Crystal structures of human caspase 6 reveal a new mechanism for intramolecular cleavage self-activation
The crystal structures of the effector caspase-6 zymogen and of Ac-VEID-CHO inhibited caspase-6 reveal that the protein can be activated and regulated through intramolecular self-cleavage.
Keywords: Alzheimer disease, apoptosis, caspase 6, cysteine protease, intramolecular cleavage
Abstract
Dimeric effectors caspase 3 and caspase 7 are activated by initiator caspase processing. In this study, we report the crystal structures of effector caspase 6 (CASP6) zymogen and N-Acetyl-Val-Glu-Ile-Asp-al-inhibited CASP6. Both of these forms of CASP6 have a dimeric structure, and in CASP6 zymogen the intersubunit cleavage site 190TEVD193 is well structured and inserts into the active site. This positions residue Asp 193 to be easily attacked by the catalytic residue Cys 163. We demonstrate biochemically that intramolecular cleavage at Asp 193 is a prerequisite for CASP6 self-activation and that this activation mechanism is dependent on the length of the L2 loop. Our results indicate that CASP6 can be activated and regulated through intramolecular self-cleavage.
Introduction
Caspases—a family of cysteine proteases that cleave substrates after an aspartate residue—are involved in apoptosis and inflammation, and are classified on the basis of their function and the length of their pro-domains (Nicholson, 1999). Long pro-domain initiator caspases (CASP8 and CASP9) undergo induced proximity activation, and then cleave and activate short pro-domain effector caspases (CASP3 and CASP7; Fuentes-Prior & Salvesen, 2004; Yan & Shi, 2005). CASP6—which is classified as an effector—is expressed as a dimeric zymogen composed of a short pro-domain, a large subunit (p20, containing the Cys 163 catalytic cysteine), an inter-subunit linker and a small subunit (p10). CASP6 is activated by proteolytic processing at Asp 23, Asp 179 and Asp 193 (Srinivasula et al, 1996). CASP6 is often activated by CASP3 rather than initiator caspases during apoptosis (Slee et al, 1999), but can also be activated in the absence of CASP3 activity (LeBlanc et al, 1999; Allsopp et al, 2000; Doostzadeh-Cizeron et al, 2000).
Active CASP6 is abundant in the neuropathological lesions of Alzheimer disease (Guo et al, 2004; Albrecht et al, 2007) and has a role in the disruption of the neuronal cytoskeleton (Klaiman et al, 2008). Furthermore, CASP6 mediates axonal degeneration in amyloid precursor protein-mediated death receptor 6 signalling (Nikolaev et al, 2009).
The molecular mechanism for effector CASP3 and CASP7 activation is well understood (Chai et al, 2001), but little is known about the mechanism for CASP6 activation. CASP6 shares 41% and 37% sequence identity with CASP3 and CASP7, respectively, but it has several unique features. Its substrate specificity is different from CASP3 and CASP7, but similar to that of the initiators CASP8 and CASP9 (Thornberry et al, 1997). The inhibitors of apoptosis family proteins—which inhibit CASP3, CASP7 and CASP9—do not inhibit CASP6 (Salvesen & Duckett, 2002). Furthermore, CASP6 undergoes self-processing and activation in vitro and in vivo (Klaiman et al, 2009).
A previous study has shown the crystal structure of a ligand-free form of active CASP6; the free active CASP6 is a dimer and exists in a latent conformation (Baumgartner et al, 2009). However, that study could not explain the above mentioned activation features of CASP6. In this study, we report the crystal structures of a CASP6 zymogen and an Ac-VEID-CHO-bound active CASP6. The results of our structural and biochemical analyses revealed a new intramolecular self-activation mechanism for CASP6.
Results And Discussion
Crystal structure analyses
To obtain well-diffracting crystals of CASP6 zymogen, catalytic residue Cys 163 was mutated to alanine and the pro-domain was deleted. This CASP6 zymogen (ΔproCASP6C163A) was expressed as a single contiguous peptide, that was homogenized and crystallized (supplementary Fig S1 online). The structures of ΔproCASP6C163A and the Ac-VEID-CHO-inhibited CASP6 were determined at 2.9 Å and 1.6 Å resolutions, and refined to R factors Rcryst/Rfree to 18.5/23.7% and 15.5/18.9%, respectively (Table 1).
Table 1. Data collection and statistics from crystallographic analysis.
| ΔproCASP6C163A | Ac-VEID-CHO-inhibited CASP6 | |
|---|---|---|
| Wavelength (Å) | 1.5418 | 1.0 |
| Space group | P 65 2 2 | P 21 |
| Cell dimension | ||
| a, b, c (Å) | 127.98, 127.98, 167.91 | 55.46, 89.59, 61.15 |
| α, β, γ (°) | 90, 90, 120 | 90, 111.67, 90 |
| Resolution (Å) | 29.9–2.9 (3.03–2.9) | 40–1.60 (1.66–1.60) |
| Rsym* (%) | 8.8 (32.0) | 8.1 (41.5) |
| Mean I/σI | 6.7 (1.6) | 11.7 (1.7) |
| Completeness (%) | 90.5 (95.4) | 93.5 (85.2) |
| Redundancy | 3.8 | 2.3 |
| Refinement | ||
| Resolution range (Å) | 29.9–2.9 | 40–1.60 |
| Number of reflections | 15,216 | 68,035 |
| Rwork‡/Rfree‡§ (%) | 18.5/23.7 | 15.5/ 18.9 |
| Average B-factors | 34.1 | 10.7 |
| r.m.s.d.∣∣ | ||
| Bond lengths (Å) | 0.009 | 0.015 |
| Bond angles (°) | 1.218 | 1.547 |
| Ramachandran plots | ||
| Most favoured | 416 | 462 |
| Allowed | 33 | 15 |
| Disallowed | 2 | 1 |
| Values in parentheses are for the highest resolution shell. | ||
| *Rsym=∑∣Iobs–Iavg∣/∑Iobs; | ||
| ‡Rwork,free=∑∣∣Fobs∣–∣Fcalc∣∣/∑∣Fobs∣; | ||
| §Rfree values are calculated for a randomly selected 5% of the data that was excluded from the refinement; | ||
| ∣∣r.m.s.d. from ideal/target geometries. Ac-VEID-CHO, N-Acetyl-Val-Glu-Ile-Asp-al. | ||
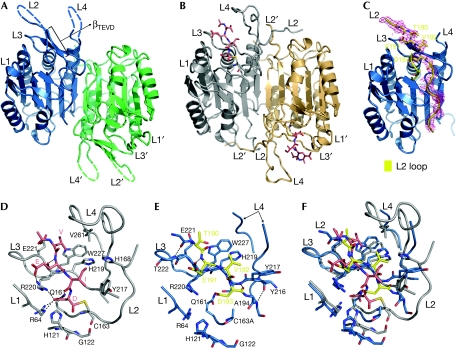
Both forms of CASP6 have the typical caspase dimer structure, with each monomer assembled into a central six-stranded β-sheet flanked by five α-helices and three small β-strands. The two monomers form a dimer by edge-to-edge interaction between their central β-sheets (Fig 1A,B). Four loops (L1–L4) in each monomer protrude from the central β-sheet to form the active site. Effector caspase activation requires cleavage in the intersubunit linker (part of the L2 loop) to release the amino-terminus of p10, which rotates about 180° to form the ‘loop bundle' with the four loops of the adjacent catalytic unit and stabilizes the substrate-binding pockets (Chai et al, 2001). The same changes are also observed in CASP6 (Fig 1A,B). Unexpectedly, in the CASP6 zymogen, the intersubunit cleavage site 190TEVD193 forms a well-defined β-strand (βTEVD), that occupies the substrate-binding cleft (Fig 1A,C).
Figure 1.
Structures of the caspase 6. (A,B) The overall dimeric structures of (A) CASP6 zymogen and (B) Ac-VEID-CHO-inhibited CASP6. The dashed lines indicate unobserved flexible residues. (C) The electron density map (2fo–fc maps) surrounding the L2 loop shown at 1.0 σ, calculated by phenix.refine. (D,E) Active sites of (D) CASP6 zymogen and (E) Ac-VEID-CHO-inhibited CASP6. The 190TEVD193 is shown in yellow and the hydrogen bonds and ‘salt bridges' are represented by black dashed lines. (F) Active sites overlay of CASP6 zymogen and Ac-VEID-CHO-inhibited CASP6. Ac-VEID-CHO, N-Acetyl-Val-Glu-Ile-Asp-al.
In the Ac-VEID-CHO-inhibited CASP6 structure, the inhibitor binds to the active site mainly through side-chain interactions and forms a covalent bond with the catalytic Cys 163, and the active site ‘loop bundle' is perfectly formed (Fig 1D). The S1 pocket is constructed from the side chains of residues Arg 64, Gln 161 and Arg 220. Arg 220 is also involved in the S3 pocket formation, which forms a ‘salt bridge' and two main-chain hydrogen bonds with the P3 glutamate of the inhibitor. The S2 pocket is composed of side chains of Tyr 217, His 168 and His 219, and the S4 pocket is formed by side chains of His 219, Glu 221, Trp 227 and Val 261. By contrast, the L2 loop of the CASP6 zymogen is intact. Residues 168–186 are flexible without clear electron density and the ‘loop bundle' is not formed. However, the βTEVD strand with flanking sequence, 189ITEVDAA195, forms an anti-parallel β-sheet with 216YYSHRET222 of the L3 loop through six main-chain hydrogen bonds binding in the substrate cleft (Fig 1E). The βTEVD strand is well structured with B-factors of 192VDAA195 that are below the average B-factors (Table 1). Side chains of Thr 190, Glu 191, Val 192 and Asp 193 fill up the S4, S3, S2 and S1 pockets (with the S1 site a little off position), respectively (Fig 1E,F). Most of the residues composing the S4, S3 and S1 pockets are already in place and these three pockets are almost identical in the zymogen and Ac-VEID-CHO-inhibited CASP6 structures. However, in the zymogen structure, one side of the S2 pocket is open without His 168, the Tyr 217 rotates about 30° towards the L4 loop. Part of the L4 loop is flexible, and barely engaged in the 190TEVD193 binding. Consequently, the 190TEVD193 is placed in the active site of the same catalytic unit and the peptide bond between Asp 193 and Ala 194 is located so close to the corresponding Cys 163 in the wild-type (WT) CASP6 that it could be attacked by Cys 163 with only minor conformational adjustment. These results indicate that CASP6 might be self-cleaved and activated intramolecularly.
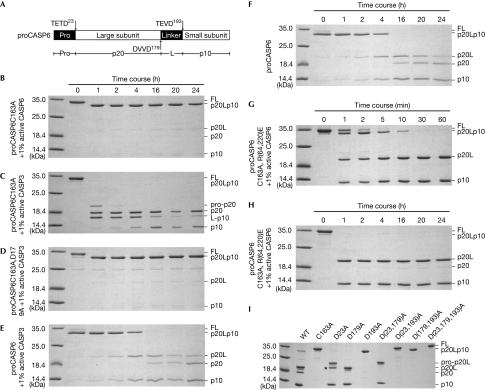
Biochemical evidence supporting intramolecular activation
The position of 190TEVD193 in the structure of the CASP6 zymogen was supported by the inability of active CASP6 (1%) to efficiently process proCASP6C163A intermolecularly at Asp 193 (Fig 2A,B). However, CASP6 activity was sufficient to process Asp 23. The cleavability of the proCASP6C163A sites was confirmed with active CASP3; it first efficiently cleaved proCASP6C163A at Asp 23 and Asp 179, generating p20 and L-p10, and then at Asp 193 to yield p10 (Fig 2C). Furthermore, CASP3 could not process proCASP6C163A,D179A at Asp 193 effectively, despite 190TEVD193 being a good CASP3 substrate site (Fig 2D). This indicates that Asp 193 was not accessible before cleavage at Asp 179, and cleavage at Asp 179 will increase the flexibility of the L2 Loop and facilitate the rotation of the N-terminus of L-p10 by 180° with the βTEVD to form the ‘loop bundle', which will release the 190TEVD193 from the active site.
Figure 2.
Biochemical evidence for caspase 6 intramolecular self-activation. (A) Schematic diagram of CASP6. (B–I) Coomassie-blue-stained gels showing proCASP6C163A cleaved by 1% active (B) CASP6 or (C) CASP3, (D) proCASP6C163AD179A cleaved by 1% active CASP3, proCASP6 incubated (E) with or (F) without 1% active CASP6, proCASP6C163A,R(64,220)E cleaved by 1% active CASP6 in (G) 1 h and (H) 24 h, and (I) self-processed activity of purified WT or CASP6 mutants. The 25-kDa unlabelled bands in (D) were a bacterial contaminated protein. The asterisk in I shows this band was pro-p20 (supplementary Figs S5 and S6 online). FL, full-length CASP6; L, intersubunit linker; p10, small subunit; p20, large subunit; pro, pro-domain; WT, wild type.
By contrast, when catalytically competent proCASP6 was incubated with or without 1% active CASP6, proCASP6 was cleaved at all the three sites (Fig 2E,F). Processing first occurred at Asp 23 and proCASP6 was completely processed into p20Lp10 within one hour. Processing continued at Asp 193, generating increasing amounts of the p20L and p10 subunits, as the level of p20Lp10 decreased beween one and four hours of incubation. Cleavage at Asp 179 occurred last, indicated by increasing levels of the p20 subunit as the p20L subunit decreased.
Furthermore, when Arg 64 and Arg 220 of the S1 pocket were mutated to glutamates, the 190TEVD193 site was excluded from the active site and became exposed and accessible. Active CASP6 cleaved proCASP6C163A,R(64,220)E efficiently at Asp 23 and Asp 193 within 1 h (Fig 2G), and cleaved slightly at Asp 179 after 16 h (Fig 2H). Interestingly, active CASP6 cleaved proCASP6C163A,R(64,220)E at Asp 23 before Asp 193, as there was no pro-p20L band observed, suggesting that 20TETD23 is a better substrate than 190TEVD193 when both sites were accessible. Cleavage at Asp 179 was inefficient because only a small amount of active CASP6 was added and DVVD is not a good substrate for CASP6.
Purified bacterial-expressed CASP6 mutant D193A allowed self-processing at Asp 23 but not at Asp 179, whereas mutants D23A and D179A allowed processing at the other two sites (Fig 2I). Similarly, CASP6D193A intermolecularly cleaved proCASP6C163A at Asp 23, but not at Asp 179 (supplementary Fig S3A online). These results confirm that Asp 193 is the first intersubunit linker site to be cleaved, and suggest that removal of the pro-domain is not essential for CASP6 self-activation.
Together, these results demonstrate that 190TEVD193 is not accessible for efficient intermolecular cleavage. During self-processing of proCASP6, cleavage occurs intermolecularly at Asp 23, then intramolecularly at Asp 193, and last at Asp 179, only after Asp 193 has been cleaved. It is also possible that cleavage at Asp 193 happens first and the resulting trace amount of pro-p20L and p10 subunits rapidly cleave proCASP6 at Asp 23. Processing of the Asp 179 site is probably through intermolecular cleavage because Asp 179 is flexible and not proximal to the intra-subunit active site of CASP6.
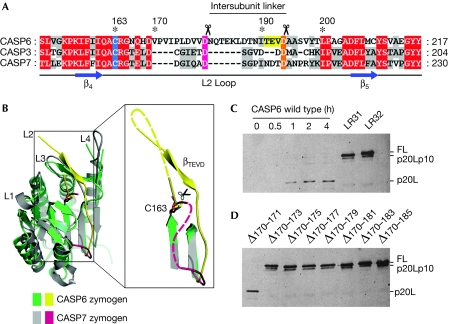
The L2 loop regulates intramolecular cleavage
Sequence alignment shows that CASP6 has a relatively long L2 loop compared with CASP3 and CASP7 (Fig 3A). Overlay of CASP7 and CASP6 zymogen structures shows that the longer L2 loop allows the intersubunit cleavage site to bind to the active site in CASP6, but not in CASP7 (Fig 3B). To determine whether the longer L2 loop facilitated intramolecular cleavage, residues 166–187 of CASP6 were replaced with the shorter L2 loop sequence 166TELDCG171 (LR31) or 166TELDCGIETA175 (LR32) of CASP3, and the recognition sequence 188NITEVD193 was retained. As 171IETD175 is a substrate of CASP6, Asp 175 was mutated to alanine in CASP6LR32 to avoid cleavage at this site. Bacterially expressed WT CASP6 underwent self-cleavage at Asp 23 and Asp 193 immediately after expression and produced p20L. By contrast, CASP6LR31 and CASP6LR32 were only slightly cleaved at Asp 23 after 4 h of expression (Fig 3C). Furthermore, deletion of only four residues of the L2 loop in CASP6 eliminated self-cleavage at Asp 193 and Asp 179, but not at Asp 23 (Fig 3D), which indicates that the mutated proteins have activity but lose the ability to intramolecularly self-cleave at Asp 193. These results indicate that the longer L2 loop of CASP6 allows the intramolecular cleavage at Asp 193 to occur first, to trigger the self-activation of CASP6.
Figure 3.
Intramolecular cleavage of caspase 6 at Asp 193 depends on the length of the L2 loop. (A) Sequence alignment of the L2 loop region for effectors. The catalytic cysteines are shadowed in blue, and the intersubunit cleavage sites indicated by scissors are in pink and orange. The 190TEVD193 of CASP6 is highlighted in yellow. (B) Overlay of CASP6 and CASP7 zymogens. The dashed lines refer to the unobserved flexible residues. (C,D) Western blot analyses of bacterially expressed wild type or mutant CASP6 showing auto-activation from (C) 0 to 4 h or (D) after 4 h.
Intramolecular cleavage is a new and unique mechanism
The CASP7 zymogen structure (Chai et al, 2001; Riedl et al, 2001) and the shorter L2 loop of both CASP7 and CASP3 do not support intramolecular cleavage of these enzymes. Furthermore, unlike CASP6—which only cleaves the pro-domain of proCASP6C163A—active CASP3 and CASP7 cleave catalytically inactive CASP3 and CASP7 at their intersubunit sites, respectively (Denault & Salvesen, 2003; Kang et al, 2008). A survey of caspases revealed that CASP1, CASP2 and CASP9 have longer L2 loops with approximately 30 residues between their catalytic cysteine and their processing sites. However, the zymogen structure of CASP1 (Elliott et al, 2009) and the report that active CASP2 processes catalytically inactive CASP2 into its active subunits (Baliga et al, 2003) do not support intramolecular self-cleavage in CASP1 and CASP2. Self-cleavage of CASP9 has been reported (Srinivasula et al, 2001), but intramolecular cleavage could not be confirmed in the absence of the zymogen structure.
Intramolecular cleavage is essential to CASP6 self-activation
CASP3 and CASP7 can process proCASP6 into its active form in vitro (Fig 2C; supplementary Fig S4A online) and in vivo. CASP8 removes the pro-domain of CASP6 without further processing the intersubunit linker sites (supplementary Fig S4B online), and CASP9 cannot process CASP6 (Srinivasula et al, 1998). However, we have observed that in well-characterized cases of Alzheimer disease and in human primary neurons, CASP6 is active in the absence of active CASP3 and CASP7 (LeBlanc et al, 1999; Guo et al, 2004; Albrecht et al, 2007), and that CASP6 undergoes self-activation in vivo (Klaiman et al, 2009). This intramolecular mechanism of activation offers a new biochemical explanation for previous observations of CASP6 self-activation in vitro and in mammalian cells (Klaiman et al, 2009). Thus, intramolecular self-cleavage is essential to initiate CASP6 activation in the absence of active CASP3 and CASP7.
Conclusions
We concluded that processing of CASP6 at the intersubunit linker Asp 193 site occurs through an intramolecular mechanism for a number of reasons. First, the crystal structure of the CASP6 zymogen shows that the intersubunit cleavage site 190TEVD193 binds to the active site of the same catalytic unit. Second, active CASP6 was unable to intermolecularly process the 190TEVD193 site in the catalytically inactive proCASP6C163A, whereas active CASP6 was able to cleave the substrate-binding pocket mutant proCASP6C163A,R(64,220)E at 190TEVD193. Third, shortening the L2 loop eliminated processing at Asp 193. We also concluded that intramolecular processing at Asp 193 is required for CASP6 self-activation. As strong CASP6 activity requires proCASP6 to be processed at either or both Asp 179 and Asp 193 sites (Klaiman et al, 2009), absence of cleavage at Asp 193 and consequently Asp 179 cannot generate active CASP6. The results also suggest that the ordered 190TEVD193 structure has a dual role in regulating CASP6 activity. We found that the 190TEVD193 inhibited CASP6 activity; the presence of 190TEVD193 at the active site physically excluded CASP6 substrates, and with an intact L2 loop, the Asp 193 site was not accessible for intermolecular cleavage. However, the peptide bond between Asp 193 and Ala 194 is located near the catalytic Cys 163, and minor conformational changes might trigger intramolecular cleavage to activate CASP6. As 190TEVD193 might still occupy the active site after cleavage at Asp 193, intramolecular processing at Asp 193 generates only partial activity and further cleavage at Asp 179 is required for full activity (Klaiman et al, 2009). Steric hindrance is not the only reason for poor initial cleavage at Asp 179. The 176DVVD179 is not an optimal CASP6 substrate, because the S4 pocket E221 repels Asp 176 and impedes CASP6 cleavage of 176DVVD179. When DVVD was mutated to TETD, the new Asp 179 site was efficiently processed by active CASP6 (supplementary Fig S4C online).
These results add to the known molecular mechanisms of effector caspase activation. High levels of CASP6 are found in Alzheimer disease (Guo et al, 2004; Albrecht et al, 2007) and it has a role in neurodegeneration (Klaiman et al, 2009; Nikolaev et al, 2009). Therefore, blocking CASP6 activation by preventing intramolecular processing might be an efficient therapeutic target for Alzheimer disease.
Methods
Mutagenesis of CASP6. The WT and mutated CASP6 constructs have been described previously (Klaiman et al, 2009). ProCASP6C163A,D179A, proCASP6C163A,R(64,220)E, L2 loop replacement and deletion mutants were generated by using overlapping PCR mutagenic oligonucleotides.
Protein preparation. The CASP6 and CASP3 constructs, provided by G. Salvesen (Burnham Institute, CA), were cloned in pET21b vector with a carboxy-terminal (His)6-tag and expressed in Escherichia coli Rosetta (DE3) strain at 18°C for 20 h. Homogeneous proteins were obtained by two-step purification; nickel-chelating column (5 ml HisTrap HP column, GE Healthcare) and gel filtration chromatography (120 ml Superdex-75, GE Healthcare). Ten millimolar dithiothreitol was added to the purified proteins. To obtain fully processed CASP6, 5 μg/μl purified WT CASP6 was incubated with 1 ng/μl active CASP3 at 4°C overnight. As the amount of CASP3 added into CASP6 was negligible compared with CASP6, fully processed CASP6 was not further purified.
Unprocessed proCASP6 was prepared according to a modified protocol (Wolan et al, 2009). ProCASP6 was expressed with a C-terminal (His)6-tag in E. coli BL21(DE3)pLysS strain at 30°C for 1 h. Cells were collected in ice-cold equilibration buffer (20 mM Tris–HCl (pH 7.5), 500 mM NaCl), sonicated and centrifuged at 50,000g for 40 min at 4°C. Soluble fractions were separated in a 1-ml HisTrap HP column (GE Healthcare) and purified proteins were transferred into equilibration buffer and concentrated to 1 μg/μl.
Crystallization and data collection. Crystals of ΔproCASP6C163A were grown by the hanging-drop vapour diffusion method. Crystals were obtained in 50 mM sodium cacodylate (pH 6.0), 7.5% w/v PEG 4000 and 1% w/v β-D-glucopyranoside at 20°C. Crystals were mounted into capillaries for data collection on a Bruker SMART 6000 CCD detector mounted on a Bruker Nonius FR591 rotating anode generator with Cu Kα radiation at 20°C. PROTEUM suite software was used to process the data.
The Ac-VEID-CHO-inhibited CASP6 crystals were grown by the sitting-drop diffusion method. Before crystallization, 0.3 mM Ac-VEID-CHO (Sigma) was added to 2 μg/μl fully processed CASP6. Crystals were obtained in 20% w/v PEG 8000, 0.2 M magnesium acetate and 0.1 M sodium cacodylate (pH 6.5) at 20°C. Diffraction data were collected on the Beamline BL6A at the KEK (the High Energy Accelerator Research Organization), Photon Factory, Tsukuba, Japan and processed by the program Mosflm (Leslie, 1992).
Structure determination and refinement. Both structures of CASP6 were determined by molecular replacement calculations with MOLREP (Vagin & Teplyakov, 1997). The structure of ΔproCASP6C163A was determined using CASP3 monomer (Protein Data Bank (PDB) ID 1CP3) as the search model, and the structure of Ac-VEID-CHO-inhibited CASP6 was determined using ΔproCASP6C163A monomer as the search model. The models were completed using COOT (Emsley & Cowtan, 2004) and refined using PHENIX (Adams et al, 2002). The data processing and refinement statistics are summarized in Table 1. The PDB access code for ΔproCASP6C163A is 3NR2 and for Ac-VEID-CHO-inhibited active CASP6 it is 3OD5.
Proteolytic processing of CASP6 variants by active caspases. The substrates—3 μM (0.1 μg/μl) purified CASP6 variants (proCASP6C163A, proCASP6C163A,D179A, proCASP6 or proCASP6C163A,R(64,220)E)—were incubated with 30 nM (1 ng/μl) active CASP6 or CASP3 (supplementary Fig S2 online) in 20 mM HEPES (pH 7.4), 50 mM NaCl, 2 mM EDTA, 0.1% CHAPS and 5 mM dithiothreitol at 37°C for 24 h. Samples were analysed by 15% SDS–PAGE.
Expression and auto-activation analysis of CASP6 variants by western blot. Expression of CASP6 variants was induced with 0.5 mM Isopropyl β-D-1-thiogalactopyranoside in E. coli Rosetta (DE3) strain at 25°C for 4 h. Cell cultures were sampled at 0, 0.5, 1, 2 and 4 h. Samples were separated by 15% SDS–PAGE, transferred to Immobilon-P polyvinylidene fluoride membranes and probed with 1:20,000 dilution of the polyclonal purified IgG against the p20 N-terminal region of CASP6 (Neomarker, Fremont), 1:5,000 dilution of secondary horseradish peroxidase-conjugated donkey anti-rabbit antibodies (GE Healthcare) and detected with enhanced chemiluminescence (GE Healthcare).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank J. Wang, J. Jodoin, D. Halawani, J. Nan, T.-M. Fu and A. Lee for valuable discussions. This study was supported by the Canadian Institutes of Health Research (CCI-85682)–National Natural Science Foundation of China (30711120581) Canada–China Joint Health Initiative and Canadian Institute for Health Research Open Operation Grant 81146. Grants from the Chinese Ministry of Science and Technology National High Technology, 863 Program (2006AA02A317) and 973 Program (2006CB806504) also supported this study.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954 [DOI] [PubMed] [Google Scholar]
- Albrecht S, Bourdeau M, Bennett D, Mufson EJ, Bhattacharjee M, LeBlanc AC (2007) Activation of caspase-6 in aging and mild cognitive impairment. Am J Pathol 170: 1200–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allsopp TE, McLuckie J, Kerr LE, Macleod M, Sharkey J, Kelly JS (2000) Caspase 6 activity initiates caspase 3 activation in cerebellar granule cell apoptosis. Cell Death Differ 7: 984–993 [DOI] [PubMed] [Google Scholar]
- Baliga BC, Colussi PA, Read SH, Dias MM, Jans DA, Kumar S (2003) Role of prodomain in importin-mediated nuclear localization and activation of caspase-2. J Biol Chem 278: 4899–4905 [DOI] [PubMed] [Google Scholar]
- Baumgartner R, Meder G, Briand C, Decock A, D'Arcy A, Hassiepen U, Morse R, Renatus M (2009) The crystal structure of caspase-6, a selective effector of axonal degeneration. Biochem J 423: 429–439 [DOI] [PubMed] [Google Scholar]
- Chai J, Wu Q, Shiozaki E, Srinivasula SM, Alnemri ES, Shi Y (2001) Crystal structure of a procaspase-7 zymogen: mechanisms of activation and substrate binding. Cell 107: 399–407 [DOI] [PubMed] [Google Scholar]
- Denault JB, Salvesen GS (2003) Human caspase-7 activity and regulation by its N-terminal peptide. J Biol Chem 278: 34042–34050 [DOI] [PubMed] [Google Scholar]
- Doostzadeh-Cizeron J, Yin S, Goodrich DW (2000) Apoptosis induced by the nuclear death domain protein p84N5 is associated with caspase-6 and NF-kappa B activation. J Biol Chem 275: 25336–25341 [DOI] [PubMed] [Google Scholar]
- Elliott JM, Rouge L, Wiesmann C, Scheer JM (2009) Crystal structure of procaspase-1 zymogen domain reveals insight into inflammatory caspase autoactivation. J Biol Chem 284: 6546–6553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Fuentes-Prior P, Salvesen GS (2004) The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem J 384: 201–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Albrecht S, Bourdeau M, Petzke T, Bergeron C, LeBlanc AC (2004) Active caspase-6 and caspase-6-cleaved tau in neuropil threads, neuritic plaques, and neurofibrillary tangles of Alzheimer's disease. Am J Pathol 165: 523–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Lee YM, Jeong YJ, Park K, Jang M, Park SG, Bae KH, Kim M, Chung SJ (2008) Large-scale preparation of active caspase-3 in E. coli by designing its thrombin-activatable precursors. BMC Biotechnol 8: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaiman G, Petzke TL, Hammond J, LeBlanc AC (2008) Targets of caspase-6 activity in human neurons and Alzheimer disease. Mol Cell Proteomics 7: 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaiman G, Champagne N, LeBlanc AC (2009) Self-activation of Caspase-6 in vitro and in vivo: Caspase-6 activation does not induce cell death in HEK293T cells. Biochim Biophys Acta 1793: 592–601 [DOI] [PubMed] [Google Scholar]
- LeBlanc A, Liu H, Goodyer C, Bergeron C, Hammond J (1999) Caspase-6 role in apoptosis of human neurons, amyloidogenesis, and Alzheimer's disease. J Biol Chem 274: 23426–23436 [DOI] [PubMed] [Google Scholar]
- Leslie AGW (1992) Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 and ESF-EACMB Newsletter on Protein Crystallography 26 [in press] [Google Scholar]
- Nicholson DW (1999) Caspase structure, proteolytic substrates, and function during apoptotic cell death. Cell Death Differ 6: 1028–1042 [DOI] [PubMed] [Google Scholar]
- Nikolaev A, McLaughlin T, O'Leary DD, Tessier-Lavigne M (2009) APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature 457: 981–989 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Riedl SJ, Fuentes-Prior P, Renatus M, Kairies N, Krapp S, Huber R, Salvesen GS, Bode W (2001) Structural basis for the activation of human procaspase-7. Proc Natl Acad Sci USA 98: 14790–14795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvesen GS, Duckett CS (2002) IAP proteins: blocking the road to death's door. Nat Rev Mol Cell Biol 3: 401–410 [DOI] [PubMed] [Google Scholar]
- Slee EA et al. (1999) Ordering the cytochrome c-initiated caspase cascade: hierarchical activation of caspases-2, -3, -6, -7, -8, and -10 in a caspase-9-dependent manner. J Cell Biol 144: 281–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasula SM, Fernandes-Alnemri T, Zangrilli J, Robertson N, Armstrong RC, Wang L, Trapani JA, Tomaselli KJ, Litwack G, Alnemri ES (1996) The Ced-3/interleukin 1beta converting enzyme-like homolog Mch6 and the lamin-cleaving enzyme Mch2alpha are substrates for the apoptotic mediator CPP32. J Biol Chem 271: 27099–27106 [DOI] [PubMed] [Google Scholar]
- Srinivasula SM, Ahmad M, Fernandes-Alnemri T, Alnemri ES (1998) Autoactivation of procaspase-9 by Apaf-1-mediated oligomerization. Mol Cell 1: 949–957 [DOI] [PubMed] [Google Scholar]
- Srinivasula SM et al. (2001) A conserved XIAP-interaction motif in caspase-9 and Smac/DIABLO regulates caspase activity and apoptosis. Nature 410: 112–116 [DOI] [PubMed] [Google Scholar]
- Thornberry NA et al. (1997) A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J Biol Chem 272: 17907–17911 [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A (1997) MOLREP: an automated program for molecular replacement. J Appl Cryst 30: 1022–1025 [Google Scholar]
- Wolan DW, Zorn JA, Gray DC, Wells JA (2009) Small-molecule activators of a proenzyme. Science 326: 853–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N, Shi Y (2005) Mechanisms of apoptosis through structural biology. Annu Rev Cell Dev Biol 21: 35–56 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.