Abstract
The history of p53 and the various interpretations of its function in cells serves as an example of how scientific paradigms can influence research fields. It also offers some interesting lessons for young scientists in search of a rewarding research topic.
The p53 gene has spent 30 years oscillating chaotically between classification as an oncogene and as a tumour suppressor. The uncertainty about its role is not owing to the incidental cloning of a mutant p53, but has been influenced by the various paradigms that have been popular during that time. The identification of both oncogenes and, shortly thereafter, tumour suppressor genes were breakthroughs that revolutionized our understanding of cancer and consequently influenced our perception of p53 and its function. The thirtieth anniversary of the discovery of p53 has been marked by the publication of several reviews about its complexity and function. In this Outlook, I discuss the history of p53 research from a more personal and epistemological point of view.
Following the model of scientific revolutions proposed by the American physicist Thomas Kuhn (1962), I analyse the effect that various paradigms have had on our understanding of p53 and how they might have unintentionally (mis)directed the approach of scientists studying the gene. I highlight lessons that can be drawn from the history of p53; both for young scientists in search of a research topic, and for experienced researchers faced with an unorthodox grant application.
My scientific career started in a laboratory adjacent to that of Pierre May—one of the discoverers of p53—at the Cancer Research Institute in Villejuif, France. Our institute had chosen to study cancer in the context of two related virus models: SV40 and polyoma virus. It is important to put this into the historical context of 1977; the viral theory of cancer was popular and the association between viral and cellular oncogenes was only beginning to emerge. Several other DNA tumour viruses such as human papilloma virus (HPV) and Epstein–Barr virus were already strongly associated with human neoplasia. In vitro and in vivo experiments had shown that both SV40 and polyoma viruses could transform cells or induce tumours in animals. The similarities between polyoma virus and SV40 allowed us to work in parallel on these viruses, and findings in one were rapidly confirmed in the other.
The history of the search for the mt protein in SV40 shows how our desire to classify, define and model can be detrimental
I vividly remember a postdoc bursting into our laboratory to announce that his friend, Michel Kress, had identified the middle T (mt) antigen of the SV40 virus: it was a 53-kDa protein. In view of my somewhat blank response, it was explained to me in learned terms that this was an important finding. It was known that the polyoma virus expressed three early antigens associated with cell transformation: the large (90 kDa), small (17 kDa) and mt (55 kDa) antigens. Therefore, the search for the mt antigen of SV40 was an important subject. However, in the 1980s it became clear that the SV40 mt antigen does not exist. In fact, Michel Kress found that the virus does not code the protein; the host cell does. He also demonstrated that the protein accumulates in the nucleus of tumour cells and associates with the T antigen of SV40. The publication of these results, and other reports by Arnold Levine and Lionel Crawford, created the field of p53 research (Kress et al, 1979; Lane & Crawford, 1979; Linzer & Levine, 1979).
History mainly remembers the virological approach that led to the discovery of p53. However, Lloyd Old's team at the Ludwig Institute for Cancer Research in New York, USA took an immunological approach and published their findings in the same year (DeLeo et al, 1979). Old showed that the humoral response of mice to some induced tumour cells was directed against a 53-kDa protein. His team found that in animals, SV40 tumours elicited an immune response specific to this protein. Crawford also described antibodies in human serum that target the 53-kDa protein in 9% of breast cancer patients. This immunological research was just as original, but the compartmentalization of research meant that scientists did not immediately recognize the link between the two sets of observations (DeLeo et al, 1979). Several years later, it was established that the protein was the same and it was imaginatively baptized p53 (Crawford, 1983).
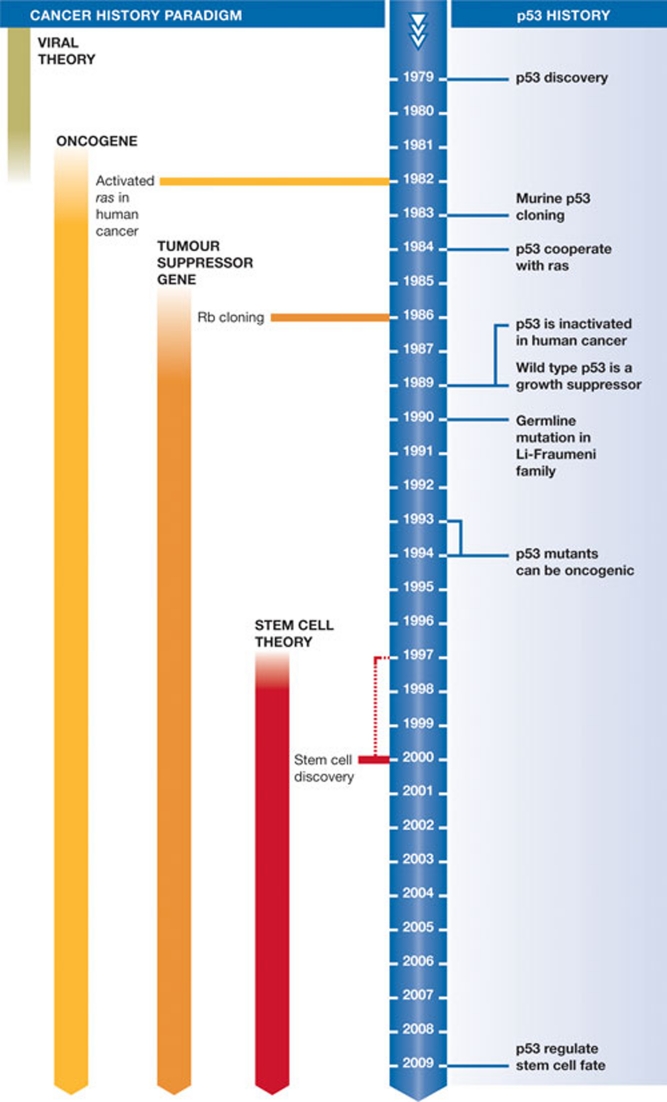
The history of the search for the mt protein in SV40 shows how our desire to classify, define and model can be detrimental. I do not discuss the relative strengths of discovery-driven science and hypothesis-driven science, but it is clear that the history of p53 research has been shaped by the different paradigms that have dominated the study of cell transformation (Fig 1).
Figure 1.
A brief timeline of p53 research and key discoveries.
When Michel Kress left for a postdoctoral fellowship in 1981 he was not allowed to continue wasting his time working on a molecule of no interest and had to change his research subject. I joined Pierre May's laboratory in 1983. A common problem for French university research is the teaching workload for young assistants, which was 180 hours a year at that time. I therefore chose to work on p53 because it was a calm, non-competitive field—about ten teams were working on it—and I thought it would allow me to balance my research and teaching responsibilities. Choosing p53 at that time did not follow the general advice for choosing a research topic, which encourages scientists to pick an area based on the interest of the outcome or identify an under-occupied niche with potential for future development.
However, from 1984–1988, the world of oncology underwent a revolution, triggered by the cloning and analysis of the first cellular oncogenes, as well as the discovery of ras mutations and their dominant role in cell transformation (Land et al, 1983). At the same time molecular biology developed new methodological approaches, some of which dominate modern molecular oncology. The rediscovery of the transfection technique, developed in 1973, enabled the discovery of oncogenes and it is one of the most widely used methodologies in biology (Weinberg, 1981).
Finally, it was also at this time that the viral theory of cancer—the ‘enemy from outside'—was abandoned in favour of the ‘enemy from within' theory: the activation of oncogenes (Cairns, 1981). As with any revolution of ideas, the story of p53 is linked to several intellectual and methodological dogmas. The cDNA of murine and human p53 were cloned in 1982 and 1983 and, in 1984, three articles published in Nature showed that transfection of p53 could cooperate with another oncogene to transform cells (Eliyahu et al, 1984; Jenkins et al, 1984; Parada et al, 1984). It therefore seemed reasonable to classify p53 as an oncogene, as this explained various observations including its accumulation in tumour cells and the fact that p53 knockout induced the arrest of cell proliferation. This shows how the oncogene-induced cancer paradigm influenced the initial view of p53.
The euphoria in 1984 was short-lived; from 1985–1988, studies of p53 began to encounter difficulties and the 131 articles published on the topic during this period did not integrate p53 into the family of oncogenes. Among these studies was the finding that both p53 alleles were inactivated in murine erythroleukaemia induced by the Friend virus (Mowat et al, 1985). Another ‘strange' observation was the high frequency of p53 gene rearrangements and deletions in human and murine osteosarcoma (Masuda et al, 1987; Chandar et al, 1992). As a result of these findings, p53 again declined in popularity. When one of my colleagues presented her thesis on p53 at the end of 1987, one member of the examination board—a prominent scientist at the INSERM (National Institute of Health and Medical Research; the French equivalent of the NIH)—told her that her scientific future would require a change of research topic to a more serious and better funded subject.
When I began my postdoctoral fellowship on p53 with Pierre May in 1983, he asked me to continue to study the oncogenic properties of p53. I was not very enthusiastic; not because I had doubts about this programme, but because I was more interested in a different system. One of my colleagues had just returned from a postdoctoral fellowship with John Gurdon and Ronald Laskey—specialists on the Xenopus laevis model organism. The study of p53 in an organism so distant from humans stimulated my imagination, as I had always been fascinated by evolution. We convinced Pierre to pursue this idea, and rapidly cloned the X. laevis p53 cDNA and gene. We also added other species to the list—trout, rat and chicken—which enabled us to propose a structural diagram of the p53 protein that was confirmed several years later by functional and crystallographic studies (Soussi et al, 1987, 1990).
During years of plenty, adventurous research topics and research that is off the beaten track have a chance of obtaining adequate funding
The other outcome of this phylogenetic study was its contribution to the transition of p53 from classification as an oncogene to a tumour suppressor. Several studies had suggested that p53 had a more complicated role than a classic oncogene (Lane & Benchimol, 1990). First, the p53 gene could be inactivated in several models of human or murine cell transformation (Mowat et al, 1985). A second study focussed on the heterogeneity of the murine and human clones used for cell transformation experiments. Our phylogenetic study of p53 had demonstrated five highly preserved domains during evolution. Examination of murine and human cDNA showed differences affecting one or two amino acids that were attributed to negligible polymorphisms. However, these differences were located in highly preserved domains, which was not compatible with evolutionary logic. All of the mammalian cDNAs had been cloned from transformed cells, whereas those used for our phylogenetic studies were taken from healthy animal tissues.
Levine's team were the first to clone nonmutant murine cDNA and show that it was devoid of oncogenic activity (Finlay et al, 1989). The simultaneous publication of the anti-proliferative property of wild-type p53 and the demonstration of inactivating mutations in human cancers eventually changed the status of p53 to a tumour suppressor gene (Baker et al, 1989; Takahashi et al, 1989). Once again, this occurred in parallel with a change in the popular paradigm in cell transformation research after tumour suppressor genes had entered the stage (Knudson, 1971; Friend et al, 1986). Indeed, the use of DNA tumour viruses was essential not only for the study of p53, but also for the study of another tumour suppressor gene, retinoblastoma protein. The observation that both cellular gene products were systematically inactivated during lytic infection through an interaction with viral proteins was a key finding for our understanding of their functions. 1989 was therefore a turning point in the history of p53 for its classification and the extent of research interest in it. In 1992 alone, 413 articles were published on p53—many more than the 250 articles that had been published during the previous decade, though less than the 1,600 articles that would be published in 2000.
Modern literature can be used to reinterpret old experiments in a new scientific framework, with the benefit of hindsight; p53 is an excellent case in point of this. An article published in 1984 showed that UV irradiation of normal cells induced the accumulation of nuclear p53 (Maltzman & Czyzyk, 1984). As the concept of the DNA damage checkpoint was not yet popular, the results of this experiment were not interpreted correctly at the time. Eight years later, Kastan and co-workers confirmed that p53 controls the cell cycle after DNA damage (Kuerbitz et al, 1992). Similarly, although Crawford and coworkers discovered that breast cancer patients developed p53 antibodies in 1982, it was 10 years before it was demonstrated that this was caused by an autoimmune reaction to overexpressed mutant p53 in tumours (Crawford et al, 1982; Lubin et al, 1995; Soussi, 1996).
Studies have recently shown that p53 can interact with RNA (Riley et al, 2007), but again, this is an old story. Caron de Fromentel and colleagues demonstrated the colocalization of p53 and heterogeneous nuclear RNA by electron microscopy as early as 1986 (Caron de Fromentel et al, 1986). There is no satisfactory explanation of the function of p53 as an RNA-binding protein yet, but this previous work will be useful when such an explanation is found and researchers can put this information together.
The understanding of the complexity of the p53 gene in humans has also been furthered by the reinterpretation of previous research. For almost 15 years, the gene was thought to have one transcript and one isoform of 393 amino acid residues. The discovery in 2005 of multiple transcripts coding for various isoforms overthrew this notion (Bourdon et al, 2005). Various publications in 1987 and 1996, however, had already suggested the presence of alternative splicing (Matlashewski et al, 1987; Flaman et al, 1996). It was only after p63 and p73 had been cloned in 1997 and their multiple isoforms had been demonstrated that the extent of the complexity of p53 was recognized.
Consciously or unconsciously, we self-censor our grant applications to be compatible with current paradigms
The benefits of re-reading literature are not limited to any particular field. When Bernard Dutrillaux published the karyotypic observation of recurrent loss of the short arm of chromosome 17 in colon cancer in 1985, it was impossible to predict that his finding also described the loss of heterozygosity of the p53 gene, which was demonstrated several years later (Muleris et al, 1985; Vogelstein et al, 1989). The process of accumulating information over many years might seem unappealing or difficult, especially as many original findings were not published in high-impact factor journals.
What lessons can we learn from this story? The first concerns scientific choices. No-one can predict the long-term impact of a scientific discovery. This works in both directions: one issue of a medical journal focusing on a subject that, like a shooting star, made a spectacular entry to the scientific stage and then rapidly disappeared would not be an accurate representation of the importance of the subject. By contrast, subjects such as p53 had humble beginnings before they became widely, and possibly excessively, covered in the scientific literature. One of the main problems in research is the arbitrary approach to defining ‘good' science and science that is ‘a waste of time'. Who could have predicted that the study of rare diseases such as Fanconi's anaemia would reveal the role of BRCA2 in breast cancer? Who could have imagined that a protein as trivial as cytochrome C is crucial in apoptosis?
A crucial factor in scientific choices is the availability of research grants. During years of plenty, adventurous topics and research that is ‘off the beaten track' have a chance of obtaining adequate funding. Unfortunately, in this period of austerity, funds have dwindled and profitability-based logic imposed by politicians has modified the behaviour of scientists. Consciously or unconsciously, we self-censor our grant applications to be compatible with current paradigms. This has been demonstrated clearly in the field of tumour virology. During the 1930s and 1940s, John Bittner could not call mouse mammary tumour virus (MMTV) a virus, because he would not have been able to get funding for his work. Instead, he called it the milk factor; emphasizing genetics and downplaying the potential role of viruses in cancer pathogenesis made his work respectable, and therefore fundable. If early p53 research had taken place in the current, highly competitive economic climate, the topic would not have found many supporters.
Another negative influence that is more difficult to quantify is the filter of peer review. This is also influenced by the popular ideas of the day, which makes publishing data that is contradictory to them difficult. Paradigm-driven selection is not a trivial problem, as it determines both the topics we choose to research and our grant applications; something which has been noticed outside the scientific microcosm. The New York Times published a somewhat melodramatic headline: ‘Grant system leads cancer researchers to play it safe', and linked our safe behaviour to a lack of innovation in the field of cancer therapy (Kolata, 2009). Although this might be an overstatement, it is true that governmental agencies tend to fund conservative projects that are supported by an abundance of preliminary results.
Our scientific reductionism has also affected funding agencies and research institutions, by limiting research to clearly defined fields
Those who sit on commissions know that atypical grant applications are often politely put aside. Our scientific reductionism has also affected funding agencies and research institutions, by limiting research to clearly defined fields. This situation must change. Research shows that no signalling pathway or research topic is limited to one particular field. There are many examples of overlap between seemingly distinct areas, something which, again, p53 typifies. Although research on p53 was initially focused on cancer, it has led to the discovery of a multigene family that includes p63 and p73. These have been implicated in genetic dysmorphic diseases and neurodegenerative diseases, respectively (McKeon & Melino, 2007). If the first member of this family to be identified was p63, for its alterations in ectrodactyly, ectodermal dysplasia syndrome including ectrodactyly, ectodermal dysplasia and cleft lip/palate (van Bokhoven & McKeon, 2002), to which field would research have been confined? How would the subsequent discoveries of p53 and p73 and their analyses have been interpreted?
Over more than 30 years of molecular biology teaching, many dogmas have crumbled: the universal nature of the genetic code, the collinearity of genes, the share of epigenetic phenomena in the regulation of transcription, the discovery of prions and the RNA regulator revolution. In his essay on the structure of scientific revolutions, Kuhn suggested that research is discontinuous and punctuated by revolutions that are linked to paradigm shifts (Kuhn, 1970). These revolutions can have a profound impact on the entire discipline—for example the Darwinian revolution—or they can influence only a specific field. The discovery of retroviruses and their ability to induce tumours, the demonstration of reverse transcription, and the identification of viral oncogene sequences led to the emergence of the ‘virus and cancer' paradigm, which formed the basis of US President Richard Nixon's ‘war on cancer' in 1971. The discovery of cellular oncogenes in the 1980s led to a major paradigm shift with its own series of excesses, as we have seen in the case of p53. The discovery of tumour suppressor genes and DNA repair genes, although not constituting a major revolution, have introduced flexibility to this paradigm, to integrate ‘orphan' concepts. Finally, even though the viral theory of cancer does not apply to all human cancers, it led to the discovery of human oncogenes and tumour suppressor genes, the development of several vaccines—including those against human papilloma virus and hepatitis B virus—and a Nobel Prize for Harald zur Hausen for his work on human papilloma virus and cervical cancer.
With the benefit of hindsight, can we discern whether we missed a trick with p53? Would it have been possible to predict the changes in the role of p53? A review of the literature reveals that there were signs that p53 was not a conventional oncogene, although these were often disregarded at the time and interpretted as exceptions to particular models. It is important to remember that the current classification of p53 as a tumour suppressor gene is not conclusive. Wild-type p53 acts as a negative regulator of cell proliferation, but mutant p53 must be considered to be an oncogene with additional functions, which can vary from one mutant to another.
Scientific revolutions in the future might radically change biology again, hopefully then we will be able to solve this puzzle and discover the true role of p53
Although p53 was discovered in oncology research, recent data show that its natural role is much broader. Ten-thousand years ago, human life expectancy was about 30 years, which meant that people usually died before they were at high-risk of cancer. In the absence of cancer-derived selection pressure, the signalling roles of the p53 family pathways had already evolved, indicating that the ‘tumour suppressor' function was not the primary activity of p53, but rather that cancer is a disastrous consequence of its deregulation. Conversely, p53 preserves its function as the ‘guardian of the genome' by monitoring all types of stress from DNA damage to hypoxic metabolism or differentiation. Recent studies show that p53 could also be an important player in female fertility, development, stem-cell division and ageing (Danilova et al, 2008; Hu et al, 2008; Puzio-Kuter & Levine, 2009; Shi et al, 2009). Clinical studies published several years ago suggested that a polymorphism in Arg 72 of p53 is linked to recurrent implantation failure in women undergoing in vitro fertilization treatment and embryo transfer (Kay et al, 2006). It has also been demonstrated that p53 regulates the expression of leukaemia inhibitory factor, a gene that plays a crucial role in blastocyst implantation (Hu et al, 2007). These two lines of evidence, as well as the observation that p53-deficient mice have reproductive problems, are encouraging for the development of a new field of investigation that could reconcile many observations from across the literature.
Elucidating the true function of wild-type p53 will require better understanding of the activity of the various isoforms of p53, how they are inter-regulated with the p63 and p73 signalling pathways and how this network is integrated into cellular homeostasis. Scientific revolutions in the future might radically change biology again, hopefully then we will be able to solve this puzzle and discover the true role of p53. I predict that the past 30 years has been the beginning of the age of reason and that p53 will have many birthdays before reaching full maturity.

Thierry Soussi
Acknowledgments
This article is dedicated to Pierre May, in memoriam. I am grateful to G. Klein, G. Selinova and K. Wiman for all of our regular discussions. Special thanks to G. Klein for critical reading of this article and showing that intellect is an unlimited force, driven by curiosity and a deep thirst for knowledge.
Footnotes
The author declares that he has no conflict of interest.
References
- Baker SJ et al. (1989) Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science 244: 217–221 [DOI] [PubMed] [Google Scholar]
- Bourdon JC, Fernandes K, Murray-Zmijewski F, Liu G, Diot A, Xirodimas DP, Saville MK, Lane DP (2005) p53 isoforms can regulate p53 transcriptional activity. Genes Dev 19: 2122–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns J (1981) The origin of human cancers. Nature 289: 353–357 [DOI] [PubMed] [Google Scholar]
- Caron de Fromentel C, Viron A, Puvion E, May P (1986) SV40 large T-antigen and transformation related protein p53 are associated in situ with nuclear RNP structures containing hnRNA of transformed cells. Exp Cell Res 164: 35–48 [DOI] [PubMed] [Google Scholar]
- Chandar N, Billig B, McMaster J, Novak J (1992) Inactivation of p53 gene in human and murine osteosarcoma cells. Br J Cancer 65: 208–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford L (1983) The 53,000-dalton cellular protein and its role in transformation. Int Rev Exp Pathol 25: 1–50 [PubMed] [Google Scholar]
- Crawford LV, Pim DC, Bulbrook RD (1982) Detection of antibodies against the cellular protein p53 in sera from patients with breast cancer. Int J Cancer 30: 403–408 [DOI] [PubMed] [Google Scholar]
- Danilova N, Sakamoto KM, Lin S (2008) p53 family in development. Mech Dev 125: 919–931 [DOI] [PubMed] [Google Scholar]
- DeLeo AB, Jay G, Appella E, Dubois GC, Law LW, Old LJ (1979) Detection of a transformation-related antigen in chemically induced sarcomas and other transformed cells of the mouse. Proc Natl Acad Sci USA 76: 2420–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliyahu D, Raz A, Gruss P, Givol D, Oren M (1984) Participation of p53 cellular tumour antigen in transformation of normal embryonic cells. Nature 312: 646–649 [DOI] [PubMed] [Google Scholar]
- Finlay CA, Hinds PW, Levine AJ (1989) The p53 proto-oncogene can act as a suppressor of transformation. Cell 57: 1083–1093 [DOI] [PubMed] [Google Scholar]
- Flaman JM, Waridel F, Estreicher A, Vannier A, Limacher JM, Gilbert D, Iggo R, Frebourg T (1996) The human tumour suppressor gene p53 is alternatively spliced in normal cells. Oncogene 12: 813–818 [PubMed] [Google Scholar]
- Friend SH, Bernards R, Rogelj S, Weinberg RA, Rapaport JM, Albert DM, Dryja TP (1986) A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature 323: 643–646 [DOI] [PubMed] [Google Scholar]
- Hu W, Feng Z, Atwal GS, Levine AJ (2008) p53: a new player in reproduction. Cell Cycle 7: 848–852 [DOI] [PubMed] [Google Scholar]
- Hu W, Feng Z, Teresky AK, Levine AJ (2007) p53 regulates maternal reproduction through LIF. Nature 450: 721–724 [DOI] [PubMed] [Google Scholar]
- Jenkins JR, Rudge K, Currie GA (1984) Cellular immortalization by a cDNA clone encoding the transformation-associated phosphoprotein p53. Nature 312: 651–654 [DOI] [PubMed] [Google Scholar]
- Kay C, Jeyendran RS, Coulam CB (2006) p53 tumour suppressor gene polymorphism is associated with recurrent implantation failure. Reprod Biomed Online 13: 492–496 [DOI] [PubMed] [Google Scholar]
- Knudson A (1971) Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci USA 68: 820–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolata G (2009) Grant system leads cancer researchers to play it safe. New York Times 24 June [Google Scholar]
- Kress M, May E, Cassingena R, May P (1979) Simian virus 40-transformed cells express new species of proteins precipitable by anti-simian virus 40 tumor serum. Virol J 31: 472–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuerbitz SJ, Plunkett BS, Walsh WV, Kastan MB (1992) Wild-type p53 is a cell cycle checkpoint determinant following irradiation. Proc Natl Acad Sci USA 89: 7491–7495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn T (1962) The Structure of Scientific Revolutions. Chicago, USA: University of Chicago Press [Google Scholar]
- Land H, Parada LF, Weinberg RA (1983) Cellular oncogenes and multistep carcinogenesis. Science 222: 771–778 [DOI] [PubMed] [Google Scholar]
- Lane DP, Benchimol S (1990) p53: oncogene or anti-oncogene? Genes Dev 4: 1–8 [DOI] [PubMed] [Google Scholar]
- Lane DP, Crawford LV (1979) T antigen is bound to a host protein in SV40-transformed cells. Nature 278: 261–263 [DOI] [PubMed] [Google Scholar]
- Linzer DI, Levine AJ (1979) Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell 17: 43–52 [DOI] [PubMed] [Google Scholar]
- Lubin R, Zalcman G, Bouchet L, Tredanel J, Legros Y, Cazals D, Hirsch A, Soussi T (1995) Serum p53 antibodies as early markers of lung cancer. Nat Med 1: 701–702 [DOI] [PubMed] [Google Scholar]
- Maltzman W, Czyzyk L (1984) UV irradiation stimulates levels of p53 cellular tumor antigen in nontransformed mouse cells. Mol Cell Biol 4: 1689–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda H, Miller C, Koeffler HP, Battifora H, Cline MJ (1987) Rearrangement of the p53 gene in human osteogenic sarcomas. Proc Natl Acad Sci USA 84: 7716–7719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlashewski G, Pim D, Banks L, Crawford L (1987) Alternative splicing of human p53 transcripts. Oncogene Res 1: 77–85 [PubMed] [Google Scholar]
- McKeon F, Melino G (2007) Fog of war: the emerging p53 family. Cell Cycle 6: 229–232 [DOI] [PubMed] [Google Scholar]
- Mowat M, Cheng A, Kimura N, Bernstein A, Benchimol S (1985) Rearrangements of the cellular p53 gene in erythroleukaemic cells transformed by Friend virus. Nature 314: 633–636 [DOI] [PubMed] [Google Scholar]
- Muleris M, Salmon RJ, Zafrani B, Girodet J, Dutrillaux B (1985) Consistent deficiencies of chromosome 18 and of the short arm of chromosome 17 in eleven cases of human large bowel cancer: a possible recessive determinism. Ann Genet 28: 206–213 [PubMed] [Google Scholar]
- Parada LF, Land H, Weinberg RA, Wolf D, Rotter V (1984) Cooperation between gene encoding p53 tumour antigen and ras in cellular transformation. Nature 312: 649–651 [DOI] [PubMed] [Google Scholar]
- Puzio-Kuter AM, Levine AJ (2009) Stem cell biology meets p53. Nat Biotechnol 27: 914–915 [DOI] [PubMed] [Google Scholar]
- Riley KJ, Ramirez-Alvarado M, Maher L Jr (2007) RNA-p53 interactions in vitro. Biochemistry 46: 2480–2487 [DOI] [PubMed] [Google Scholar]
- Shi H et al. (2009) Winter temperature and UV are tightly linked to genetic changes in the p53 tumor suppressor pathway in Eastern Asia. Am J Hum Genet 84: 534–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soussi T (1996) The humoral response to the tumor-suppressor gene-product p53 in human cancer: implications for diagnosis and therapy. Immunol Today 17: 354–356 [DOI] [PubMed] [Google Scholar]
- Soussi T, Caron de Fromentel C, May P (1990) Structural aspects of the p53 protein in relation to gene evolution. Oncogene 5: 945–952 [PubMed] [Google Scholar]
- Soussi T, Caron de Fromentel C, Mechali M, May P, Kress M (1987) Cloning and characterization of a cDNA from Xenopus laevis coding for a protein homologous to human and murine p53. Oncogene 1: 71–78 [PubMed] [Google Scholar]
- Takahashi T, Nau MM, Chiba I, Birrer MJ, Rosenberg RK, Vinocour M, Levitt M, Pass H, Gazdar AF, Minna JD (1989) p53: a frequent target for genetic abnormalities in lung cancer. Science 246: 491–494 [DOI] [PubMed] [Google Scholar]
- van Bokhoven H, McKeon F (2002) Mutations in the p53 homolog p63: allele-specific developmental syndromes in humans. Trends Mol Med 8: 133–139 [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Fearon ER, Kern SE, Hamilton SR, Preisinger AC, Nakamura Y, White R (1989) Allelotype of colorectal carcinomas. Science 244: 207–211 [DOI] [PubMed] [Google Scholar]
- Weinberg RA (1981) Use of transfection to analyze genetic information and malignant transformation. Biochim Biophys Acta 651: 25–35 [DOI] [PubMed] [Google Scholar]