Abstract
MRI is increasingly used to evaluate cartilage in tissue constructs, explants, and animal and patient studies. However, while mean values of MR parameters, including T1, T2, magnetization transfer rate km, apparent diffusion coefficient ADC, and the dGEMRIC-derived fixed charge density, correlate with tissue status, the ability to classify tissue according to these parameters has not been explored. Therefore, the sensitivity and specificity with which each of these parameters was able to distinguish between normal and trypsin- degraded, and between normal and collagenase-degraded, cartilage explants were determined. Initial analysis was performed using a training set to determine simple group means to which parameters obtained from a validation set were compared. T1 and ADC showed the greatest ability to discriminate between normal and degraded cartilage. Further analysis with k-means clustering, which eliminates the need for a priori identification of sample status, generally performed comparably. Use of fuzzy c-means (FCM) clustering to define centroids likewise did not result in improvement in discrimination. Finally, a FCM clustering approach in which validation samples were assigned in a probabilistic fashion to control and degraded groups was implemented, reflecting the range of tissue characteristics seen with cartilage degradation.
Keywords: cartilage, degradation, sensitivity, specificity
Introduction
A number of MRI parameters, including T1, T2, magnetization transfer ratio (MTR) and apparent rate (km), self-diffusion coefficient (ADC), and T1 in the presence of gadolinium (the dGEMRIC index), have been used to characterize cartilage (1–3). These parameters all exhibit, to a varying degree, sensitivity to cartilage pathology, and in certain cases a degree of specificity for particular matrix components and ultrastructure. Analysis of the means of the parameter of interest between control and experimentally degraded ex vivo cartilage, or between normal and osteoarthritic cartilage within living subjects, indicates whether there is a statistically significant variation in that parameter between groups.
In such experiments, T1, T2, and ADC tend to increase, while MTR, km, and the dGEMRIC index decrease, with matrix degradation. MTR and km exhibit enhanced specificity to changes in the collagen component (3,4), while the dGEMRIC index is preferentially specific to proteoglycan (1,2). However, although relationships between MR parameters and cartilage pathology have been well-documented, the sensitivity and specificity of these parameters have not been evaluated in detail.
Osteoarthritis (OA) is characterized by loss or modification of the matrix components of cartilage (5), so that degradation with trypsin and collagenase are frequently used as pathomimetic interventions. Trypsin, acting primarily to digest proteoglycan, corresponds to the loss of this component seen in the early stages of OA. Collagenase produces a more mixed degradation; while it cleaves the collagen molecule, this loss of tissue framework potentiates proteoglycan loss as well.
We sought to evaluate the sensitivity and specificity of several commonly used MRI outcome measures to experimental cartilage degeneration. The parameters examined were T1, T2, magnetization transfer rate km, self-diffusion coefficient ADC, and fixed charge density (FCD) as determined by the dGEMRIC technique. Our initial evaluation was based on the notion that classification of an unknown sample based on any particular single parameter would be performed by comparing the parameter value found for that sample, to the overall means of that parameter in the control and digested sample groups. This provides an objective measure of the accuracy of what is in effect a widely-used conventional approach to tissue classification. In addition, we implemented three alternative classification procedures based on cluster analysis. The first of these was classification based on comparison of a sample’s parameter values with control and degraded cluster centroids defined by k-means clustering (KMC), with k, the number of clusters, set equal to 2, representing control and degraded groups. While both the arithmetic means and KMC approaches would be expected to yield similar results, the KMC algorithm does not require a priori knowledge of the classification status of training set samples. KMC provides a potentially more robust analytic framework for classification at the expense of forming cluster centroids that are potentially contaminated by the presence of samples that are misclassified by the algorithm. As a second alternative to classification based on simple means, we assigned samples to either control or degraded groups based on cluster centroids defined using fuzzy c-means (FCM) clustering, again with c = 2. While the FCM clustering algorithm used in this way limits the influence of data outliers on the definition of cluster centroids, it nevertheless still provides only a binary assignment of validation set samples to one group or another. However, the primary utility of FCM clustering is through an implementation that assigns samples to clusters in a probabilistic fashion; this is particularly suitable for applications in which a range of tissue characteristics can be expected, such as in osteoarthritis. Finally, therefore, we implemented a FCM clustering approach in which cluster centroids were defined through FCM as above, and the membership strength of each validation set sample to a given cluster was assigned using a graded weighting function.
Methods
Sample Preparation
Bovine nasal cartilage (BNC) disks (8 mm diameter) were excised from the nasal septa of 5–6 month old calves (T. D. Morris, Inc., Reisterstown, MD). Disks were threaded onto a hollow tube through a central 2.5-mm diameter hole. Multiple samples were inserted into each well of a susceptibility-matched four-well ULTEM sample holder and bathed in Dulbecco’s phosphate buffered saline (DPBS) (pH 7.5 +/− 0.1) (6). After imaging, degradation was induced by incubating two different sets of samples in DPBS to which 1mg/ml trypsin (Sigma-Aldrich, St. Louis, MO) or 30 units/ml collagenase type II (Worthington Biochemical Corp., Lakewood, NJ) had been added at 37 °C in a 5% CO2 atmosphere. The durations of the degradation protocols were 18 hours for trypsin and 20 hours for collagenase; these degradation protocols have been found in our laboratory to result in severe degradation in BNC as confirmed by biochemical evaluation. A total of 40 control, 40 trypsin-digested, and 40 collagenase-digested samples were studied. Two separate analyses were performed, using i) 80 samples consisting of the control group plus the trypsin-digested group, and ii) 80 samples consisting of that same control group plus the collagenase-digested group.
MRI Measurements
Imaging was performed using a 9.4T/105-mm Bruker DMX spectrometer (Bruker Biospin GmbH, Rheinstetten, Germany), using a 30-mm proton birdcage resonator. Sample temperature was maintained at 4.0 ± 0.1 °C. Data were acquired from sagittal slices (thickness = 0.5 mm) through the center of each of the four wells in the sample holder, permitting all samples within the holder (typically numbering ~6) to be imaged simultaneously. A 64-echo CPMG sequence with TE/TR = 12.8 ms/ 5 s was used for T2 measurements. T1 mapping was performed with a progressive saturation spin-echo sequence with TE = 12.8 ms and TR varying from 100 ms to 15 s in 12 steps. MT data were obtained using this spin-echo sequence preceded by a 6 kHz off-resonance saturation pulse of amplitude B1 = 12 µT and duration tp incremented from 0.1 to 4.6 s in 8 steps. Measurements of ADC were performed by incorporating a pair of identical gradient pulses of δ = 5 ms duration on either side of the 180° refocusing pulse, with a fixed interval Δ = 13.8 ms between pulse centers. Gradient strength was incremented from 0 to 320 mT/m in 8 steps. We found that diffusion images obtained with these parameters did not show geometric distortion. After the above measurements were performed, 2mM Gd-DTPA2− (Magnevist; Berlex Laboratories, Wayne, NJ) was added to the DPBS buffer. Full equilibration of Gd-DTPA2− within the samples was confirmed by repeat T1 mapping over a period of approximately 12 hours, with a maximum TR = 5s. FCD was then calculated from T1 measurements made in the absence and presence of Gd-DTPA2− (7). Other parameters included BW = 50 kHz, FOV = 4.0 × 1.5 cm, matrix size 256 × 128, resolution 156 × 117 µm (vertical × horizontal) and NEX = 2. Signal intensity was averaged over all pixels in a region of interest (ROI) covering an entire BNC disk. T1, T2, km and MTR, and ADC were then calculated by individually fitting mean ROI intensities as a function of TR, TE, off-resonance saturation pulse length (8), and b, a function of diffusion-sensitizing gradient strength and duration, respectively, to appropriate 3-parameter monoexponential functions incorporating offsets from baseline. We define MTR = 1 − Mss/M0, where Mss is the steady-state magnetization after application of a long-time saturation pulse, while km = MTR/T1sat. Fits were performed using the Bruker-supplied ParaVision software package. Values were calculated independently for every sample and averaged.
Assessment of Sensitivity and Specificity Based on Assignments to Particular Clusters
Assignments based on arithmetic means
Sensitivity and specificity were calculated for each MRI parameter as follows. 53 (approximately 2/3) out of the total of 80 samples composed of the control plus trypsin- or collagenase-degraded group were randomly selected as a training set (9). After determination of parameter means for the control and degraded samples in this training set, the remaining 27 samples, used as a validation set, were each assigned to the group for which the group mean was closest to that sample's value. The sensitivity and specificity of this procedure was calculated according to:
and
To account for the possibility of an unusually favorable or unfavorable random selection of the training set, the above procedure was repeated for 100 independent selections of the training set, with the results then averaged and reported.
In addition to classification of the validation set, after establishment of group means, classification was also performed on the training set itself as a rough indicator of the maximal ability of arithmetic means to distinguish between the control and degraded groups.
Assignments based on k-means and fuzzy c-means clustering centroids
The analysis of sensitivity and specificity for each MR parameter was performed in a fashion identical to that described above, with the exception that cluster centroids as defined by KMC (10–12) and as defined by FCM clustering (13) were used to determine the classification of the samples in the validation set. The clustering procedures with k = 2 and c = 2 were performed using the statistics and fuzzy logic tool packages in Matlab (The MathWorks, Inc. Natick, MA), respectively.
Probabilistic Assignments to Clusters
After definition of cluster centroids in the training set by FCM clustering, membership probabilities of the samples in the validation set were calculated using a standard metric. The probability mik of membership of sample xi in cluster k was taken as:
where d is the distance between the parameter value for sample xi and the k-th centroid Ck, and where the summation is over all centroids Cj (14).
Statistical Analysis
Data are reported as mean ± SD, with statistical significance taken as p < 0.05.
Results
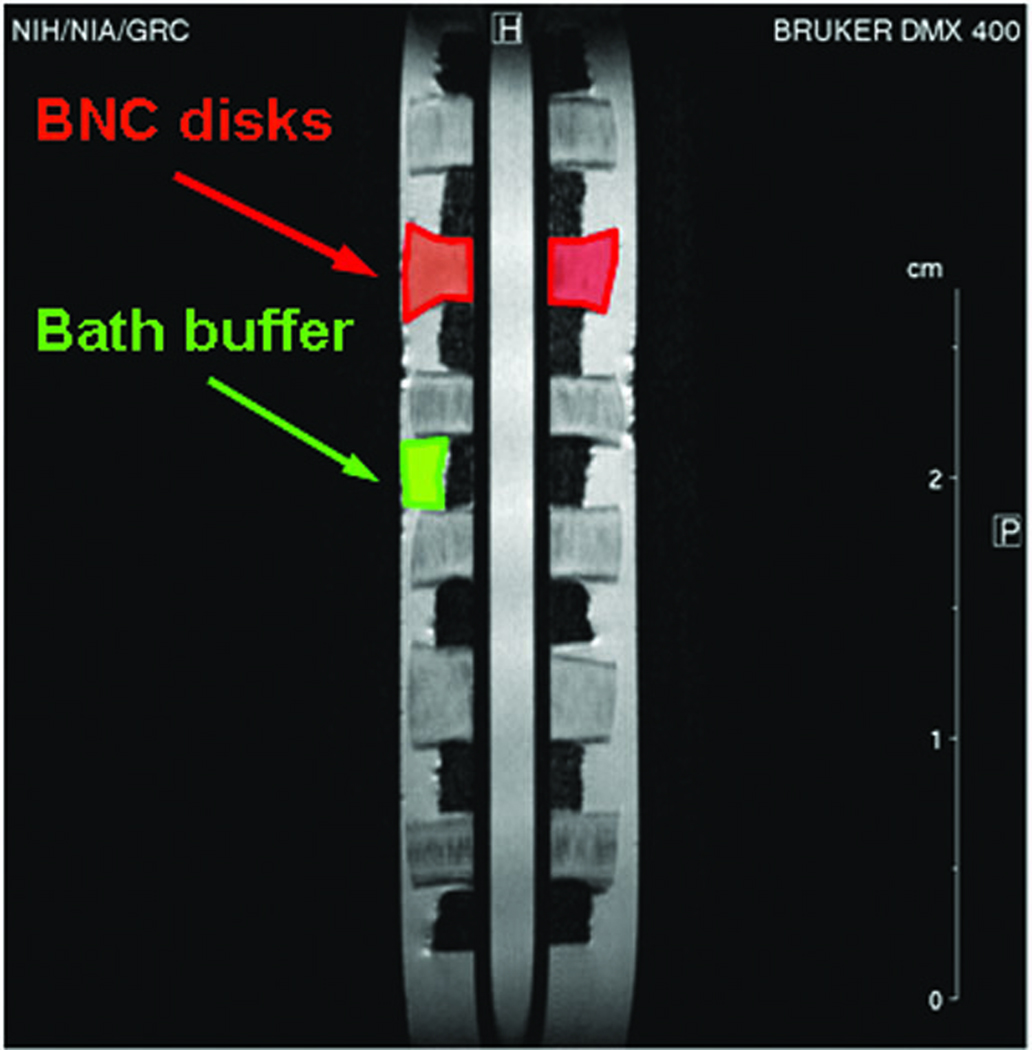
Figure 1 shows a T2 weighted image of one well of the sample holder with the six BNC disks threaded onto a thin central tube. ROI's were readily defined within the cartilage.
Figure 1.
T2-weighted MR image of one well of the sample holder. Data were acquired using a 64-echo CPMG pulse sequence with TE = 12.8 ms and a constant TR of 5 s at 4 °C. The cartilage disks are clearly delineated from surrounding bath fluid. Similar images were simultaneously acquired for the other wells of the same sample holder at each time point, which permitted direct comparison of the effects of the various bath fluids. A typical ROI is indicated by the outline and arrow; the ROI's determined using the T2-weighted images were applied to all parameters investigated.
Table 1 shows the parameter mean values. There were statistically significant differences between the control and enzymatically digested groups in most cases. This fact by itself does not indicate the degree to which a given parameter can be used to assign samples of unknown status to non-degraded and degraded categories, motivating the use of the sensitivity and specificity analysis outlined above.
Table 1.
MRI parameters in non- and enzymatically- digested BNC samples
| T1 (ms) | T2 (ms) | km (s−1) | ADC (× 10−4 mm2/s) | FCD (mM) | |
|---|---|---|---|---|---|
| Control | 1208 ± 101 | 55.0 ± 11.1 | 0.87 ± 0.22 | 9.29 ± 1.00 | 201.8 ± 33.8 |
| 18-hr Trypsin | 1527 ± 107* | 66.5 ± 10.8* | 0.64 ± 0.16* | 11.34 ± 0.63* | 74.9 ± 58.0* |
| 20-hr Collagenase | 1448 ± 129* | 57.9 ± 10.6 | 0.72 ± 0.21* | 10.39 ± 0.71* | 175.1 ± 46.1* |
* p < 0.05 for control vs. trypsin- or control vs. collagenase-digested samples.
Each of the 3 sample sets consisted of 40 samples
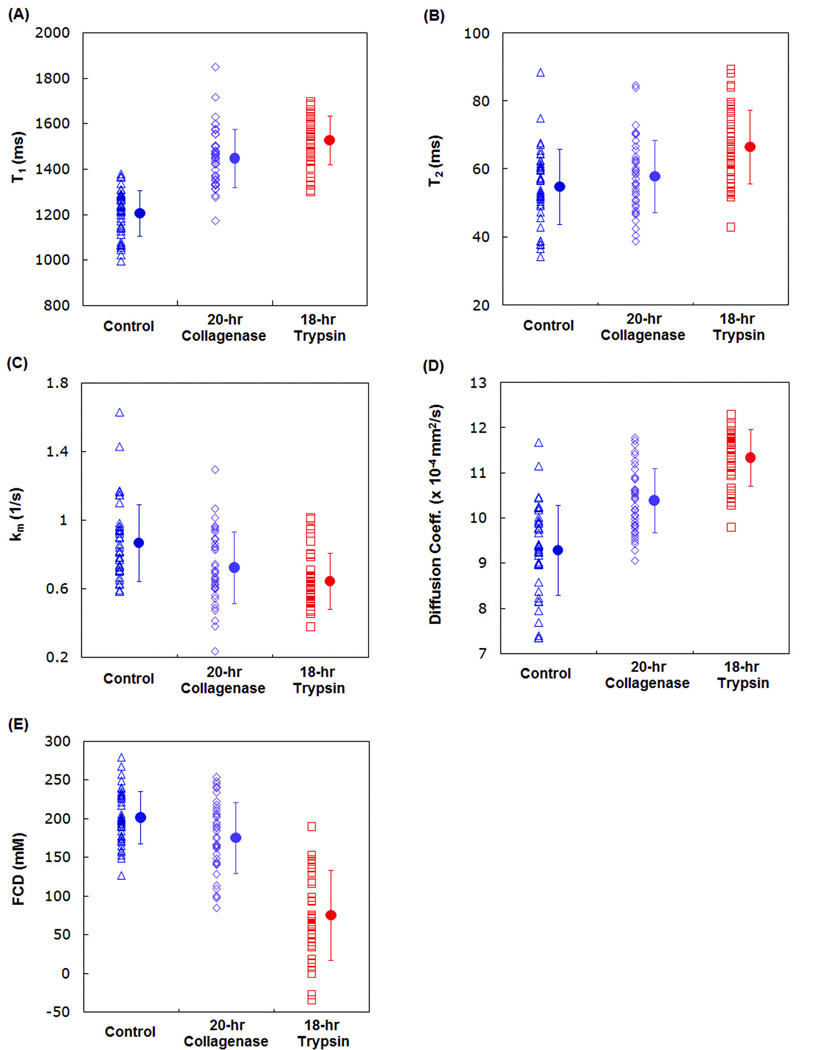
Figure 2 shows the values obtained for each sample for each of the five MRI parameters evaluated. For each parameter, overlap between the groups is clearly seen, in spite of the generally statistically significant group mean differences obtained in these relatively severe degradation protocols. It is this overlap which limits the sensitivity and specificity of the parameters.
Figure 2.
Plots of the values of MRI modalities influenced by enzymatic depletion in BNC disk samples. The triangle symbols in the left column of each panel represent the measured values in control set, the diamond symbols in the trypsin-degraded set, and the square symbols in the collagnease-degraded set. The solid circle with error bar in each column depicts the mean and standard deviation of the indicated parameter. Data variation in (A) T1, (B)T2, (C) km, (D) translational diffusion coefficient, ADC, and (E) fixed charge density, FCD is readily appreciated.
Table 2A shows the results of the sensitivity and specificity analyses performed with respect to simple arithmetic means. The analyses for the training sets, from which the mean values guiding the classification were determined, and the corresponding validation sets, were compared. A paired t-test with sample size taken as 100 was used, as the distributions of the sensitivities and specificities over the random selections of training and validation data sets were generally Gaussian for each parameter, each of the 100 selections of training sets completely defined a corresponding validation set, and each entry in Table 2A is the average of values obtained using 100 random selections of the training set. Results were further corrected using the Bonferroni method for multiple comparisons. We found no significant differences between the results for the training sets and the corresponding validation sets. This indicates that the classification scheme is not highly sensitive to the random choice of data subset. We see that for trypsin degradation, classification according to both T1 and ADC values achieved excellent sensitivity and specificity of approximately 90% or greater in the validation set. For collagenase digestion, classification according to T1 was the most successful, with sensitivity and specificity above 80%, while diffusion was again the second-best discriminator. Interestingly, while T2 and km are often regarded as having a particular sensitivity to collagen content, classification according to these parameters was substantially less accurate for collagenase digestion than for trypsin digestion. However, FCD as determined by the dGEMRIC procedure performed better in the trypsin digestion protocol than in the collagenase protocol, consistent with its greater sensitivity to GAG than to collagen. However, direct comparison of the trypsin and collagenase protocols is problematic, given the preferential action of these two enzymes on different macromolecules.
Table 2.
Univariate analysis of control vs. enzymatically degraded BNC samples
| (A) Using training set means for classification | |||||
|---|---|---|---|---|---|
| Category | Parameter | Training set | Validation set | ||
|
|
|||||
| Sensitivity | Specificity | Sensitivity | Specificity | ||
| Normal vs. 18-hr trypsin |
T1 | 0.873 ± 0.032 | 0.956 ± 0.031 | 0.902 ± 0.074 | 0.944 ± 0.065 |
| T2 | 0.657 ± 0.049 | 0.744 ± 0.052 | 0.692 ± 0.132 | 0.764 ± 0.135 | |
| km | 0.765 ± 0.039 | 0.620 ± 0.049 | 0.793 ± 0.086 | 0.625 ± 0.124 | |
| ADC | 0.945 ± 0.032 | 0.885 ± 0.032 | 0.933 ± 0.073 | 0.896 ± 0.093 | |
| FCD | 0.816 ± 0.035 | 0.974 ± 0.018 | 0.814 ± 0.105 | 0.977 ± 0.035 | |
| Normal vs. 20-hr collagenase |
T1 | 0.837 ± 0.048 | 0.906 ± 0.031 | 0.814 ± 0.110 | 0.891 ± 0.068 |
| T2 | 0.580 ± 0.044 | 0.600 ± 0.051 | 0.616 ± 0.123 | 0.573 ± 0.110 | |
| km | 0.633 ± 0.048 | 0.551 ± 0.048 | 0.611 ± 0.105 | 0.553 ± 0.133 | |
| ADC | 0.734 ± 0.052 | 0.706 ± 0.042 | 0.746 ± 0.118 | 0.683 ± 0.135 | |
| FCD | 0.586 ± 0.044 | 0.679 ± 0.053 | 0.546 ± 0.131 | 0.684 ± 0.132 | |
| (B) Using training set k-means cluster centroids for classification | |||||
|---|---|---|---|---|---|
| Category | Parameter | Training set | Validation set | ||
|
|
|||||
| Sensitivity | Specificity | Sensitivity | Specificity | ||
| Normal vs. 18-hr trypsin |
T1 | 0.861 ± 0.042 | 0.995 ± 0.017 | 0.892 ± 0.076 | 0.991 ± 0.027 |
| T2 | 0.494 ± 0.111 | 0.877 ± 0.099 | 0.511 ± 0.149 | 0.890 ± 0.112 | |
| km | 0.826 ± 0.046 | 0.481 ± 0.096 | 0.846 ± 0.082 | 0.470 ± 0.127 | |
| ADC | 0.962 ± 0.036 | 0.816 ± 0.066 | 0.967 ± 0.049 | 0.820 ± 0.099 | |
| FCD | 0.727 ± 0.056 | 0.999 ± 0.006 | 0.732 ± 0.106 | 1.000 ± 0.000 | |
| Normal vs. 20-hr collagenase |
T1 | 0.753 ± 0.088 | 0.941 ± 0.044 | 0.729 ± 0.122 | 0.930 ± 0.061 |
| T2 | 0.580 ± 0.072 | 0.612 ± 0.091 | 0.603 ± 0.119 | 0.579 ± 0.133 | |
| km | 0.685 ± 0.070 | 0.473 ± 0.088 | 0.681 ± 0.120 | 0.466 ± 0.145 | |
| ADC | 0.801 ± 0.114 | 0.660 ± 0.090 | 0.799 ± 0.129 | 0.624 ± 0.152 | |
| FCD | 0.540 ± 0.064 | 0.720 ± 0.063 | 0.511 ± 0.122 | 0.729 ± 0.110 | |
In all cases, 100 random selections of a training set consisting of 53 randomly-selected samples, with the remaining 27 samples comprising the validation set, were used to determine sensitivity and specificity.
Table 2B shows the results of classification with respect to k-means clustering centroids. Overall results were generally similar to those obtained using arithmetic means. Again, the sensitivity and specificity for each parameter and both degradations were similar for the training set and validation set. The specificity of T2 for classification of the trypsin-degraded samples was 89.0% when based on k-means centroids, while it was 76.4% using arithmetic means. In contrast, the sensitivity achieved for T2 using the k-means approach was 51.1% which was lower than the value of 69.2% obtained using arithmetic means.
We also performed classification in which cluster centroids were defined in accordance with FCM clustering, with each validation sample then assigned to the cluster with the closer centroid. The results (not shown) were virtually indistinguishable from the KMC results, indicating that the small change in cluster centroid location resulting from use of FCM clustering rather than KMC did not lead to any improvement in classification.
Results of implementation of FCM clustering with probabalistic assignments to clusters are shown in Table 3. For this, sensitivity and specificity of the aggregate test results are no longer the appropriate outcome measures. Instead, results for each sample in the validation set must be displayed individually. The Table shows the probabilities of membership of validation samples to clusters; the clusters were defined using a single random selection of 53 total digested and control samples, with the 27 remaining samples forming the validation set. Assignments can be seen to vary among samples within a given treatment group (control, trypsin-degraded, or collagenase-degraded). As noted, the FCM clustering analysis, avoiding definite cluster assignments in favor of probabilities, is motivated by recognition of the fact that there will be a continuous variation of parameter values both within the group of control samples and within the group of degraded samples. Nevertheless, means of the assignment probabilities are shown illustrative purposes, and indicate the average degree to which samples were assigned to groups according to the various parameters studied. It is of note that T1 was again the most accurate classifier in terms of these averages, showing a high degree of average probability in assigning control and degraded samples to the appropriate group. Classification based on the other parameters was of limited accuracy due to much wider variation in the measured values. Interestingly, FCD was a very poor classifier for the mixture of control and collagenase-degraded samples, but performed much better for the mixture of control and trypsin-degraded samples. This is consistent with the known primary action of typsin to digest proteoglycans, which are the primary contributor to FCD, while collagenase has a large effect on both the proteoglycan and collagen components of cartilage matrix.
Table 3.
Cluster membership probabilities for validation set samples using fuzzy c-means clustering
| (A) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Treatment | T1 | T2 | km | ADC | FCD | |||||
| Normal | Degraded | Normal | Degraded | Normal | Degraded | Normal | Degraded | Normal | Degraded | |
| Control | 0.98 | 0.02 | 0.19 | 0.81 | 0.97 | 0.03 | 0.40 | 0.60 | 0.73 | 0.27 |
| 0.97 | 0.03 | 0.44 | 0.56 | 0.27 | 0.73 | 0.06 | 0.94 | 1.00 | 0.00 | |
| 0.98 | 0.02 | 0.99 | 0.01 | 1.00 | 0.00 | 0.66 | 0.34 | 0.93 | 0.07 | |
| 0.91 | 0.09 | 0.93 | 0.07 | 0.71 | 0.29 | 0.58 | 0.42 | 0.95 | 0.05 | |
| 0.93 | 0.07 | 1.00 | 0.00 | 0.11 | 0.89 | 0.63 | 0.37 | 1.00 | 0.00 | |
| 0.80 | 0.20 | 0.32 | 0.68 | 0.00 | 1.00 | 0.86 | 0.14 | 0.78 | 0.22 | |
| 0.99 | 0.01 | 0.25 | 0.75 | 0.12 | 0.88 | 0.67 | 0.33 | 0.84 | 0.16 | |
| 1.00 | 0.00 | 0.25 | 0.75 | 0.87 | 0.13 | 0.71 | 0.29 | 1.00 | 0.00 | |
| 0.88 | 0.12 | 0.49 | 0.51 | 0.52 | 0.48 | 0.86 | 0.14 | 0.92 | 0.08 | |
| 0.91 | 0.09 | 0.11 | 0.89 | 0.49 | 0.51 | 0.67 | 0.33 | 0.99 | 0.01 | |
| 0.97 | 0.03 | 0.50 | 0.50 | 0.52 | 0.48 | 0.63 | 0.37 | 1.00 | 0.00 | |
| 0.52 | 0.48 | 0.07 | 0.93 | 0.01 | 0.99 | 1.00 | 0.00 | 0.96 | 0.04 | |
| 0.67 | 0.33 | 0.79 | 0.21 | 0.00 | 1.00 | 0.38 | 0.62 | 0.98 | 0.02 | |
| Average | 0.88 | 0.12 | 0.49 | 0.51 | 0.43 | 0.57 | 0.62 | 0.38 | 0.93 | 0.07 |
| Trypsin | 0.81 | 0.19 | 0.34 | 0.66 | 1.00 | 0.00 | 0.32 | 0.68 | 0.13 | 0.87 |
| 0.01 | 0.99 | 0.08 | 0.92 | 0.00 | 1.00 | 0.38 | 0.62 | 0.00 | 1.00 | |
| 0.02 | 0.98 | 0.00 | 1.00 | 0.06 | 0.94 | 0.43 | 0.57 | 0.05 | 0.95 | |
| 0.08 | 0.92 | 0.51 | 0.49 | 0.02 | 0.98 | 0.37 | 0.63 | 0.64 | 0.36 | |
| 0.04 | 0.96 | 0.15 | 0.85 | 0.08 | 0.92 | 0.43 | 0.57 | 0.00 | 1.00 | |
| 0.05 | 0.95 | 0.04 | 0.96 | 0.08 | 0.92 | 0.40 | 0.60 | 0.77 | 0.23 | |
| 0.01 | 0.99 | 0.00 | 1.00 | 0.02 | 0.98 | 0.41 | 0.59 | 0.71 | 0.29 | |
| 0.10 | 0.90 | 0.95 | 0.05 | 0.01 | 0.99 | 0.30 | 0.70 | 0.99 | 0.01 | |
| 0.01 | 0.99 | 0.31 | 0.69 | 0.02 | 0.98 | 0.31 | 0.69 | 0.01 | 0.99 | |
| 0.07 | 0.93 | 0.09 | 0.91 | 0.05 | 0.95 | 0.41 | 0.59 | 0.12 | 0.88 | |
| 0.18 | 0.82 | 0.93 | 0.07 | 0.42 | 0.58 | 0.01 | 0.99 | 0.58 | 0.42 | |
| 0.02 | 0.98 | 0.16 | 0.84 | 0.08 | 0.92 | 0.40 | 0.60 | 0.10 | 0.90 | |
| 0.08 | 0.92 | 0.08 | 0.92 | 0.06 | 0.94 | 0.42 | 0.58 | 0.06 | 0.94 | |
| 0.00 | 1.00 | 0.57 | 0.43 | 0.00 | 1.00 | 0.35 | 0.65 | 0.00 | 1.00 | |
| Average | 0.10 | 0.90 | 0.30 | 0.70 | 0.14 | 0.86 | 0.35 | 0.65 | 0.30 | 0.70 |
| (B) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Treatment | T1 | T2 | km | ADC | FCD | |||||
| Normal | Degraded | Normal | Degraded | Normal | Degraded | Normal | Degraded | Normal | Degraded | |
| Control | 0.97 | 0.03 | 0.24 | 0.76 | 0.93 | 0.07 | 0.33 | 0.67 | 0.00 | 1.00 |
| 0.96 | 0.04 | 0.10 | 0.90 | 0.18 | 0.82 | 0.03 | 0.97 | 0.77 | 0.23 | |
| 0.97 | 0.03 | 0.85 | 0.15 | 0.98 | 0.02 | 0.92 | 0.08 | 0.17 | 0.83 | |
| 0.89 | 0.11 | 0.97 | 0.03 | 0.73 | 0.27 | 0.69 | 0.31 | 0.30 | 0.70 | |
| 0.91 | 0.09 | 0.89 | 0.11 | 0.06 | 0.94 | 0.85 | 0.15 | 0.85 | 0.15 | |
| 0.74 | 0.26 | 0.05 | 0.95 | 0.00 | 1.00 | 0.54 | 0.46 | 0.00 | 1.00 | |
| 0.99 | 0.01 | 0.02 | 0.98 | 0.07 | 0.93 | 0.94 | 0.06 | 0.03 | 0.97 | |
| 1.00 | 0.00 | 0.03 | 0.97 | 0.78 | 0.22 | 1.00 | 0.00 | 0.92 | 0.08 | |
| 0.84 | 0.16 | 0.13 | 0.87 | 0.40 | 0.60 | 0.52 | 0.48 | 0.14 | 0.86 | |
| 0.88 | 0.12 | 0.00 | 1.00 | 0.37 | 0.63 | 0.95 | 0.05 | 1.00 | 0.00 | |
| 0.96 | 0.04 | 0.13 | 0.87 | 0.40 | 0.60 | 0.85 | 0.15 | 0.88 | 0.12 | |
| 0.41 | 0.59 | 0.00 | 1.00 | 0.02 | 0.98 | 0.03 | 0.97 | 0.97 | 0.03 | |
| 0.58 | 0.42 | 0.41 | 0.59 | 0.00 | 1.00 | 0.01 | 0.99 | 0.99 | 0.01 | |
| Average | 0.85 | 0.15 | 0.29 | 0.71 | 0.38 | 0.62 | 0.59 | 0.41 | 0.54 | 0.46 |
| Collagenase | 0.86 | 0.14 | 0.81 | 0.19 | 1.00 | 0.00 | 0.22 | 0.78 | 0.18 | 0.82 |
| 0.37 | 0.63 | 1.00 | 0.00 | 0.95 | 0.05 | 1.00 | 0.00 | 0.01 | 0.99 | |
| 0.50 | 0.50 | 0.75 | 0.25 | 0.93 | 0.07 | 0.11 | 0.89 | 0.05 | 0.95 | |
| 0.03 | 0.97 | 0.02 | 0.98 | 0.08 | 0.92 | 0.33 | 0.67 | 0.10 | 0.90 | |
| 0.64 | 0.36 | 0.97 | 0.03 | 0.96 | 0.04 | 0.01 | 0.99 | 1.00 | 0.00 | |
| 0.59 | 0.41 | 0.99 | 0.01 | 0.96 | 0.04 | 1.00 | 0.00 | 0.98 | 0.02 | |
| 0.11 | 0.89 | 0.35 | 0.65 | 0.00 | 1.00 | 0.28 | 0.72 | 0.01 | 0.99 | |
| 0.03 | 0.97 | 0.02 | 0.98 | 0.00 | 1.00 | 0.03 | 0.97 | 0.98 | 0.02 | |
| 0.02 | 0.98 | 0.15 | 0.85 | 0.03 | 0.97 | 0.01 | 0.99 | 0.22 | 0.78 | |
| 0.23 | 0.77 | 0.57 | 0.43 | 0.63 | 0.37 | 0.32 | 0.68 | 1.00 | 0.00 | |
| 0.72 | 0.28 | 0.96 | 0.04 | 0.75 | 0.25 | 0.41 | 0.59 | 0.68 | 0.32 | |
| 0.02 | 0.98 | 0.68 | 0.32 | 0.00 | 1.00 | 0.87 | 0.13 | 0.96 | 0.04 | |
| 0.13 | 0.87 | 0.98 | 0.02 | 0.08 | 0.92 | 0.55 | 0.45 | 0.26 | 0.74 | |
| 0.00 | 1.00 | 0.93 | 0.07 | 0.04 | 0.96 | 0.14 | 0.86 | 0.56 | 0.44 | |
| Average | 0.30 | 0.70 | 0.66 | 0.34 | 0.46 | 0.54 | 0.38 | 0.62 | 0.50 | 0.50 |
For both Table 3A and 3B, out of the two groups of 80 samples as defined in the text, 53 randomly-selected samples were used to define cluster centroids using FCM clustering, and the remaining 27 samples were then assigned in a probabilistic fashion to control or degraded clusters.
Discussion
Early changes in cartilage associated with OA are characterized by degradation and potential loss of matrix components (5), followed by decrease in cartilage volume (15). To date, there are no well-established methods to achieve early detection of this process, which is an established goal towards development of therapeutic interventions. Given the noninvasive nature of MRI studies, interest has been attached to differences in mean values of several MRI parameters, including T1, T2, MTR, km, and ADC as they relate to tissue status (1–3,16,17). Cartilage tissue status can be altered through degradation by pathomimetic enzymes, including trypsin and collagenase as used in the present study. However, even with significant mean differences being observed in MRI parameters of control as compared to degraded samples, for any given parameter there is still a large overlap between groups (Figure 2). Thus, the issue of sensitivity and specificity of the various parameters becomes of interest.
As expected, the ability of each parameter investigated to distinguish control from degraded samples was related to the amount by which its mean value changed under the degradation protocols, with a greater change indicating a generally greater sensitivity and specificity, and the standard deviation of the measurements, with a smaller standard deviation being more favorable. We interpret the magnitude of the change in mean parameter values to reflect the average extent of matrix degradation, while the standard deviations are related to the homogeneity of matrix composition in control samples, and, in addition, to the uniformity of enzymatic action in degraded samples. In spite of the limitations in using pathomimetic enzymatic degradation as a model for disease, evaluation of classification sensitivity and specificity is meaningful because such differences and variations are observed as well in healthy and diseased cartilage.
The studies presented here were performed at a field strength substantially greater than those used for clinical studies. It has been well-documented that T1's increase with field strength. Similarly, field strength has a known, though smaller, effect on T2 values. An excellent discussion of these phenomena in the context of brain imaging has been previously presented (18). It has been pointed out that with increasing B0 and increasing T1's, the difference in tissue T1's may decrease, but the overall-contrast-to-noise ratio may be maintained or even increased, given the increase in signal-to-noise ratio (SNR) at higher field. Thus, the applicability of our results to studies at lower field will depend upon several factors, including the unknown dependence upon field strength of the difference in T1's between control and degraded cartilage, and SNR of the imaging protocols implemented. Similar comments apply to cartilage T2 values, although the dependence on field strength is more complex (18).
Sample temperature in our study was maintained at 4.0 °C during MR data acquisition in order to attenuate any degradation, while clinical studies are performed at body temperature of approximately 37 °C. To our knowledge, the difference between cartilage tissue relaxation times at 9.4 T as a function of temperature has not been studied. However, any such effect would be expected to be small, given the relatively weak dependence of relaxation times on temperatures in this range. Similar comments apply to ADC, which increases weakly with temperature.
Matrix inhomogeneity in the control and degraded samples can be estimated in several ways. Matrix inhomogeneity is reflected by inhomogeneity in the T2 weighted images shown in Fig. 1, and is also evident in typical histological sections of control and degraded BNC when stained with alcian blue for GAG or with picrosirius red for collagen (not shown). More quantitatively, Fourier transform imaging spectroscopy of BNC subjected to a trypsin degradation protocol resulted in a spatial distribution of cartilage matrix constituents with a coefficient of variation (CV) of 24% (collagen) and 43% (proteoglycan) relative to their averaged amounts (19). In contrast, biochemical analysis of BNC samples prepared in the same way as for those in the present study demonstrated a CV of less than 10% for GAG content; this value increased to 20% for collagenase digestion and 40% for trypsin degration. In terms of average matrix composition, these samples lost 10% of their GAG content upon digestion with collagenase, and 73% under trypsin degradation.
The maximum parameter CV's resulting from the fits of MR data to appropriate functional forms were, for T1, T2, km, and ADC, 4%, 6%, 16% and 11%, respectively, with much smaller values for the mean errors over all analyses taken together. As these values are on the order of or smaller than observed sample inhomogeneities, we conclude that errors due to finite SNR and potentially incorrect data models are negligible in this study.
We conclude from these observations that the sample changes due to degradation are larger than both the expected inhomogeneity within control samples and the variation between control samples; that is, degradation produces changes larger than those attributable to biologic variability. This indicates that alterations in MR parameters are potentially sufficient to distinguish between control and degraded samples in this study. Clearly, however, given the intra-sample variability, a classification approach in which maps of MR parameters, as opposed to global characterization of entire samples, may demonstrate superior performance. This approach, which is akin to segmentation, would be complicated by the loss of sensitivity inherent in localized as compared to bulk analyses.
A related point is that the bulk-sample approach undertaken in the present study does not address the sensitivity and specificity of measurements of MR parameter variation, such as the gradient in T2 values that is observed from deep to superficial cartilage and which may be a marker for pathology. Such studies would require discriminant analysis based on spatially localized parameter measurements.
Analysis through use of arithmetic means
As expected, both trypsin- and collagenase-induced matrix degradation caused increases in the mean values of T1,T2, and diffusivity, and decreases in mean km and dGEMRIC index (3). There was a statistically significant change in the mean values of all of these parameters with both enzymatic degradation protocols, with the exception of the minimal increase in T2 under collagenase digestion. This was consistent with the relatively low sensitivity displayed by T2 in both classification schemes, which we attribute to sample inhomogeneity and to non-uniformity of trypsin-induced depletion of proteoglycans (20,21). In contrast, the large changes in T1 resulting from degradation, combined with relatively small standard deviations, led to the high degree of sensitivity and specificity of that parameter to distinguish control from degraded samples. This is consistent with T1 being relatively insensitive to minor sample inhomogeneities and small variations in degradation. A particularly large mean change in MRI-determined FCD was seen under trypsin digestion, with that value decreasing by nearly a factor of three. This is consistent with the well-known specificity of the gadolinium exclusion technique for the proteoglycan component of the matrix. However, the large standard deviation associated with this measurement limited its sensitivity to degradation. Under collagenase digestion, the mean value of the FCD changed by only 13%; this, combined with the large standard deviation, was reflected in the poor ability of that parameter to characterize collagenase degradation. In contrast, although the change in the mean value of ADC after trypsin digestion was small, the small standard deviations in that parameter for both the control and the degraded tissue resulted in a high degree of sensitivity and specificity for ADC. However, with a smaller change in mean value and a somewhat larger standard deviation, ADC performed much more poorly in the collagenase digestion. This is analogous to the results for T1. Furthermore, differences in the variance between non-degraded and degraded groups influenced the relationship between sensitivity and specificity for a given parameter. As an example, the variance of FCD was larger in the trypsin-degraded group than the control group. Therefore, a degraded sample would be more likely to be assigned to the control group than conversely; that is, the specificity of this parameter is greater than its sensitivity. Similarly, the variance of km was smaller in the trypsin degraded group than the control group, so that this parameter exhibited greater sensitivity than specificity.
In the context of a larger survey of diagnostics for cartilage degradation, Kiviranta et al. determined cut off values of T1 in the presence of Gd-DTPA2− and of T2 that maximized the sum of sensitivity and specificity in distinguishing between non-degenerative and spontaneously degenerated bovine patellae as defined by Mankin scores (22). They found that these two parameters gave identical performance. This approach uses sensitivity and specificity themselves to determine a parameter cut-off value between tissue groups. In contrast, we have implemented objective classification techniques, including k-means clustering and FCM clustering, which do not require knowledge of tissue status to perform classification. Only after sample assignments were made according to pre-established procedures were sensitivity and specificity calculated. It is unclear how the results of Kiviranta et al. would translate into analysis of a test set of unknowns based on cut off values established from a training set.
Analysis through use of k-means clustering and fuzzy c-means clustering
Unlike classification based on mean values, KMC and FCM clustering do not require a priori knowledge of the status of training set samples. In addition, the centroids resulting from these two clustering approaches are less influenced by data outliers than are mean values. Therefore, we compared classification based on means with classification based on cluster centroids defined through use of KMC and FCM clustering, respectively, finding that the sensitivity and specificity of all three approaches was essentially indistinguishable.
This study was carried out on cartilage samples which had been subjected to relatively severe degradation protocols in order to examine the basic performance of MRI parameters in classification. With more subtle changes, such as in the early stages of osteoarthritis, more extensive overlap between normal and depleted cartilage data sets, and therefore possibly more limited discrimination ability, can be expected as compared to the present study.
The three clustering and discriminant analyses described above all yield binary results, that is, designation of a sample as being either in the control or degraded group. However, a gradation in degradation was seen in these samples and is characteristic of degenerative cartilage in general. Therefore, use of a clustering approach in which the training set centroid is defined by a FCM clustering algorithm and further, each validation set sample is assigned in a probabalistic fashion to control and degraded groups jointly, may be more appropriate. The sample-by-sample results we have obtained with this approach are consistent with this range of degradation. Although this approach necessarily does not designate whether an assignment is correct or incorrect, it is interesting to note that T1 was again the most accurate classifier based on average probability of assignment of validation set samples to the control or degraded groups.
In summary, even given statistically significant changes in mean parameter values under enzymatic digestion of cartilage, individual MRI parameters demonstrated very different sensitivity and specificity to degradation, with T1 exhibiting the most favorable characteristics for both collagenase and trypsin digestions. Classification accuracy based on training set mean values, k-means clustering centroids, and FCM clustering centroids was similar. Probablistic assignment through FCM clustering was also implemented successfully. We conclude that formal assessment of test performance as applied here to uni-parametric MRI characterization of cartilage may be an important component in the development of high-quality cartilage assessment protocols.
Acknowledgement
This work was supported by the Intramural Research Program of the NIH, National Institute on Aging.
References
- 1.Burstein D, Bashir A, Gray ML. MRI techniques in early stages of cartilage disease. Investigative Radiology. 2000;35(10):622–638. doi: 10.1097/00004424-200010000-00008. [DOI] [PubMed] [Google Scholar]
- 2.Mlynarik V, Sulzbacher I, Bittsansky M, Fuiko R, Trattnig S. Investigation of apparent diffusion constant as an indicator of early degenerative disease in articular cartilage. Journal of Magnetic Resonance Imaging. 2003;17(4):440–444. doi: 10.1002/jmri.10276. [DOI] [PubMed] [Google Scholar]
- 3.Laurent D, Wasvary J, Yin J, Rudin M, Pellas TC, O'Byrne E. Quantitative and qualitative assessment of articular cartilage in the goat knee with magnetization transfer imaging. Magn Reson Imaging. 2001;19(10):1279. doi: 10.1016/s0730-725x(01)00433-7. [DOI] [PubMed] [Google Scholar]
- 4.Fishbein KW, Gluzband YA, Kaku M, Ambia-Sobhan H, Shapses SA, Yamauchi M, Spencer RG. Effects of formalin fixation and collagen cross-linking on T-2 and magnetization transfer in bovine nasal cartilage. Magn Reson Med. 2007;57(6):1000–1011. doi: 10.1002/mrm.21216. [DOI] [PubMed] [Google Scholar]
- 5.Buckwalter JA, Mankin HJ. Articular cartilage.1. Tissue design and chondrocyte-matrix interactions. Journal of Bone and Joint Surgery-American Volume. 1997;79A(4):600–611. [Google Scholar]
- 6.Doty FD, Entzminger G, Yang YA. Magnetism in high-resolution NMR probe design. I: General methods. Concepts Magn Resonance. 1998;10(3):133–156. [Google Scholar]
- 7.Bashir AA, Gray MML, Burstein DD. Gd-DTPA2− as a measure of cartilage degradation. Magn Reson Med. 1996;36(5):665. doi: 10.1002/mrm.1910360504. [DOI] [PubMed] [Google Scholar]
- 8.Hajnal JV, Baudouin CJ, Oatridge A, Young IR, Bydder GM. Design and Implementation of Magnetization Transfer Pulse Sequences for Clinical Use. Journal of Computer Assisted Tomography. 1992;16(1):7–18. doi: 10.1097/00004728-199201000-00003. [DOI] [PubMed] [Google Scholar]
- 9.Stone M. Cross-Validation And Multinomial Prediction. Biometrika. 1974;61(3):509–515. [Google Scholar]
- 10.Hartigan J. Clustering Algorithms. New York: Wiley; 1975. [Google Scholar]
- 11.MacQueen J. Some methods for classification and analysis of multivariate observations; Proceedings 5th Berkeley Symposium on Mathematics, Statistics and Probability; 1967. pp. 281–297. [Google Scholar]
- 12.Hartigan JA, Wong MA. Algorithm AS 136: A K-Means Clustering Algorithm. Applied Statistics. 1979;28(1):100–108. [Google Scholar]
- 13.Bezdek JC, Ehrlich R, Full W. FCM - The Fuzzy C-Means Clustering-Algorithm. Comput Geosci. 1984;10(2–3):191–203. [Google Scholar]
- 14.Suckling J, Sigmundsson T, Greenwood K, Bullmore ET. A modified fuzzy clustering algorithm for operator independent brain tissue classification of dual echo MR images. Magn Reson Imaging. 1999;17(7):1065–1076. doi: 10.1016/s0730-725x(99)00055-7. [DOI] [PubMed] [Google Scholar]
- 15.Poole AR. An introduction to the pathophysiology of osteoarthritis. Frontiers in Bioscience. 1999;4:d662–d670. doi: 10.2741/poole. [DOI] [PubMed] [Google Scholar]
- 16.Menezes NM, Gray ML, Hartke JR, Burstein D. T-2 and T-1, MRI in articular cartilage systems. Magn Reson Med. 2004;51(3):503–509. doi: 10.1002/mrm.10710. [DOI] [PubMed] [Google Scholar]
- 17.Nissi MJ, Toyras J, Laasanen MS, Rieppo J, Saarakkala S, Lappalainen R, Jurvelin JS, Nieminen MT. Proteoglycan and collagen sensitive MRI evaluation of normal and degenerated articular cartilage. Journal of Orthopaedic Research. 2004;22(3):557–564. doi: 10.1016/j.orthres.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 18.de Graaf RA, Brown PB, McIntyre S, Nixon TW, Behar KL, Rothman DL. High magnetic field water and metabolite proton T-1 and T-2 relaxation in rat brain in vivo. Magn Reson Med. 2006;56(2):386–394. doi: 10.1002/mrm.20946. [DOI] [PubMed] [Google Scholar]
- 19.Potter K, Kidder LH, Levin IW, Lewis EN, Spencer RGS. Imaging of collagen and proteoglycan in cartilage sections using Fourier transform infrared spectral imaging. Arthritis Rheum. 2001;44(4):846–855. doi: 10.1002/1529-0131(200104)44:4<846::AID-ANR141>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 20.Moody HR, Brown CP, Bowden JC, Crawford RW, McElwain DLS, Oloyede AO. In vitro degradation of articular cartilage: does trypsin treatment produce consistent results? J Anat. 2006;209(2):259–267. doi: 10.1111/j.1469-7580.2006.00605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nieminen MT, Toyras J, Rieppo J, Hakumaki JM, Silvennoinen J, Helminen HJ, Jurvelin JS. Quantitative MR microscopy of enzymatically degraded articular cartilage. Magn Reson Med. 2000;43(5):676–681. doi: 10.1002/(sici)1522-2594(200005)43:5<676::aid-mrm9>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 22.Kiviranta P, Toyras J, Nieminen MT, Laasanen MS, Saarakkala S, Nieminen HJ, Nissi MJ, Jurvelin JS. Comparison of novel clinically applicable methodology for sensitive diagnostics of cartilage degeneration. Eur Cells Mater. 2007;13:46–55. doi: 10.22203/ecm.v013a05. [DOI] [PubMed] [Google Scholar]