Summary
The plant steroid hormones brassinosteroids (BRs) play an important role in a wide range of developmental and physiological processes. How BR signaling regulates diverse processes remains unclear. To understand the molecular details of BR responses, we have performed a proteomic study of BR-regulated proteins in Arabidopsis using two-dimensional difference gel electrophoresis (2-D DIGE) coupled with liquid chromatography-tandem mass spectrometry (LC-MS/MS). We identified 42 BR-regulated proteins, which are predicted to play potential roles in BR regulation of specific cellular processes, such as signaling, cytoskeleton rearrangement, vesicle trafficking, and biosynthesis of hormones and vitamins. Analyses of the BR insensitive mutant bri1-116 and BR hypersensitive mutant bzr1-1D identified 5 proteins (PATL1, PATL2, THI1, AtMDAR3 and NADP-ME2) affected by both BR-treatment and in the mutants, suggesting their importance in BR action. Selected proteins were further studied using insertion knockout mutants or immunoblotting. Interestingly, about 80% of the BR-responsive proteins were not identified in previous microarray studies, and direct comparison between protein- and RNA changes in BR mutants revealed a very weak correlation. RT-PCR analysis of selected genes revealed gene-specific kinetic relationships between RNA and protein responses. Furthermore, BR-regulated posttranslational modification of BiP2 protein was detected as spot shifts in 2-D DIGE. This study provides novel insights into the molecular networks that link BR signaling to specific cellular and physiological responses.
Keywords: Brassinosteroid, 2-D DIGE, proteomics, steroid, Arabidopsis
INTRODUCTION
Brassinosteroids (BRs) are growth-promoting steroid hormones that regulate multiple physiological and developmental processes in plants. While BR was initially identified based on its growth-promoting activity, subsequent physiological and genetic studies revealed additional functions of BRs in regulating a wide range of processes, including source/sink relationship, photosynthesis, responses to abiotic and biotic stresses, senescence, seed germination, photomorphogenesis, and flowering (1). BR-deficient or insensitive mutant plants exhibit a wide range of phenotypes, which include dwarfism, curly and dark-green leaves, delayed flowering and senescence, reduced fertility, photomorphogenesis in the dark, and altered vascular development (2–5). Molecular genetic studies have elucidated a BR signal transduction pathway. However, the molecular links between the BR signaling pathway and diverse developmental and physiological processes remain unclear.
Unlike its animal counterparts, which are perceived mostly by nuclear receptors (6), BR is perceived at the plasma membrane by a leucine-rich repeat receptor-like kinase (LRR-RLK), BRI1. BR binding to the extracellular domain of BRI1 activates its kinase activity (7,8) and induces dimerization with and activation of another LRR-RLK, BAK1 (9–12). Activation of the receptor kinases initiates a signaling cascade that leads to dephosphorylation and activation of the transcription factors BZR1 and BZR2 (also known as BES1) (13,14), likely through inhibiting the GSK3-like kinase BIN2 or activating the phosphatase BSU1 (15,16). Unphosphorylated BZR1 and BZR2 bind to promoter DNA and regulate BR-responsive gene expression (17–19).
A large number of BR-regulated genes have been identified by microarray studies (14,20–24). These genes are implicated in mediating specific cellular and physiological responses, such as cell wall modification, cytoskeleton, hormone synthesis, and various metabolic processes. While these changes at the RNA level might cause similar changes at the protein level, very few proteins have been directly shown to be BR regulated. An interesting observation made in some of these microarray studies is that BR-regulated genes tend to respond slowly to the hormone treatment with small fold change (around 2-fold), whereas most auxin-regulated genes were detected within three hours of auxin treatment with changes up to 10 fold (22,24). It has been proposed that BR might induce more robust changes at the protein level by regulating translational or posttranslational processes (24). Furthermore, discrepancies between responses at RNA and protein levels have been observed in yeast, mouse and human cells when microarray data of RNA expression were compared with data of proteomic profiling (25–28). Studies that combine protein expression mapping with transcript expression profiling are still uncommon in plants (29). Thus, a proteomic study of BR-responsive proteins is likely to advance our understanding of the molecular basis of BR responses as well as posttranscriptional regulation in plants.
Various methods have been developed for quantitative analysis of proteomic changes, based on quantitation with either mass spectrometry or two-dimensional gel electrophoresis (2-DE) (30). In traditional 2-DE, comparison between samples is made between gels and suffers from gel-to-gel reproducibility. Such methods have been used to identify a small number of BR-regulated proteins in rice (31) and pathogen elicitor-regulated phosphoproteins in Arabidopsis cell culture (32). In 2-D DIGE, protein samples are labeled with size- and charge-matched but spectrally distinct fluorescent dyes (CyDye DIGE fluor Cy2, Cy3 or Cy5), and multiple samples are mixed and then separated in the same 2-DE gel (33,34). Perfectly matching spot map images obtained from the same 2-DE gel allow for accurate quantitation of relative protein abundance of the spots. The protein identities of spots of interest are subsequently determined by mass spectrometry. 2-D DIGE has been used widely in quantitative proteomic studies in a wide range of biological and disease systems (35–42). In plants, 2-D DIGE has been used to identify proteins regulated by UV-B (43), salt and osmotic stresses (44), cold stress (45), fungal elicitors (46), and gibberellins (47).
Here we have applied 2-D DIGE coupled with mass spectrometry to identify BR-regulated proteins in Arabidopsis. Both BR treatment and BR-signaling mutant samples were analyzed. We identified forty-two BR-responsive proteins. Most of these protein have not been shown to be BR-responsive at the RNA level in microarray studies, and overall there is a poor correlation between the proteomic and microarray data. Comparison between the proteomes of BR signaling mutants and wild type showed that a small number of the BR-responsive proteins were affected in BR signaling mutants, suggesting their important functions in BR response. Selected proteins were further studied using RT-PCR, immunoblotting, and T-DNA knockout mutants, which confirmed the proteomic data. Furthermore, 2-D DIGE results revealed BR-regulation of BiP2 phosphorylation. The results provide valuable insight into the protein changes underlying BR-induced cellular and physiological responses.
Experimental Procedures
Plant materials and growth conditions
The BR-deficient mutant det2-1 (48), BR insensitive mutant bri1-116 (4), transgenic plants expressing the mutant bzr1-1D gene fused to cyan fluorescence protein (mBZR1-CFP) (49) were in the Arabidopsis thaliana Columbia ecotype background. The bri1-116 mutant seedlings were selected from heterozygous population based on their dwarf phenotype, because homozygous bri1-116 plants are sterile. Arabidopsis seeds were sterilized in 100% bleach for 10 min. After extensive washing with sterile water, seeds were added to liquid growth media (1/2 Murashige and Skoog basal salt mixture, 1.5% sucrose, pH 5.7) at a ratio of 50 mg seeds per 250 ml media. Seeds were cold treated for 2 days to synchronize germination, and grown under continuous light for 5 days on a shaker at a speed of 90 rpm. Brassinolide (BL) or equal volume of solvent (80% ethanol, V/V) was added to the culture to start the BL- or mock treatment.
Measurement of fresh weight and hypocotyl length
Total weight of 30 seedlings was measured on a balance-beam scale and average fresh weight per seedling was calculated. For each data point, 6 measurements were performed. Hypocotyl length was determined with image analysis software ImageJ 1.34s (http://rsb.info.nih.gov/ij/). At least 25 seedlings were measured for each data point.
Extraction of total proteins for 2-D DIGE
Total proteins were extracted with the phenol-methanol method (50) with modifications. Approximately 0.1 g plant tissues were mixed with 3 volume of SDS extraction buffer (100 mM Tris-HCl, pH 8.0, 2% SDS, 1% β-Mercaptoethanol, 5 mM EGTA, 10 mM EDTA), heated for 10 min at 65 °C, and then centrifuged at 20,000 g for 20 min. The supernatant was mixed with an equal volume of ice-cold phenol (Tris buffered, pH 7.5–7.9), and centrifuged at 20,000 g for 15 min at 4 °C to separate phenol and aqueous phases. The upper aqueous phase was removed leaving the interface intact, and the phenol phase was extracted twice with 50 mM Tris-HCl, pH 8.0, then mixed with 5 volumes of cold 0.1 M ammonium acetate in methanol and left at –20 °C overnight to precipitate proteins. After centrifugation at 20,000 g for 20 min, the protein pellet was washed three times with 1 ml cold 0.1 M ammonium acetate in methanol and once with ethanol, and then resuspended in lysis buffer (7M urea, 2M thiourea, 4% CHAPS). After centrifugation, the supernatant was transferred to a new tube and the protein concentration was determined with Bio-Rad protein assay (Bio-Rad, Hercules, CA) using BSA as a standard.
Immunoblotting
Proteins were separated on Laemmli SDS-PAGE gels (51), transferred to a nitrocellulose membrane, and probed with different antibodies followed by detection with ECL Plus Western Detection System (GE Healthcare, Piscataway, NJ). The polyclonal PATL1 and PATL2 antisera were produced using synthetic tetravalent multiantigenic peptides as described in Li et al (52) based on the 30 N-terminal residues.
Protein CyDye labeling
Proteins were labeled with DIGE specific Cy3 or Cy5 according to the manufacture’s instructions (GE healthcare, Piscataway, NJ) with modifications. Briefly, after adjusting pH to 8.5, 30 µg proteins were mixed with 50 picomole of CyDye and incubated on ice in the dark for at least 30 min. The reaction was stopped by addition of 0.5 µl of 10 mM lysine and incubated on ice for 10 min.
Two-dimensional gel electrophoresis
Pairs of Cy3- and Cy5-labeled protein samples were mixed together. The DIGE sample buffer (7M urea, 2M thiourea, 4% CHAPS, 20 mM DTT and 0.5% IPG buffer (GE Healthcare)) was added to bring the volume to 450 µl, and the samples were then applied to 24 cm Immobiline Drystrips (GE Healthcare). IEF was carried out on an Ettan IPGphor II (GE healthcare) at 20°C with maximum 50 µA per strip and the following setting: rehydration for 2 hr and then 40 V for 10 hr, 500 and 1000 V each for 1 hr, gradient increase to 8000 V in 3 hr, and remaining at 8000V until reaching the desired total Vhr (70 K for pH 4–7 and 140 K for pH 4.5–5.5 or pH 5.3–6.5 IPG strips). After IEF, IPG strips were equilibrated in equilibration buffer (6 M urea, 30% w/v glycerol, 2% SDS, and 50 mM Tris-HCl, pH 8.0) first with 0.5% DTT, and then with 2% iodoacetamide each for 15 min. The strips were then transferred to 12.5% SDS-PAGE gels for the second dimension electrophoresis using the Ettan Dalt Twelve gel system (GE healthcare). SDS-PAGE was run at 1 W per gel for 2 hr and then 3W per gel for 12–16 hr until the bromophenol blue dye front reached the gel end.
Image scanning
The gels were scanned using a Typhoon 8600 scanner (GE Healthcare). Cy3 labeled sample was scanned using a 532 nm laser and a 580/30 nm band pass emission filter, while Cy5 labeled sample was scanned with a 633 nm laser and 670/30 nm band pass emission filter. The photomultiplier tube was set to ensure that the maximum pixel intensity was between 60,000 and 90,000 pixels. Post-electrophoretic staining with deep purple (GE Healthcare), SYPRO Ruby (Bio-Rad) fluorescent dyes were performed essentially according to the manufacturer’s instructions, and subsequent scanning was performed using the same setting as Cy3 labeled sample.
2-D DIGE data analysis using the DeCyder software
The DIGE images were analyzed using DeCyder v6.5 software as described in the user manual (GE Healthcare). Briefly, the differential in-gel analysis module was used for spot detection, spot volume quantification and volume ratio normalization of different samples in the same gel. Then, the biological variation analysis module was used to match protein spots among different gels and to identify protein spots that exhibit significant difference. Manual editing was performed in the biological variation analysis module to ensure that spots were correctly matched between different gels, and were not contaminated with artifacts, such as streaks or dusts. Differentially expressed spots were selected based on the p-value of t-test (p<0.05), and presence on at least two of the three replicates, including presence on the reverse labeled one, and peak height of over 100 on at least two gels.
Spot picking
Approximately 800 µg of protein was labeled with 40 picomole Cy5 dye, and separated with 2-DE. SDS-PAGE gels were stained with deep purple dye (GE Healthcare) or SYPRO Ruby dye (Bio-Rad). After scanning the gels with Typhoon 8600, spots of interests were selected with DeCyder software, and picked by an Ettan spot picker (GE Healthcare).
In-gel tryptic digestion, mass spectrometry, and database searching
Gel spots of interest were subjected to in-gel digestion (http://ms-facilit.ucsf.edu/ingel.html) with trypsin (porcine, side-chain protected, Promega). Briefly, specific protein spots were washed twice with 50% acetonitrile in 25 mM ammonium bicarbonate (NH4HCO3), vacuum dried, rehydrated in 8 µl of digestion buffer (10 ng µl−1 trypsin in 25 mM NH4HCO3), and covered with the minimum volume NH4HCO3. After overnight digestion at 37 °C, peptides were extracted twice with a solution containing 50% acetonitrile and 5% formic acid. The extracted digests were vacuum-dried and resuspended in 5 µl 0.1% formic acid in water.
The digests were analyzed by capillary HPLC-tandem mass spectrometry. The separation was performed with C18 PepMap 75 µm ×150 mm column (LC Packings, Sunnyvale, CA) used on either an Ultimate HPLC system linked with a FAMOS auto sampler (LC Packings, San Francisco, CA) or an Agilent 1100 series HPLC system equipped with an auto sampler (Agilent Technologies, Palo Alto, CA). Solvent A was 0.1% formic acid in water, and B was 0.1 % formic acid in acetonitrile, and a flow rate of ∼330 nL/min was employed. One µl of the digests was injected at 5% B, then the organic content of the mobile phase was increased linearly to 50% B over 30 min. The column effluent was directed to either a QSTAR-Pulsar or QSTAR-XL tandem mass spectrometer (Applied Biosystems/MDS Sciex, Toronto, CA). Throughout the chromatographic separation, a one-second MS acquisition was followed by a three second collision-induced-dissociation (CID) acquisition for computer-selected precursor ions in information-dependent acquisition mode. The collision energy was set according to the m/z value and charge state of the precursor ion. In some cases, the LC/MS/MS analyses were performed on an LTQ-FT mass spectrometer (Thermo Electron, San Jose, CA) where the instrument alternated between acquiring a full MS scan, a zoom scan and then a subsequent MS/MS spectrum.
The CID spectra were submitted for NCBInr database searching using the Mascot search engine version 2.0 (http://www.matrixscience.com). The maximum number of missed enzyme cleavages per peptide was set at two. Variable modifications selected for searching include oxidation of methionine, N-terminal acetylation, N-terminal pyroglutamate, and modification of cysteine by carbamidomethylation or propionamide. In some cases phosphorylation was also taken into consideration for queries. The CID spectra of the identified peptides were manually inspected. All protein identifications were matched by Mascot with scores that exceeded their 95% confidence threshold. All protein spots were verified to contain at least one peptide unique to the assigned protein.
RNA analysis
Total RNA was extracted from liquid-nitrogen ground seedlings using TRIzol reagent (Invitrogen, Carlsbad, CA). First strand cDNA was synthesized from total RNA with SuperScript II reverse transcriptase (Invitrogen) and Oligo (dT)15 primers. Reactions of quantitative RT-PCR were performed with iQ Syber Green Supermix (Bio-Rad) on a PTC-200 Peltier Thermal Cylcer Chromo 4 (Bio-Rad) with the following settings: 95 °C for 10 min, then 45 cycles of 94 °C for 15 sec, 62 °C for 30 sec and 72 °C for 30 sec. Primers were designed to ensure that at least one primer per gene spanned the junction of two adjoining exons so that no genomic DNA would be amplified. The primers used for each gene are: ACC oxidase (At1g62380, 5’-GACCAACTTGAGGTGATAACCAA and 5’-GCGAAGTAGCTGGTGAGATCTC-3’), THI1 (At5g54770, 5’-CACCGAGAATGGGACCAACT-3’ and TTCCAACGAGAGTTCCGTCAA-3’), allene oxide cyclase (At3g25770, 5’-GACCAAGCAAAGTTCAAGAACTG-3’ and 5’-CGAGATCTCCGAGACCGAACA-3’), S-adenosylmethionine synthetase (At3g17390, 5’-CTCCTTTTAAGGTTTCTTTCGTGA-3’ and 5’-GGTCTTGTTCAAGGCAAGCATC-3’). The reference gene is an ubiquitin-conjugating enzyme gene (At5g56150, 5’-CAAATCCAAAACCCTAGAAACCGAA-3’ and 5’-ATCTCCCGTAGGACCTGCACTG-3’). For each data point, three PCR replicates from different biological samples were performed.
RESULTS
Identification of BR-regulated proteins by 2-D DIGE and mass spectrometry
To obtain optimal separation of Arabidopsis proteins by 2-DE, we have tested several protein isolation protocols, including TCA/acetone (53), methanol/chloroform (54), and the phenol-methanol method (50). The phenol-methanol method consistently yielded highly resolved 2-DE gels and was used in this study. Typically, approximately 2,300 protein spots were resolved in each gel. To determine technical variations, we first analyzed protein samples prepared in parallel from the same tissue sample in a 2-D DIGE gel (e.g. Supplementary Figure 1A). Four pairs of samples from two independent preparations were analyzed and the 2x standard deviation (SD) for the normalized protein volume ratio varied from 16% to 18%. We also compared proteins of biological repeat samples by 2-D DIGE to determine combined biological and technical variations. Both wild type (Columbia-0) and BR deficient or insensitive mutants were used, and the 2x SD from five repeats varied from 18% to 23%. We thus used a spot abundance ratio of <0.81 or >1.23 and p<0.05 as a threshold to identify differentially expressed proteins in subsequent studies.
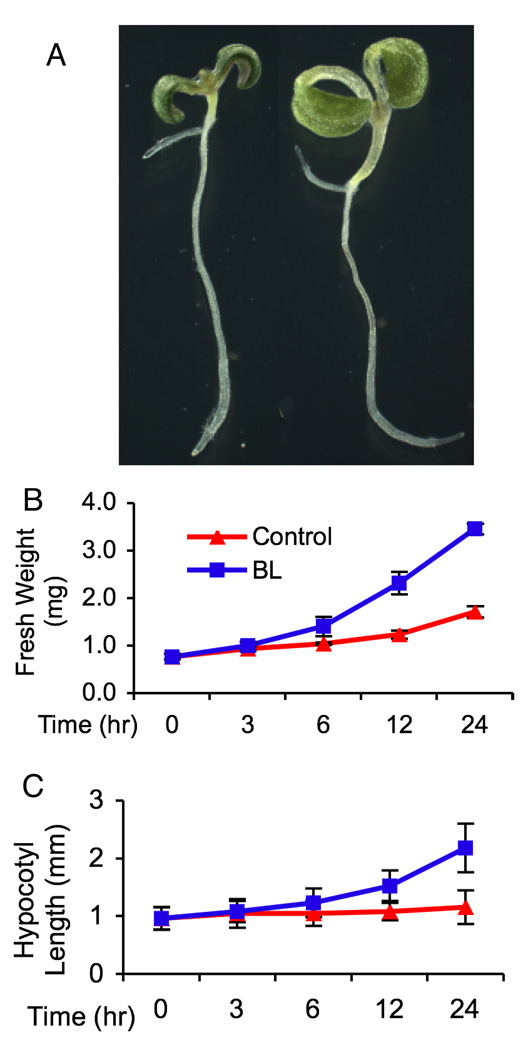
It was reported that most BR-regulated genes exhibited stronger response to BR in the BR-deficient det2 seedlings than in the wild type (20). To reduce the effect of endogenous hormone on BR treatment and to compare the proteomic data with the published microarray results, we chose det2 as plant materials. Seedlings were grown in continuous light condition to minimize the effect of circadian clock, as described in the microarray study (20). We treated det2 seedlings with 100 nM brassinolide (BL, the most active BR) or mock solution for up to 24 hr. BL induced growth becomes significant after 6 hr, as measured by both average fresh weight and hypocotyl length. The size of seedlings nearly doubled after 24 hr BR treatment (Figure 1B and 1C). We compared each pair of treated and control samples using 2-D DIGE to identify BR-regulated proteins. Triplicate pairs of BL and mock treated samples were analyzed using pH 4–7 IPG strips of 24 cm. A total of 2407 spots were detected and analyzed. No protein spot with significant differential expression (average spot ratio <0.81 or >1.23, p<0.05) was detected after 0.5 (data not shown) or 3 hr BL treatment (Supplementary Figure 2). Six BR-induced protein spots were detected after 6 hr BL treatment (Supplementary Figure 2), which include both PATL1 and PATL2 (Peterman et al., 2004), and a MutT/nudix family protein (GFG1) (55). Fifteen BR-induced and 3 BR-repressed protein spots were detected after 12 hr BL treatment (Supplementary Figure 2). After 24 hr of BL treatment, 82 proteins were up regulated and 21 were down regulated (Figure 2, Table 1). Similar results were obtained when 10 nM BL was applied or when wild type Columbia-0 was used as plant materials (data not shown).
Figure 1. BL induces seedling growth.
(A) Representatives of Arabidopsis det2 seedling treated with 100 nm BL (right) or mock solution (left) for 24 hr. (B and C) Kinetics of seedling growth measured by change of fresh weight per seedling (B) and hypocotyl length (C) during the course of BL treatment. Results are shown as means (±standard deviation).
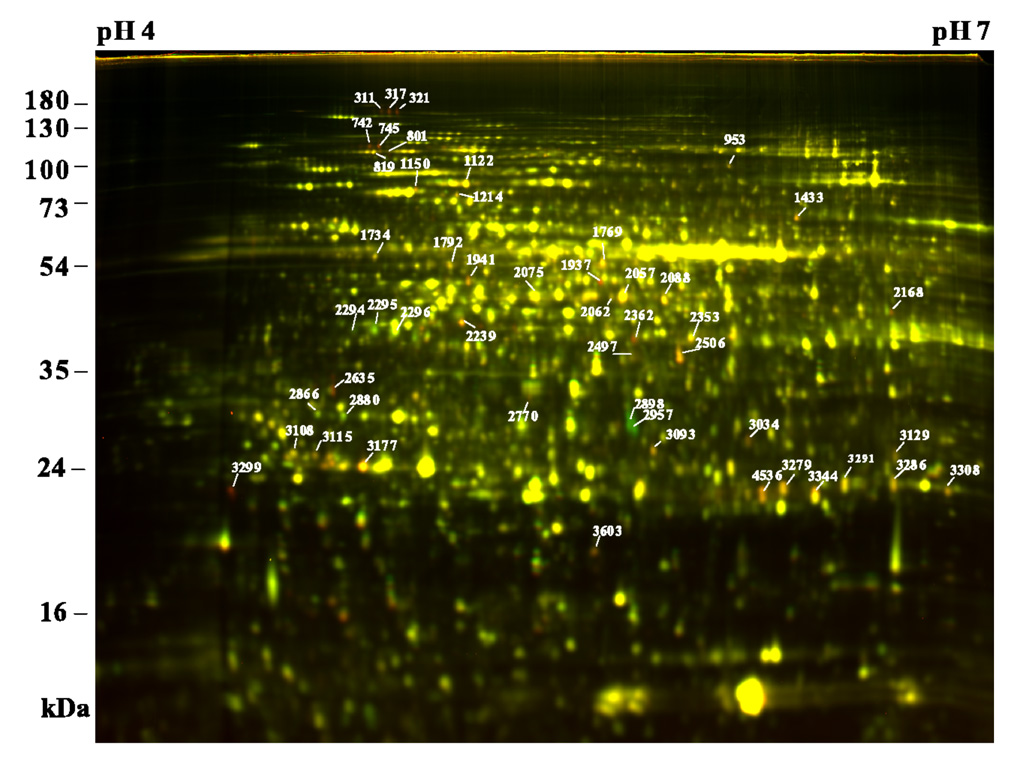
Figure 2. 2-D DIGE analysis of BR-regulated proteins.
Proteins of det2 seedlings treated with 100 nM BL for 24 hr (Cy5, red) were compared with those of mock treated samples (Cy3, green) by 2-D DIGE using 24 cm pH 4–7 IPG strips and 12.5% SDS-PAGE gel. Proteins increased by BL appear red, and those repressed by BR appear green, while those unaffected show yellow. Protein spots of interest are marked with spot numbers and their identities are shown on Table 1.
Table 1.
BR-regulated proteins. Average protein level ratios of BR-treated/untreated (positive) or BR-untreated/BR-treated (negative) and the t-test p-values are calculated from three repeat experiments of the 24 hr time point.
| Spot | Gene Locus | Protein name | Mascot Score |
6-hr ratio |
6-hr p-value |
12-hr ratio |
12-hr p-value |
24-hr ratio |
24-hr p-value |
|---|---|---|---|---|---|---|---|---|---|
| Secretion and membrane trafficking | |||||||||
| 311 | At1g22530 a | Sec14 cytosolic factor (PATL2) | 486 | 2.16 | 3.0E-2 | 1.72 | 1.4E-2 | 2.69 | 1.5E-2 |
| 317 | At1g22530 a | Sec14 cytosolic factor (PATL2) | 407 | 2.13 | 2.1E-3 | 2.07 | 6.3E-4 | 2.78 | 6.6E-3 |
| 321 | At1g22530 a | Sec14 cytosolic factor (PATL2) | 473 | 1.88 | 3.9E-2 | 2.04 | 1.5E-3 | 2.87 | 8.8E-3 |
| 742 | At1g72150 | Sec14 cytosolic factor (PATL1) | 336 | 1.45 | 6.0E-3 | 1.81 | 6.5E-2 | 3.22 | 8.2E-6 |
| 745 | At1g72150 | Sec14 cytosolic factor (PATL1) | 967 | 1.97 | 1.8E-2 | 2.19 | 2.0E-4 | 3.51 | 3.0E-5 |
| 801 | At1g30690 a | Sec14 cytosolic factor (PATL4) | 364 | 2.16 | 3.0E-2 | 1.21 | 2.3E-1 | 1.86 | 5.0E-3 |
| Signal protein | |||||||||
| 2770 | At2g41100 b | calmodulin-related protein 3 (TCH3) | 589 | 1.00 | 1.0E+0 | 1.05 | 5.0E-2 | 1.59 | 7.7E-3 |
| 3108 | At5g10450 | 14-3-3 protein GF14 lambda (GRF6) | 527 | 1.06 | 1.4E-1 | 1.09 | 1.0E-2 | 1.27 | 6.6E-4 |
| 3115 | At5g65430 | 14-3-3 protein GF14 kappa (GRF8) | 118 | 1.05 | 5.6E-3 | 1.11 | 1.2E-1 | 1.38 | 3.1E-5 |
| 3299 | At2g41090 a,b | calmodulin-like protein (CABP-22) | 163 | 1.01 | 7.2E-1 | 1.78 | 2.7E-2 | 5.33 | 5.6E-4 |
| Hormone and vitamin biosynthesis | |||||||||
| 2168 | At1g76680 a | 12-oxophytodienoate reductase (OPR1) | 404 | −1.03 | 5.8E-1 | 1.35 | 6.3E-2 | 1.52 | 4.6E-3 |
| 2295 | At1g62380 | ACC oxidase (ACO2 ) | 356 | −1.02 | 9.0E-2 | −1.13 | 3.5E-4 | −1.55 | 6.3E-4 |
| 2296 | At1g62380 | ACC oxidase (ACO2 ) | 491 | −1.04 | 1.1E-2 | 1.18 | 2.9E-1 | −1.32 | 3.0E-2 |
| 3603 | At3g25770 | allene oxide cyclase (AOC2) | 112 | 1.04 | 7.3E-1 | 1.15 | 1.2E-3 | 1.67 | 1.6E-3 |
| 2898 | At5g54770 | thiazole biosynthetic enzyme (THI1) | 431 | 1.01 | 9.2E-1 | −1.25 | 6.2E-2 | −3.16 | 1.9E-3 |
| 2957 | At5g54770 | thiazole biosynthetic enzyme (THI1) | 390 | 1.02 | 7.3E-1 | −1.44 | 1.4E-2 | −3.37 | 2.0E-2 |
| Cytoskeleton | |||||||||
| 1734 | At5g44340 b | tubulin beta-4 chain (TUB4) | 490 | 1.14 | 2.9E-1 | 1.05 | 1.7E-2 | 1.42 | 2.5E-2 |
| 1792 | At4g14960 b | tubulin alpha-6 chain (TUA6) | 1016 | 1.16 | 5.5E-2 | 1.14 | 1.2E-1 | 1.39 | 8.5E-3 |
| 2075 | At3g18780 | actin2 (ACT2) | 1248 | 1.08 | 8.3E-3 | 1.13 | 1.1E-2 | 1.34 | 1.3E-2 |
| Heat shock proteins (chaperones) | |||||||||
| 819 | At4g24190 | SHEPHERD (GRP94) | 1271 | 1.14 | 3.4E-1 | 1.19 | 4.8E-2 | 1.36 | 2.1E-3 |
| 1122 | At5g42020 | luminal binding protein 2 (BiP-2) | 1956 | 1.16 | 4.9E-2 | 1.24 | 1.1E-2 | 1.58 | 3.0E-3 |
| 1150 | At5g02490 a | heat shock cognate (HSC70-2) | 962 | −1.02 | 7.0E-1 | 1.10 | 4.3E-3 | 1.69 | 1.0E-2 |
| 1214 | At3g12580 a | heat shock protein 70 | 880 | 1.12 | 7.7E-2 | 1.29 | 4.8E-2 | 1.47 | 9.9E-3 |
| Redox related proteins | |||||||||
| 1941 | At3g09940 | monodehydroascorbate reductase MDAR3 |
377 | 1.03 | 2.6E-1 | 1.02 | 3.7E-1 | 1.59 | 4.0E-3 |
| 2294 | At1g76180 | dehydrin ERD14 | 188 | −1.13 | 9.0E-2 | 1.09 | 1.4E-1 | −1.34 | 2.2E-3 |
| 2353 | At2g47470 | thioredoxin family protein | 454 | 1.09 | 2.6E-1 | 1.18 | 2.9E-1 | 1.30 | 3.2E-2 |
| 2497 | At1g54040 | kelch repeat-containing protein | 372 | −1.02 | 3.5E-1 | 1.70 | 1.1E-2 | 1.53 | 1.3E-2 |
| 2506 | At1g54040 | kelch repeat-containing protein | 908 | 1.01 | 9.6E-1 | 1.03 | 9.1E-1 | 1.87 | 2.1E-6 |
| 2635 | At4g12720 a | MutT/nudix family protein (GFG1) | 296 | 1.41 | 1.7E-2 | 1.86 | 1.9E-3 | 2.89 | 3.2E-2 |
| 3034 | At1g10370 | glutathione S-transferase (GSTU17) | 164 | 1.07 | 4.4E-1 | −1.42 | 1.4E-2 | 1.67 | 6.4E-6 |
| 3093 | At4g30530 | defense-related protein | 376 | 1.10 | 2.3E-1 | 1.18 | 1.8E-2 | 1.59 | 1.2E-5 |
| 3129 | At5g20630 | germin-like protein (GER3) | 174 | 1.08 | 3.0E-2 | 1.22 | 2.4E-2 | 1.48 | 1.4E-2 |
| 3279 | At4g02520 b | glutathione S-transferase (GSTF2) | 211 | 1.66 | 8.0E-2 | −1.03 | 7.9E-1 | 1.70 | 4.0E-4 |
| 3286 | At2g30860 b | glutathione S-transferase (GSTF9) | 234 | −1.10 | 2.2E-1 | 1.04 | 6.3E-1 | 1.34 | 5.7E-5 |
| 3291 | At4g02520 b | glutathione S-transferase(GSTF2) | 929 | −1.05 | 5.4E-1 | −1.05 | 5.3E-1 | 1.33 | 4.8E-3 |
| 3308 | At1g02920 a,b | glutathione S-transferase (GSTF7) | 356 | −1.04 | 5.1E-1 | −1.01 | 8.8E-1 | 1.43 | 2.0E-4 |
| 3344 | At2g47730 b | glutathione S-transferase 6 (GST6) | 328 | −1.02 | 7.4E-1 | 1.15 | 3.1E-1 | 1.52 | 2.9E-3 |
| 4536 | At1g02930b | glutathione S-transferase 1 (GST1) | 177 | −1.07 | 3.7E-1 | 1.30 | 2.9E-2 | 1.88 | 9.5E-4 |
| Metabolism | |||||||||
| 953 | At3g46970 | alpha-glucan phosphorylase (PHS2) | 78 | 1.12 | 1.1E-1 | 1.13 | 8.3E-3 | 1.34 | 4.3E-5 |
| 1433 | At5g11670 | malate oxidoreductase (NADP-ME2) | 550 | 1.03 | 1.2E-1 | 1.05 | 1.9E-1 | 1.65 | 2.6E-3 |
| 1769 | At3g23810a | S-adenosyl-L-homocysteine hydrolase | 652 | 1.11 | 1.6E-1 | 1.21 | 4.9E-2 | 1.63 | 2.6E-4 |
| 2057 | At3g17390 b | S-adenosylmethionine synthetase | 826 | 1.13 | 1.1E-3 | 1.17 | 1.6E-1 | 1.46 | 5.4E-5 |
| 2062 | At1g08200b | putative UDP-D-apiose synthetase | 662 | 1.15 | 5.8E-2 | 1.21 | 1.8E-4 | 1.39 | 1.6E-3 |
| 2088 | At4g23600 | coronatine-responsive tyrosine aminotransferase |
1265 | 1.04 | 4.6E-1 | 1.09 | 1.4E-1 | 1.46 | 3.0E-5 |
| 2239 | At5g37600 | glutamine synthetase | 460 | 1.09 | 3.8E-2 | 1.29 | 3.1E-2 | 1.89 | 3.1E-4 |
| 2362 | At3g44860 | SAM:carboxyl methyltransferase | 218 | 1.07 | 7.2E-1 | 1.02 | 8.5E-1 | 1.59 | 9.3E-3 |
| Others | |||||||||
| 1937 | At1g57720 | elongation factor 1B–gamma | 612 | −1.01 | 7.8E-1 | −1.02 | 6.5E-1 | 3.10 | 2.4E-2 |
| 2866 | At3g53460 | 29 kDa chloroplast ribonucleoprotein | 603 | 1.02 | 8.4E-1 | 1.03 | 6.1E-1 | −1.24 | 6.1E-4 |
| 2880 | At3g53460 | 29 kDa chloroplast ribonucleoprotein | 613 | −1.01 | 8.1E-1 | 1.05 | 4.0E-2 | −1.34 | 8.3E-3 |
| 3177 | At1g50670 | OTU-like cysteine protease | 83 | 1.66 | 8.0E-2 | 1.47 | 2.8E-1 | 2.22 | 2.0E-3 |
Spots of interest were picked with a robotic spot picker from post-electrophoretically stained preparative gels, in-gel trypsin digested, and protein identities analyzed with LC-MS/MS. Multiple protein identities were obtained in some of the protein spots in crowded gel areas. In this case, 2-D DIGE was performed using narrow pH range IPG strips (pH 4.5–5.5 or pH 5.3–6.5), and spots were picked again based on differential expression and relative locations. A total of 50 BR-regulated protein spots were successfully identified (Table 1), and they represent products of 42 unique genes. Some of the gene products were identified in multiple spots, which are likely caused by posttranslational modifications that shift the mobility in 2-DE.
These 42 genes can be classified into several functional groups (Table 1). These include three cytoskeleton proteins, several proteins involved in hormone or vitamin biosynthesis, proteins with signaling/regulatory roles, proteins involved in vesicle trafficking and cell plate formation.
Analysis of proteomic changes in the bri1-116 and bzr1-1D mutants
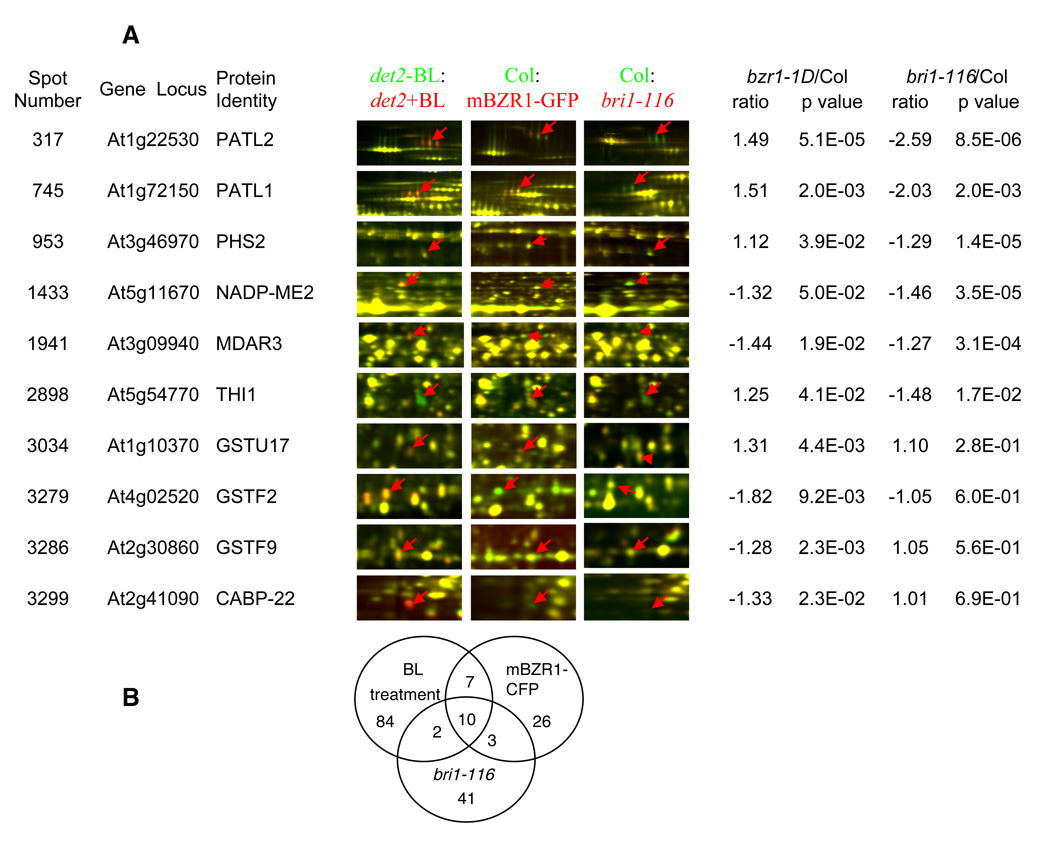
We expect that the BR-regulated proteins with important roles in plant growth will be affected both by BR treatment and in BR signaling mutants. We thus studied the proteomic changes caused by the BR insensitive bri1-116 and the BR hypersensitive bzr1-1D mutations (Figure 3A). The bri1-116 mutant contains a nonsense mutation at Gln583 in the extracellular domain of the BRI1 resulting in the absence of the BR receptor in the plasma membrane and is considered a null mutant (7,56). Direct comparison between bri1-116 and wild type by 2-D DIGE revealed 31 spots up regulated and 25 down regulated in bri1-116 (Figure 3A, B). Twelve BR-regulated spots containing products of seven genes, PATL1, PATL2, THI1 (57), malate oxidoreductase (NADP-ME2) (58), alpha-glucan phosphorylase PHS2 (59), monodehydroascorbate reductase MDAR3 (60) and GFG1 (55), were affected in bri1-116 (Figure 3A, Supplementary Table 1). Interestingly, THI1 was down regulated by both BR treatment and the bri1-116 mutation, possibly because of the increased BR level in the bri1 mutant (61).
Figure 3. Proteomic changes in the bri1-116 or bzr1-1D mutant.
(A) Representative proteins regulated by both BL treatment and bri1-116 or bzr1-1D mutations. Protein samples from bri1-116 or a transgenic line expressing mBZR1-CFP (bzr1-1D) (Cy5 labeled, red) were compared with wild type samples (Cy3, green) by 2-D DIGE. The representative images showing the affected spots and the relative ratios of protein abundance are listed. In the superimposed 2-D DIGE images, protein spots up regulated by BL, bri1-116, or bzr1-1D appear red, those down regulated appear green, and those unaffected are yellow. (B) Venn diagram of protein spots regulated by BL treatment, the bri1-116 mutation, or bzr1-1D mutation.
The dominant bzr1-1D mutation activates downstream BR responses and promotes feedback inhibition of BR biosynthesis (17). Transgenic plants expressing the mutant bzr1-1D gene fused to the cyan fluorescence protein (mBZR1-CFP) (49) show nearly identical phenotypes to the bzr1-1D mutant and were used to study the proteomic changes caused by the bzr1-1D mutation (Fig 3A). A total of 19 up-regulated and 27 down-regulated protein spots were found in the mBZR1-CFP line compared with wild type. Consistent with the dual roles of BZR1 in positively regulating BR responses and negatively regulating BR synthesis (17), 7 BR-induced protein spots containing products of four genes (PATL1, PATL2, BiP2, and GSTU17 (62)) were up-regulated in the mBZR1-CFP line, and 6 BR-induced protein spots containing products of 5 genes (GSTF2, GSTF9, NADP-ME2, MDAR3, and a calmodulin-like protein CABP-22 (63)) were down-regulated in the transgenic line (Figure 3A). The BR-repressed THI1 was up regulated in mBZR1-CFP plants. Interestingly, only 5 BR-regulated proteins (PATL1, PATL2, THI1, MDAR3 and NADP-ME2, found in 10 protein spots) were affected by BR treatment and the bri1-116 and bzr1-1D mutations, suggesting their importance in BR action.
Confirmation of BR-regulated proteins
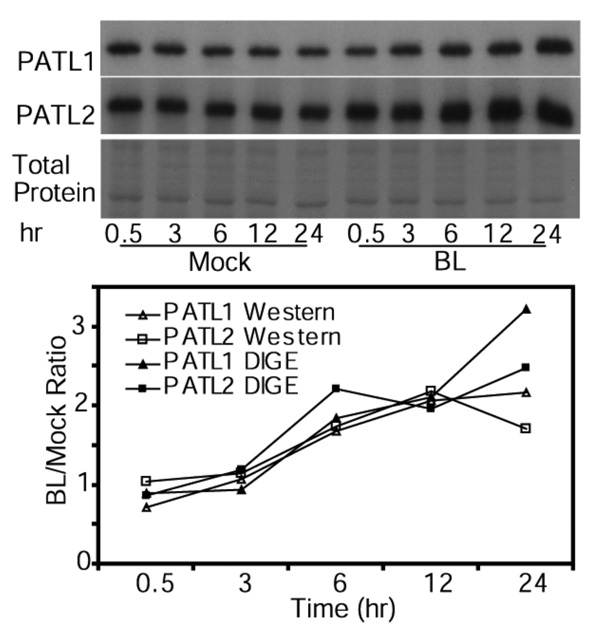
To confirm some of the results of 2-D DIGE and mass spectrometry, BR regulation of PATL1 and PATL2 was studied by immunoblotting. Protein extracts of mock- and BR-treated det2 samples were separated by SDS-PAGE, blotted, and detected by antibodies raised against each protein (64). The results confirm that BR induces both PATL1 and PATL2 (Figure 4). The kinetics of protein abundance change determined by immunoblot were very similar to those measured by 2-D DIGE (Figure 4), which demonstrates that 2-D DIGE yields accurate measurements of differential protein expression.
Figure 4. Immunoblot analysis of PATL1 and PATL2 expression in response to BL treatment.
Immunoblot of mock- and BR-treated samples was probed first with anti-PATL1 antibody, striped, and then probed with anti-PATL2 antibody. A duplicate SDS-PAGE gel stained by Commassie blue R-250 is shown. The graph shows the PATL1 and PATL2 protein levels quantified using either immunoblots or 2-D DIGE. Immunoblot data were quantitated using ImageQuant 5.2 and 2-D DIGE data were quantitated using the DeCyder 6.5 software.
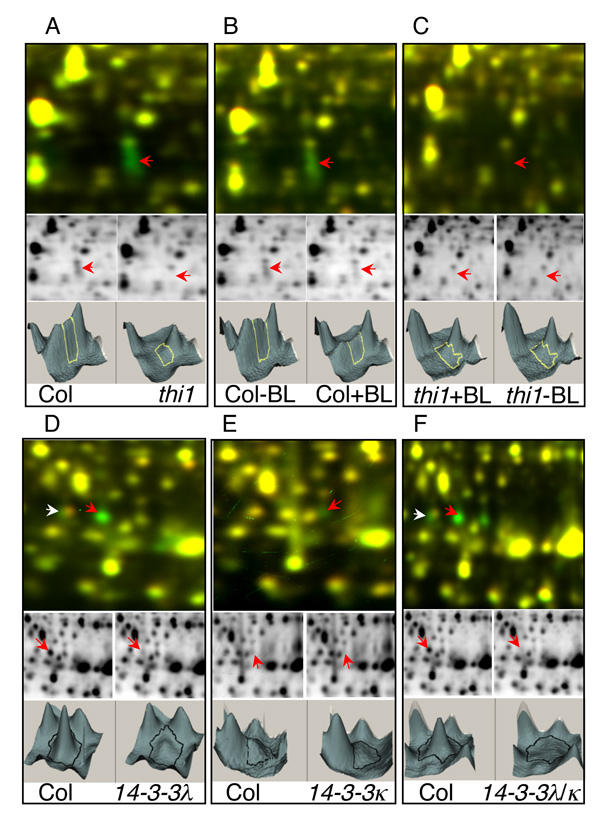
Another way to confirm the protein identity is to show that the 2-DE spot is absent when the coding gene is mutated by a T-DNA insertion. We have obtained T-DNA knockout lines for THI1, 14-3-3κ, and 14-3-3λ. Direct comparison between wild type and these mutants showed the disappearance of the corresponding spots in the mutants, confirming the correct protein identification by LC-MS/MS. THI1 was identified in spots 2898 and 2957 (Figure 2) by LC-MS/MS. A T-DNA insertion mutant, Salk_086269, in which the T-DNA was inserted into the first exon of THI1, was obtained from the Arabidopsis stock center. This thi1 mutant has similar phenotypes to those reported for the tz-201 allele, which has yellow true leaves and requires thiamine for survival (65). Comparing the protein profile between the thi1 and wild type plants showed that THI1 protein spots were absent in the knockout mutant (Figure 5A). BR treatment of the thi1 mutant did not change abundance of any spot in the corresponding gel area (Figure 5B and 5C). These data indicate that THI1 was correctly identified as the BR-regulated protein spots.
Figure 5. Confirmation of protein identity by 2-D DIGE analyses of T-DNA knockout mutant of thi1 (A–C), and single and double mutants of 14-3-3λ and 14-3-3κ (D–F).
In each panel, the top image shows the Cy3 (green) and Cy5 (red) overlay, in the middle are the individual Cy3 (left) or Cy5 (right) images, and at the bottom are the 3D view of the middle images. As expected, spots 2898 (red arrow) and 2957 are absent in the thi1 mutant compared to the wild type (A). BL treatment reduced the signal levels of both spots in wild type (B) but not in thi1 (C). Disappearance of spots in the T-DNA mutants confirms the spots 3062 (white arrow) and 3108 (red arrow) as 14-3-3λ (D, F), and spot 3115 as 14-3-3κ (E, F).
14-3-3 proteins are ubiquitous eukaryotic proteins that bind specifically to phosphoserine or phosphothreonine protein motifs and modulate diverse cellular processes (66). In this study, 14-3-3λ (spot 3108) and 14-3-3κ (spot 3115) were found to be up regulated by BR using 2-D DIGE. We obtained T-DNA knockout mutants of 14-3-3λ and 14-3-3κ and generated a double mutant. 2-D DIGE analysis showed that both proteins disappeared in the T-DNA knockout mutants, which confirmed the spot identities (Figure 5D–F). Spot 3062 was found to contain 14-3-3 λ and two additional proteins by mass spectrometry, and 2-D DIGE showed that only part of the spot exhibited reduced level in the T-DNA mutant (Figure 5D).
Comparing BR responses at the protein and mRNA levels
Since large numbers of early BR-regulated genes have been identified by microarray studies (14,20–24), we compared the BR-regulated proteins identified in this study with the BR-regulated RNAs identified in the microarray studies. The det2 mutant was used in three of the five studies. Six of the 42 BR-regulated proteins were identified in one or more of the previous microarray studies (Table 1), which include PATL2, PATL4, CABP-22, 12-oxophytodienoate reductase (OPR1), HSP70 (At3g12580), and an S-adenosyl-L-homocysteine hydrolase. The small overlap between proteomic and microarray data may be due to the different time of BR treatments used in these studies. These microarray studies used 3 hr or less BR treatments and the proteins identified in our study only showed response after 6 hr BR treatment. We then compared our proteomic data with a microarray study that included longer BR treatments (22). This study used the Affymetrix array that contains 8200 genes (about 1/3 of the genome) and also uses det2 mutants. We found 22 of the 42 genes identified in our study were present in this array but only 6 of them (27%) were identified as BR-regulated RNAs, which correspond to PATL4, CABP-22, 12-oxophytodienoate reductase (OPR1), HSC70-2, MutT/nudix family protein, and GSTF7. Taken together, only 9 of the 42 BR-regulated proteins were identified by previous microarray analysis (Table 1), indicating that our proteomic study yielded mostly novel information.
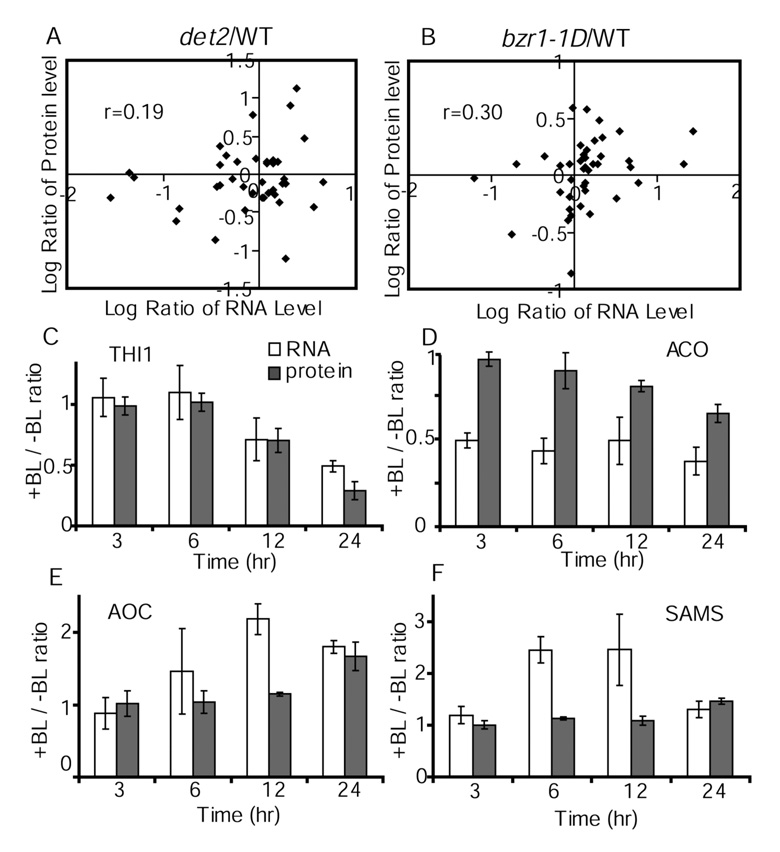
We then compared protein with RNA changes caused by the det2 or bzr1-1D mutation, which causes BR deficiency or BR-hypersensitivity, respectively. We have performed microarray analyses and identified transcripts differentially expressed in det2 and bzr1-1D compared to wild type (17). The average ratios of RNA expression between the mutant and wild type for the 42 genes in Table 1 were compared to the protein ratios (Figure 6A, 6B). The RNA data showed only moderate correlations to the protein data for both mutants (correlation coefficient for det2 r=0.19, bzr1-1D: r=0.30). For example, the PATL2 protein was significantly reduced in det2 (0.72 fold) and increased in bzr1-1D (1.49 fold), but their mRNA levels showed no significant changes (0.91 and 1.11 fold, respectively) in the mutants.
Figure 6. Comparison between changes of protein and mRNA levels caused by BR mutants (A, B) or BR treatment (C–F).
The 2-D DIGE data of Table 1 and microarray data were used to compare the ratios (log2) of protein versus mRNA expression levels between det2 (A) or bzr1-1D (B) and wild type. (C to F) The ratios of RNA (open bars) or protein (closed bars) abundance between samples treated with BL for various time and un-treated samples are shown for THI1 (panel C), ACC oxidase (ACO, panel D), allene oxide cyclase (AOC, panel E), and S-adenosylmethionine synthetase (SAMS, panel F). Protein levels were determined by 2-D DIGE analysis, and the RNA levels were quantified using real-time RT-PCR. Error bars indicate standard deviation.
To determine whether some of the new BR responsive proteins are regulated at the posttranscriptional level, we performed quantitative RT-PCR analysis of RNA expression in the same tissues analyzed by 2-D DIGE. We analyzed RNA expression of THI1 (At5g54770), ACC oxidase (At1g62380), S-adenosylmethionine synthetase (At3g17390), and allene oxide cyclase (At3g25770), which were not identified as BR responsive genes in previous microarray studies (24). However, RT-PCR analysis showed that these 4 genes responded to BR at the RNA level (Figure 6C–F). The RNA of THI1 showed a similar kinetic change as its protein (Figure 6C), whereas the other three genes showed earlier response and larger fold change at the RNA level than those at the protein level (Figure 6D–F). These data indicate that the relationship between RNA change and protein change is gene-specific and that the discrepancies between microarray and proteomic data can be due to differences between RNA and protein in their kinetics of accumulation/turn-over.
BR regulates phosphorylation of BiP2
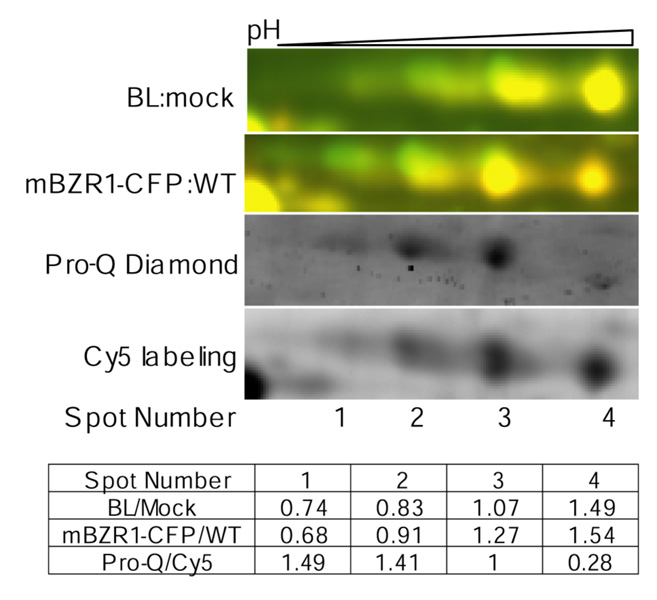
One advantage of 2-D DIGE over traditional 2-DE is its ability to detect subtle changes of spot mobility caused by changes in posttranslational modifications, such as protein phosphorylation, which alter the pI of the protein. In this study, we found that BiP2, a luminal binding protein, was regulated post-translationally by both BR treatment and bzr1-1D mutation. As in Figure 7, spots 1 to 4 were all identified by mass spectrometry as the BiP2 protein. However, the abundance of spot 1 (more acidic and lower pI) was reduced and spot 4 (higher pI) was increased after BR treatment and in the mBZR1-CFP transgenic line. To determine whether the BiP2 protein in different spots was differentially phosphorylated, proteins were minimally labeled with Cy5, separated by 2-DE, and then stained with Pro-Q Diamond stain, which specifically detects phosphorylated proteins. Comparing ratios of the Pro-Q Diamond signals to the Cy5 signals indicated that the more acidic spots were more phosphorylated than the less acidic ones (Figure 7). These results suggest that both BR and the bzr1-1D mutation decrease BiP2 phosphorylation.
Figure 7. BR regulates BiP2 phosphorylation.
BR treatment and the bzr1-1D mutation affect phosphorylation of BiP2. From top panel to bottom in: Overlaid 2-D DIGE images of BR treated (Cy5, red) compared with untreated (Cy3, green) det2 samples, 2-D DIGE image of transgenic line expressing mBZR1-CFP (Cy5, red) compared with wild type (Cy3), and Pro-Q Diamond stain and Cy5 images from the same 2-DE gel, and the table showing the quantified data. A higher ratio of Pro-Q diamond to Cy5 signal indicates increased phosphorylation. The Pro-Q/Cy5 ratio was normalized to spot 3.
DISCUSSION
2-D DIGE coupled with mass spectrometry is a powerful quantitative proteomic approach for identifying proteins that change during a biological response. Using this approach, here we identified a large number of BR-regulated proteins. Analyses of proteomic changes caused by genetic mutations that inactivate or activate the BR pathway have identified a small subgroup of the BR-regulated proteins as candidates for proteins with important roles in growth regulation. Interestingly, most of these BR-responsive proteins were not identified in previous microarray studies, and the changes of protein expression levels showed a weak correlation to the changes of their RNA. Furthermore, BR-regulation of BiP2 phosphorylation was detected as spot shift in the 2-D DIGE gels. The results thus provide novel insight into the downstream targets of BR that potentially mediate specific cellular and physiological responses.
We have been able to reproducibly generate highly resolved 2-D DIGE images from Arabidopsis tissue using a protocol based on the phenol/methanol method of protein sample preparation (50). Efficient hormone treatment was achieved by growing seedlings in liquid medium, and protein degradation and modification were minimized by extracting proteins directly from frozen tissues using an SDS buffer. Phenol extraction and methanol precipitation effectively remove lipids and nucleic acids that interfere with 2-DE. With such an optimized method of sample preparation, we were able to separate over 2000 protein spots in a single 2-DE gel, and up to 5000 proteins in multiple gels using overlapping narrow pH range IPG strips (data not shown). The sensitivity of the CyDye labeling method is very high and many detected spots were of low abundance and could not be identified by LC-MS/MS analysis. What limits the number of proteins detected in 2-D DIGE appears to be the spatial resolution of the gel rather than the sensitivity of the fluorescence labeling. Because only about 2000 protein spots can be resolved in a 24×20 cm 2-DE gel, there is a great deal of spot overlap and only the most abundant proteins are detected and quantified.
Spot overlap not only reduces the number of proteins quantified, but also poses a potential problem for unambiguous protein identification. How serious this problem is has been debated recently (67,68). Based on the sizes of the whole gel area and of each spot, Campostrini et al. (67) proposed that most spots would each contain multiple proteins, and such spot overlap causes errors in protein identification and is a serious problem in 2-DE analysis. Hunsucker and Duncan argued that the problem is not as severe as it appears, mostly because of the huge dynamic range of protein abundance in the cells (68). When several proteins of very different abundance are present in one spot, the one with the highest level will dominate both spot quantification and MS identification, resulting in correct protein identification. However, when multiple proteins of similar levels are present in one spot, assigning the correct protein identity to the expression data can be challenging. When more than one protein is found with similar level of confidence in one spot by mass spectrometry analysis, a secondary experiment would be required to confirm the protein identity. We have used knockout mutants of thi1, 14-3-3λ and 14-3-3κ to evaluate the problem of spot overlap. By directly comparing the mutant with wild type using 2-D DIGE, we not only confirmed our protein identifications but also revealed that spot 3062 indeed contained 14-3-3λ and other proteins. For some other spots that yielded ambiguous protein identity (Mascot scores within 2 fold difference) we have re-picked spots from 2-D DIGE gels using narrow pH range IPG strips, which greatly reduce spot overlap (data not shown). We also confirmed the BR regulation of PATL1 and PATL2 by immunoblot analysis. Overall, our study indicates that when a dominant protein identity is obtained in a spot, the data is of high confidence.
Most of the BR regulated proteins identified in this study were not identified by previous proteomic studies. In a recent proteomic study of BR responses in the rice lamina joints and roots using traditional 2-DE, a total of 21 BR-regulated proteins were identified (31). Four of these proteins, including tubulin, dihydroflavonol reductase, glutathione S-transferase, and calmodulin, share similarity with BR-regulated proteins identified in our study. A proteomic study of mung bean epicotyl identified 17 proteins that were down regulated by chilling but up regulated by BR (69). A homolog and an ortholog of these proteins, alpha tubulin and SAM synthetase, respectively, were found in the current study. The small overlap between these studies might be due to different methods of protein extraction, different treatment time, or different species used. It appears that at least some BR actions are conserved in higher plants, and the larger data set of Arabidopsis BR-responsive proteins identified in this study is likely to have broad implications for understanding BR actions in other higher plants.
About 80% of the BR-regulated proteins identified here were not reported in previous microarray studies. The correlation between changes of RNA- and protein-levels observed in det2 or bzr1-1D mutants is also very weak, and different kinetics of RNA and protein changes were observed for specific proteins. Because we did not perform MS analysis of the BRunregulated proteins, we cannot tell whether any BR-regulated RNAs lead to no change in the protein level. It is unclear how much of the discrepancy between RNA and protein data was due to different sensitivities of the methods, different biological samples analyzed, meaningless RNA changes that lead to no protein change, or posttranscriptional and posttranslational regulation. Similar discrepancies between RNA and protein profiling have been also observed in other biological systems (25–28). The discrepancy suggests that proteomic data is more relevant to biological responses than microarray data, because proteins, not RNAs, are the functional products of these genes. The data generated in this study are therefore very valuable for understanding BR-responses at the molecular levels.
Many BR-regulated proteins identified in this study potentially mediate BR-regulation of specific cellular and physiological processes, which include intracellular signaling, cytoskeleton, secretion and vesicle trafficking, and biosynthesis of other hormones. Proteins with predicted role in signal transduction or cellular regulation include two calmodulin-like proteins and two 14-3-3 proteins. It was shown recently that calmodulin is involved in regulation of BR biosynthesis by interacting with the BR biosynthetic enzyme DWF1 (70). BR has been shown to affect intracellular calcium fluxes (71). The BR-regulated calmodulin might mediate feedback regulation of BR biosynthesis as well as other BR-regulated metabolism and cellular responses. 14-3-3 proteins are signaling proteins that bind specifically to phosphorylated proteins. They play important regulatory roles in a wide range of cellular processes including cell division, signal transduction, transcription, and metabolism (72). There are 15 genes encoding 14-3-3 proteins in Arabidopsis (73–75). Both 14-3-3λ and 14-3-3κ were up regulated by BR. Although the fold changes were mild, the changes in protein amount can be significant given the high abundance of this family of proteins (73). Neither single nor double mutants of the two 14-3-3s show an obvious phenotype under normal growth conditions (data not shown), suggesting redundant function of the 14-3-3 family members. Interestingly, 14-3-3λ interacted with SERK1 (a close homolog of BAK1) in yeast two-hybrid (76), and 14-3-3ν was identified together with BRI1 and BAK1 in a SERK1-immunoprecipitated complex (77). 14-3-3 proteins also interact with the barley homolog of BAK1 in yeast two-hybrid assays (78). We have shown that 14-3-3 proteins interact with phosphorylated BZR1 to inhibit its nuclear localization (79,80). Since 14-3-3s modulate BR signaling by interacting with primary BR signaling components such as BZR1 and BAK1, the BR-induced accumulation of 14-3-3s is likely to contribute to feedback regulation of BR signaling.
We found three BR-induced cytoskeleton proteins, including actin 2 (At3g18780), tubulin alpha-6 chain (At4g14960), and tubulin beta-4 chain (At5g44340). Interestingly, different actin and tubulin genes, ACT11, TUB1, and TUB8, showed BR-induced expression in previous microarray studies (24). Our finding that several tubulin genes are BR-induced is particularly interesting in light of studies of the BR-deficient mutant bul1, which exhibits defects in microtubule organization that can be rescued by BR treatment (81).
Sec14 proteins play an essential role in controlling signaling interfaces between lipid metabolism and membrane trafficking (82–85). There are 31 members of Sec14-like proteins in Arabidopsis and they are divided into several groups (86). Three Sec14-like proteins (PATL1, 2, and 4) are induced by BR treatment and PATL1 and PATL2 are also affected in bri1-116 and bzr1-1D mutants, suggesting an important function of these proteins in BR-mediated growth response. Interestingly a phylogenetic analysis of plant PATLs revealed that PATL1 and PATL2 belong to the same clade, consistent with a common function (87). AtSFH1p, a member of the Sec14-nodulin domain family, is essential for root hair tip growth and was suggested to function as a regulator of polarized membrane growth in plants (86,88). Although the precise function of the PATLs in Arabidopsis remains to be elucidated, there is evidence that supports a role in vesicle trafficking and cell wall formation. Vesicle cosedimentation assays indicated binding of PATL1 to phosphoinositides (64). Immunolocalization studies showed prominent localization of PATL1 to the expanding cell plate at late telophase, where dynamic vesicle trafficking and vesicle fusion occur (64). Further studies of the BR responsive PATL proteins will shed light on how vesicle trafficking is regulated by BR and contributes to BR induced cell elongation.
BR-induced heat shock protein/chaperone expression has been proposed to contribute to increased thermal tolerance (89). Several molecular chaperones were induced by BR treatment in this study. These include Shepherd (SHD)/CLAVATA formation protein, luminal binding protein 2 (BiP2), heat shock cognate 70 kDa protein 2, and an HSP70. SHD is an ortholog of GRP94, an ER-resident HSP90-like protein. Genetic studies of the Arabidopsis shd mutant indicated that SHD is required for function of the CLV signaling pathway, which include a peptide hormone and a receptor kinase with a similar structure to BRI1 (90). It was proposed that SHD mediates the folding or complex formation of CLV proteins (90). BiP is also an ER molecular chaperone. In mammals, BiP and GRP94 interact sequentially with immunoglobin to facilitate its folding in the ER (91). In Arabidopsis, it has been shown that BiP is up regulated during defense responses to mediate folding and secretion of PR proteins (92), and enhanced BiP accumulation increases tolerance to water stress (93). The mammalian BiP is regulated by phosphorylation, and it is generally accepted that its unmodified form constitutes the biologically active species (94). Our observation that unphosphorylated form of BiP2 is increased by BR treatment suggests that BR might up regulate BiP2 activity. Up regulation of BiP2 and SHD by BR is likely to facilitate folding and secretion of cell wall proteins that mediate cell expansion as well as increase stress tolerance.
BR regulates biosynthesis or metabolism of other hormonal and regulatory molecules. An ACC oxidase (At1g62380), which converts ACC to ethylene, was down regulated by BR treatment at both the RNA and protein levels. This observation supports a crosstalk between BR and ethylene actions. It is also possible that reduced ACC oxidase accumulation helps funnel more methyl group from ethylene production to other methylation processes to promote growth since methylation is involved in the synthesis of diverse compounds such as phospholipids, pectin and lignin. In support of this hypothesis, there is increased accumulation of several enzymes involved in the S-adenosyl-L-methionine cycle, which include S-adenosyl-L-methionine:carboxyl methyltransferase, S-adenosylmethionine synthetase and S-adenosyl-L-homocysteine hydrolase. Two jasmonic acid (JA) biosynthetic proteins, allene oxide cyclase (At3g25770) and 12-oxophytodienoate reductase 1 (At1g76680, OPR1), were found to be up regulated by BR. The RNA level of the 12-oxophytodienoate reductase 2 (OPR2) was shown to be up regulated by BR in microarray studies (24). Thus, BR is likely to induce JA biosynthesis. It is interesting to note that in tomato, the BRI1 receptor kinase perceives both BR and systemin, a peptide signal involved in wounding response (95–97), and systemin induces JA biosynthesis to activate wounding responses. Furthermore, overexpression of systemin increases seedling stem elongation in a JA-dependent manner (96–98). Therefore, it is possible that BR induces JA biosynthesis to promote cell elongation and defense responses.
Surprisingly, only a small number of BR-regulated proteins were affected in BR mutants. When total protein of BR oversensitive bzr1-1D, which shows slightly dwarf phenotype in the light (contrary to BR over-expressing lines), approximately 40% of bzr1-1D regulated protein spots were affected by BL treatment. Four BL-induced proteins (PATL1, PATL2, BiP2, and GSTU17) were up regulated, and 5 BL-induced proteins (AtGSTF2, AtGSTF9, NADP-ME2, MDAR3, and a CABP-22) were down regulated in the transgenic line, in agreement with a dual role of BZR1 in brassinosteroid homeostasis and growth responses (17). When total proteins of the bri1 mutant, which have severe growth phenotypes, were compared to wild type, a total of 56 of the over 2000 protein spots showed differential levels between bri1 and wild type. Of the 42 BR-responsive proteins, only 6 were affected in the bri1 mutant. These results suggest that these six proteins, PATL1, PATL2, THI1, NADP-ME2, PHS2, MDAR3 and GFG1, play important roles in BR regulation of cell growth. In support of this conclusion, the T-DNA knockout mutant of GFG1 has been shown to have a dwarf phenotype with reduced cell size (55). THI1 is involved in biosynthesis of thiamine (vitamin B1). Thiamine pyrophosphate is a key cofactor for several enzymes involved in carbon metabolism, such as glycolysis, citric acid cycle, and the pentose phosphate cycle. The thi1 mutant is seedling lethal (99), and lacks chlorophyll content. Since BL treatment reduced chlorophyll content, it is likely BL down regulates chlorophyll content by repressing THI1.
While the BR-responsive proteins identified this study provide insight into BR regulation of specific cellular processes, no early BR-responsive proteins or BR-signaling components were detected by 2-D DIGE analysis of total protein samples. This is likely due to two reasons. First, it has been shown that most BR-induced RNAs respond slowly to BR treatment with mild fold changes (22,23). As the change in protein abundance is expected to lag behind that of RNA, the proteins regulated at the transcript level would mostly show slow response to BR treatment. Second, the early BR response proteins might be of low abundance and not detected in 2-D DIGE. Indeed, several BR signal transduction components have been shown to be phosphorylated or dephosphorylated within about 10 min of BR treatment (49). Apparently detection of these signaling proteins of low abundance by 2-D DIGE would require enrichment by prefractionation.
Supplementary Material
Supplementary Figure 1. Evaluation of the reproducibility of sample preparation and 2-D DIGE.
Supplementary Figure 2. Time course of proteomic responses to BR treatment.
Supplementary Table 1: BR-regulated proteins and their expression in response to BR treatment and in the bri1-116 mutant and mBZR1-CFP transgenic plants.
Acknowledgement
We thank Dr. Zuxiong Chen (Stanford University) for assistance with image analysis. T-DNA knockout mutants were generated by Dr. Joe Ecker at the Salk Institute and provided by the Arabidopsis Stock Center. This work is supported by grants from the U.S. Department of Energy (DE-FG02-04ER15525) to Z.Y.W. and A.L.B., the National Institute of General Medical Sciences (R01GM066258) to Z.Y.W., and US Department of Agriculture (NRICGP 2001-35304-10903) to T.K.P.. J.M.G is supported by a training grant from National Institutes of Health (5T32GM007276). The UCSF Mass Spectrometry Facility (A.L. Burlingame, Director) is supported by the Biomedical Research Technology Program of the National Center for Research Resources, NIH NCRR RR01614, RR012961 and RR019934.
Abbreviations
- BR
brassinosteroid
- BL
brassinolide
- GSK3
glycogen synthase kinase 3
- LRR-RLK
leucine-rich repeat receptor-like kinase
- JA
jasmonic acid
References
- 1.Clouse SD, Sasse JM. BRASSINOSTEROIDS: Essential Regulators of Plant Growth and Development. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:427–451. doi: 10.1146/annurev.arplant.49.1.427. [DOI] [PubMed] [Google Scholar]
- 2.Li J, Nagpal P, Vitart V, McMorris TC, Chory J. A role for brassinosteroids in light-dependent development of Arabidopsis. Science. 1996;272:398–401. doi: 10.1126/science.272.5260.398. [DOI] [PubMed] [Google Scholar]
- 3.Clouse SD, Langford M, McMorris TC. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111:671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 5.Szekeres M, Nemeth K, Koncz-Kalman Z, Mathur J, Kauschmann A, Altmann T, Redei GP, Nagy F, Schell J, Koncz C. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- 6.Thummel CS, Chory J. Steroid signaling in plants and insects - common themes, different pathways. Genes & Development. 2002;16:3113–3129. doi: 10.1101/gad.1042102. [DOI] [PubMed] [Google Scholar]
- 7.Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature. 2001;410:380–383. doi: 10.1038/35066597. [DOI] [PubMed] [Google Scholar]
- 8.Kinoshita T, Cano-Delgado A, Seto H, Hiranuma S, Fujioka S, Yoshida S, Chory J. Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature. 2005;433:167–171. doi: 10.1038/nature03227. [DOI] [PubMed] [Google Scholar]
- 9.Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110:213–222. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- 10.Nam KH, Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. 2002;110:203–212. doi: 10.1016/s0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- 11.Wang XL, Li XQ, Meisenhelder J, Hunter T, Yoshida S, Asami T, Chory J. Autoregulation and homodimerization are involved in the activation of the plant steroid receptor BRI1. Developmental Cell. 2005;8:855–865. doi: 10.1016/j.devcel.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 12.Russinova E, Borst JW, Kwaaitaal M, Cano-Delgado A, Yin Y, Chory J, de Vries SC. Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1) Plant Cell. 2004;16:3216–3229. doi: 10.1105/tpc.104.025387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang ZY, Nakano T, Gendron J, He JX, Chen M, Vafeados D, Yang YL, Fujioka S, Yoshida S, Asami T, et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Developmental Cell. 2002;2:505–513. doi: 10.1016/s1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- 14.Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 15.Li J, Nam KH. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science. 2002;295:1299–1301. doi: 10.1126/science.1065769. [DOI] [PubMed] [Google Scholar]
- 16.Mora-Garcia S, Vert G, Yin Y, Cano-Delgado A, Cheong H, Chory J. Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 2004;18:448–460. doi: 10.1101/gad.1174204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He JX, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, Wang ZY. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307:1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell. 2005;120:249–259. doi: 10.1016/j.cell.2004.11.044. [DOI] [PubMed] [Google Scholar]
- 19.Vert G, Chory J. Downstream nuclear events in brassinosteroid signalling. Nature. 2006;441:96–100. doi: 10.1038/nature04681. [DOI] [PubMed] [Google Scholar]
- 20.Goda H, Shimada Y, Asami T, Fujioka S, Yoshida S. Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 2002;130:1319–1334. doi: 10.1104/pp.011254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mussig C, Fischer S, Altmann T. Brassinosteroid-regulated gene expression. Plant Physiol. 2002;129:1241–1251. doi: 10.1104/pp.011003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S. Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 2004;134:1555–1573. doi: 10.1104/pp.103.034736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nemhauser JL, Mockler TC, Chory J. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2004;2:E258. doi: 10.1371/journal.pbio.0020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vert G, Nemhauser JL, Geldner N, Hong FX, Chory J. Molecular mechanisms of steroid hormone signaling in plants. Annual Review of Cell and Developmental Biology. 2005;21:177–201. doi: 10.1146/annurev.cellbio.21.090704.151241. [DOI] [PubMed] [Google Scholar]
- 25.Griffin TJ, Gygi SP, Ideker T, Rist B, Eng J, Hood L, Aebersold R. Complementary profiling of gene expression at the transcriptome and proteome levels in Saccharomyces cerevisiae. Mol Cell Proteomics. 2002;1:323–333. doi: 10.1074/mcp.m200001-mcp200. [DOI] [PubMed] [Google Scholar]
- 26.Ideker T, Thorsson V, Ranish JA, Christmas R, Buhler J, Eng JK, Bumgarner R, Goodlett DR, Aebersold R, Hood L. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292:929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- 27.Tian Q, Stepaniants SB, Mao M, Weng L, Feetham MC, Doyle MJ, Yi EC, Dai H, Thorsson V, Eng J, et al. Integrated genomic and proteomic analyses of gene expression in Mammalian cells. Mol Cell Proteomics. 2004;3:960–969. doi: 10.1074/mcp.M400055-MCP200. [DOI] [PubMed] [Google Scholar]
- 28.Huber M, Bahr I, Kratzschmar JR, Becker A, Muller EC, Donner P, Pohlenz HD, Schneider MR, Sommer A. Comparison of proteomic and genomic analyses of the human breast cancer cell line T47D and the antiestrogen-resistant derivative T47D–r. Mol Cell Proteomics. 2004;3:43–55. doi: 10.1074/mcp.M300047-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Rossignol M, Peltier JB, Mock HP, Matros A, Maldonado AM, Jorrin JV. Plant proteome analysis: a 2004–2006 update. Proteomics. 2006;6:5529–5548. doi: 10.1002/pmic.200600260. [DOI] [PubMed] [Google Scholar]
- 30.Lilley KS, Dupree P. Methods of quantitative proteomics and their application to plant organelle characterization. J Exp Bot. 2006;57:1493–1499. doi: 10.1093/jxb/erj141. [DOI] [PubMed] [Google Scholar]
- 31.Konishi H, Komatsu S. A proteomics approach to investigating promotive effects of brassinolide on lamina inclination and root growth in rice seedlings. Biol Pharm Bull. 2003;26:401–408. doi: 10.1248/bpb.26.401. [DOI] [PubMed] [Google Scholar]
- 32.Peck SC, Nuhse TS, Hess D, Iglesias A, Meins F, Boller T. Directed proteomics identifies a plant-specific protein rapidly phosphorylated in response to bacterial and fungal elicitors. Plant Cell. 2001;13:1467–1475. doi: 10.1105/tpc.13.6.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Unlu M, Morgan ME, Minden JS. Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis. 1997;18:2071–2077. doi: 10.1002/elps.1150181133. [DOI] [PubMed] [Google Scholar]
- 34.Tonge R, Shaw J, Middleton B, Rowlinson R, Rayner S, Young J, Pognan F, Hawkins E, Currie I, Davison M. Validation and development of fluorescence two-dimensional differential gel electrophoresis proteomics technology. Proteomics. 2001;1:377–396. doi: 10.1002/1615-9861(200103)1:3<377::AID-PROT377>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 35.Weeks ME, Sinclair J, Jacob RJ, Saxton MJ, Kirby S, Jones J, Waterfield MD, Cramer R, Timms JF. Stress-induced changes in the Schizosaccharomyces pombe proteome using two-dimensional difference gel electrophoresis, mass spectrometry and a novel integrated robotics platform. Proteomics. 2005;5:1669–1685. doi: 10.1002/pmic.200401241. [DOI] [PubMed] [Google Scholar]
- 36.Yan JX, Devenish AT, Wait R, Stone T, Lewis S, Fowler S. Fluorescence two-dimensional difference gel electrophoresis and mass spectrometry based proteomic analysis of Escherichia coli. Proteomics. 2002;2:1682–1698. doi: 10.1002/1615-9861(200212)2:12<1682::AID-PROT1682>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 37.Hu Y, Wang G, Chen GY, Fu X, Yao SQ. Proteome analysis of Saccharomyces cerevisiae under metal stress by two-dimensional differential gel electrophoresis. Electrophoresis. 2003;24:1458–1470. doi: 10.1002/elps.200390188. [DOI] [PubMed] [Google Scholar]
- 38.Henkel C, Roderfeld M, Weiskirchen R, Scheibe B, Matern S, Roeb E. Identification of fibrosis-relevant proteins using DIGE (difference in gel electrophoresis) in different models of hepatic fibrosis. Z Gastroenterol. 2005;43:23–29. doi: 10.1055/s-2004-813911. [DOI] [PubMed] [Google Scholar]
- 39.Czupalla C, Mansukoski H, Pursche T, Krause E, Hoflack B. Comparative study of protein and mRNA expression during osteoclastogenesis. Proteomics. 2005;5:3868–3875. doi: 10.1002/pmic.200402059. [DOI] [PubMed] [Google Scholar]
- 40.Beckner ME, Chen X, An J, Day BW, Pollack IF. Proteomic characterization of harvested pseudopodia with differential gel electrophoresis and specific antibodies. Lab Invest. 2005;85:316–327. doi: 10.1038/labinvest.3700239. [DOI] [PubMed] [Google Scholar]
- 41.Alfonso P, Nunez A, Madoz-Gurpide J, Lombardia L, Sanchez L, Casal JI. Proteomic expression analysis of colorectal cancer by two-dimensional differential gel electrophoresis. Proteomics. 2005;5:2602–2611. doi: 10.1002/pmic.200401196. [DOI] [PubMed] [Google Scholar]
- 42.Seike M, Kondo T, Fujii K, Yamada T, Gemma A, Kudoh S, Hirohashi S. Proteomic signature of human cancer cells. Proteomics. 2004;4:2776–2788. doi: 10.1002/pmic.200300795. [DOI] [PubMed] [Google Scholar]
- 43.Casati P, Zhang X, Burlingame AL, Walbot V. Analysis of leaf proteome after UV-B irradiation in maize lines differing in sensitivity. Mol Cell Proteomics. 2005;4:1673–1685. doi: 10.1074/mcp.M500173-MCP200. [DOI] [PubMed] [Google Scholar]
- 44.Ndimba BK, Chivasa S, Simon WJ, Slabas AR. Identification of Arabidopsis salt and osmotic stress responsive proteins using two-dimensional difference gel electrophoresis and mass spectrometry. Proteomics. 2005;5:4185–4196. doi: 10.1002/pmic.200401282. [DOI] [PubMed] [Google Scholar]
- 45.Amme S, Matros A, Schlesier B, Mock HP. Proteome analysis of cold stress response in Arabidopsis thaliana using DIGE-technology. J Exp Bot. 2006;57:1537–1546. doi: 10.1093/jxb/erj129. [DOI] [PubMed] [Google Scholar]
- 46.Chivasa S, Hamilton JM, Pringle RS, Ndimba BK, Simon WJ, Lindsey K, Slabas AR. Proteomic analysis of differentially expressed proteins in fungal elicitor-treated Arabidopsis cell cultures. J Exp Bot. 2006;57:1553–1562. doi: 10.1093/jxb/erj149. [DOI] [PubMed] [Google Scholar]
- 47.Komatsu S, Zang X, Tanaka N. Comparison of two proteomics techniques used to identify proteins regulated by gibberellin in rice. J Proteome Res. 2006;5:270–276. doi: 10.1021/pr0502929. [DOI] [PubMed] [Google Scholar]
- 48.Chory J, Nagpal P, Peto CA. Phenotypic and Genetic Analysis of det2, a New Mutant That Affects Light-Regulated Seedling Development in Arabidopsis. Plant Cell. 1991;3:445–459. doi: 10.1105/tpc.3.5.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He JX, Gendron JM, Yang Y, Li J, Wang ZY. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci U S A. 2002;99:10185–10190. doi: 10.1073/pnas.152342599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hurkman WJ, Tanaka CK. Solubilization of Plant Membrane-Proteins for Analysis by Two-Dimensional Gel-Electrophoresis. Plant Physiology. 1986;81:802–806. doi: 10.1104/pp.81.3.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 52.Li Y, Kandasamy MK, Meagher RB. Rapid isolation of monoclonal antibodies. Monitoring enzymes in the phytochelatin synthesis pathway. Plant Physiol. 2001;127:711–719. [PMC free article] [PubMed] [Google Scholar]
- 53.Granier F. Extraction of plant proteins for two-dimensional electrophoresis. Electrophoresis. 1988;9:712–718. doi: 10.1002/elps.1150091106. [DOI] [PubMed] [Google Scholar]
- 54.Wessel D, Flugge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 55.Jambunathan N, Mahalingam R. Analysis of Arabidopsis growth factor gene 1 (GFG1) encoding a nudix hydrolase during oxidative signaling. Planta. 2006;224:1–11. doi: 10.1007/s00425-005-0183-y. [DOI] [PubMed] [Google Scholar]
- 56.Friedrichsen DM, Joazeiro CA, Li J, Hunter T, Chory J. Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 2000;123:1247–1256. doi: 10.1104/pp.123.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Machado CR, de Oliveira RL, Boiteux S, Praekelt UM, Meacock PA, Menck CF. Thi1, a thiamine biosynthetic gene in Arabidopsis thaliana, complements bacterial defects in DNA repair. Plant Mol Biol. 1996;31:585–593. doi: 10.1007/BF00042231. [DOI] [PubMed] [Google Scholar]
- 58.Wheeler MC, Tronconi MA, Drincovich MF, Andreo CS, Flugge UI, Maurino VG. A comprehensive analysis of the NADP-malic enzyme gene family of Arabidopsis. Plant Physiol. 2005;139:39–51. doi: 10.1104/pp.105.065953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zeeman SC, Thorneycroft D, Schupp N, Chapple A, Weck M, Dunstan H, Haldimann P, Bechtold N, Smith AM, Smith SM. Plastidial alpha-glucan phosphorylase is not required for starch degradation in Arabidopsis leaves but has a role in the tolerance of abiotic stress. Plant Physiol. 2004;135:849–858. doi: 10.1104/pp.103.032631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lisenbee CS, Lingard MJ, Trelease RN. Arabidopsis peroxisomes possess functionally redundant membrane and matrix isoforms of monodehydroascorbate reductase. Plant J. 2005;43:900–914. doi: 10.1111/j.1365-313X.2005.02503.x. [DOI] [PubMed] [Google Scholar]
- 61.Noguchi T, Fujioka S, Choe S, Takatsuto S, Yoshida S, Yuan H, Feldmann KA, Tax FE. Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 1999;121:743–752. doi: 10.1104/pp.121.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wagner U, Edwards R, Dixon DP, Mauch F. Probing the diversity of the Arabidopsis glutathione S-transferase gene family. Plant Mol Biol. 2002;49:515–532. doi: 10.1023/a:1015557300450. [DOI] [PubMed] [Google Scholar]
- 63.Ling V, Zielinski RE. Isolation of an Arabidopsis cDNA sequence encoding a 22 kDa calcium-binding protein (CaBP-22) related to calmodulin. Plant Mol Biol. 1993;22:207–214. doi: 10.1007/BF00014929. [DOI] [PubMed] [Google Scholar]
- 64.Peterman TK, Ohol YM, McReynolds LJ, Luna EJ. Patellin1, a novel Sec14-like protein, localizes to the cell plate and binds phosphoinositides. Plant Physiol. 2004;136:3080–3094. doi: 10.1104/pp.104.045369. discussion 3001–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Papini-Terzi FS, Galhardo RS, Farias LP, Menck CF, Van Sluys MA. Point mutation is responsible for Arabidopsis tz-201 mutant phenotype affecting thiamin biosynthesis. Plant Cell Physiol. 2003;44:856–860. doi: 10.1093/pcp/pcg104. [DOI] [PubMed] [Google Scholar]
- 66.Darling DL, Yingling J, Wynshaw-Boris A. Role of 14-3-3 proteins in eukaryotic signaling and development. Curr Top Dev Biol. 2005;68:281–315. doi: 10.1016/S0070-2153(05)68010-6. [DOI] [PubMed] [Google Scholar]
- 67.Campostrini N, Areces LB, Rappsilber J, Pietrogrande MC, Dondi F, Pastorino F, Ponzoni M, Righetti PG. Spot overlapping in two-dimensional maps: a serious problem ignored for much too long. Proteomics. 2005;5:2385–2395. doi: 10.1002/pmic.200401253. [DOI] [PubMed] [Google Scholar]
- 68.Hunsucker SW, Duncan MW. Is protein overlap in two-dimensional gels a serious practical problem? Proteomics. 2006;6:1374–1375. doi: 10.1002/pmic.200500521. [DOI] [PubMed] [Google Scholar]
- 69.Huang B, Chu CH, Chen SL, Juan HF, Chen YM. A proteomics study of the mung bean epicotyl regulated by brassinosteroids under conditions of chilling stress. Cell Mol Biol Lett. 2006;11:264–278. doi: 10.2478/s11658-006-0021-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Du L, Poovaiah BW. Ca2+/calmodulin is critical for brassinosteroid biosynthesis and plant growth. Nature. 2005;437:741–745. doi: 10.1038/nature03973. [DOI] [PubMed] [Google Scholar]
- 71.Allen GJ, Chu SP, Schumacher K, Shimazaki CT, Vafeados D, Kemper A, Hawke SD, Tallman G, Tsien RY, Harper JF, et al. Alteration of stimulusspecific guard cell calcium oscillations and stomatal closing in Arabidopsis det3 mutant. Science. 2000;289:2338–2342. doi: 10.1126/science.289.5488.2338. [DOI] [PubMed] [Google Scholar]
- 72.Tzivion G, Avruch J. 14-3-3 proteins: active cofactors in cellular regulation by serine/threonine phosphorylation. J Biol Chem. 2002;277:3061–3064. doi: 10.1074/jbc.R100059200. [DOI] [PubMed] [Google Scholar]
- 73.Sehnke PC, Rosenquist M, Alsterfjord M, DeLille J, Sommarin M, Larsson C, Ferl RJ. Evolution and isoform specificity of plant 14-3-3 proteins. Plant Mol Biol. 2002;50:1011–1018. doi: 10.1023/a:1021289127519. [DOI] [PubMed] [Google Scholar]
- 74.Rosenquist M, Alsterfjord M, Larsson C, Sommarin M. Data mining the Arabidopsis genome reveals fifteen 14-3-3 genes. Expression is demonstrated for two out of five novel genes. Plant Physiol. 2001;127:142–149. doi: 10.1104/pp.127.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.DeLille JM, Sehnke PC, Ferl RJ. The arabidopsis 14-3-3 family of signaling regulators. Plant Physiol. 2001;126:35–38. doi: 10.1104/pp.126.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rienties IM, Vink J, Borst JW, Russinova E, Vries SC. The Arabidopsis SERK1 protein interacts with the AAA-ATPase AtCDC48, the 14-3-3 protein GF14lambda and the PP2C phosphatase KAPP. Planta. 2004 doi: 10.1007/s00425-004-1447-7. [DOI] [PubMed] [Google Scholar]
- 77.Karlova R, Boeren S, Russinova E, Aker J, Vervoort J, de Vries S. The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 Protein Complex Includes BRASSINOSTEROID-INSENSITIVE1. Plant Cell. 2006;18:626–638. doi: 10.1105/tpc.105.039412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schoonheim PJ, Veiga H, da Costa Pereira D, Friso G, van Wijk KJ, de Boer AH. A Comprehensive Analysis of the 14-3-3 Interactome in Barley Leaves Using a Complementary Proteomics and Two-hybrid Approach. Plant Physiol. 2006 doi: 10.1104/pp.106.090159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gampala SS, Kim TW, He JX, Tang W, Deng Z, Bai MY, Guan S, Lalonde S, Sun Y, Gendron JM, et al. An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev Cell. 2007;13:177–189. doi: 10.1016/j.devcel.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bai MY, Zhang LY, Gampala SS, Zhu SW, Song WY, Chong K, Wang ZY. Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc Natl Acad Sci U S A. 2007;104:13839–13844. doi: 10.1073/pnas.0706386104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Catterou M, Dubois F, Schaller H, Aubanelle L, Vilcot B, Sangwan-Norreel BS, Sangwan RS. Brassinosteroids, microtubules and cell elongation in Arabidopsis thaliana. II. Effects of brassinosteroids on microtubules and cell elongation in the bul1 mutant. Planta. 2001;212:673–683. doi: 10.1007/s004250000467. [DOI] [PubMed] [Google Scholar]
- 82.Li X, Xie Z, Bankaitis VA. Phosphatidylinositol/phosphatidylcholine transfer proteins in yeast. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2000;1486:55–71. doi: 10.1016/s1388-1981(00)00048-2. [DOI] [PubMed] [Google Scholar]
- 83.Cockcroft S. Phosphatidylinositol transfer proteins: a requirement in signal transduction and vesicle traffic. Bioessays. 1998;20:423–432. doi: 10.1002/(SICI)1521-1878(199805)20:5<423::AID-BIES9>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 84.Routt SM, Bankaitis VA. Biological functions of phosphatidylinositol transfer proteins. Biochem Cell Biol. 2004;82:254–262. doi: 10.1139/o03-089. [DOI] [PubMed] [Google Scholar]
- 85.Phillips SE, Vincent P, Rizzieri KE, Schaaf G, Bankaitis VA, Gaucher EA. The diverse biological functions of phosphatidylinositol transfer proteins in eukaryotes. Crit Rev Biochem Mol Biol. 2006;41:21–49. doi: 10.1080/10409230500519573. [DOI] [PubMed] [Google Scholar]
- 86.Vincent P, Chua M, Nogue F, Fairbrother A, Mekeel H, Xu Y, Allen N, Bibikova TN, Gilroy S, Bankaitis VA. A Sec14p–nodulin domain phosphatidylinositol transfer protein polarizes membrane growth of Arabidopsis thaliana root hairs. J Cell Biol. 2005;168:801–812. doi: 10.1083/jcb.200412074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Peterman T, Sequeira A, Samia J, Lunde E. Molecular cloning and characterization of patellin1, a novel sec14-related protein, from zucchini (Cucurbita pepo) J Plant Physiol. 2006 doi: 10.1016/j.jplph.2006.01.009. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 88.Bohme K, Li Y, Charlot F, Grierson C, Marrocco K, Okada K, Laloue M, Nogue F. The Arabidopsis COW1 gene encodes a phosphatidylinositol transfer protein essential for root hair tip growth. Plant J. 2004;40:686–698. doi: 10.1111/j.1365-313X.2004.02245.x. [DOI] [PubMed] [Google Scholar]
- 89.Dhaubhadel S, Chaudhary S, Dobinson KF, Krishna P. Treatment with 24-epibrassinolide, a brassinosteroid, increases the basic thermotolerance of Brassica napus and tomato seedlings. Plant Mol Biol. 1999;40:333–342. doi: 10.1023/a:1006283015582. [DOI] [PubMed] [Google Scholar]
- 90.Ishiguro S, Watanabe Y, Ito N, Nonaka H, Takeda N, Sakai T, Kanaya H, Okada K. SHEPHERD is the Arabidopsis GRP94 responsible for the formation of functional CLAVATA proteins. Embo J. 2002;21:898–908. doi: 10.1093/emboj/21.5.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Melnick J, Dul JL, Argon Y. Sequential interaction of the chaperones BiP and GRP94 with immunoglobulin chains in the endoplasmic reticulum. Nature. 1994;370:373–375. doi: 10.1038/370373a0. [DOI] [PubMed] [Google Scholar]
- 92.Wang D, Weaver ND, Kesarwani M, Dong X. Induction of protein secretory pathway is required for systemic acquired resistance. Science. 2005;308:1036–1040. doi: 10.1126/science.1108791. [DOI] [PubMed] [Google Scholar]
- 93.Alvim FC, Carolino SM, Cascardo JC, Nunes CC, Martinez CA, Otoni WC, Fontes EP. Enhanced accumulation of BiP in transgenic plants confers tolerance to water stress. Plant Physiol. 2001;126:1042–1054. doi: 10.1104/pp.126.3.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Freiden PJ, Gaut JR, Hendershot LM. Interconversion of three differentially modified and assembled forms of BiP. Embo J. 1992;11:63–70. doi: 10.1002/j.1460-2075.1992.tb05028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Szekeres M. Brassinosteroid and systemin: two hormones perceived by the same receptor. Trends Plant Sci. 2003;8:102–104. doi: 10.1016/S1360-1385(03)00010-4. [DOI] [PubMed] [Google Scholar]
- 96.Wang ZY, He JX. Brassinosteroid signal transduction--choices of signals and receptors. Trends Plant Sci. 2004;9:91–96. doi: 10.1016/j.tplants.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 97.Howe GA, Ryan CA. Suppressors of systemin signaling identify genes in the tomato wound response pathway. Genetics. 1999;153:1411–1421. doi: 10.1093/genetics/153.3.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li C, Liu G, Xu C, Lee GI, Bauer P, Ling HQ, Ganal MW, Howe GA. The tomato suppressor of prosystemin-mediated responses2 gene encodes a fatty acid desaturase required for the biosynthesis of jasmonic acid and the production of a systemic wound signal for defense gene expression. Plant Cell. 2003;15:1646–1661. doi: 10.1105/tpc.012237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lichtenthaler HK. The 1-Deoxy-D-Xylulose-5-Phosphate Pathway Of Isoprenoid Biosynthesis In Plants. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:47–65. doi: 10.1146/annurev.arplant.50.1.47. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Evaluation of the reproducibility of sample preparation and 2-D DIGE.
Supplementary Figure 2. Time course of proteomic responses to BR treatment.
Supplementary Table 1: BR-regulated proteins and their expression in response to BR treatment and in the bri1-116 mutant and mBZR1-CFP transgenic plants.