin, smooth muscle actin (SMA), α-sarcomeric actin (α-SA), and CD31 in undifferentiated high-grade pleomorphic sarcoma (UPS), leiomyosarcoma (LS), rhabdomyosarcoma (RS), and angiosarcoma (AS). AS features endothelial nuclear atypia (arrow). Hematoxylin and eosin (left column images), and ABC Elite method with hematoxylin counterstaining (right column images). Calibration bar = 50 μm (all large images), 20 μm (all insets). B: Gating of STS cells positive (double pos, 17%) or negative (double neg, 6%) for F4/80 (PE) and CD11b (FITC) during FACS based isolation. C: Detection of Cre-loxP–mediated deletion of the Rb gene by PCR in F4/80+CD11b+ (+) and F4/80−CD11b− (−) cell subpopulations isolated by FACS from primary STS. loxP, DNA with floxed gene, S1 and S2 indicate representative individual tumor samples. See Figure 1 for description of PCR fragments.
According to immunohistochemical analysis significant areas of desmin and either smooth muscle actin or α-sarcomeric actin were detected in several pleomorphic sarcomas which were respectively classified as leiomyosarcoma and rhabdomyosarcoma. However, 70% of sarcomas (18 of 27) and 33% (1 of 3) in p53loxP/loxPRbloxP/loxP and p53loxP/loxP mice, respectively, did not demonstrate any significant (more than 5%) differentiation with an exception of procollagen type I, a marker of collagen producing cells, such as fibroblasts. Additionally, sarcomas frequently contained a significant number of cells expressing the macrophage marker F4/80 (Figure 2A). However, these cells did not have neoplastic morphology. Consistently, macrophage population selected by fluorescence-activated cell sorting with antibodies to F4/80 and CD11b, contained only traces of Cre-loxP mediated deletion of the Rb gene (Figure 2, B and C), likely due to phagocytosis of neoplastic cells and/or presence of contaminating neoplastic cells.
CXCR4 Overexpression Contributes to Invasive Properties of STS
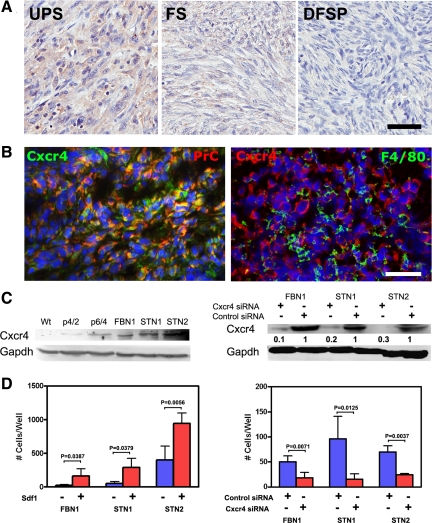
It has been reported that the expression of CXCR4 in human STSs is associated with poor prognosis and metastasis,34 particularly in malignant nonround cell tumors such as UPS and leiomyosarcoma. In agreement with these observations, we determined high expression of CXCR4 in human UPS (MFH) and leiomyosarcoma (Table 2). Notably, we observed that expression of CXCR4 was high in UPS, moderate in fibrosarcoma, and negligible in dermatofibrosarcoma protuberans (Figure 3A).
Table 2.
Expression of CXCR4 in Human Soft Tissue Tumors
| Tumor type | Frequency of tumors with specific level of staining, %*
|
|||
|---|---|---|---|---|
| − | + | ++ | +++ | |
| UPS | 26.7 (4/15) | 20 (3/15) | 46.7 (7/15) | 6.7 (1/15) |
| Fibrosarcoma | 28.6 (2/7) | 42.9 (3/7) | 28.6 (2/7) | 0 (0/7) |
| DFSP† | 100 (5/5) | 0 (0/5) | 0 (0/5) | 0 (0/5) |
| Neurofibrosarcoma | 50 (2/4) | 25 (1/4) | 25 (1/4) | 0 (0/4) |
| Liposarcoma | 18.8 (3/16) | 50 (8/16) | 31.3 (5/16) | 0 (0/16) |
| Leiomyosarcoma | 18.2 (2/11) | 18.2 (2/11) | 45.5 (5/11) | 18.2 (2/11) |
| Rhabdomyosarcoma | 0 (0/1) | 100 (1/1) | 0 (0/1) | 0 (0/1) |
| Hemangiosarcoma | 0 (0/1) | 0 (0/1) | 0 (0/1) | 100 (1/1) |
| M. Schwannoma | 100 (1/1) | 0 (0/1) | 0 (0/1) | 0 (0/1) |
Numbers in parentheses indicate number of particular tumors with specific level of CXCR4 expression of total number of neoplasms of the same type.
Expression CXCR4 is significantly lower in dermatofibrosarcoma protuberance (DFSP) compared with UPS (Fisher’s exact test, P = 0.0081) and leiomyosarcoma (Fisher’s exact test, P = 0.0048).
Figure 3.
CXCR4 overexpression contributes to invasive properties of STS. A: CXCR4 expression (brown color) in human undifferentiated, high-grade sarcoma (UPS), fibrosarcoma (FS), and dermatofibrosarcoma protuberans (DFSP). Note preferential expression of CXCR4 in UPS. ABC Elite method with hematoxylin counterstaining. B: Colocalization (yellow color) of Cxcr4 expression in cells containing procollagen type I (PrC) but not F4/80. Double immunofluorescence with DAPI counterstaining. A and B: Calibration bar = 50 μm. C, Left: Cxcr4 expression in wild-type (Wt) and p53loxP/loxPRbloxP/loxP fibroblasts two (p4/2) and four (p6/4) passages after exposure to AdCMVCre, and in established cell lines FBN1 (passage 43), STN1 and STN2 (both passage 39). Right: Cxcr4 expression after exposure of cell lines FBN1, STN1, and STN2 to Cxcr4 or control siRNA. D: Effects of Sdf1 (left) and siRNA-mediated knockdown of Cxcr4 (right) on cell invasion in Matrigel assay. All values are the mean ± SD, n = 4 (FVBN1 and STN1) or n = 3 (STN2). Control siRNA, scrambled siRNA.
Consistent with observations in human STS, mouse sarcomas expressed Cxcr4 according to immunostaining and Western blot analyses (Figure 3, B and C). The majority of Cxcr4 expressing cells also expressed procollagen type I but not macrophage marker F4/80. Levels of CXCR4 were negligible in primary fibroblast cell cultures but increased after inactivation of p53 and Rb. To test whether Cxcr4 plays role in sarcoma progression, Matrigel invasion assays were performed with STS cell lines STN1 and STN2 as well as p53 and Rb-deficient fibroblastic cell line FBN1. Cells of all lines showed increased invasion after addition of Cxcr4 ligand Sdf-1 (Figure 3D). At the same time, siRNA-mediated knockdown of Cxcr4 significantly reduced invasion activity of these cells (Figure 3D).
Connective Tissue Cells Targeted with Adenovirus
To establish which dermal cells were targeted by adenovirus, expression of EGFP was detected within 1 to 14 days after subcutaneous injection of adenovirus expressing Cre-EGFP (AdCMVCre-EGFP). EGFP expression in subcutaneous tissues was strongest at 1 and 3 days after infection. According to cell phenotyping, 39% of fibroblasts (procollagen type I+), 44% (F4/80+) or 42% (Iba-1+) of macrophages, 33% of smooth muscle cells (smooth muscle actin+), and 16% of endothelial cells (CD31+) were labeled with AdCMVCre-EGFP (Table 3 and Figure 4A).
Table 3.
Percentage of EGFP-Expressing Cells in Dermal Cell Populations 3 Days after AdCMVCre-EGFP Infection
| Cell marker | Pro-collagen type I | F4/80 | Iba-1 | Smooth muscle actin | CD31 |
|---|---|---|---|---|---|
| Mean±SD (%)* | 38.88 ± 18.77 | 44.33 ± 14.38 | 42.28 ± 6.53 | 32.8 ± 33.37 | 15.66 ± 26.46 |
At least 50 DAPI-stained cells per field were counted in 10 fields at magnification ×400.
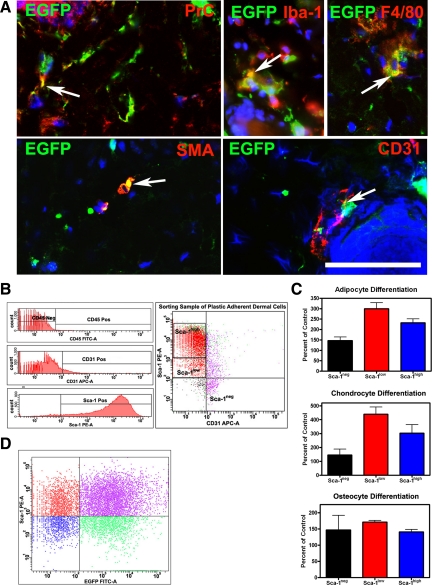
Figure 4.
Identification of connective tissue cells infected with AdCMVCre-EGFP adenovirus. A: Colocalization (yellow color, arrows) of EGFP and fibroblastic (procollagen type I, PrC), macrophage (F4/80), smooth muscle (smooth muscle actin, SMA), or endothelial (CD31) markers. Calibration bar = 50 μm. B: Expression of CD45, CD31, and Sca-1 in plastic adherent dermal cells (left). Sorting gates for isolation of Sca-1high, Sca-1low, and Sca-1neg cell populations after negative selection for CD45 and CD31 (right). C: Sca-1low cells have a greater potential for differentiation into adipocytes (P = 0.0076), chondrocytes (P = 0.0139), and osteocytes (P = 0.0006) compared with Sca-1high population. Additionally, Sca-1low cells have greater adipocyte (P = 0.0001) and chondrocyte (P = 0.0001) potential compared with Sca-1neg cells. Values (mean ± SD, n = 4) represent percentage of controls without differentiation medium. All experiments were repeated at least twice and subjected to unpaired t-tests with two-tailed P values. D: Flow cytometry analysis of expression of Sca-1 (y axis) and EGFP (x axis) 1 week after infection of dermal plastic adherent cells with adenovirus expressing EGFP.
To test whether adenovirus also infected MSC, this population was isolated from dissociated dermal cells by strict plastic adhesion followed by negative selection for CD45, a marker for cells of hematopoietic origin, including hematopoietic stem cells, and for endothelial cell marker CD31 and positive selection for Sca-1 expression (Figure 4B). Although all plastic adherent cells had a potential for differentiation toward adipogenic and osteogenic differentiation,35 our studies demonstrated that further selection for Sca-1low fraction results in additional enrichment for cells with enhanced ability to form adipocytes, chondrocytes and osteocytes (Figure 4C). All cell subpopulations of plastic adherent cells were equally well infected with AdCMVEGFP (Figure 4D).
Local Origin of STS
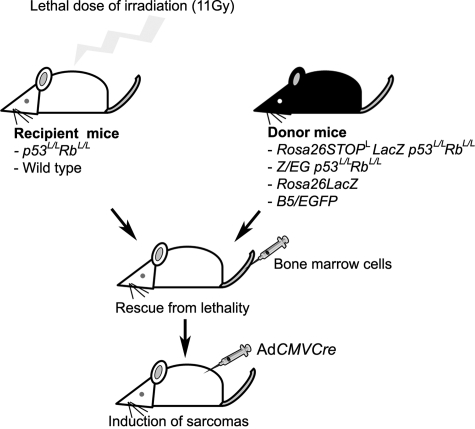
To discriminate between bone marrow and local dermal cell of origin of STS, irradiation chimeras reconstituted with donor-derived bone marrow cells isolated from mice with reporter transgenes were generated (Figure 5).
Figure 5.
Identification of soft tissue sarcoma cellular origin in irradiation chimeras. p53loxP/loxPRbloxP/loxP (p53L/LRbL/L) or wild-type mice were exposed to lethal dose (11 Gy) of irradiation, followed by rescue with bone marrow cells derived from the Rosa26StoploxPLacZ (Rosa26StopLLacZ) or the Z/EG reporter mice containing either wild-type or floxed p53 and Rb. Soft tissue sarcomas were induced by the subcutaneous AdCMVCre injection 14 days after bone marrow reconstitution.
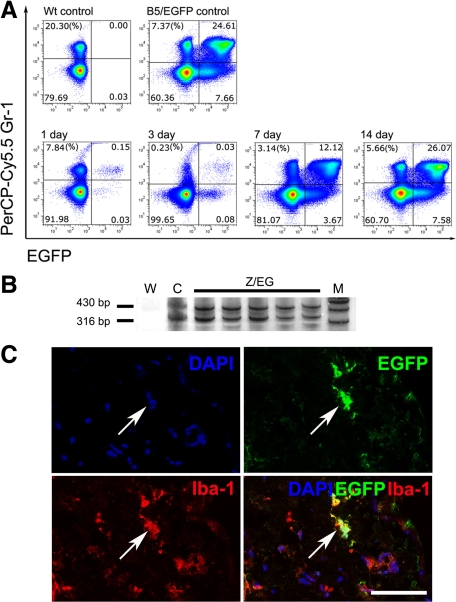
To evaluate a proportion of donor bone marrow cells, bone marrow cells and the dermal cells were evaluated for EGFP expression at 1, 3, 7, and 14 days after irradiation and transplantation of bone marrow cells from B5/EGFP reporter mice. Reduced numbers of cells expressing Gr-1, a marker for granulocytes and monocytes, and limited EGFP expression was detected at 1 and 3 days after irradiation followed by immediate bone marrow transplantation (Figure 6A). Both cell populations increased by day 7 and were within normal range for a B5/EGFP reporter mouse. These results were consistent with detection of EGFP transgene copy numbers similar to those in B5/EGFP mice by multiplex PCR (Figure 6B). At the same time, 24% of Iba-1+ macrophages (98.5 ± 29, Mean ± SD, out of 410 ± 86.3 counted) expressed EGFP 14 days postirradiation (Figure 6C).
Figure 6.
Assessment of bone marrow rescue in irradiation chimeras. A: Efficiency of reconstitution of bone marrow cells according to coexpression of donor-derived EGFP and granulocyte/monocyte marker Gr-1 between 1 and 14 days after irradiation and transplantation of bone marrow from B5/EGFP mice (lower row). Wt and B5/EGFP, Gr-1/EGFP profiles of bone marrow cells from wild-type and B5/EGFP mice, respectively (upper row). B: Detection of ACTBStoploxPEGFP transgene (430 bp) by PCR analysis of blood from chimeras that were transplanted with bone marrow cells from the Z/EG reporter mice. The 316-bp DNA fragment diagnostic of floxed p53 was used as internal reference control. (W, water control; C, DNA from a mouse carrying the Z/EG reporter, as well as floxed p53 and Rb genes; M, marker). Silver staining of acrylamide gels. C: Coexpression of EGFP and macrophage marker Iba-1 in dermal cells 14 days after irradiation and transplantation of bone marrow cell–derived B5/EGFP donor mice. DAPI counterstaining. Arrows indicate the same area in all images. Calibration bar = 50 μm.
To determine whether any STS developed from bone marrow–derived cells, sarcomas were initiated by a single subcutaneous injection of AdCMVCre into irradiated p53loxP/loxPRbloxP/loxP (n = 29) and wild-type mice (n = 9) reconstituted with bone marrow from mice carrying floxed p53 and Rb as well as Rosa26StoploxPLacZ or Z/EG reporter constructs. In these reporter constructs, expression of β-galactosidase or EGFP, respectively, is possible only after deletion of a STOP codon flanked by loxP sites. Additionally, another group of p53loxP/loxPRbloxP/loxP mice (n = 4) was reconstituted with bone marrow of mice carrying constitutively expressed EGFP (B5/EGFP) or LacZ (Rosa26LacZ) transgene. In parallel with collecting tumors, blood from each chimera was collected and genotyped by PCR analysis to confirm the reconstitution with donor cells.
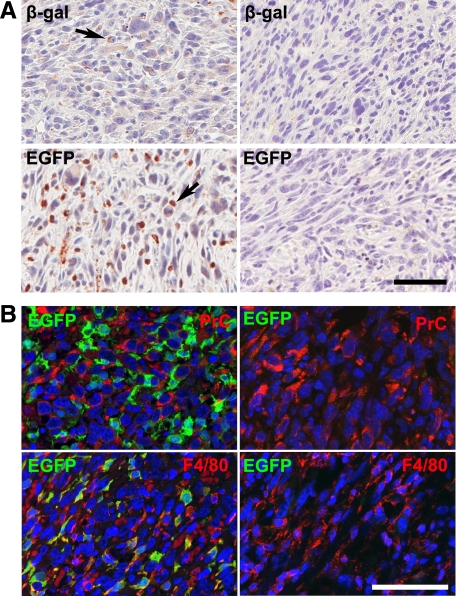
All chimeric mice with host p53loxP/loxPRbloxP/loxP genotype (n = 33) but none of wild-type (n = 9) developed sarcomas independently of the type of donor mouse strain. The sarcomas were diagnosed as UPS similar to the majority of sarcomas associated with p53 and Rb deficiency. The immunohistochemistry results showed that tumors from control chimeras rescued with bone marrow of mice carrying constitutively expressed EGFP (B5/EGFP) or LacZ (Rosa26LacZ) transgene contained numerous cells expressing EGFP or β-galactosidase (Figure 7A). These cells did not have atypical cytological features and expressed macrophage marker F4/80 but no procollagen type I (Figure 7B). At the same time, sarcomas developed from chimeras that contain bone marrow cells from the Z/EG (n = 19) or the Rosa26STOPloxPLacZ (n = 10) reporter mice did not show any GFP or β-galactosidase expression (Figure 7A). Thus none of the 38 mice developed STS from bone marrow with floxed p53 and Rb. Taken together with complete reconstitution of bone marrow cells and 24% reconstitution of the dermal macrophage population, these results indicate that STS associated with p53 and Rb inactivation most likely originate from the local resident cells as opposed to bone marrow derived cells, such as macrophages.
Figure 7.
Detection of bone marrow–derived cells in soft tissue sarcomas. A: Expression of β-galactosidase (β-gal, arrow) and EGFP (arrow) in sarcomas initiated by a single SC AdCMVCre injection into lethally irradiated p53loxP/loxPRbloxP/loxP mice transplanted with bone marrow cells from Rosa26LacZ (upper left panel) and B5/EGFP reporter mice (lower left panel) but not in those with Rosa26StoploxPLacZ (upper right panel) or a Z/EG (lower right panel) bone marrow cells. ABC Elite method with hematoxylin counterstaining. B: Expression of EGFP, procollagen type I (PrC), and F4/80 markers in sarcomas of p53loxP/loxPRbloxP/loxP irradiation chimeras reconstituted with bone marrow cells from a B5/EGFP (left) and Z/EG reporter mice (right). Note coexpression (yellow color) of EGFP and F4/80 in sarcoma of mouse reconstituted with B5/EGFP bone marrow. Double immunofluorescence with DAPI counterstaining. Calibration bar: 50 μm (A and B).
MSC Have Enhanced Potential for Malignant Transformation
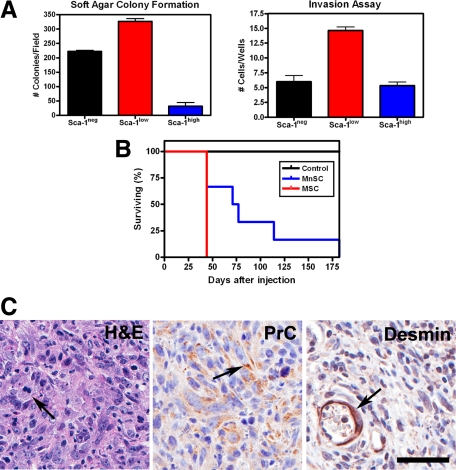
To investigate whether dermal MSC could be a target for malignant transformation in our model, these cells were subjected to a series of transformation-related assays after AdCMVCre-mediated inactivation of p53 and Rb. First, the ability of the three plastic adherent dermal cell populations to form colonies in soft agar was compared. Interestingly, Sca-1low cells showed an increase in the number of colonies when compared to the other 2 plastic adherent dermal cell populations (Figure 8A). Secondly, invasion ability of transformed plastic adherent dermal cells among Sca-1neg, Sca-1low, and Sca-1high populations was tested using a Matrigel invasion chamber. Sca-1low cells once again exhibited the greatest potential for malignant transformation compared to the other two plastic adherent dermal cell populations (Figure 8A). Finally, tumorigenicity of MSC was tested after their subcutaneous transplantation into SCID mice. Sca-1low cells formed tumors faster (Figure 8B). Importantly, tumors were UPS histologically similar to conditionally induced sarcomas in p53loxP/loxPRbloxP/loxP mice (Figure 8C). These tumors expressed procollagen type I but no other diagnostic markers in neoplastic cells, including desmin (Figure 8C). These results indicate that Sca-1low multipotent dermal stem/progenitor cells are a preferential target for malignant transformation associated with p53 and Rb deficiency.
Figure 8.
Neoplastic potential of dermal mesenchymal stem cells (MSC) after conditional inactivation of p53 and Rb. A: Compared with Sca-1neg and Sca-1high subpopulations, the Sca-1low MSC cells have significantly higher propensity for forming colonies in soft agar (left panel, mean ± SD, n = 3, P < 0.0001) and invasion in Matrigel chamber (right panel, mean ± SD, n = 3, Sca-1low versus Sca-1neg, P = 0.0002; Sca-1low versus Sca-1high, P < 0.0001). B: Accelerated formation of tumors after SC transplantation of MSC (median survival 44 days, n = 3) compared with mesenchymal non-stem cells (MnSC, median survival 74 days, n = 6; P = 0.0376). C: Undifferentiated high-grade pleomorphic sarcoma with mitotic figures (H&E, arrow) and expression of procollagen type I (PrC, arrow) formed after MSC implantation. Desmin is detected only in smooth muscle cells of blood vessels (arrow). Left panel, Hematoxylin and eosin, lower, middle, and right panels, ABC Elite method with hematoxylin counterstaining. Calibration bar = 50 μm.
Discussion
STS are relatively uncommon neoplasms and constitute less than 1% of all cancers, with an estimated 10,660 new STSs diagnoses and 3820 related deaths in 2009.36 However, lack of identifiable precursor lesions and high heterogeneity of these neoplasms, as well as incomplete understanding of ontogenesis and phenotypical plasticity of normal connective tissue cells represent a continuous challenge for diagnosis, which is reflected in continuous shift in STS classifications. Therefore, development of mouse models which accurately recapitulate both morphological features and frequent genetic alterations of human STS is of critical importance. The mouse model of STS reported here addresses this need because histology of induced sarcomas is similar to those of human STS. Furthermore, mutations in p53 and Rb are the most common genetic alterations occurring in sarcomas with complex genotype, such as UPS, fibrosarcoma, and leiomyosarcoma.
Similarity between human STS and sarcomas in our mouse model was further confirmed by observation of expression of CXCR4 which is a receptor of chemokine CXCL12 (SDF-1α). It has been previously reported that signaling through CXCR4 plays an important role in progression of a broad variety of malignancies, including breast cancers,37 prostate cancers,38 malignant melanomas,37,39 gastric cancer,40 osteosarcomas,41 and rhabdomyosarcomas.42 In agreement with a previous report that expression of CXCR4 is associated with poor prognosis of STS34 we observed that expression of CXCR4 is significantly higher in high-grade sarcomas, such as UPS and leiomyosarcomas as compared to low-grade dermatofibrosarcoma and fibrosarcoma. Furthermore, according to our Matrigel invasion assays, stimulation of Cxcr4 by addition of Sdf-1 increased invasion of sarcoma cells, while siRNA-mediated knockdown of Cxcr4 significantly reduced their invasion. Taken together, these results demonstrate that the CXCR4/SDF-1 axis is an important mediator in sarcoma progression. It has been reported that CXCR4 expression is repressed by p53.43 Our observations of increased Cxcr4 expression in fibroblasts after p53 and Rb inactivation is consistent with that report and indicate that inactivation of p53 during sarcoma initiation may predetermine some of the advanced metastatic traits.
Diagnosis of UPS, also known as MFH, is a matter of a particular controversy. Given the lack of specific immunohistochemical markers and firm diagnostic criteria of this neoplasm, it is not surprising that the frequency of UPS/MFH diagnosis ranges from 5% to 70%.3,4,44 MFH was originally described by A. P. Stout and colleagues45,46,47 as a neoplasm consisting of histiocyte-like and fibroblast-like cells. Some initial studies performed on human cells and tissues, as well as in transplantation experiments, have supported an origin of these sarcomas from bone marrow–derived cells of the mononuclear phagocyte system, that is histiocytes/macrophages.46,48,49,50,51,52,53 However, later observations based on advanced ultrastructural and immunohistochemical characterization of neoplastic cells, as well as studies of sequential evaluation of sarcoma formation in rats and mice, favor a view that UPS/MFH originate from fibroblastic or mesenchymal precursor/stem cells.2,54,55,56,57 These studies have also demonstrated that UPS/MFH frequently contain a large number of nonneoplastic infiltrating macrophages.57,58,59,60 Consistent with those reports we observed that cells expressing macrophage markers F4/80 and/or Iba1 do not have neoplastic morphology and lack Cre-loxP–mediated recombination.
Some UPS may represent other STS that exhibit high degree of phenotypical plasticity during tumor progression.61,62,63,64,65 However, findings of MFH-specific genomic alterations66,67,68 and gene expression profiles69,70,71 indicate that diagnosis of UPS/MFH corresponds to a distinct nosological entity and may reflect development of these neoplasms from MSC.
Recent progress in isolation of MSC and their potential use in therapeutic approaches72,73 have renewed interest in assessment of transformation potential of these cells. By using human bone marrow MSC Matushansky et al5 demonstrated that the expression pattern of these cells correlates with that of UPS/MFH. Furthermore, they demonstrated that MSC immortalized with SV-40 large T antigen recapitulated UPS/MFH morphology after subcutaneous transplantation into nude mice. Consistently, formation of UPS was reported after transplantation of bone marrow–derived mouse MSC treated with 2-methylcholanthrene.7 Intriguingly, it has been reported that according to cell lineage tracing experiments in irradiation chimeras, spontaneous fibrosarcomas arise from bone marrow–derived cells in the mouse.74
To evaluate a possibility of bone marrow origin of STS in our model, we have performed irradiation chimera experiments in which donor and recipient cells carried floxed copies of p53 and Rb. Given 24% reconstitution of dermal macrophage population in irradiation chimeras, 70% frequency of UPS in our model and near equal infectivity of macrophages and fibroblasts by adenovirus, one would expect that sarcomas in six to seven mice could arise from macrophages. However, none of 38 mice developed STS from bone marrow with floxed p53 and Rb. These results provide the direct experimental evidence that UPS/MFH do not arise from macrophages and indicate that initiation of sarcomas from bone marrow–derived cells is unlikely.
To test whether STS may preferentially arise from local resident MSC after p53 and Rb inactivation we have isolated dermal mesenchymal stem/progenitors cells based on their adherence to plastic and expression of Sca-1 marker (Ly-6A/E) in combination with negative selection for CD45 and CD31. Sca-1 has been shown to be an effective marker for stem cells in many different tissues, including MSC.75,76,77,78,79 Interestingly, in our experiments the Sca-1low population was enriched for multipotent cells. It was reported that Sca-1low population was more enriched for stem cells than the expected Sca-1high population in the mammary epithelial cells80 and low levels of positive staining for Sca-1 were detected in spermatogonial stem cells.81 Similar to these observations we also found that prolonged culturing of cells, before staining and FACS, induced expression of Sca-1 (our unpublished observations). Therefore, it is quite possible that the culture step necessary to select for plastic adherence upregulated Sca-1 expression in the PAD cells, throwing off the Sca-1 expression levels that would be expected based on the majority of studies that sort cells before any culturing.75,82
By using isolated dermal MSC enriched in the Sca-1low fraction we were able to demonstrate their increased transformation potential, as comported to more differentiated connective tissue cells of the dermis, after inactivation of p53 and Rb. Interestingly, transformation of dermal MSC resulted in formation of UPS, consistent with previous reports which used bone marrow MSC.5,7 At the same time, Rodriguez et al35 reported formation of fibrosarcomas with areas of adipose tissue differentiation after subcutaneous injection of MSC derived from adipose tissue of p21−/− p53−/− mice into immunodeficient mice. These variations in tumor type may reflect either different differentiation potential of MSC derived from different locations or gene-dependent differentiation potential.
Recent observations indicate that p53 and Rb tumor suppressors have important functions in stem cells. p53 has been shown to mediate the onset of senescence of endothelial progenitor cells, to negatively regulate proliferation and survival of neural stem cells, to induce differentiation of embryonic stem cells, and to control efficiency of induced pluripotent stem cell generation.83,84,85 Rb and its pathway are involved in control of stem cell cycle progression, senescence and tissue homeostasis.86 It has been reported that fibroblasts lacking Rb form spheres and become cancer stem cells.87 Notably, p16/Rb and ARF/p53 pathways are extensively connected and may be coordinately regulated by Bmi-1, which is required for maintenance and self-renewal of hematopoietic and neural stem and progenitor cells.88 Consistently it has been demonstrated that combined p53 and Rb deficiency results in prostate cancer arising from the prostate stem cell compartment.33 Further studies of established STS model will address the specific molecular and cellular mechanisms responsible for cooperation of p53 and Rb in control of MSC and their malignant transformation. It also remains to be addressed whether sarcomas with distinct differentiation features, such as leiomyosarcoma and angiosarcoma, originate from MSC or more committed progenitor cells which are less prone to transformation after p53 and Rb inactivation. It is of interest that according to comparative genomic hybridization studies, UPS/MFH share similarities with leiomyosarcomas,70,89 consistent with frequent observations of areas of smooth muscle differentiation in UPS, including our model.
In conclusion, we have established a genetically engineered mouse model of the STS by temporally and spatially controlled p53 and Rb inactivation in connective tissue cells of the dermis. We have also demonstrated that STS develop locally, most likely from MSC. This mouse model of STS will serve as a valuable basis for future studies of STS pathogenesis, particularly of those for sarcomas with complex genotypes. This model also provides new opportunities for exploration of targeted therapies and screening for novel therapeutic agent to treat the disease.
Acknowledgments
We thank Dr. Anton Berns (Netherlands Cancer Institute, Amsterdam, the Netherlands) for the generous gift of the p53loxP/loxP and RbloxP/loxP mice, Dr. Margaret Colleen McEntee (Cornell College of Veterinary Medicine) for helping with mice irradiation, James Smith III, (Biomedical Sciences Flow Cytometry Core Laboratory) for help with FACS experiments, Dr. Kyung-Chul Choi for his contributions to the initial stages of this project, David C. Corney for critical reading of the manuscript, and members of Nikitin lab for helpful comments.
Footnotes
Address reprint requests to Alexander Yu. Nikitin, M.D., Ph.D., Department of Biomedical Sciences, Cornell University, T2 014A VRT Campus Road, Ithaca, NY 14853-6401. E-mail: an58@cornell.edu.
Supported by grants BK21, Korea and Graduate Research Assistantship awarded by the College of Veterinary Medicine, Cornell University (J.C.) and RR17595 (National Institutes of Health/National Center for Research Resources), CA96823 (National Institutes of Health/National Cancer Institute), and C023050 (New York State Stem Cell Science) awards (A.Y.N.).
References
- Collini P, Sorensen PH, Patel S, Blay JY, Issels RD, Maki RG, Eriksson M, del Muro XG. Sarcomas with spindle cell morphology. Semin Oncol. 2009;36:324–337. doi: 10.1053/j.seminoncol.2009.06.007. [DOI] [PubMed] [Google Scholar]
- Mackall CL, Meltzer PS, Helman LJ. Focus on sarcomas. Cancer Cell. 2002;2:175–178. doi: 10.1016/s1535-6108(02)00132-0. [DOI] [PubMed] [Google Scholar]
- Guillou L, Aurias A: Soft tissue sarcomas with complex genomic profiles. Virchows Arch 2009, Nov 13 [Epub ahead of press] [DOI] [PubMed] [Google Scholar]
- Weiss SW, Goldblum JR. Enzinger & Weiss’s Soft Tissue Tumors. Weiss SW, Goldblum JR, editors. Philadelphia: Mosby, Elsevier; 2008:pp1–1258. [Google Scholar]
- Matushansky I, Hernando E, Socci ND, Mills JE, Matos TA, Edgar MA, Singer S, Maki RG, Cordon-Cardo C. Derivation of sarcomas from mesenchymal stem cells via inactivation of the Wnt pathway. J Clin Invest. 2007;117:3248–3257. doi: 10.1172/JCI31377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasso R, Augello A, Carida M, Postiglione F, Tibiletti MG, Bernasconi B, Astigiano S, Fais F, Truini M, Cancedda R, Pennesi G. Development of sarcomas in mice implanted with mesenchymal stem cells seeded onto bioscaffolds. Carcinogenesis. 2009;30:150–157. doi: 10.1093/carcin/bgn234. [DOI] [PubMed] [Google Scholar]
- Li Q, Hisha H, Takaki T, Adachi Y, Li M, Song C, Feng W, Okazaki S, Mizokami T, Kato J, Inaba M, Hosaka N, Maki M, Ikehara S. Transformation potential of bone marrow stromal cells into undifferentiated high-grade pleomorphic sarcoma. J Cancer Res Clin Oncol. 2010;136:829–838. doi: 10.1007/s00432-009-0723-0. [DOI] [PubMed] [Google Scholar]
- Tolar J, Nauta AJ, Osborn MJ, Panoskaltsis Mortari A, McElmurry RT, Bell S, Xia L, Zhou N, Riddle M, Schroeder TM, Westendorf JJ, McIvor RS, Hogendoorn PC, Szuhai K, Oseth L, Hirsch B, Yant SR, Kay MA, Peister A, Prockop DJ, Fibbe WE, Blazar BR. Sarcoma derived from cultured mesenchymal stem cells. Stem Cells. 2007;25:371–379. doi: 10.1634/stemcells.2005-0620. [DOI] [PubMed] [Google Scholar]
- Li H, Fan X, Kovi RC, Jo Y, Moquin B, Konz R, Stoicov C, Kurt-Jones E, Grossman SR, Lyle S, Rogers AB, Montrose M, Houghton J. Spontaneous expression of embryonic factors and p53 point mutations in aged mesenchymal stem cells: a model of age-related tumorigenesis in mice. Cancer Res. 2007;67:10889–10898. doi: 10.1158/0008-5472.CAN-07-2665. [DOI] [PubMed] [Google Scholar]
- Stratton MR, Moss S, Warren W, Patterson H, Clark J, Fisher C, Fletcher CD, Ball A, Thomas M, Gusterson BA, Cooper CS. Mutation of the p53 gene in human soft tissue sarcomas: association with abnormalities of the RB1 gene. Oncogene. 1990;5:1297–1301. [PubMed] [Google Scholar]
- Walkley CR, Qudsi R, Sankaran VG, Perry JA, Gostissa M, Roth SI, Rodda SJ, Snay E, Dunning P, Fahey FH, Alt FW, McMahon AP, Orkin SH. Conditional mouse osteosarcoma, dependent on p53 loss and potentiated by loss of Rb, mimics the human disease. Genes Dev. 2008;22:1662–1676. doi: 10.1101/gad.1656808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman SD, Calo E, Landman AS, Danielian PS, Miller ES, West JC, Fonhoue BD, Caron A, Bronson R, Bouxsein ML, Mukherjee S, Lees JA. Metastatic osteosarcoma induced by inactivation of Rb and p53 in the osteoblast lineage. Proc Natl Acad Sci USA. 2008;105:11851–11856. doi: 10.1073/pnas.0805462105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin PP, Pandey MK, Jin F, Raymond AK, Akiyama H, Lozano G. Targeted mutation of p53 and Rb in mesenchymal cells of the limb bud produces sarcomas in mice. Carcinogenesis. 2009;30:1789–1795. doi: 10.1093/carcin/bgp180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch DG, Dinulescu DM, Miller JB, Grimm J, Santiago PM, Young NP, Nielsen GP, Quade BJ, Chaber CJ, Schultz CP, Takeuchi O, Bronson RT, Crowley D, Korsmeyer SJ, Yoon SS, Hornicek FJ, Weissleder R, Jacks T. A spatially and temporally restricted mouse model of soft tissue sarcoma. Nat Med. 2007;13:992–997. doi: 10.1038/nm1602. [DOI] [PubMed] [Google Scholar]
- Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29:418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- Marino S, Vooijs M, van Der Gulden H, Jonkers J, Berns A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 2000;14:994–1004. [PMC free article] [PubMed] [Google Scholar]
- Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- Chai Y, Jiang X, Ito Y, Bringas P, Jr, Han J, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- Jiang X, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cardiac neural crest. Development. 2000;127:1607–1616. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- Hadjantonakis AK, Gertsenstein M, Ikawa M, Okabe M, Nagy A. Generating green fluorescent mice by germline transmission of green fluorescent ES cells. Mech Dev. 1998;76:79–90. doi: 10.1016/s0925-4773(98)00093-8. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Flesken-Nikitin A, Choi KC, Eng JP, Shmidt EN, Nikitin AY. Induction of carcinogenesis by concurrent inactivation of p53 and Rb1 in the mouse ovarian surface epithelium. Cancer Res. 2003;63:3459–3463. [PubMed] [Google Scholar]
- Anderson RD, Haskell RE, Xia H, Roessler BJ, Davidson BL. A simple method for the rapid generation of recombinant adenovirus vectors. Gene Ther. 2000;7:1034–1038. doi: 10.1038/sj.gt.3301197. [DOI] [PubMed] [Google Scholar]
- Choi J, Burns AA, Williams RM, Zhou Z, Flesken-Nikitin A, Zipfel WR, Wiesner U, Nikitin AY. Core-shell silica nanoparticles as fluorescent labels for nanomedicine. J Biomed Opt. 2007;12:064007. doi: 10.1117/1.2823149. [DOI] [PubMed] [Google Scholar]
- Cui YZ, Hisha H, Yang GX, Fan TX, Jin T, Li Q, Lian Z, Ikehara S. Optimal protocol for total body irradiation for allogeneic bone marrow transplantation in mice. Bone Marrow Transplant. 2002;30:843–849. doi: 10.1038/sj.bmt.1703766. [DOI] [PubMed] [Google Scholar]
- McDonald TP, Lange RD, Congdon CC, Toya RE. Effect of hypoxia, irradiation, and bone marrow transplantation on erythropoietin levels in mice. Radiat Res. 1970;42:151–163. [PubMed] [Google Scholar]
- Mohr U. International Classification of Rodent Tumours. Mohr U, editor. Berlin: Heidelberg, Springer; The Mouse. 2001:pp1–474. [Google Scholar]
- Zhou Z, Flesken-Nikitin A, Corney DC, Wang W, Goodrich DW, Roy-Burman P, Nikitin AY. Synergy of p53 and Rb deficiency in a conditional mouse model for metastatic prostate cancer. Cancer Res. 2006;66:7889–7898. doi: 10.1158/0008-5472.CAN-06-0486. [DOI] [PubMed] [Google Scholar]
- Matoso A, Zhou Z, Hayama R, Flesken-Nikitin A, Nikitin AY. Cell lineage-specific interactions between Men1 and Rb in neuroendocrine neoplasia. Carcinogenesis. 2008;29:620–628. doi: 10.1093/carcin/bgm207. [DOI] [PubMed] [Google Scholar]
- Wilson L, Fathke C, Isik F. Tissue dispersion and flow cytometry for the cellular analysis of wound healing. Biotechniques. 2002;32:548–551. doi: 10.2144/02323st07. [DOI] [PubMed] [Google Scholar]
- Corney DC, Hwang CI, Matoso A, Vogt M, Flesken-Nikitin A, Godwin AK, Kamat AA, Sood AK, Ellenson LH, Hermeking H, Nikitin AY. Frequent downregulation of miR-34 family in human ovarian cancers. Clin Cancer Res. 2010;16:1119–1128. doi: 10.1158/1078-0432.CCR-09-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corney DC, Flesken-Nikitin A, Godwin AK, Wang W, Nikitin AY. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67:8433–8438. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Flesken-Nikitin A, Nikitin AY. Prostate cancer associated with p53 and Rb deficiency arises from the stem/progenitor cell-enriched proximal region of prostatic ducts. Cancer Res. 2007;67:5683–5690. doi: 10.1158/0008-5472.CAN-07-0768. [DOI] [PubMed] [Google Scholar]
- Oda Y, Tateishi N, Matono H, Matsuura S, Yamamaoto H, Tamiya S, Yokoyama R, Matsuda S, Iwamoto Y, Tsuneyoshi M. Chemokine receptor CXCR4 expression is correlated with VEGF expression and poor survival in soft-tissue sarcoma. Int J Cancer. 2009;124:1852–1859. doi: 10.1002/ijc.24128. [DOI] [PubMed] [Google Scholar]
- Rodriguez R, Rubio R, Masip M, Catalina P, Nieto A, de la Cueva T, Arriero M, San Martin N, de la Cueva E, Balomenos D, Menendez P, Garcia-Castro J. Loss of p53 induces tumorigenesis in p21-deficient mesenchymal stem cells. Neoplasia. 2009;11:397–407. doi: 10.1593/neo.81620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- Muller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN, Barrera JL, Mohar A, Verastegui E, Zlotnik A. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- Taichman RS, Cooper C, Keller ET, Pienta KJ, Taichman NS, McCauley LK. Use of the stromal cell-derived factor-1/CXCR4 pathway in prostate cancer metastasis to bone. Cancer Res. 2002;62:1832–1837. [PubMed] [Google Scholar]
- Scala S, Ottaiano A, Ascierto PA, Cavalli M, Simeone E, Giuliano P, Napolitano M, Franco R, Botti G, Castello G. Expression of CXCR4 predicts poor prognosis in patients with malignant melanoma. Clin Cancer Res. 2005;11:1835–1841. doi: 10.1158/1078-0432.CCR-04-1887. [DOI] [PubMed] [Google Scholar]
- Yasumoto K, Koizumi K, Kawashima A, Saitoh Y, Arita Y, Shinohara K, Minami T, Nakayama T, Sakurai H, Takahashi Y, Yoshie O, Saiki I. Role of the CXCL12/CXCR4 axis in peritoneal carcinomatosis of gastric cancer. Cancer Res. 2006;66:2181–2187. doi: 10.1158/0008-5472.CAN-05-3393. [DOI] [PubMed] [Google Scholar]
- Oda Y, Yamamoto H, Tamiya S, Matsuda S, Tanaka K, Yokoyama R, Iwamoto Y, Tsuneyoshi M. CXCR4 and VEGF expression in the primary site and the metastatic site of human osteosarcoma: analysis within a group of patients, all of whom developed lung metastasis. Mod Pathol. 2006;19:738–745. doi: 10.1038/modpathol.3800587. [DOI] [PubMed] [Google Scholar]
- Strahm B, Durbin AD, Sexsmith E, Malkin D. The CXCR4-SDF1alpha axis is a critical mediator of rhabdomyosarcoma metastatic signaling induced by bone marrow stroma. Clin Exp Metastasis. 2008;25:1–10. doi: 10.1007/s10585-007-9094-6. [DOI] [PubMed] [Google Scholar]
- Mehta SA, Christopherson KW, Bhat-Nakshatri P, Goulet RJ, Jr, Broxmeyer HE, Kopelovich L, Nakshatri H. Negative regulation of chemokine receptor CXCR4 by tumor suppressor p53 in breast cancer cells: implications of p53 mutation or isoform expression on breast cancer cell invasion. Oncogene. 2007;26:3329–3337. doi: 10.1038/sj.onc.1210120. [DOI] [PubMed] [Google Scholar]
- Engellau J, Anderson H, Rydholm A, Bauer HC, Hall KS, Gustafson P, Akerman M, Meis-Kindblom J, Alvegard TA, Nilbert M. Time dependence of prognostic factors for patients with soft tissue sarcoma: a Scandinavian Sarcoma Group Study of 338 malignant fibrous histiocytomas. Cancer. 2004;100:2233–2239. doi: 10.1002/cncr.20254. [DOI] [PubMed] [Google Scholar]
- Kauffman SL, Stout AP. Histiocytic tumors (fibrous xanthoma and histiocytoma) in children. Cancer. 1961;14:469–482. doi: 10.1002/1097-0142(199005/06)14:3<469::aid-cncr2820140304>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Ozzello L, Stout AP, Murray MR. Cultural characteristics of malignant histiocytomas and fibrous xanthomas. Cancer. 1963;16:331–344. doi: 10.1002/1097-0142(196303)16:3<331::aid-cncr2820160307>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- O'Brien JE, Stout AP. Malignant fibrous xanthomas. Cancer. 1964;17:1445–1455. doi: 10.1002/1097-0142(196411)17:11<1445::aid-cncr2820171112>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Yumoto T, Morimoto K. Experimental approach to fibrous histiocytoma. Acta Pathol Jpn. 1980;30:767–778. doi: 10.1111/j.1440-1827.1980.tb00972.x. [DOI] [PubMed] [Google Scholar]
- Shirasuna K, Sugiyama M, Miyazaki T. Establishment and characterization of neoplastic cells from a malignant fibrous histiocytoma: a possible stem cell line. Cancer. 1985;55:2521–2532. doi: 10.1002/1097-0142(19850601)55:11<2521::aid-cncr2820551102>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Strauchen JA, Dimitriu-Bona A. Malignant fibrous histiocytoma: expression of monocyte/macrophage differentiation antigens detected with monoclonal antibodies. Am J Pathol. 1986;124:303–309. [PMC free article] [PubMed] [Google Scholar]
- Hagari Y, Yumoto T. Experimental tumors of myxoid malignant fibrous histiocytoma and hyaluronic acid production. Acta Pathol Jpn. 1987;37:975–988. doi: 10.1111/j.1440-1827.1987.tb00447.x. [DOI] [PubMed] [Google Scholar]
- Yamate J, Tajima M, Togo M, Shibuya K, Ihara M, Kudow S. Heterogeneity of cloned cell lines established from a transplantable rat malignant fibrous histiocytoma. Jpn J Cancer Res. 1991;82:298–307. doi: 10.1111/j.1349-7006.1991.tb01846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder SW, Said JW, Shintaku IP, Pinkus GS. A histiocyte-specific marker in the diagnosis of malignant fibrous histiocytoma: use of monoclonal antibody KP-1 (CD68) Am J Clin Pathol. 1992;97:759–763. doi: 10.1093/ajcp/97.6.759. [DOI] [PubMed] [Google Scholar]
- Fu YS, Gabbiani G, Kaye GI, Lattes R. Malignant soft tissue tumors of probable histiocytic origin (malignant fibrous histiocytomas): general considerations and electron microscopic and tissue culture studies. Cancer. 1975;35:176–198. doi: 10.1002/1097-0142(197501)35:1<176::aid-cncr2820350123>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Hoffman MA, Dickersin GR. Malignant fibrous histiocytoma: an ultrastructural study of eleven cases. Hum Pathol. 1983;14:913–922. doi: 10.1016/s0046-8177(83)80166-x. [DOI] [PubMed] [Google Scholar]
- Nikitin AY, Rajewsky MF, Pozharisski KM. Development of malignant fibrous histiocytoma induced by 7,12-dimethylbenz[a]anthracene in the rat: characterization of early atypical cells. Virchows Arch B Cell Pathol Incl Mol Pathol. 1993;64:151–159. doi: 10.1007/BF02915108. [DOI] [PubMed] [Google Scholar]
- Takeya M, Yamashiro S, Yoshimura T, Takahashi K. Immunophenotypic and immunoelectron microscopic characterization of major constituent cells in malignant fibrous histiocytoma using human cell lines and their transplanted tumors in immunodeficient mice. Lab Invest. 1995;72:679–688. [PubMed] [Google Scholar]
- Roessner A, Vassallo J, Vollmer E, Zwadlo G, Sorg C, Grundmann E. Biological characterization of human bone tumors. X. The proliferation behavior of macrophages as compared to fibroblastic cells in malignant fibrous histiocytoma and giant cell tumor of bone. J Cancer Res Clin Oncol. 1987;113:559–562. doi: 10.1007/BF00390865. [DOI] [PubMed] [Google Scholar]
- Kato T, Takeya M, Takagi K, Takahashi K. Chemically induced transplantable malignant fibrous histiocytoma of the rat. Analyses with immunohistochemistry, immunoelectron microscopy and [3H]thymidine autoradiography. Lab Invest. 1990;62:635–645. [PubMed] [Google Scholar]
- Takeya M, Yoshimura T, Leonard EJ, Kato T, Okabe H, Takahashi K. Production of monocyte chemoattractant protein-1 by malignant fibrous histiocytoma: relation to the origin of histiocyte-like cells. Exp Mol Pathol. 1991;54:61–71. doi: 10.1016/0014-4800(91)90044-x. [DOI] [PubMed] [Google Scholar]
- Brooks JJ. The significance of double phenotypic patterns and markers in human sarcomas. A new model of mesenchymal differentiation Am J Pathol. 1986;125:113–123. [PMC free article] [PubMed] [Google Scholar]
- Dehner LP. Regressing atypical histiocytosis: the controversy continues. Arch Dermatol. 1988;124:319–321. doi: 10.1001/archderm.124.3.319. [DOI] [PubMed] [Google Scholar]
- Katenkamp D. Cellular heterogeneity. Explanation for changing of tumor phenotype and biologic behavior in soft tissue sarcomas. Pathol Res Pract. 1988;183:698–705. doi: 10.1016/S0344-0338(88)80055-4. [DOI] [PubMed] [Google Scholar]
- Fletcher CD. Pleomorphic malignant fibrous histiocytoma: fact or fiction? A critical reappraisal based on 159 tumors diagnosed as pleomorphic sarcoma. Am J Surg Pathol. 1992;16:213–228. [PubMed] [Google Scholar]
- Hollowood K, Fletcher CD. Malignant fibrous histiocytoma: morphologic pattern or pathologic entity? Semin Diagn Pathol. 1995;12:210–220. [PubMed] [Google Scholar]
- Meltzer PS, Jankowski SA, Dal Cin P, Sandberg AA, Paz IB, Coccia MA. Identification and cloning of a novel amplified DNA sequence in human malignant fibrous histiocytoma derived from a region of chromosome 12 frequently rearranged in soft tissue tumors. Cell Growth Differ. 1991;2:495–501. [PubMed] [Google Scholar]
- Mairal A, Terrier P, Chibon F, Sastre X, Lecesne A, Aurias A. Loss of chromosome 13 is the most frequent genomic imbalance in malignant fibrous histiocytomas. A comparative genomic hybridization analysis of a series of 30 cases. Cancer Genet Cytogenet. 1999;111:134–138. doi: 10.1016/s0165-4608(98)00227-1. [DOI] [PubMed] [Google Scholar]
- Mandahl N, Heim S, Willen H, Rydholm A, Eneroth M, Nilbert M, Kreicbergs A, Mitelman F. Characteristic karyotypic anomalies identify subtypes of malignant fibrous histiocytoma. Genes Chromosomes Cancer. 1989;1:9–14. doi: 10.1002/gcc.2870010104. [DOI] [PubMed] [Google Scholar]
- Nielsen TO, West RB, Linn SC, Alter O, Knowling MA, O'Connell JX, Zhu S, Fero M, Sherlock G, Pollack JR, Brown PO, Botstein D, van de Rijn M. Molecular characterisation of soft tissue tumours: a gene expression study. Lancet. 2002;359:1301–1307. doi: 10.1016/S0140-6736(02)08270-3. [DOI] [PubMed] [Google Scholar]
- Segal NH, Pavlidis P, Antonescu CR, Maki RG, Noble WS, DeSantis D, Woodruff JM, Lewis JJ, Brennan MF, Houghton AN, Cordon-Cardo C. Classification and subtype prediction of adult soft tissue sarcoma by functional genomics. Am J Pathol. 2003;163:691–700. doi: 10.1016/S0002-9440(10)63696-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazziola C, Cordani N, Wasserman B, Carta S, Colombatti A, Perris R. Malignant fibrous histiocytoma: a proposed cellular origin and identification of its characterizing gene transcripts. Int J Oncol. 2003;23:343–351. [PubMed] [Google Scholar]
- Toma JG, Akhavan M, Fernandes KJ, Barnabe-Heider F, Sadikot A, Kaplan DR, Miller FD. Isolation of multipotent adult stem cells from the dermis of mammalian skin. Nat Cell Biol. 2001;3:778–784. doi: 10.1038/ncb0901-778. [DOI] [PubMed] [Google Scholar]
- Mata A, Boehm C, Fleischman AJ, Muschler G, Roy S. Growth of connective tissue progenitor cells on microtextured polydimethylsiloxane surfaces. J Biomed Mater Res. 2002;62:499–506. doi: 10.1002/jbm.10353. [DOI] [PubMed] [Google Scholar]
- Li WT, Leu YC. Effects of low level red-light irradiation on the proliferation of mesenchymal stem cells derived from rat bone marrow. Conf Proc IEEE Eng Med Biol Soc. 2007;2007:5830–5833. doi: 10.1109/IEMBS.2007.4353673. [DOI] [PubMed] [Google Scholar]
- Xin L, Lawson DA, Witte ON. The Sca-1 cell surface marker enriches for a prostate-regenerating cell subpopulation that can initiate prostate tumorigenesis. Proc Natl Acad Sci USA. 2005;102:6942–6947. doi: 10.1073/pnas.0502320102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meirelles Lda S, Nardi NB. Murine marrow-derived mesenchymal stem cell: isolation, in vitro expansion, and characterization. Br J Haematol. 2003;123:702–711. doi: 10.1046/j.1365-2141.2003.04669.x. [DOI] [PubMed] [Google Scholar]
- Oh H, Chi X, Bradfute SB, Mishina Y, Pocius J, Michael LH, Behringer RR, Schwartz RJ, Entman ML, Schneider MD. Cardiac muscle plasticity in adult and embryo by heart-derived progenitor cells. Ann NY Acad Sci. 2004;1015:182–189. doi: 10.1196/annals.1302.015. [DOI] [PubMed] [Google Scholar]
- Welm BE, Tepera SB, Venezia T, Graubert TA, Rosen JM, Goodell MA. Sca-1(pos) cells in the mouse mammary gland represent an enriched progenitor cell population. Dev Biol. 2002;245:42–56. doi: 10.1006/dbio.2002.0625. [DOI] [PubMed] [Google Scholar]
- Sun S, Guo Z, Xiao X, Liu B, Liu X, Tang PH, Mao N. Isolation of mouse marrow mesenchymal progenitors by a novel and reliable method. Stem Cells. 2003;21:527–535. doi: 10.1634/stemcells.21-5-527. [DOI] [PubMed] [Google Scholar]
- Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, Li HI, Eaves CJ. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- Guan K, Nayernia K, Maier LS, Wagner S, Dressel R, Lee JH, Nolte J, Wolf F, Li M, Engel W, Hasenfuss G. Pluripotency of spermatogonial stem cells from adult mouse testis. Nature. 2006;440:1199–1203. doi: 10.1038/nature04697. [DOI] [PubMed] [Google Scholar]
- Kim CF, Jackson EL, Woolfenden AE, Lawrence S, Babar I, Vogel S, Crowley D, Bronson RT, Jacks T. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Stingl J, Caldas C. Molecular heterogeneity of breast carcinomas and the cancer stem cell hypothesis. Nat Rev Cancer. 2007;7:791–799. doi: 10.1038/nrc2212. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Yin X, Qin H, Zhu F, Liu H, Yang W, Zhang Q, Xiang C, Hou P, Song Z, Liu Y, Yong J, Zhang P, Cai J, Liu M, Li H, Li Y, Qu X, Cui K, Zhang W, Xiang T, Wu Y, Zhao Y, Liu C, Yu C, Yuan K, Lou J, Ding M, Deng H. Two supporting factors greatly improve the efficiency of human iPSC generation. Cell Stem Cell. 2008;3:475–479. doi: 10.1016/j.stem.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Feng B, Ng JH, Heng JC, Ng HH. Molecules that promote or enhance reprogramming of somatic cells to induced pluripotent stem cells. Cell Stem Cell. 2009;4:301–312. doi: 10.1016/j.stem.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Galderisi U, Cipollaro M, Giordano A. The retinoblastoma gene is involved in multiple aspects of stem cell biology. Oncogene. 2006;25:5250–5256. doi: 10.1038/sj.onc.1209736. [DOI] [PubMed] [Google Scholar]
- Liu Y, Clem B, Zuba-Surma EK, El-Naggar S, Telang S, Jenson AB, Wang Y, Shao H, Ratajczak MZ, Chesney J, Dean DC. Mouse fibroblasts lacking RB1 function form spheres and undergo reprogramming to a cancer stem cell phenotype. Cell Stem Cell. 2009;4:336–347. doi: 10.1016/j.stem.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valk-Lingbeek ME, Bruggeman SW, van Lohuizen M. Stem cells and cancer; the polycomb connection. Cell. 2004;118:409–418. doi: 10.1016/j.cell.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Derre J, Lagace R, Nicolas A, Mairal A, Chibon F, Coindre JM, Terrier P, Sastre X, Aurias A. Leiomyosarcomas and most malignant fibrous histiocytomas share very similar comparative genomic hybridization imbalances: an analysis of a series of 27 leiomyosarcomas. Lab Invest. 2001;81:211–215. doi: 10.1038/labinvest.3780229. [DOI] [PubMed] [Google Scholar]