Crystal structure of zinc-finger domain of Nanos and its functional implications
The crystal structure of the RNA-binding domain of Nanos, which adopts a novel zinc-finger structure, reveals that the conserved basic surface is indispensable for the interaction with mRNA.
Keywords: crystal structure, germ cell, Nanos, RNA-binding protein, translational regulation
Abstract
Nanos is an RNA-binding protein that is involved in the development and maintenance of germ cells. In combination with Pumilio, Nanos binds to the 3′ untranslated region of a messenger RNA and represses its translation. Nanos has two conserved Cys-Cys-His-Cys zinc-finger motifs that are indispensable for its function. In this study, we have determined the crystal structure of the zinc-finger domain of zebrafish Nanos, for the first time revealing that Nanos adopts a novel zinc-finger structure. In addition, Nanos has a conserved basic surface that is directly involved in RNA binding. Our results provide the structural basis for further studies to clarify Nanos function.
Introduction
Translational control of messenger RNAs (mRNAs) is crucial in developmental processes including cell division, cell-fate determination and embryonic axis establishment in early embryogenesis. Most types of translational control are mediated by a sequence in the 3′ untranslated region (3′-UTR) and are achieved by the interaction of various regulatory factors such as RNA-binding proteins (Kuersten & Goodwin, 2003). Nanos is a highly conserved RNA-binding protein in higher eukaryotes and functions as a key regulatory protein in translational control using a 3′-UTR during the development and maintenance of germ cells. The nanos gene was first identified as a maternal gene crucial for posterior pattern formation in the Drosophila melanogaster embryo (Lehmann & Nusslein-Volhard, 1991). In combination with Pumilio, Nanos represses the translation of maternal hunchback mRNA in the early Drosophila embryo, thereby governing abdominal segmentation (Murata & Wharton, 1995; Wharton et al, 1998). Nanos and Pumilio also have a variety of functions in the primary germ cells (PGCs). Nanos is essential for the development of PGCs (Kobayashi et al, 1996); PGCs lacking Nanos or Pumilio enter mitosis prematurely, fail to migrate to the somatic gonad, undergo apoptosis and fail to maintain stem cell identity in adults (Lin & Spradling, 1997; Asaoka-Taguchi et al, 1999; Asaoka & Lin, 2004; Hayashi et al, 2004; Wang & Lin, 2004). One of the regulatory targets of Nanos and Pumilio in PGCs is thought to be Cyclin B mRNA (Asaoka-Taguchi et al, 1999), Pumilio and Nanos directly bind to an element in the 3′-UTR to repress its translation (Kadyrova et al, 2007).
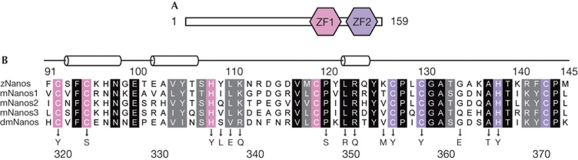
Nanos is widespread in higher eukaryotes and comprises a non-conserved amino-terminus and highly conserved carboxy-terminal regions (Fig 1). The C-terminal region has two conserved Cys-Cys-His-Cys (CCHC)-type zinc-finger motifs (Fig 1A) that are indispensable for Nanos function (Curtis et al, 1997). The motif pattern is Cx2Cx12Hx10Cx7Cx2Cx7Hx4C (‘x' indicates any amino acid; Fig 1B). The CCHC motif of a zinc finger is known to have a zinc knuckle structure, as observed in human immunodeficiency virus (HIV) nucleocapsid protein (NC). Although HIV NC, which is an RNA-binding protein, has two zinc knuckles, the C and H spacing of both tandem zinc knuckle motifs is Cx2Cx4Hx4C (Henderson et al, 1988; Summers et al, 1990; Gitti et al, 1996). Thus, the zinc-finger motif of Nanos is distinct from the symmetric motif and the Nanos zinc-finger domain is expected to adopt a new structure.
Figure 1.
Domain architecture and sequence alignment of Nanos. (A) Domain architecture of zebrafish Nanos composed of 159 amino-acid residues. The two CCHC zinc-finger motifs, ZF1 and ZF2, are indicated. (B) Sequence alignment of the zinc-finger domain of Nanos from zebrafish (z), mouse (m) and Drosophila melanogaster (dm). Identical and homologous residues are highlighted by a black and grey background, respectively. The CCHC motifs in ZF1 and ZF2 are highlighted by a pink and purple background, respectively. The secondary structure of zNanos is also shown above the sequence. Missense mutations causing an abdominal or oogenesis nanos phenotype in Drosophila are shown below the sequence (Arrizabalaga & Lehmann, 1999). CCHC, Cys-Cys-His-Cys; ZF, zinc-finger.
Selective genetic screens have revealed that the CCHC motifs are essential for Drosophila Nanos to function correctly (Arrizabalaga & Lehmann, 1999). The CCHC motifs of D. melanogaster Nanos (dmNanos) have been identified as potential zinc-binding sites (Curtis et al, 1997). It has been reported that zebrafish Nanos (zNanos) has a significant role in both the development of the germ line and oogenesis (Koprunner et al, 2001; Draper et al, 2007). Mammals have several Nanos paralogues; for example, three mouse Nanos (Nanos1, Nanos2 and Nanos3) have been identified. Nanos1 is expressed in the central nervous system (Haraguchi et al, 2003). Nanos2 and Nanos3 are expressed in embryonic germ cells, and a deficiency in these genes results in a loss of germ cells (Tsuda et al, 2003). Although studies have revealed important functions of Nanos, neither the atomic structure of Nanos nor the structural basis of the interaction between Nanos and RNA has been reported. In this study, we present the first, to our knowledge, crystal structure of the zinc-finger domain of zNanos (residues 59–159; zNanos59–159), which includes the two conserved zinc-finger motifs. Our study also reveals that the two CCHC motifs actually bind zinc ions and that the zinc-finger domain of Nanos adopts a novel structure. Furthermore, we reveal a conserved basic surface that is responsible for RNA binding.
Results And Discussion
Structure of Nanos zinc-finger domain
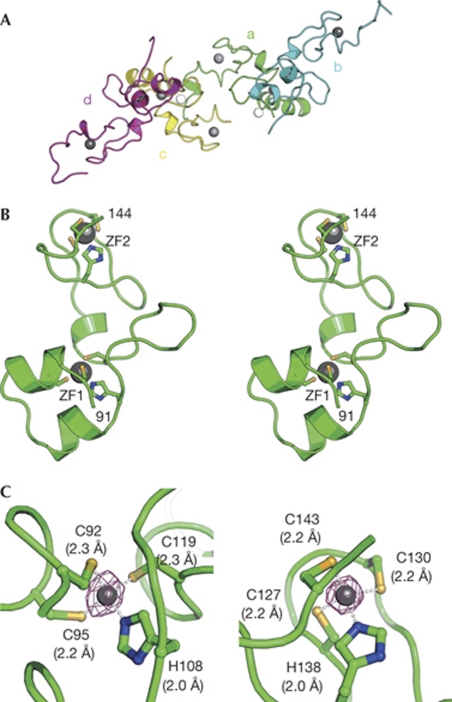
Four zNanos59–159 molecules are present in the crystallographic asymmetric unit (Fig 2A). Although the structures of the N- and C-terminal regions of the four molecules in the asymmetric unit are variable, those of residues 91–144 superimpose well, with an RMSD value of 0.48 Å for the Cα atoms, indicating that the four structures of residues 91–144 are essentially identical and this region is a structural core of zNanos59–159 (supplementary Fig S1A online). We therefore describe the core structure (residues 91–144), hereafter termed zNanos91–144, of the A-molecule as a representative structure. The structure of zNanos91–144 is composed of two independent zinc-finger (ZF) lobes—the N-terminal ZF1 (92–119) and the C-terminal ZF2 (127–143)—which are connected by a linker helix (120–126; Figs 1B,2B). These lobes create a large cleft (Fig 3A,B). Zinc ions in ZF1 and ZF2 are bound to the CCHC motif by tetrahedral coordination (Fig 2C). The CCHC zinc finger of Nanos is unique and has not been observed in other CCHC zinc finger proteins. In fact, a search for homologous whole or partial structures, including ZF1 and ZF2 of zNanos91–144, using the DALI server (Holm et al, 2008) revealed no similar structure, indicating that zNanos91–144 adopts a novel structure. The CCHC-type zinc-finger motif that binds to RNA is known to have a zinc knuckle structure, as found in HIV NC. The zinc knuckle is composed of two ligands from a short β-sheet (knuckle) and two more from a short helix or loop. Thus, the structures of ZF1 and ZF2 of zNanos are distinct from the typical zinc knuckle structure and thereby distinct from that of HIV NC (De Guzman et al, 1998; Matsui et al, 2007).
Figure 2.
Structure of the zebrafish Nanos zinc-finger domain. (A) The four zNanos molecules (a, b, c and d) in the asymmetric unit, shown by green, cyan, yellow and magenta ribbon representations, respectively. Grey and white spheres indicate zinc ions in the CCHC motif and the molecular interface (supplementary Fig S1B online), respectively. (B) Structure of the zNanos zinc-finger domain, shown by a green ribbon representation in stereo view. Only the core region, residues 91–144, is shown. The residue numbers are labelled at the amino- and carboxy-terminal ends, 91 and 144, respectively. Zinc ions in ZF1 and ZF2 are shown by grey spheres. Residues in the CCHC motifs bound to the zinc ion are shown by stick representation. (C) Close-up views of the zinc-finger motifs. Structures of ZF1 and ZF2 are shown in the left and right panels, respectively. The zinc ion is shown by a grey sphere. Residues bound to the zinc ion are shown by stick representation. The purple cage shows the electron density (10 σ) of the zinc ion generated by difference anomalous Fourier calculation using Zn-Peak data at 2.5 Å resolution. Interactions of the zinc ion with the CCHC are shown by white dots and the bond lengths are also given in parentheses. CCHC, Cys-Cys-His-Cys; ZF, zinc-finger; zNanos, zebrafish Nanos.
Figure 3.
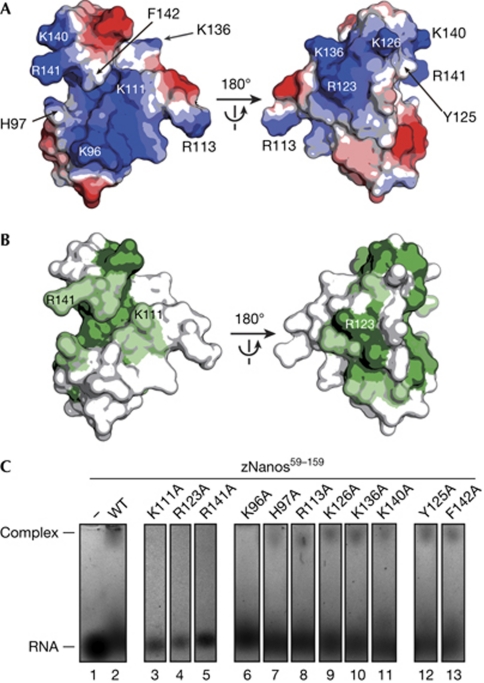
Structural basis of the interaction between zebrafish Nanos and RNA. (A) Electrostatic potential of zNanos91–144 depicted on the molecular surface. Blue and red surfaces indicate positive and negative potential, respectively. The orientation in the left panel is the same as that in Fig 2B. Substituted residues to alanine in EMSA are indicated. (B) Sequence conservation of Nanos shown on the molecular surface of zNanos91–144. Green and light green surfaces indicate identical and homologous residues, respectively. (C) Interaction between zNanos59–159 and ssRNA by EMSA. Lanes 1, 2 and 3–13 are RNA alone, RNA+zNanos59–159 wild-type and RNA+zNanos59–159 mutants, respectively. EMSA, electrophoresis mobility shift assay; ssRNA, single-stranded RNA; WT, wild-type; zNanos, zebrafish Nanos.
zNanos59–159 forms a dimer, and two dimers are present in the asymmetric unit (Fig 2A; supplementary Fig S2A online). To clarify the assembly of Nanos in solution, we performed sedimentation velocity analysis in an ultracentrifugation experiment. This revealed that dimer formation depends on the protein concentration (supplementary Fig S2B online). The buried solvent-accessible surfaces of the AB and CD dimers are 1,514 and 1,483 Å2, respectively. Water molecules are observed throughout the dimer interface (supplementary Fig S2A online) and most of the interactions between the Nanos monomers are formed by water molecules. Taken together with the above results, these observations suggest that the Nanos dimer would not be functional.
Structural basis of interaction between Nanos and RNA
It has been shown that Nanos binds to RNA with no sequence specificity (Curtis et al, 1997), suggesting that electrostatic interactions with the phosphate backbone of RNA are crucial. Consistent with this idea, calculation of electrostatic potential revealed that zNanos91–144 has a large basic region on the surface (Fig 3A) comprising mostly conserved residues (Fig 3B), implying that it is involved in the interaction with RNA. To investigate this, we prepared alanine mutants and tested the interaction between zNanos59–159 and single-stranded RNA (ssRNA) by using electrophoresis mobility shift assay (EMSA). We selected Lys 111, Arg 123 and Arg 141 as conserved basic residues. These correspond to Arg 339, Arg 351 and Lys 369 of dmNanos, respectively (Fig 1B). Lys 96, His 97, Arg 113, Lys 126, Lys 136 and Lys 140 were selected as non-conserved basic residues. These residues contribute to the positive surface potential (Fig 3A). A structural study of TIS11d—which has tandem CCCH zinc fingers and binds to the AU-rich element of mRNA—has revealed that aromatic residues are involved in stacking interactions with the base moieties of ssRNA (Hudson et al, 2004). Stacking interactions between aromatic residues and base moieties have also been observed in single-stranded DNA-binding proteins (Bochkarev et al, 1997; Raghunathan et al, 2000). Thus, we selected Tyr 125 and Phe 142 as conserved and solvent-exposed aromatic residues. In this EMSA, we used a 10-mer ssRNA derived from the 3′-UTR of Cyclin B mRNA (Kadyrova et al, 2007). Initially, we confirmed that wild-type zNanos59–159 binds to this RNA (Fig 3C, lane 2). As expected, Lys 11A, Arg 123A or Arg 141A substitution significantly impaired the interaction between zNanos59–159 and the RNA (Fig 3C, lane 3–5). Interestingly, mutation of Arg 339 or Arg 351 of dmNanos—which corresponds to Lys 111 or Arg 123 of zNanos, respectively—abrogates Nanos function, resulting in two strong abdominal and oogenesis nanos phenotypes (Arrizabalaga & Lehmann, 1999). Thus, our results reveal that those nanos phenotypes are attributable to defects in RNA-binding activity. Although mutation in Lys 369 of dmNanos—which corresponds to Arg 141 of zNanos—has not been reported in genetic studies, our results suggest that this mutation causes a strong nanos phenotype owing to defects in RNA-binding activity. By contrast, zNanos59–159 with a mutation in a non-conserved basic residue maintained its interaction with RNA, except for the K96A mutant (Fig 3C, lane 6–11). Furthermore, Y125A and F142A mutants also retained binding activity. These results indicate that the electrostatic interactions between the basic residues and the phosphate backbone of RNA are crucial in formation of the Nanos–RNA complex. This contrasts with the situation for TIS11d or single-stranded DNA-binding proteins; Lys 96, Lys 111 and Arg 141 are in close proximity and provide a large basic surface (Fig 3A, left panel), implying that this could be a major RNA-binding site. Furthermore, Arg 123—which is located on the backside of Lys 96, Lys 111 and Arg 141—also contributes to RNA binding.
Sequence preference in Nanos binding to 3′-UTR of Cyclin B mRNA has been demonstrated (Kadyrova et al, 2007). Thus, we performed EMSA using RNA lacking the preferential sequence (supplementary Fig S3A online). The assays showed that zNanos59–159 binds to RNA with no sequence specificity. Furthermore, we tested whether the RNA-binding domain (residues 822–1206) of zebrafish Pumilio (zPumilio822–1206) affects the interaction between zNanos59–159 and RNA (supplementary Fig S3B online). We found that zNanos59–159 binds to RNA lacking the preferential sequence in the presence of zPumilio822–1206, suggesting that zPumilio822–1206 does not affect RNA binding by zNanos59–159.
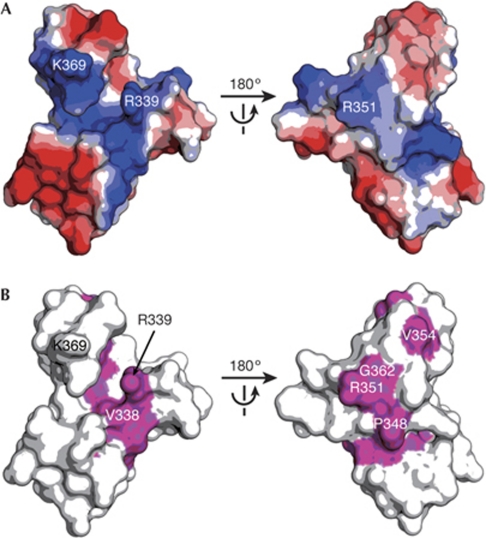
Missense mutations in the zinc-finger domain causing a strong abdominal or oogenesis nanos phenotype have been reported in Drosophila (Fig 1B; Arrizabalaga & Lehmann, 1999). To investigate the structure of those mutations causing a deficiency in Nanos function, we built a homology model of the zinc-finger domain (residues 319–372) of dmNanos (dmNanos319–372; Fig 4). The homology model shows that dmNanos319–372 also has a large basic surface, and the distribution of positive potential is similar to that of zNanos91–144 (Figs 3A,4A). Mapping the missense mutations that cause the nanos phenotype in Drosophila on the molecular surface of dmNanos319–372 revealed the mechanism of dysfunction (Fig 4). Mutations in the CCHC motif (C320Y, C323S, H336Y, C355Y, C358Y and H366Y) are likely to destabilize the structure. In addition, the mutations P348S, L350R, V354M and A365T are likely to affect maintenance of the structure, because Leu 350, Val 354 and Ala 365 are buried in the interior. The EMSA results showed that Lys 111 and Arg 123 of zNanos contribute to the ionic interactions with RNA (Fig 3C). Thus, mutations in the Drosophila counterparts, R339Q and R351Q, might change the charge distribution of the molecular surface. Furthermore, the V338E and G362E mutations might also change the basic properties of the surface, because Val 338 and Gly 362 are positioned adjacent to Arg 339 and Arg 351 (Fig 4B).
Figure 4.
Homology model of the Drosophila Nanos zinc-finger domain. (A) Electrostatic potential of dmNanos319–372 depicted on the molecular surface. Blue and red surfaces indicate positive and negative potential, respectively. (B) Missense mutations causing a strong phenotype in D. melanogaster, shown in magenta on the molecular surface of dmNanos319–372. dmNanos, Drosophila melanogaster Nanos.
In this study, we have determined the first, to our knowledge, crystal structure of the Nanos zinc-finger domain, which adopts a novel structure. We have demonstrated the structural basis of the interaction between Nanos and RNA, whereby Nanos binds to ssRNA with no sequence specificity and the electrostatic interactions made by the conserved basic residues of Nanos function predominantly in RNA binding. Furthermore, our structure elucidates the effects of mutations that cause the nanos phenotype in Drosophila at an atomic level. Our results provide the structural basis for further studies to clarify Nanos function. However, the detailed interaction between Nanos and RNA, and the mechanism by which it functionally collaborates with Pumilio in 3′-UTR-mediated translational repression remains unclear. To address these issues, further structural studies of Nanos in complex with RNA and/or Pumilo will be required.
Methods
Structure determination. The purification and crystallization of zNanos59–159 has been described previously (Hashimoto et al, 2009). X-ray data were collected on beamline BL5A with a Quantum 315 charge-coupled device detector (Area Detector Systems Corporation) at the Tsukuba Photon Factory. All diffraction data were processed with the program HKL2000 (Otwinowski & Minor, 1997). The crystal structure of zNanos59–159 was determined by using a single anomalous diffraction method using intrinsic zinc atoms with the programs SOLVE (Terwilliger & Berendzen, 1999) and RESOLVE (Terwilliger, 2000). Model building and fitting were carried out with the programs O (Jones et al, 1991) and COOT (Emsley & Cowtan, 2004). The structure was refined with the programs CNS (Brunger et al, 1998) and REFMAC (Murshudov et al, 1997), and validated with the program PROCHECK (Laskowski et al, 1993). Crystallographic statistics are given in Table 1. Secondary structures are defined by the program PyMOL. The final coordinates and the structure factors have been deposited in the Protein Data Bank Japan (PDBj; 3ALR). The homology model of dmNanos319–372 was built by the program MODELLER (Marti-Renom et al, 2000) using the zNanos91–144 structure.
Table 1. Data collection, phasing and refinement statistics.
| Zn-Peak (λ=1.2818 Å) | Native (λ=1.0000 Å) | |
|---|---|---|
| Crystallographic data | ||
| Space group | P63 | P63 |
| Cell dimensions | ||
| a=b (Å) | 100.96 | 100.96 |
| c (Å) | 71.54 | 71.57 |
| γ (°) | 120 | 120 |
| Resolution range (Å) | 20.00–2.50 (2.59–2.50) | 50.00–2.10 (2.18–2.10) |
| Observations | 56,941 | 244,320 |
| Unique | 13,866 | 23,796 |
| Rmerge (%) | 0.079 (0.154) | 0.082 (0.300) |
| Completeness (%) | 95.9 (70.8) | 97.7 (80.5) |
| 〈I〉/σ〈I〉 | 11.8 (6.5) | 15.7 (5.1) |
| Phasing statistics | ||
| Resolution range (Å) | 20.00–3.00 | |
| 〈m〉/Z-score from SOLVE | 0.32/28.15 | |
| Heavy atoms found | 10 | |
| Refinement statistics | ||
| Resolution range (Å) | 20.00–2.10 | |
| Protein atoms | 1,985 | |
| Zinc ions | 10 | |
| Water molecules | 109 | |
| R/Rfree | 0.165/0.208 | |
| RMSD bond distances (Å)/angles (°) | 0.016/1.476 | |
| Ramachandran statistics | 92.1%/7.9%/0% | |
| Mean B value (Å2) | 46.52 | |
| Protein Data Bank ID | 3ALR | |
| Values in parentheses are for the highest resolution shell. Ramachandran statistics indicate the fraction of residues in the most favoured, allowed and disallowed regions, respectively. | ||
RNA-binding assay. To perform EMSA, an alanine mutation was introduced into zNanos59–159 by using QuikChange (Stratagene). The glutathione S-transferase-fused zNanos59–159 mutant was overexpressed, purified with GS4B resin (GE Healthcare) and digested with HRV3C protease (Hashimoto et al, 2009). The reaction mixture was applied to a HiTrap Q column (GE Healthcare) equilibrated with 50 mM Tris–HCl (pH 9.0) and 150 mM NaCl. The mutant was collected in a flow-through fraction and concentrated in a storage buffer containing 50 mM HEPES–NaOH (pH 7.4), 100 mM NaCl and 10% glycerol. A 10-mer of ssRNA, 5′-GACUAUUUGU-3′, was used in this assay (supplementary Table S1 online). RNA and zNanos59–159 were mixed in storage buffer and incubated for 1 h on ice. The concentrations of RNA and zNanos59–159 in the mixed solution were 13.3 and 26.7 μM, respectively. The solutions were separated by electrophoresis at 4 °C on a 1% agarose gel at 100 V for 45 min in a conventional Tris–acetate–ethylenediaminetetra acetic acid buffer. The gel was stained with SYBR Gold (Invitrogen), and bands were detected using an LAS4000 charge-coupled device image analyser with a blue-light transilluminator (GE Healthcare).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank the beamline staff of Photon Factory for their kind support. This study was supported by grants from KAKENHI, the Protein 3000 Project and the Target Protein Research Programs to M.S., T.S. and H.H. from the Japanese Ministry of Education, Culture, Sports, Science and Technology.
Footnotes
The authors declare that they have no conflict of interest.
References
- Arrizabalaga G, Lehmann R (1999) A selective screen reveals discrete functional domains in Drosophila Nanos. Genetics 153: 1825–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asaoka M, Lin H (2004) Germline stem cells in the Drosophila ovary descend from pole cells in the anterior region of the embryonic gonad. Development 131: 5079–5089 [DOI] [PubMed] [Google Scholar]
- Asaoka-Taguchi M, Yamada M, Nakamura A, Hanyu K, Kobayashi S (1999) Maternal Pumilio acts together with Nanos in germline development in Drosophila embryos. Nat Cell Biol 1: 431–437 [DOI] [PubMed] [Google Scholar]
- Bochkarev A, Pfuetzner RA, Edwards AM, Frappier L (1997) Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature 385: 176–181 [DOI] [PubMed] [Google Scholar]
- Brunger AT et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Curtis D, Treiber DK, Tao F, Zamore PD, Williamson JR, Lehmann R (1997) A CCHC metal-binding domain in Nanos is essential for translational regulation. EMBO J 16: 834–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF (1998) Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science 279: 384–388 [DOI] [PubMed] [Google Scholar]
- Draper BW, McCallum CM, Moens CB (2007) Nanos1 is required to maintain oocyte production in adult zebrafish. Dev Biol 305: 589–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Gitti RK, Lee BM, Walker J, Summers MF, Yoo S, Sundquist WI (1996) Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science 273: 231–235 [DOI] [PubMed] [Google Scholar]
- Haraguchi S, Tsuda M, Kitajima S, Sasaoka Y, Nomura-Kitabayashi A, Kurokawa K, Saga Y (2003) Nanos1: a mouse nanos gene expressed in the central nervous system is dispensable for normal development. Mech Dev 120: 721–731 [DOI] [PubMed] [Google Scholar]
- Hashimoto H, Kawaguchi S, Hara K, Nakamura K, Shimizu T, Tamaru Y, Sato M (2009) Purification, crystallization and initial X-ray diffraction study of the zinc-finger domain of zebrafish Nanos. Acta Crystallogr Sect F Struct Biol Cryst Commun 65: 959–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Hayashi M, Kobayashi S (2004) Nanos suppresses somatic cell fate in Drosophila germ line. Proc Natl Acad Sci USA 101: 10338–10342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson LE, Sowder RC, Copeland TD, Benveniste RE, Oroszlan S (1988) Isolation and characterization of a novel protein (X-ORF product) from SIV and HIV-2. Science 241: 199–201 [DOI] [PubMed] [Google Scholar]
- Holm L, Kaariainen S, Rosenstrom P, Schenkel A (2008) Searching protein structure databases with DaliLite v.3. Bioinformatics 24: 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE (2004) Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat Struct Mol Biol 11: 257–264 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Kadyrova LY, Habara Y, Lee TH, Wharton RP (2007) Translational control of maternal Cyclin B mRNA by Nanos in the Drosophila germline. Development 134: 1519–1527 [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Yamada M, Asaoka M, Kitamura T (1996) Essential role of the posterior morphogen nanos for germline development in Drosophila. Nature 380: 708–711 [DOI] [PubMed] [Google Scholar]
- Koprunner M, Thisse C, Thisse B, Raz E (2001) A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev 15: 2877–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuersten S, Goodwin EB (2003) The power of the 3′ UTR: translational control and development. Nat Rev Genet 4: 626–637 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26: 283–291 [Google Scholar]
- Lehmann R, Nusslein-Volhard C (1991) The maternal gene nanos has a central role in posterior pattern formation of the Drosophila embryo. Development 112: 679–691 [DOI] [PubMed] [Google Scholar]
- Lin H, Spradling AC (1997) A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development 124: 2463–2476 [DOI] [PubMed] [Google Scholar]
- Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A (2000) Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct 29: 291–325 [DOI] [PubMed] [Google Scholar]
- Matsui T, Kodera Y, Endoh H, Miyauchi E, Komatsu H, Sato K, Tanaka T, Kohno T, Maeda T (2007) RNA recognition mechanism of the minimal active domain of the human immunodeficiency virus type-2 nucleocapsid protein. J Biochem 141: 269–277 [DOI] [PubMed] [Google Scholar]
- Murata Y, Wharton RP (1995) Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell 80: 747–756 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in the oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Raghunathan S, Kozlov AG, Lohman TM, Waksman G (2000) Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat Struct Biol 7: 648–652 [DOI] [PubMed] [Google Scholar]
- Summers MF, South TL, Kim B, Hare DR (1990) High-resolution structure of an HIV zinc fingerlike domain via a new NMR-based distance geometry approach. Biochemistry 29: 329–340 [DOI] [PubMed] [Google Scholar]
- Terwilliger TC (2000) Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr 56: 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J (1999) Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr 55: 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda M, Sasaoka Y, Kiso M, Abe K, Haraguchi S, Kobayashi S, Saga Y (2003) Conserved role of nanos proteins in germ cell development. Science 301: 1239–1241 [DOI] [PubMed] [Google Scholar]
- Wang Z, Lin H (2004) Nanos maintains germline stem cell self-renewal by preventing differentiation. Science 303: 2016–2019 [DOI] [PubMed] [Google Scholar]
- Wharton RP, Sonoda J, Lee T, Patterson M, Murata Y (1998) The Pumilio RNA-binding domain is also a translational regulator. Mol Cell 1: 863–872 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.