Trichoplein/mitostatin regulates endoplasmic reticulum–mitochondria juxtaposition
The authors report the identification of the putative tumor suppressor trichoplein/mitostatin as a novel component of the interface between mitochondria and endoplasmic reticulum. Here it interacts with the mitofusin 2 to negatively regulate the tethering between these organelles.
Keywords: mitochondria, endoplasmic reticulum, trichoplein, mitofusin, apoptosis
Abstract
Trichoplein/mitostatin (TpMs) is a keratin-binding protein that partly colocalizes with mitochondria and is often downregulated in epithelial cancers, but its function remains unclear. In this study, we report that TpMs regulates the tethering between mitochondria and endoplasmic reticulum (ER) in a Mitofusin 2 (Mfn2)-dependent manner. Subcellular fractionation and immunostaining show that TpMs is present at the interface between mitochondria and ER. The expression of TpMs leads to mitochondrial fragmentation and loosens tethering with ER, whereas its silencing has opposite effects. Functionally, the reduced tethering by TpMs inhibits apoptosis by Ca2+-dependent stimuli that require ER–mitochondria juxtaposition. Biochemical and genetic evidence support a model in which TpMs requires Mfn2 to modulate mitochondrial shape and tethering. Thus, TpMs is a new regulator of mitochondria–ER juxtaposition.
Introduction
Mitochondria are dynamic organelles; their location within the cytoplasm is controlled by the concerted action of organelle fusion/fission, movement along microtubuli and anchoring at defined cytoskeletal sites, including intermediate filaments (Anesti & Scorrano, 2006). This organization probably affects the juxtaposition between mitochondria and the endoplasmic reticulum (ER)—a structural feature that is required for mitochondrial lipid biosynthesis and delivery, for efficient mitochondrial Ca2+ uptake and for the amplification of apoptosis (Hayashi et al, 2009).
The molecular basis of ER–mitochondria juxtaposition is starting to be unravelled, as researchers can now purify the mitochondria-associated membranes (MAMs)—patches of ER attached to the outer mitochondrial membrane (OMM; Vance, 1990). The ER Ca2+ channel inositol triphosphate receptor (Szabadkai et al, 2006), enzymes of the lipid biosynthetic pathway (Stone & Vance, 2000), chaperones (Szabadkai et al, 2006; Hayashi & Su, 2007) and kinases (Simmen et al, 2005) are enriched in MAMs and these proteins can indirectly regulate juxtaposition between ER and mitochondria (Simmen et al, 2005; Hayashi & Su, 2007). The first structural tether identified between the two organelles was the mitochondria-shaping protein mitofusin 2 (Mfn2), ablation of which in mammalian cells loosens the interaction between ER and mitochondria (de Brito & Scorrano, 2008). Conversely, in yeast a multiprotein complex was retrieved (Kornmann et al, 2009).
Trichoplein/mitostatin (TpMs) shows weak homology to trichohyalin, plectin and myosin and was originally reported to bind to keratins 8 and 18 (Nishizawa et al, 2005), which are the major intermediate filaments in epithelial cells (Fuchs, 1996). In addition, the genomic locus of TpMs (12q24) is a hotspot of allele deletion in several cancers; it partly colocalizes with mitochondria and regulates apoptosis (Vecchione et al, 2009). These results have prompted a more detailed analysis of its biological function. In this study, we report that TpMs is a new MAM protein interacting with Mfn2 to regulate mitochondrial morphology and tethering with the ER.
Results
TpMs localizes at the mitochondria–ER interface
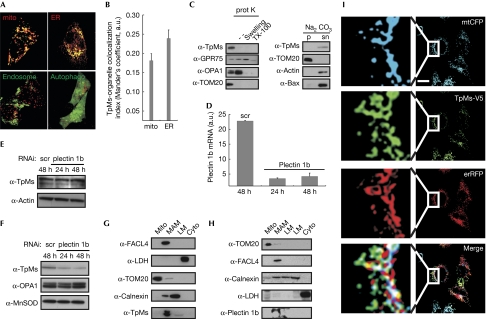
Endogenous or expressed TpMs shows a punctuate pattern, partly overlapping with mitochondria (Nishizawa et al, 2005; Vecchione et al, 2009). As TpMs lacks a canonical mitochondrial targeting sequence or a clear transmembrane domain for insertion into the mitochondrial membrane, we analysed the subcellular localization of a TpMs–green fluorescent protein (GFP) chimaera. Visual inspection and a quantitative analysis showed that TpMs–GFP partly colocalized with mitochondria and ER, but not with autophagosomes or late endosomes—two other organelles with shapes that are reminiscent of the TpMs puncta (Fig 1A,B). TpMs was retrieved in a crude mitochondrial fraction from HeLa cells that also contains ER and MAMs as contaminants (Fig 1C). When these crude mitochondria were treated with proteinase K, TpMs was completely degraded, similarly to the OMM marker translocase of outer membrane 20 (TOM20). After alkaline carbonate extraction, TpMs was retrieved in the supernatant, similarly to the loosely attached Bax and actin, whereas the integral protein TOM20 remained in the membranous pellet (Fig 1C). Thus, TpMs is loosely attached to the OMM. To map the domain of TpMs required for its mitochondrial localization, we fused fragments of increasing length and the trichohyalin–plectin homology domain of TpMs to GFP. Live confocal imaging and subcellular fractionation revealed that only full-length TpMs was clearly localised to the mitochondria (supplementary Fig S1A,B online).
Figure 1.
Trichoplein/mitostatin is enriched at the endoplasmic reticulum–mitochondria interface. (A) Upper panels: representative confocal images of HeLa cells co-transfected with TpMs–GFP (green) and mtRFP (mito, red) or erRFP (ER, red). Lower panels: confocal images of HeLa cells co-transfected with TpMs–V5 and empty vector (left) or YFP-LC3 (autophago, right), fixed after 24 h and immunostained with TRITC-conjugated anti-V5 (red) and with an FITC-conjugated cM6PR antibody (endosomes, green) in the left panel. Scale bar, 20 μm. (B) Mean±s.e. (n=3) of interaction data from (A). (C) Left: crude HeLa mitochondria (5 mg/ml) were incubated where indicated with proteinase K (100 μg/ml). Mitochondria were in isolation buffer (Frezza et al, 2007) or in 20 mM HEPES (pH 7.4; swelling) or in 0.1% Triton X-100. Proteins (25 μg) separated by SDS–PAGE were immunoblotted with the indicated antibodies. Right: crude HeLa mitochondria were incubated in 0.1 M Na2CO3 (pH 11.3; 30 min; 4°C). After centrifugation, proteins (25 μg) from pellet (p) and supernatant (sn) separated by SDS–PAGE were immunoblotted with the indicated antibodies. (D) Real-time PCR of plectin 1b levels from HeLa cells transfected with the indicated siRNA. (E) Proteins (20 μg) from HeLa cells transfected as indicated were separated by SDS–PAGE and immunoblotted using the indicated antibodies. (F) Mitochondria were isolated at indicated times from 5 × 108 HeLa cells transfected as indicated and proteins (25 μg) were separated by SDS–PAGE and immunoblotted. (G,H) Proteins (40 μg) from Percoll-purified subcellular fractions of mouse liver, were separated by SDS–PAGE and immunoblotted. (I) Representative confocal images of HeLa cells co-transfected with the indicated plasmids. After 24 h cells were fixed and immunostained with FITC-conjugated V5 antibody. Boxed areas are magnified × 9. Scale bar, 20 μm. White in the merge image: overlap of the three channels. Cyto, cytosol; ER, endoplasmic reticulum; ER-RFP, ER-targeted red fluorescent protein; FITC, fluorescein isothiocyanate; GFP, green fluorescent protein; GRP75, glucose regulatory protein 75; LC3, microtuble-associated protein 1 light chain 3; LM, light membranes; MAM, mitochondria-associated membrane; mito, mitochondria; mtRFP, matrix red fluorescent protein; SDS–PAGE, sodium dodecyl sulphate–polyacrylamide gel electrophoresis; scr, scrambled; siRNA, small interfering RNA; TpMs, trichoplein/mitostatin; TRITC, tetramethyl rhodamine iso-thiocyanate; YFP, yellow fluorescent protein.
TpMs has a plectin homology domain, suggesting that the mitochondrial plectin (plectin 1b; Winter et al, 2008) could mediate its association with mitochondria. Efficient downregulation of plectin 1b by small interfering RNA (siRNA; Fig 1D) reduced the mitochondrial levels of TpMs (Fig 1F), without affecting the total amount of TpMs (Fig 1E). Several biochemical approaches failed, however, to show a direct interaction between TpMs and plectin 1b (data not shown), raising the question of whether plectin 1b uses keratin as a ‘bridge' to direct TpMs to the OMM. Immunoprecipitation confirmed an interaction between TpMs and keratins, and immunofluorescence showed that TpMs was present between mitochondria and keratins in HeLa cells. However, levels of TpMs did not affect the organization of keratins or other cytoskeletal components (supplementary Fig S2 online). In mouse embryonic fibroblasts (MEFs) that do not express keratins 8 or 18 (data not shown) the distribution of fragments of and full-length TpMs–GFP expressed was similar to that observed in HeLa (supplementary Fig S1C online). This excludes a role for keratin in the association of TpMs with mitochondria. The localization pattern of TpMs (Fig 1A,B) resembled that of the MAM component Mfn2 (de Brito & Scorrano, 2008). When we probed MAMs purified from mouse liver for TpMs and plectin 1b, these proteins were observed almost exclusively (Fig 1G,H). Immunofluorescence confirmed that expressed TpMs–V5 was localized at puncta overlapping with both ER and mitochondria (Fig 1I). In conclusion, TpMs is localized at the interface between mitochondria and ER.
TpMs regulates ER–mitochondria tethering
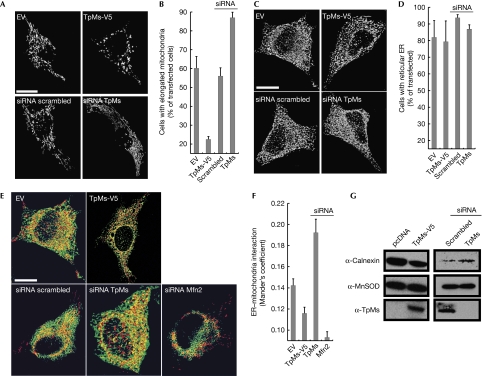
The subcellular location of TpMs prompted us to analyse its role in mitochondrial, ER morphology and tethering. Immunoblotting showed efficient siRNA-mediated downregulation and expression of TpMs (V5-tagged or untagged; supplementary Fig S3 online). Changes in the levels of TpMs affected mitochondrial morphology; less TpMs was associated with mitochondrial elongation, whereas overexpressed TpMs caused mitochondrial fragmentation (Fig 2A,B). The pro-fission effect of TpMs was replicated in MEFs, ruling out a role for keratin (supplementary Fig S1D online) and was not associated with increased levels of the pro-fission protein Drp1 on mitochondria (data not shown). Conversely, TpMs did not alter the ER structure, as shown by three-dimensional reconstructions of ER-targeted yellow fluorescent protein (de Brito & Scorrano, 2008; Fig 2C,D). A confocal semiquantitative assay of ER–mitochondria juxtaposition (de Brito & Scorrano, 2008) showed that tethering was reduced when we increased the levels of TpMs (Fig 2E,F). Accordingly, association of ER with mitochondria isolated from HeLa cells was also inversely proportional to levels of TpMs (Fig 2G). Finally, the phenotype could be reproduced in prostate LnCaP cells stably expressing TpMs and in MEFs (data not shown), thereby excluding the possibility that it was a consequence of transient transfection, or that it requires keratin. In conclusion, TpMs negatively regulates tethering of ER to mitochondria.
Figure 2.
Trichoplein/mitostatin regulates mitochondrial shape and mitochondria–endoplasmic reticulum juxtaposition. (A) Representative confocal images of HeLa cells co-transfected with mtRFP and the indicated plasmids. Scale bar, 20 μm. (B) Mean±s.e.m. (n=3) of morphometric analysis from (A). (C) Representative three-dimensional reconstructions of ER HeLa cells co-transfected with ER-YFP and the indicated plasmids. Scale bar, 20 μm. (D) Mean±s.e.m. (n=3) of morphometric analysis from (C). (E) Representative three-dimensional reconstructions of ER and mitochondria in HeLa cells co-transfected with mtRFP, ER-YFP and the indicated plasmids. Yellow, organelles are closer than around 270 nm. Scale bar, 20 μm. (F) Mean±s.e. (n=5) of interaction data from (E). (G) A total of 25 μg of proteins from mitochondria isolated after 24 (left) or 48 h (right) from HeLa cells (5 × 108), transfected as indicated, were analysed by SDS–PAGE and immunoblotting. ER, endoplasmic reticulum; ER-RFP, ER-targeted red fluorescent protein; Mfn2, mitofusin 2; MtRFP, matrix red fluorescent protein; SDS–PAGE, sodium dodecyl sulphate–polyacrylamide gel electrophoresis; siRNA, small interfering RNA; TpMs, trichoplein/mitostatin; YFP, yellow fluorescent protein.
TpMs regulates ER–mitochondria tethering via Mfn2
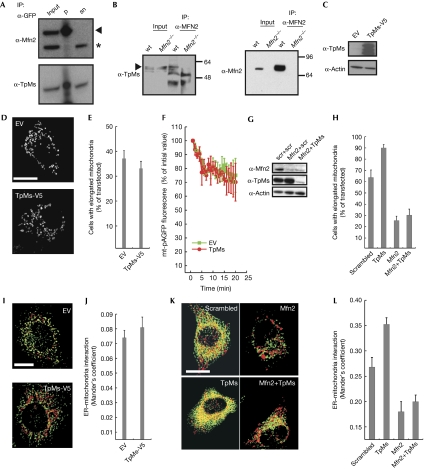
The localization and function of TpMs suggested a possible interaction between it and Mfn2. Mfn2–GFP expressed in HeLa cells pulled down TpMs (Fig 3A) similarly to endogenous Mfn2 in wild type, but not in Mfn2−/− MEFs (Fig 3B). We therefore analysed whether TpMs interacted genetically with Mfn2. TpMs was efficiently expressed in Mfn2−/− MEFs (Fig 3C), but did not further fragment mitochondria (Fig 3D,E) or inhibit mitochondrial fusion, as measured previously (Karbowski et al, 2004; Fig 3F). Efficient silencing of Mfn2 in HeLa cells (Fig 3G) resulted in the expected fragmentation, whereas silencing of TpMs (Fig 3G) alone caused mitochondrial elongation (Fig 3H). When TpMs and Mfn2 were efficiently silenced together (Fig 3G), mitochondrial elongation was not detected (Fig 3H). We then checked whether the effect of TpMs on ER–mitochondria juxtaposition required Mfn2; the expression of TpMs–V5 in Mfn2−/− MEFs did not decrease the already limited tethering any further (Fig 3I,J). In HeLa cells, levels of juxtaposition were increased by silencing TpMs and decreased by ablation of Mfn2 alone or in combination with TpMs (Fig 3K,L). In conclusion, TpMs interacts physically and genetically with Mfn2 to regulate mitochondrial morphology and tethering with the ER.
Figure 3.
Trichoplein/mitostatin physically and functionally interacts with mitofusin 2. (A) Proteins (300 μg) from pre-cleared lysates of HeLa cells transfected with Mfn2–GFP were immunoprecipitated as indicated and analysed by SDS–PAGE and immunoblotting. Input and supernatant are diluted 1:10. (B) Proteins (1 mg) from pre-cleared lysates of wt and Mfn2−/− MEFs were immunoprecipitated as indicated and analysed by SDS–PAGE and immunoblotting. Input and supernatant are diluted 1:10. (C) Proteins (20 μg) from Mfn2−/− MEFs 24 h after transfection with the indicated plasmids were analysed by SDS–PAGE and immunoblotting. (D) Representative confocal images of Mfn2−/− MEFs co-transfected for 24 h with mtRFP and the indicated plasmids. Scale bar, 20 μm. (E) Mean±s.e. (n=3) of morphometric analysis of mitochondrial shape from (D). (F) Mitochondrial fusion assay of Mfn2−/− MEFs co-transfected with mtRFP, mt-pAGFP and the indicated plasmids. Data are mean±s.e. (n=3). (G) HeLa cells were co-transfected with the indicated siRNA and after 48 h cells were lysed and protein (20 μg) was analysed by SDS–PAGE and immunoblotting using the indicated antibodies. (H) Mean±s.e. (n=3) of morphometric analysis of mitochondrial shape from HeLa cells transfected with the indicated siRNA and mtRFP. (I) Three-dimensional reconstructions of ER and mitochondria in Mfn2−/− MEFs co-transfected with mtRFP, ER-YFP and the indicated plasmids. Yellow, organelles are closer than about 270 nm. Scale bar, 20 μm. (J) Mean±s.e. (n=3) of interaction data from (G). (K) Three-dimensional reconstructions of ER and mitochondria in HeLa cells co-transfected with mtRFP, ER-YFP and the indicated siRNA. Yellow indicates that organelles are closer than around 270 nm. Scale bar, 20 μm. (L) Mean±s.e. (n=5) of interaction data from (K). ER, endoplasmic reticulum; ER-RFP, ER-targeted red fluorescent protein; EV, empty vector; GFP, green fluorescent protein; MEFs, mouse embryonic fibroblasts; Mfn2, mitofusin 2; mtRFP, matrix red fluorescent protein; pAGFP, green fluorescent protein; SDS–PAGE, sodium dodecyl sulphate–polyacrylamide gel electrophoresis; siRNA, small interfering RNA; wt, wild type; YFP, yellow fluorescent protein.
TpMs selectively inhibits Ca2+-dependent cell death
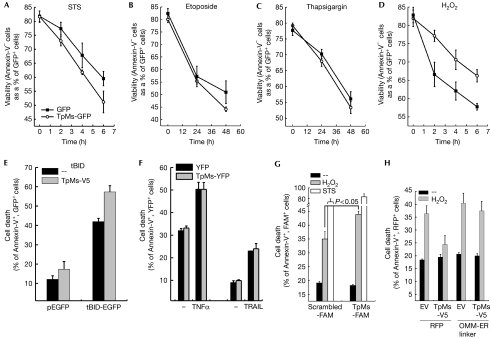
To address whether TpMs affected ER–mitochondria communication, we tested whether it influenced the apoptotic response to death stimuli that require Ca2+ transfer between the two organelles (Scorrano et al, 2003). The expression of TpMs–V5 marginally sensitized HeLa cells to apoptosis by a panoply of intrinsic stimuli (Fig 4A–C,E), whereas it had no effect on the extrinsic stimuli tumour-necrosis-factor-related apoptosis-inducing ligand and tumour necrosis factor-α (Fig 4F). Conversely, it specifically conferred resistance to H2O2, which requires ER–mitochondria Ca2+ transfer to cause death (Scorrano et al, 2003) in HeLa cells (Fig 4D) and MEFs (supplementary Fig S4 online). When TpMs was silenced, death by H2O2, but not by the other stimuli tested, was accordingly increased (Fig 4G). As TpMs fragments mitochondria—a condition that also blunts death due to Ca2+-dependent stimuli (Szabadkai et al, 2004)—we tested whether an artificial tether that increases the juxtaposition between the organelles (Csordas et al, 2006) could revert the effect of TpMs on apoptosis by H2O2. Cells co-expressing TpMs and the artificial tether were no longer resistant to H2O2-induced death (Fig 4H). In conclusion, TpMs selectively inhibits death by stimuli that require Ca2+ transfer from ER to mitochondria.
Figure 4.
Trichoplein/mitostatin protects cells from Ca2+-dependent apoptosis. (A–D) Viability of HeLa cells transfected as indicated and treated for the indicated times with staurosporine (STS, 2 μm, A), etoposide (5 μm, B), thapsigargin (1 μm, C) and H2O2 (1 mM, D). (E) Cell death of HeLa cells 48 h after transfection with the indicated and plasmids. (F) Cell death of HeLa cells transfected as indicated and treated with TNF-α (25 ng/ml plus 10 μg/ml cycloheximide) or TRAIL (50 ng/ml) for 16 h. In (A–F), data are represented as mean±s.e. (n=5). (G) Cell death of HeLa cells transfected as indicated and treated after 48 h with H2O2 or STS for 6 h. Data are represented as mean±s.e. (n=3). (H) Cell death of HeLa cells co-transfected as indicated and treated after 24 h with H2O2 for a duration of 4 h. Data are represented as mean±s.e. (n=6). EGFP, enhanced green fluorescent protein; ER, endoplasmic reticulum; EV, empty vector; FAM, carboxyfluorescein; OMM, outer mitochondrial membrane; RFP, red fluorescent protein; tBID, truncated BID; TNF, tumour necrosis factor; TpMs, trichoplein/mitostatin; TRAIL, tumour-necrosis-factor-related apoptosis-inducing ligand; YFP, yellow fluorescent protein.
Discussion
In this study we identify TpMs as a new protein localized at the mitochondria–ER interface that regulates the interaction between the two organelles. TpMs was originally described as a keratin-binding protein (Nishizawa et al, 2005), but it partly localizes to mitochondria and is downregulated in several tumours (Vecchione et al, 2009). TpMs associates with the OMM, enriched at the points of juxtaposition between mitochondria and ER. The association of TpMs with mitochondria is reduced when plectin 1b, a mitochondrial isoform of the large cytolinker plectin (Winter et al, 2008), is downregulated. Accordingly, plectin 1b is also retrieved in MAMs. It is therefore tempting to speculate that MAMs are linked to the cytoskeleton.
Despite the physiological relevance of the interface between mitochondria and ER (Hayashi et al, 2009), only a few proteins have so far been retrieved at this site. Most of them are chaperones or have an active role in signalling, with the exception of Mfn2, which acts as a structural tether (de Brito & Scorrano, 2008). In this study, we expand this group to include a protein that can interact with the cytoskeleton. However, our data indicate that keratins are dispensable for the function of TpMs in mitochondrial shape and tethering to the ER. Conversely, biochemical and genetic data point to a model in which TpMs interacts with Mfn2 and requires it to modulate mitochondrial morphology and tethering.
TpMs is downregulated in several human tumours (Vecchione et al, 2009) and higher levels of this protein are associated with a slight increase in apoptosis. Why does TpMs sensitize the cell to death? High levels of TpMs do not cause mitochondrial depolarization or latent dysfunction (data not shown). It is possible that it effects could be related to mitochondrial fragmentation, associated with a greater tendency to apoptosis. However, TpMs protects against H2O2 that requires Ca2+ transfer from the ER to mitochondria. This indicates that the role of TpMs is as a negative modulator of ER–mitochondria juxtaposition and extends the evidence supporting the importance of inter-organellar juxtaposition in cell death (Csordas et al, 2006).
In conclusion, TpMs—a cytoskeleton-binding protein that is downregulated in cancer—modulates ER–mitochondria juxtaposition in an Mfn2-dependent manner. This indicates that the functions of Mfn2 can be modulated by the levels of its partners.
Methods
Molecular biology. Mitochondrial red fluorescent protein (mtRFP), mitochondrial cyan fluorescent protein, ER-targeted red fluorescent protein, Mfn2–GFP and ER-targeted yellow fluorescent protein have been described previously (Cipolat et al, 2004; de Brito & Scorrano, 2008). pcDNA-DEST47-TpMs (TpMs–GFP) and pcDNA/V5-DEST-TpMs (TpMs–V5) were generated by standard cloning. Yellow fluorescent protein-microtubule-associated protein 1 light chain 3 was provided by T. Yoshimori (Japan). The following siRNAs were used: TpMs, 5′-GGAGGUGGAGGCGACCAAATT-3′; plectin-1b, 5′-GGGGCAUCGGCAGGCAAAGUU-3′ (Ambion, UK). Carboxyfluorescein-conjugated siRNA TpMs were obtained from Qiagen; final concentration was 100 nM.
Real-time PCR. Total RNA was isolated from HeLa cells by using the QuickPrepTM Kit (Amersham Biosciences). RNA was reverse transcribed using SuperScriptTM III Reverse Transcriptase (Invitrogen) and subsequent reverse transcriptase PCR amplification (Applied Biosystems 7000 Fast System) of a 200-bp fragment of the first exon of mouse plectin 1b was performed using the primers 5′-GAGCAAGATGAACGAGACCGTGTG-3′ and 5′-TCCAGGGCAATCTGCACATTCTGC-3′. As a control, a fragment of mouse β-actin complementary DNA was amplified using the forward primers 5′-CTGGCTCCTAGCACCATGAAGAT-3′ and reverse 5′-GGTGGACAGTGAGGCCAGGAT-3′.
Cell culture. Simian vacuolating virus 40-transformed wild-type and Mfn2−/− MEFs and HeLa cells were cultured and transfected as described previously (de Brito & Scorrano, 2008).
Imaging. Confocal imaging of live cells expressing fluorescent proteins was performed using a Nikon Eclipse TE300 inverted microscope equipped with a spinning-disk PerkinElmer Ultraview LCI as described previously (de Brito & Scorrano, 2008). For immunofluorescence, HeLa cells transfected as indicated were processed, stained with rabbit anti-ciM6PR (serum 8738, 1:200), mouse anti-V5 (1:200, Invitrogen), goat anti-mouse or anti-rabbit fluorescein isothiocyanate- or tetramethyl rhodamine iso-thiocyanate-conjugated IgG (1:500; Invitrogen) and imaged as described previously (de Brito & Scorrano, 2008).
For mitochondrial fusion, 2 × 105 cells—at 24 h after co-transfection with mtRFP, mito-photoactivatable green fluorescent protein (pAGFP) and the indicated constructs—were placed on the stage of a laser scanning microscope (TCS SP5, Leica). Using the LasAF software (Leica), regions of interest were manually defined and pAGFP was photoactivated in one z-plane using the 405-nm laser line (100%) with a × 63, 1.4 numerical aperture objective. Frames of pAGFP and mtRFP fluorescence were acquired at every minute using a 488-nm laser line. Mean pAGFP fluorescence in the photoactivated region was measured using Multi Measure plugin of ImageJ and normalized for mtRFP fluorescence (National Institutes of Health, Bethesda). In each experiment, 40 cells were analysed per condition.
Morphometric and contact analysis. Morphometric analysis of mitochondria and ER was performed with Imagetool 3.0 as described previously (de Brito & Scorrano, 2008). In each experiment, 120 cells were scored per condition. Analysis of mitochondria–ER interaction was performed as described previously (de Brito & Scorrano, 2008). In each experiment, 50 cells were scored per condition. The number of independent experiments is reported in the figure legends. Interaction analysis between the objects was performed using Manders' colocalization coefficient (Manders et al, 1993).
Biochemistry. Whole-cell lysates were prepared by disrupting 106 cells in lysis buffer (0.025 M Tris, (pH 7.4), 0.025 M NaCl, 1% Triton X-100 (Sigma), 0.1% sodium dodecyl sulphate (Sigma) and 0.5 mM ethylene glycol tetraacetic acid) supplemented with complete protease-inhibitor mixture (Sigma). For immunoprecipitation, Protein-G was incubated with mouse GFP antibody (1:15, Invitrogen) or rabbit Mfn2 antibody (1:20; for 3 h at 4°C) and subsequently (overnight at 4°C) with the indicated cell lysates prepared in lysis buffer.
Subcellular fractionation of cells and Percoll purification of MAMs were performed as described previously (Frezza et al, 2007; de Brito & Scorrano, 2008).
The following antibodies were used: GFP (1:200; Invitrogen), actin (1:30,000, Chemicon); MnSOD (1:8,000, Stressgen), TOM20 (1:4,000, Santa Cruz Biotechnology), lactate dehydrogenase (1:2,000, Rockland), V5 (1:1,000, Invitrogen), optic atrophy 1 (1:1,000, BD Biosciences), glucose regulatory protein 75 (1:1,000, Santa Cruz Biotechnology); fatty acyl-CoA ligase 4 (FACL4; 1:1,000, Santa Cruz Biotechnology); calnexin (1:1,000, Stressgen), TpMs (1:100; Vecchione et al, 2009) and Mfn2 (1:1,000, Abnova).
Proteinase K assay and carbonate extraction were performed as described previously (Dimmer et al, 2008).
Cell death. Apoptosis was measured 24 h (unless noted) after transfection of 2 × 105 cells with the indicated plasmids as described previously (Frezza et al, 2006). The number of independent experiments is reported in the figure legend.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
L.S. is a senior Telethon scientist of the Dulbecco-Telethon Institute. This study was supported by Telethon Italy; Associazione Italiana per la Ricerca sul Cancro, Italy; and Swiss National Foundation grant 31-118171, OncoSuisse.
Footnotes
The authors declare that they have no conflict of interest.
References
- Anesti V, Scorrano L (2006) The relationship between mitochondrial shape and function and the cytoskeleton. Biochim Biophys Acta 1757: 692–699 [DOI] [PubMed] [Google Scholar]
- Cipolat S, de Brito OM, Dal Zilio B, Scorrano L (2004) OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc Natl Acad Sci USA 101: 15927–15932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csordas G, Renken C, Varnai P, Walter L, Weaver D, Buttle KF, Balla T, Mannella CA, Hajnoczky G (2006) Structural and functional features and significance of the physical linkage between ER and mitochondria. J Cell Biol 174: 915–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Brito OM, Scorrano L (2008) Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 456: 605–610 [DOI] [PubMed] [Google Scholar]
- Dimmer KS, Navoni F, Casarin A, Trevisson E, Endele S, Winterpacht A, Salviati L, Scorrano L (2008) LETM1, deleted in Wolf Hirschhorn syndrome is required for normal mitochondrial morphology and cellular viability. Hum Mol Genet 17: 201–214 [DOI] [PubMed] [Google Scholar]
- Frezza C et al. (2006) OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell 126: 177–189 [DOI] [PubMed] [Google Scholar]
- Frezza C, Cipolat S, Scorrano L (2007) Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat Protoc 2: 287–295 [DOI] [PubMed] [Google Scholar]
- Fuchs E (1996) The cytoskeleton and disease: genetic disorders of intermediate filaments. Annu Rev Genet 30: 197–231 [DOI] [PubMed] [Google Scholar]
- Hayashi T, Su TP (2007) Sigma-1 receptor chaperones at the ER–mitochondrion interface regulate Ca2+ signaling and cell survival. Cell 131: 596–610 [DOI] [PubMed] [Google Scholar]
- Hayashi T, Rizzuto R, Hajnoczky G, Su TP (2009) MAM: more than just a housekeeper. Trends Cell Biol 19: 81–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M, Arnoult D, Chen H, Chan DC, Smith CL, Youle RJ (2004) Quantitation of mitochondrial dynamics by photolabeling of individual organelles shows that mitochondrial fusion is blocked during the Bax activation phase of apoptosis. J Cell Biol 164: 493–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, Walter P (2009) An ER–mitochondria tethering complex revealed by a synthetic biology screen. Science 325: 477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manders EM, Verbeek FJ, Aten JA (1993) Measurement of co-localisation of objects in dual-colour confocal images. J Microsc 169: 375–382 [DOI] [PubMed] [Google Scholar]
- Nishizawa M, Izawa I, Inoko A, Hayashi Y, Nagata K, Yokoyama T, Usukura J, Inagaki M (2005) Identification of trichoplein, a novel keratin filament-binding protein. J Cell Sci 118: 1081–1090 [DOI] [PubMed] [Google Scholar]
- Scorrano L, Oakes SA, Opferman JT, Cheng EH, Sorcinelli MD, Pozzan T, Korsmeyer SJ (2003) BAX and BAK regulation of endoplasmic reticulum Ca2+: a control point for apoptosis. Science 300: 135–139 [DOI] [PubMed] [Google Scholar]
- Simmen T, Aslan JE, Blagoveshchenskaya AD, Thomas L, Wan L, Xiang Y, Feliciangeli SF, Hung CH, Crump CM, Thomas G (2005) PACS-2 controls endoplasmic reticulum–mitochondria communication and Bid-mediated apoptosis. EMBO J 24: 717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SJ, Vance JE (2000) Phosphatidylserine synthase-1 and -2 are localized to mitochondria-associated membranes. J Biol Chem 275: 34534–34540 [DOI] [PubMed] [Google Scholar]
- Szabadkai G, Simoni AM, Chami M, Wieckowski MR, Youle RJ, Rizzuto R (2004) Drp-1-dependent division of the mitochondrial network blocks intraorganellar Ca2+ waves and protects against Ca2+-mediated apoptosis. Mol Cell 16: 59–68 [DOI] [PubMed] [Google Scholar]
- Szabadkai G, Bianchi K, Varnai P, De SD, Wieckowski MR, Cavagna D, Nagy AI, Balla T, Rizzuto R (2006) Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J Cell Biol 175: 901–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance JE (1990) Phospholipid synthesis in a membrane fraction associated with mitochondria. J Biol Chem 265: 7248–7256 [PubMed] [Google Scholar]
- Vecchione A et al. (2009) MITOSTATIN, a putative tumor suppressor on chromosome 12q24.1, is downregulated in human bladder and breast cancer. Oncogene 28: 257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter L, Abrahamsberg C, Wiche G (2008) Plectin isoform 1b mediates mitochondrion–intermediate filament network linkage and controls organelle shape. J Cell Biol 181: 903–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.