SUMMARY
PR-Set7/Set8 is a cell cycle-regulated enzyme that monomethylates lysine 20 of histone H4 (H4K20). Set8 and monomethylated H4K20 are virtually undetectable during G1 and S phases of the cell cycle but increase in late S and in G2. We identify CRL4Cdt2 as the principal E3 ubiquitin ligase responsible for Set8 proteolytic degradation in the S-phase of the cell cycle, which requires Set8-PCNA interaction. Inactivation of the CRL4-Cdt2-PCNA-Set8 degradation axis results in (a) DNA damage and the induction of tumor suppressor p53 and p53-transactivated pro-apoptotic genes, (b) delayed progression through G2 phase of the cell cycle due to activation of the G2/M check-point, (c) specific repression of histone gene transcription and depletion of the histone proteins, and (d) repression of E2F1-dependent gene transcription. These results demonstrate a central role of CRL4Cdt2-dependent cell cycle regulation of Set8 for the maintenance of a stable epigenetic state essential for cell viability.
Keywords: Epigenetics, Histone methylation, Gene silencing, PR-Set7, Set8, Cullins, Cdt2, PCNA, Ubiquitylation
INTRODUCTION
Nucleosomes, the fundamental units of chromosomes, are subject to a diverse array of post-translational modifications (PTMs) that regulate various chromosomal and genetic transactions. Among these PTMs, the methylation of histone H4, occurs exclusively on lysine 4 (H4K20), and is involved in gene silencing (Fang et al., 2002; Nishioka et al., 2002), heterochromatin formation (Sautel et al., 2007; Schotta et al., 2004), DNA damage checkpoint control (Sanders et al., 2004) and regulation of mitosis (Julien and Herr, 2004; Karachentsev et al., 2005). The mono-methylation of H4K20 (H4K20me1) is carried out by PR-Set7/Set8 (Set8 hereafter) (Fang et al., 2002; Nishioka et al., 2002; Xiao et al., 2005). The subsequent di- and tri-methylation of H4K20 (H4K20me2 and H4K20me3 respectively) are catalyzed by Suv4-20h1 and Suv4-20h2 (Yang and Mizzen, 2009; Yang et al., 2008). H4K20me1 is cell-cycle regulated with its expression paralleling that of Set8, being low in S-phase, beginning to accumulate in late S-phase and G2 and peaking in mitosis (Yang and Mizzen, 2009). How Set8 protein fluctuates during the cell cycle and the biological significance of such cell-cycle regulation remain unclear.
In this study, we show that the cullin 4 E3 ubiquitin ligase complex, in colloboration with the DCAF (DDB1 and Cul4 associated factor) and substrate specificity factor Cdt2 (CRL4Cdt2) targets Set8 for ubiquitylation and degradation. The polyubiquitylation and subsequent degradation of many CRL4Cdt2 substrates require that these substrates interact with PCNA through conserved PCNA-interacting peptides (PIP boxes). PCNA, the ring-shaped clamp protein, first recognized for its role as a processivity factor for DNA polymerase δ, is emerging in a new role of promoting the ubiquitylation of substrates by CRL4Cdt2. CRL4Cdt2 promotes the degradation of Set8 in a reaction dependent on Set8-PCNA interaction to maintain low levels of H4K20me1 during S phase of the cell cycle. The CRL4-Cdt2-PCNA-dependent degradation of Set8 is essential for cell cycle progression because it ensures the proper chromatin structure essential for gene expression and chromatin condensation.
RESULTS
Set8 Contains a Specialized PCNA-Interacting Peptide (PIP) Box Common to CRL4Cdt2 Substrates
Many proteins interact with PCNA via a conserved PCNA-interacting peptide (PIP-box), with the consensus sequence: Q-X-X-(I/L/M)-X-X-(F/Y)-(F/Y) (Moldovan et al., 2007). Besides the residues at positions 1, 4, 7 and 8 that are critical for PCNA binding (Moldovan et al., 2007), CRL4Cdt2 substrates contain conserved threonine and aspartic acid residues at positions 5 and 6 respectively, as well as a basic residue 4 amino acids c-terminal of the PIP box (position +4) that are essential for substrate degradation (Figure 1A and (Havens and Walter, 2009; Senga et al., 2006)). A second basic residue at position +3 or +5 (or both) is also common to these substrates (Figure 1A). We hypothesized that other CRL4Cdt2 substrates would include PCNA-interacting proteins with this specialized PIP box and identified Set8 as a potential substrate. Set8 is a known PCNA-interacting protein with a well-conserved PIP box (amino acids 178–185 in human Set8) which contains elements essential for targeted degradation by CRL4Cdt2, namely Thr182, Asp183, Arg188 as well as Arg189 at positions 5, 6, +3 and +4 respectively (Figure 1B).
Figure 1.
Association of Set8 with PCNA via a CRL4Cdt2 substrate-specific PIP box triggers its degradation. (A) Sequence alignment of the PIP box of various substrates of CRL4Cdt2 Red, canonical residues essential for PCNA interaction; blue, residues common to CRL4Cdt2 substrates, and green, frequent residues in these substrates. (B) Sequence alignment of Set8 PIP box from different species. (C) Schematic of Set8 protein with its two PIP boxes. (D) GST pull-down experiment showing PIP box 2 of Set8b, but not PIP box 1, is essential for direct Set8-PCNA interaction. Coomassie stained gel of the purified Gst and Gst-Set8b proteins shown on left for loading control. (E) Immuno-fluorescence images of U2OS showing that endogenous Set8 is detected in PCNA-foci positive (S-phase) cells only when proteasomes are inactivated by MG132. Red arrows indicate the cell magnified below. (F) Quantitation of data in (E). (G) Similar to E, except that Myc-Set8b or Myc-Set8b ΔPIP2 were ectopically expressed and detected by the anti-Myc epitope antibody in the absence of MG132. (H) Half-life measurement of Set8b or Set8bΔPIP2 proteins in U2OS cells following cycloheximide (CHX) treatment. Immunoblots for Myc epitope. β-actin shows equal protein loading. (I) Similar to (H), but measures the half-life of endogenous Set8 protein in U2OS cells depleted of PCNA by si-RNA. Immunoblots for endogenous Set8 and PCNA. si-RNA targeting luciferase (si-GL2) is a negative control.
Identification of a Novel Splice Variant of Set8 that Represents the Dominant Form of Set8
The Set8 protein runs as a doublet on standard SDS-PAGE (Figure 2A, Figure 4E). Primers designed to amplify the reported human Set8 ORF (1059 bp) from U2OS cells (a human osteosarcoma derived cell-line), amplified a second isoform (969 bp) encoding a protein of 322 amino acids that is missing amino acids 10–40 within Set8 Exon 2 (Figure S1A). This represents a splice variant (Set8b) to be distinguished from the reported 355 residue long Set8a protein (Accession number, HQ259683). Ectopically expressed Set8b corresponded to the faster-migrating, and most abundant form of Set8, whereas Set8a corresponded to the slower-migrating form (Figure S1B). PCR of cDNA from U2OS or the human lung carcinoma cells, H1299 confirms that Set8b is the dominant spliced isoform of Set8 in these cells (Figure S1C, and data not shown). Depletion of Set8 by siRNA decreases H4K20me1 and induces DNA damage as these cells accumulate γH2AX and Rad51 foci (Figure S2C, D and (Jorgensen et al., 2007; Tardat et al., 2007)). Stable expression of Set8b resistant to si-Set8-2 restored H4K20me1 (Figure S2C) and prevented the induction of γH2AX (Figure S2D), demonstrating that Set8b is a functional form of Set8 in vivo.
Figure 2.
Low Set8 levels in the cell-cycle coincides with high CRL4 activity and PCNA mediates Set8-Cdt2 co-localization in vivo. (A and B) Top: FACS analysis of U2OS cells showing the synchronization of cells by nocodazole or double thymidine block (DTB), respectively, and subsequent release. Bottom: proteins extracted from the synchronized cells shown above were immunoblotted with antibodies specific for the indicated proteins. *: cross-reacting unrelated band. (C and D) Immuno-fluorescence images of U2OS showing the localization of endogenous Cdt2 and myc-tagged wt Set8b (C) or Set8bΔPIP2 (D) in cells treated without or with MG132. White arrow: a representative cell, which is magnified below.
Figure 4.
CRL4Cdt2 promotes Set8 degradation after UV. (A) UV irradiation of U2OS cells decreases total and chromatin-bound Set8. Western blot of Set8 after treating U2OS with various doses of UV as indicated. Actin serves as a loading control. (B, C, D) Degradation of Set8 following UV requires Cul4 and DDB1 (B), Cdt2 (C) and PCNA (D). Western blots of Set8 and the indicated protein from untreated or UV-irradiated U2OS cells transfected with the indicated siRNAs. The level of Set8 protein relative to actin and normalized to the non-UV irradiated sample is displayed below each lane. (E) Set8b interaction with PCNA is required for its degradation following UV. Western blot of 293T cell lysates showing that ectopic wt Set8b, but not Set8bΔPIP2 is degraded via the proteasome.
Association of Set8 with PCNA via PIP box 2 Triggers its Proteolytic Degradation via the Proteasome
Set8 associates with PCNA via the above mentioned PIP box (amino acids 178–185 in human Set8a: PIP2) (Jorgensen et al., 2007), as well as though a separate more N-terminally located PIP box (amino acids 51–58: PIP1) (Figure 1C) (Huen et al., 2008). GST-tagged wt Set8b (and Set8a, data not shown) interacted with recombinant PCNA (Figure 1D), and mutations in PIP2 (with N178, L181, F183 and Y184 all converted to alanine: Set8bΔPIP2), but not in PIP1 (with Q51, I54, Y57, and M58 all converted to alanine: Set8bΔPIP1) completely disrupted PCNA binding (Figure 1D). Importantly, the stable expression of exogenous si-RNA-resistant Set8bΔPIP1, like wt Set8b, supports cell proliferation, restores H4K20me1 and prevents DNA damage upon depletion of endogenous Set8 (Figure S2B, C, D). Thus, Set8 associates with PCNA via PIP box 2, and PIP box 1 is dispensable for the H4K20 mono-methyl transferase function of Set8.
Over-expressed Set8 protein co-localizes with PCNA (Huen et al., 2008; Jorgensen et al., 2007). However, endogenous Set8 was detected only in cells negative for PCNA foci, and thus outside S phase (Figure 1E, -MG132, and (Tardat et al., 2007)). Endogenous (Figure 1E, F) and ectopically-expressed (Figure 1G, and data not shown) Set8 became visible in the same cells as PCNA foci upon treatment of cells with the proteasome inhibitor MG132. In contrast, Set8bΔPIP2 was co-expressed in cells that are positive for PCNA foci without MG132 treatment (Figure 1G). Similar results were obtained from cells arrested in early S-phase by hydroxyurea treatment (Figure S2A). Thus, disruption of the Set8b-PCNA interaction enabled Set8b to persist in S phase cells.
Half-life measurements showed that Set8bΔPIP2 and Set8aΔPIP2 proteins were highly stable compared to their wt Set8 counterparts (Figure 1H, and data not shown), demonstrating that Set8-PCNA association dramatically reduces Set8 protein stability. Furthermore, depletion of PCNA from U2OS by siRNA results in the accumulation of cells in S-phase (data not shown), and significantly increased Set8 protein (Figure 1I) due to a prolonged protein half-life (Figure 1I, Figure S3C), despite the fact that Set8 protein is normally virtually undetectable in S-phase cells (Figure 1E, Figure 2). Thus, PCNA, Set8-PCNA interaction and proteasome activity are all required for the degradation of Set8 in S phase.
Low Set8 and H4K20me1 Expression in S-phase Coincides with High CRL4Cdt2 Activity
U2OS cells synchronized by nocodazole (mitosis) (Figure 2A) or double thymidine block (DTB: G1/S) (Figure 2B) were released from these blocks and harvested at various time points. Cell-cycle distribution at each time-point was measured by FACS analysis and the expression of cyclin E2 and cyclin A2 (Figure 2A, B). The Set8 protein begins to decrease at 6 hr post-release from mitosis (late G1-early S-phase), and is already decreased in cells arrested at the early S phase by DTB (Figure 2A, B). Thus the decrease of the protein begins at the onset of S phase. Set8 protein did not appear again until late S-phase (between 8 and 10 hrs post-release from DTB), and was maximal in late G2 and in mitosis (Figure 2A, B). Because the reduction of Set8 in late G1 and during most of S-phase was recapitulated with ectopically expressed wt Set8b expressed from a heterologous promoter (see Figure 5E, middle panel, below), and Set8 protein became visible in S-phase cells by MG132 treatment (Figure 1E), the reduction in S phase is not due to transcriptional down-regulation of Set8.
Figure 5.
Failure to degrade Set8 via the CRL4Cdt2-PCNA pathway inhibits G2/M cell cycle progression and cell proliferation. (A) Expression of Set8bΔPIP2 but not a catalytically inactive Set8bΔPIP2 (Set8bΔPIP2_R265G/D338A) or wt Set8 inhibits the proliferation of U2OS cells. Day 0 post-selection corresponds to 48 hrs after retroviral transduction. (B) Phase contrast images of U2OS cells taken on day four post-transduction. Expression of Set8bΔPIP2 causes an increase in cellular and nuclear size with flattening of cells. (C) FACS analysis of U2OS cells shown in (B). Percentages of cells in the various cell cycle stages are indicated. Mean ± standard deviation of three independent transductions. (D) FACS analysis of U2OS cells transduced with retroviruses expressing the indicated proteins and synchronized (48 hrs post-transduction) by DTB block and release at various time points, in the presence of nocodazole. (E) Western blotting of lysates of U2OS shown in (D). Set8bΔPIP2 expression delays progression into mitosis as indicated by the delayed appearance of H3S10 phosphorylation. The immunoblots of protein lysates from the various cells for a given antibody were exposed for the same time to demonstrate differences in the abundance of the various proteins.
The expression of H4K20me1 through the cell-cycle mirrored that of Set8 (Figure 2A, B). Interestingly, H4K20me2 and H4K20me3 expression in cells released from nocodazole block trailed that of Set8 and H4K20me1. Thus, the expression of these chromatin marks are cell-cycle-regulated as well and are most likely dependent on Set8 first mono-methylating H4K20. Interestingly, the expression pattern of Set8 and H4K20me1 was very similar to that of Cdt1 and p21 (Figure 2A, B), two cell cycle-regulated substrates for the CRL4Cdt2 ubiquitin ligase. Importantly, when wt Set8b was stabilized by MG132, it co-localized with Cdt2 foci (Figure 2C). On the other hand, Set8bΔPIP2 did not co-localize with Cdt2 with or without MG132 treatment despite their co-expression in the same cells (Figure 2D), indicating that the Set8-Cdt2 co-localization in MG132-treated cells required Set8-PCNA interaction. Together, these results suggest that CRL4Cdt2 and PCNA may be responsible for maintaining low levels of Set8 in S phase.
Set8 is a Direct Substrate of CRL4Cdt2
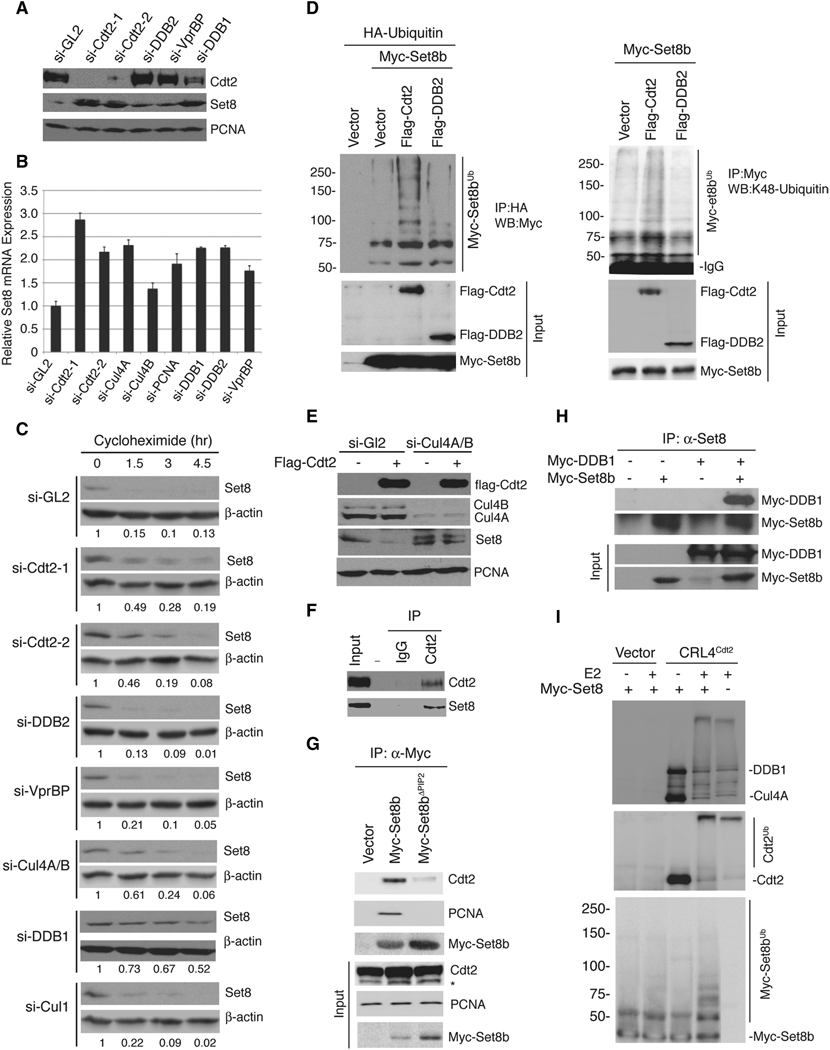
Depletion of U2OS cells of Cdt2 using two different siRNAs significantly increased Set8 protein (Figure 3A). Similar results were obtained in HeLa as well as the human colon cancer HCT116 cells (data not shown). In control siRNA-transfected cells (si-GL2, targeting the firefly luciferase gene), the half-life of Set8 is <1 hr, while in cells depleted of Cdt2, Set8 half-life is ~ 1.5 hr (Figure 3C, S3A, B). The down-regulation of Cul4A/B or DDB1 similarly stabilized Set8 protein (Figure 3C, S3A, B, C).
Figure 3.
Set8 is a direct substrate for CRL4Cdt2. (A) Western blotting of Set8 protein in cells depleted of the indicated proteins by siRNA. (B) Quantitative RT-PCR of Set8 mRNA in control (si-GL2) or cells depleted of the indicated proteins by siRNA, expressed relative to β-actin mRNA. Average ± standard deviation of three separate PCR reactions. (C) Set8 protein half-life is increased only in cells depleted of various components of the CRL4Cdt2 complex but not other related proteins. Half-life measurement of Set8 protein in control cells and in cells depleted of various proteins as indicated. 50 ug of protein from si-GL2 transfected cells or 20 ug of other samples were loaded to normalize for the starting point of Set8 expression. β-actin is loading control. The ratio of Set8 protein normalized to actin relative to the starting point (0 hr of cycloheximide treatment) is indicated below each lane. The efficiency of knockdown is shown in figure S3A. The rate of Set8 degradation is also displayed in figure S3B, C. (D) Left; Cdt2 stimulates Set8b ubiquitylation in vivo. Western blotting of Set8 expressing the indicated proteins in 293T cells. Immunoprecipitated ubiquitylated proteins (anti-HA) from MG132-treated cells were Western-blotted for Myc-Set8. Expression of the overexpressed proteins is shown below. Right, similar to the panel on left without overexpressing HA-ubiquitin. Western blot of immunoprecipitated Set8 proteins (anti-myc) with anti-K-48 ubiquitin-specific antibody. (E) Destabilization of Set8b by Cdt2 requires Cul4A/B. Western blotting of endogenous Set8 shows that ectopic Cdt2 decreases co-transfected Set8b only in control 293T but not in cells depleted of Cul4A/B by siRNA. The upper band in the anti-Set8 immunoblot, which appears after Cul4 si-RNA, represents either the stabilized Set8a isoform of Set8 protein or a modified form of Set8b, because it disappears upon co-silencing endogenous Set8 with siRNA (data not shown). (F) Endogenous Set8 interacts with endogenous Cdt2 in vivo in cells treated with MG132. Western blot analysis of Cdt2 and Set8 in control (IgG) or anti-Cdt2 immunoprecipitates. (G) Wt-Set8b but not Set8bΔPIP2 interacts with endogenous PCNA and endogenous Cdt2. Western blot of the anti-myc immunoprecipitates from cells expressing the indicated proteins. *: cross-reacting unrelated band. (H) DDB1 interacts with Set8. Western blot of the anti-Set8 immunoprecipitate from 293T cells expressing the indicated proteins. (I) CRL4Cdt2 promotes Set8 ubiquitylation in vitro. Incubation of immuno-purified CRL4Cdt2 (shown by Western in top two panels) with Set8b in an in vitro ubiquitin ligase assay increased the formation of Set8 ubiquitylated species (bottom panel). Auto-ubiquitylation of Cdt2 is also observed (middle panel).
Down-regulation of DDB2 and VprBP, two other DCAFs, which associate with CRL4, also resulted in an increased Set8 protein albeit to a lesser extent (Figure 3A). In these cases, however, the half-life of Set8 protein was not increased (Figure 3C, S3A, B, C). Depletion of all of these DCAFs, including Cdt2, resulted in an increase in Set8 mRNA (Figure 3B) and thus the induction of Set8 after depletion of Cdt2 is partly due to the stabilization of Set8 protein and partly due to the up-regulation of Set8 transcription. Depletion of the other DCAFs, on the other hand, does not stabilize Set8 protein, and the induction is solely due to increase in Set8 transcription. Thus, CRL4Cdt2 is unique among the cullins and DCAFs in being required for the degradation of Set8 protein.
To test whether CRL4Cdt2 is sufficient to down-regulate Set8, we over-expressed various cullins or DCAFs along with Set8b in 293T cells. Co-expression of Cul4A, Cul4B or Cdt2, but not Cul1 or DDB2, specifically decreased the protein level of co-expressed Set8b (Figure S3D), and this effect was prevented by MG132 (Figure S3E). Additionally, Cdt2, but not DDB2, enhanced lysine 48-specific polyubiquitylation of Set8 in vivo (Figure 3D) and decreased the half-life of co-expressed Set8b protein (Figure S3F). Importantly, the down-regulation of endogenous Set8 by Cdt2 was prevented in cells depleted of Cul4A/B (Figure 3E), demonstrating that Cdt2 targets Set8 for degradation via its association with the CRL4 ubiquitin ligase complex. Consistent with the co-localization of Cdt2 with Set8 in MG132-treated cells (Figure 2C), we found that endogenous Set8 protein interacts with endogenous Cdt2 (Figure 3F) and this interaction was dependent on PCNA as Cdt2 only interacted with wt Set8b, but not with Set8bΔPIP2 (Figure 3G). In addition, both Cul4A (not shown) and DDB1 (Figure 3H) specifically interact with Set8 protein when ectopically expressed. Finally, immuno-purified CRL4Cdt2 promoted Set8b polyubiquitylation in vitro (Figure 3I). From these data, we conclude that Set8 is a direct substrate for CRL4 ubiquitin ligase in normal unperturbed cells during the S-phase of the cell cycle.
CRL4Cdt2 Promotes Set8 Degradation Following UV Irradiation
CRL4Cdt2 ubiquitylates and promotes the degradation of at least three of its substrates (Cdt1, p21 and Spd1) not only during the S-phase of the cell cycle, but also after DNA damage (Abbas et al., 2008; Higa et al., 2003; Hu et al., 2004; Liu et al., 2005; Nishitani et al., 2008; Ralph et al., 2006; Senga et al., 2006). Set8 is also down-regulated after DNA damage although this down-regulation has been suggested to occur concurrent with the down-regulation of Set8 transcription (Shi et al., 2007). However, an active degradation of Set8 protein following DNA damage has not been ruled out. UV irradiation resulted in a dose-dependent down-regulation of total and chromatin-bound Set8 protein (Figure 4A). UV irradiation also down-regulated wt Set8b, expressed from a heterologous promoter and this was blocked by MG132 (Figure 4E), suggesting that the proteasomal degradation of Set8 plays a role in its down-regulation post-UV irradiation. Depletion of Cul4, DDB1 or Cdt2, but not other cullins or DCAFs, from U2OS cells by siRNA, stabilized Set8 protein and prevented UV-induced Set8 down-regulation (Figure 4B, C and data not shown). Similar results were obtained in HCT116 cells (data not shown). Furthermore, Set8 degradation following UV irradiation is also dependent on PCNA (Figure 4D) and Set8-PCNA interaction (Figure 4E), as Set8ΔPIP2 was not degraded in response to UV. Thus, Set8 is also a substrate of this same ubiquitin ligase in a PCNA-dependent reaction following DNA damage induced by UV irradiation.
Inactivation of the CRL4-Cdt2-PCNA-dependent Set8 Degradation Inhibits Cell Proliferation, but Does Not Interfere with S-phase Progression
U2OS stably expressing Set8b using retroviral transduction proliferated in culture with similar kinetics as mock-infected cells (Figure 5A). In contrast, cells expressing Set8bΔPIP2 did not proliferate after a couple of doublings from the initial selection with puromycin (48 hrs post-transduction) (Figure 5A). The Set8bΔPIP2 mRNA was not over-expressed relative to wt Set8b (Figure S4A), demonstrating that the proliferation defect was due to increased stability of Set8bΔPIP2 protein (Figure 1H). Similar results were obtained in H1299 cells (Figure S4B), in HeLa cells and in HCT116 cells (data not shown). Mutation of two residues essential for Set8 catalytic activity, Arg 265 and Asp 338 to Gly and Ala respectively (Set8bΔPIP2_R265G/D338A) (Nishioka et al., 2002; Shi et al., 2007), alleviated growth inhibition by Set8bΔPIP2 (Figure 5A, B), demonstrating that the histone-methyltransferase activity of stable Set8 is responsible for the growth inhibition.
Set8bΔPIP2 but not catalytically inactive Set8bΔPIP2_R265G/D338A caused a marked enlargement of cells and nuclei, mostly visible 4 days post-transduction (Figure 5B, Figure S4C, D). Set8bΔPIP2 decreases cells with G1 DNA content while increasing cells with >4N DNA content (Figure 5C) indicative of re-replication, a result that may contribute to the enlarged nuclei of these cells (Figure S4C, D, S5B and (Zhu et al., 2004)). No re-replication was seen in wt Set8b- or mock-transduced U2OS cells. In addition, 20% of Set8bΔPIP2-expressing cells underwent apoptosis to produce cells with sub G1 DNA content (Figure 5C and see below). Expression of Set8a ΔPIP2 in U2OS cells produced similar results (Figure S4E and data not shown).
Set8 and Set8-PCNA interaction have been proposed to be important for S-phase progression (Huen et al., 2008; Jorgensen et al., 2007). Because Set8bΔPIP2 does not interact with PCNA (Figure 1D, Figure 3G, and (Jorgensen et al., 2007)), we tested whether it interferes with S-phase progression. U2OS cells were synchronized at the G1/S transition after transduction and before Set8bΔPIP2-expression completely suppressed proliferation (Figure 5D). Release from the G1/S block into nocodazole showed that Set8bΔPIP2 expressing cells progressed normally through S phase (Figure 5D). Furthermore, these cells incorporated bromodeoxyuridine (BrdU) with the same kinetics as control cells (Figure S5A, B).
Like endogenous Set8, ectopic wt Set8b protein decreased substantially as cells entered and progressed through S-phase (Figure 5E, middle panel). On the other hand, Set8bΔPIP2 remained stable throughout S-phase (Figure 5E, right panel), confirming the requirement of PCNA-Set8 interaction for the S-phase-specific degradation of Set8. Although S phase progression was not impaired, cells expressing Set8bΔPIP2 reached mitosis much later than control or wt Set8b-expressing cells as indicated by the delay in the accumulation of phosphorylated Ser 10 of histone H3, a marker for entry into mitosis (Figure 5E). Consistently, when the G1/S blocked cells were released into a medium without nocodazole, control and wt Set8b-expressing cells left G2 to enter G1 at 13–16 hr, while cells expressing Set8bΔPIP2 were significantly delayed (Figure S5C, D). Thus, Set8bΔPIP2 expression does not interfere with S-phase progression but delays progression through G2 into mitosis.
Inactivation of the CRL4-Cdt2-PCNA-dependent Set8 Degradation Increases H4K20me2, and the Repressive Mark, H4K20me3
Although Set8bΔPIP2 remained stable during S-phase, global H4K20me1 remained low during S-phase and did not increase significantly at the end of S-phase as seen in mock-or wt Set8b-expressing cells (Figure 5E). Instead, these cells contained abundant H4K20me3 during S-phase despite the lower expression of histone H4 (Figure 5E, Figure 6A). In addition, we did not see the induction of total histone H4 normally seen as cells transit S phase and synthesize new histones. This will be described further later. The discrepancy between elevated Set8bΔPIP2 and low H4K20me1 levels in S-phase can be explained if in each S phase newly synthesized histone H4 is first mono-methylated and then converted to H4K20me2 and/or H4K20me3. Thus, the decrease in newly synthesized H4 and the conversion of pre-existing H4K20me1 to H4K20me2 and H4K20me3 could explain the decrease of H4K20me1 in cells expressing Set8bΔPIP2.
Figure 6.
Deregulated Set8 expression increases H4K20me2 and H4K20me3, induces DNA damage and activates the p53 tumor suppressor pathway. (A) H4K20me1 is increased initially but is converted to H4K20me2 and H4K20me3 in cells containing stable Set8. Western blot analysis of U2OS lysates, 24 and 48 hours following transduction with mock, wt Set8b, or Set8bΔPIP2-expressing retroviruses. (B) DNA damage-induced accumulation of phosphorylated p53 (Ser-15) and γH2AX and activated checkpoint pathways in cells expressing stable Set8. Rest as in (A). (C) RNA level of various p53-regulated genes in U2OS cells transduced with the indicated retroviruses as determined by quantitative RT-PCR. Data are represented relative to the expression of β-actin mRNA. (D) Similar to (C), but shows the induction of p53-regulated genes p21, FAS and PUMA in Cdt2-depleted (si-Cdt2) U2OS cells. (C, D) show the average ± standard deviation of three separate PCR reactions.
Indeed, cells expressing Set8bΔPIP2, showed an early increase in H4K20me1 one day post-transduction, but this returned to basal levels on the second day and was accompanied instead, by an increase in H4K20me2/3. Depletion of the Suv4-20h1/2 HMTs by siRNA reduced H4K20me2 and H4K20me3 in all cells, including Set8bΔPIP2-expressing cells (Figure 6A, S6B, C). Thus, (a) Set8-PCNA association is not essential for Set8 to mono-methylate H4K20 and (b) the low level of H4K20me1, despite the presence of stable Set8bΔPIP2, is at least partially due to its conversion to the di- and tri-methylated state through the activity of the HMTs Suv4-20h1/2 in the absence of new histone H4 protein synthesis.
Inactivation of CRL4-Cdt2-PCNA-dependent Set8 Degradation Activates p53
Unlike wt Set8b, the expression of Set8bΔPIP2, but not catalytically inactive Set8bΔPIP2_R265G/D338A, induced the tumor suppressor p53 and its downstream target p21 (Figure 6B, C). The stabilization of p53 is likely due the accumulation of DNA damage as p53 was phosphorylated at Ser15 in these cells (Figure 6B). There was a subtle but reproducible induction of γH2AX in Set8bΔPIP2 -transduced cells (Figure 6B). Besides p21, Set8bΔPIP2 expression up-regulated several, but not all, p53 pro-apoptotic genes such as Fas, PUMA and PIG3 (Figure 6C), likely contributing to the apoptosis seen in Figure 5C. Depletion of Cdt2 not only stabilized endogenous Set8 (Figure 3A, B), but also induced the same set of p53 target genes (Figure 6D). These results indicate that the degradation of endogenous Set8 through CRL4-Cdt2-PCNA is critical for preventing DNA damage and the activation of p53. Consistent with the delay of progression through the G2 of the cell cycle (Figure 5E, S5C, D), the expression of Set8bΔPIP2 activated the G2/M check-point as indicated by the induction of phosphorylated Tyr 15 of CDK1 (Figure 6B).
Collectively the data in this and the preceding sections indicate that stabilization of Set8 in S phase cells leads to an increase in repressive H4K20me3, DNA damage and activation of p53 and G2 checkpoint pathways. This explains the failure to enter mitosis and the increased apoptosis.
Failure to Degrade Set8 Inhibits Transcription from E2F1-Regulated and Histone Genes
To further understand the functional consequence of the failure to degrade Set8, we performed a whole-genome microarray analysis of mRNA expression of U2OS cells five days after transduction with retroviruses expressing either wt Set8b or Set8bΔPIP2. We analyzed these cells five days post transduction to enrich for genes that may be affected by the stable expression of Set8 and the consequent increase of H4K20me3. Expression of many genes was significantly altered in Set8bΔPIP2- compared to wt Set8b- or mock-transduced cells (Figure S7A, and Table S1). Set8bΔPIP2 inhibits various E2F1-regulated genes including cyclin E2, cyclin A2, CDC25A, geminin, MCM7 and Cdt1. In fact, with the exception of Rad51, E2F1-regulated genes were among the most repressed by stable Set8 (Figure S7B). Quantitative RT-PCR confirmed the inhibition of several of these genes (Figure 7A and data not shown). Importantly, full repression of E2F1-regulated genes was dependent on Set8 catalytic activity (see Set8bΔPIP2_R265G/D338A in Figure 7A).
Figure 7.
Inactivation of the CRL4-Cdt2-PCNA-Set8 pathway represses transcription of E2F1-regulated genes and histone genes and causes chromatin decompaction. (A and B) RNA level of some E2F1-regulated genes (A) and the indicated histone genes (B) expressed relative to β-actin mRNA as determined by quantitative RT-PCR. (C) Similar to (B), and represents the relative expression of histone genes in U2OS cells depleted of the indicated proteins by siRNA. (D) Chromatin immunoprecipitation (ChIP) analysis of the indicated histone promoters shows the enrichment of the H4K20me3 chromatin mark at various promoters in cells transduced with viruses expressing the indicated Set8 proteins or control pMSCV vector. The data represent the PCR-amplification from the H4K20me3 immunoprecipitates normalized to those from total histone H4 immunoprecipitates. (A, B, C &D) show the average ± standard deviation of three separate PCR reactions. (E) Ponceau-S staining of U2OS cell lysates after SDS-PAGE, showing the reduction of histone proteins upon the expression of Set8bΔPIP2. Also shown is a Western blot that shows that reduction of individual histone proteins in Set8bΔPIP2-expressing cells. (F) Cells expressing Set8bΔPIP2 exhibit decondensed chromatin. Micrococcal nuclease digestion of nuclei of cells expressing the indicated proteins shows the loss of nucleosomal pattern in nuclei from Set8bΔPIP2 -expressing cells. (G) Summary model depicting the deregulated gene expression and biological consequences of the failure to degrade Set8 via the CRL4Cdt2-PCNA during the S-phase of the cell cycle.
The second class of genes that were significantly repressed in Set8bΔPIP2-expressing cells was the various histone genes (Table S1). Quantitative RT-PCR of mRNA confirmed the repression of all four histone genes H2A, H2B, H3 and H4 as well as the linker histone H1 by stable Set8 (Figure 7B). These histone mRNAs were all significantly decreased by 48 hrs post transduction before the cells seized to proliferate (Figure S7E). By that time the cells are in G2 phase (Figure S6A). During the normal G2 phase of the cell cycle, the steady state levels of histone mRNA are known to decrease substantially due to destabilization of the mRNA (Marzluff et al., 2008 and references therein). In contrast, we did not see any destabilization of the histone H4 mRNA transcript in Set8bΔPIP2-expressing cells at this point (Figure S7F), suggesting that the decrease in histone mRNA was due to transcriptional changes and not secondary to the accumulation of cells at G2 phase of the cell cycle. Importantly, expression of wt Set8b (and not just Set8bΔPIP2) from a high-titer virus, produced the same repressive effect on histone gene transcription (Figure S7C, D). This result demonstrates that it is the abundance and activity of Set8bΔPIP2, and not a function of Set8 that may be dependent on Set8-PCNA interaction, which causes this deregulated gene expression.
Using gene-specific primers, we found that Set8bΔPIP2 expression repressed several but not all histone subtypes (Figure S7G and H, and data not shown). The dramatic repression of histone genes by Set8bΔPIP2 resulted in a near-total disappearance of all five histone proteins on SDS-PAGE (Figure 7E). Similar results were obtained in H1299 cells (data not shown). Set8bΔPIP2 also caused a modest (two-fold) repression of the histone variants H2AZ and H2AX (Figure S7G), perhaps explaining why phospho-H2AX levels are elevated but not highly so in cells expressing Set8bΔPIP2 (Figure 6B). Surprisingly, expression of Set8bΔPIP2_R265G/D338A also repressed histone gene expression and decreased histone proteins but to a lesser extent (Figure 7B). Since chromatin- modifying enzymes often work as part of stable protein complexes, it is not unexpected that a catalytically dead, but stabilized, Set8b may have dominant negative phenotypes created by titration of other components of the transcription complexes.
To test whether endogenous Set8 plays a role in histone gene transcription, we monitored the expression of histone mRNA in cells depleted of Cdt2 or Cul4A by si-RNA, where endogenous Set8 is stabilized. Figure 7C shows that the same set of histone genes repressed by Set8bΔPIP2 were also repressed, albeit to a lesser extent, in the absence of Cdt2 or Cul4A, indicating that the CRL4-Cdt2-PCNA-dependent Set8 degradation is critical for preventing the repression of histone genes during the normal cell cycle.
Failure to Degrade Set8 Results in Repressive Chromatin Modifications at the Histone Gene Promoters and Decompaction of Chromatin
Because Set8bΔPIP2 expression is associated with a significant increase in the repressive chromatin mark H4K20me3, we suspected that promoters of genes repressed by Set8bΔPIP2 may be selectively enriched for this chromatin mark. In deed, chromatin immunoprecipitation (ChIP) with H4K20me3-specific antibody at various histone gene promoters prior to the disappearance of histone proteins (four days after transduction) indicates that Set8bΔPIP2, but not catalytically inactive Set8bΔPIP2_R265G/D338A, induced H4K20me3 at many, but not all histone promoters (Figure7D). Because H4K20me3 is associated with constitutive heterochromatin (Schotta et al., 2004), we suggest that histone gene repression is mediated, at least in part, through the Set8-dependent mono-methylation H4K20 and its subsequent conversion to H4K20me3 and gene silencing.
The remarkable loss of histone proteins in cells expressing stable Set8 made us wonder whether the cells suffer from significant chromatin decompaction. Micrococcal nuclease digestion of U2OS nuclei expressing Set8bΔPIP2 showed a significant loss of nucleosomal digestion patterns (Figure 7F). Thus Set8bΔPIP2 results in a massive DNA decompaction that is likely contributing to the increased nuclear size, deregulation of gene expression, DNA damage and difficulty in progressing through mitosis.
DISCUSSION
CRL4Cdt2 Ubiquitin Ligase Complex: A master Regulator of Cell Cycle Progression
The CRL4Cdt2 ubiquitin ligase complex is a key regulator of cell cycle progression by promoting the degradation of proteins, such as the replication initiation protein Cdt1 (reviewed in Jackson and Xiong, 2009) and the CDK inhibitor p21 (Abbas et al., 2008; Kim et al., 2008; Nishitani et al., 2008), whose expression interfere with normal S-phase progression. The current study identifies Set8 as a CRL4Cdt2 substrate whose degradation during S-phase is required to express sufficiently high levels of histone proteins necessary for cells to progress through G2 phase of the cell cycle in preparation for mitosis. CRL4Cdt2 also promotes the degradation of Set8 following UV irradiation. Because the down-regulation of Set8 following DNA damage is important for the enhanced transactivation functions of p53 (Shi et al., 2007), we propose that CRL4Cdt2 may enhance responsiveness to DNA damage by accelerating Set8 down-regulation following DNA damage. However, the activation of p53 seen in cells with stable Set8 indicates that although Set8 can modulate p53, it does not completely suppress its activity.
Like Cdt1 and p21, the degradation of Set8 in S-phase or after DNA damage is dependent on its ability to interact with PCNA via a specialized PIP box. Consistent with previous finding (Jorgensen et al., 2007), we found that Set8 interacts with PCNA through PIP box2 as mutations in critical residues within this PIP box abolished PCNA-binding in vivo and in vitro. The other PIP box of Set8 (PIP box 1) however, seems to be dispensable for the ability of Set8 to promote H4K20me1 in vivo. However, it is possible that PIP box 1 may be functional in other respects, but it is not de-stabilizing Set8 or relieving the defect in cell proliferation when PIP2 is absent.
Previous studies suggest that Set8-PCNA interaction is necessary for S-phase progression and that this depended on the ability of Set8 to maintain the bulk of H4K20me1 and H4K20me3 (Huen et al., 2008; Jorgensen et al., 2007; Tardat et al., 2007). Because the loss of Set8 is accompanied by extensive DNA damage, it may indirectly interfere with S-phase progression. Using synchronization and biochemical assays as well as immuno-fluorescence experiments, we demonstrate that S-phase cells were naturally low in Set8 expression and forced expression of Set8b, using Set8b mutant proteins incapable of associating with PCNA in S-phase, delayed progression of cells through G2/M, inhibited cell proliferation and caused massive chromatin decompaction, but did not significantly affect S-phase progression (Figure 5).
Set8bΔPIP2-expressing cells also exhibited DNA damage. Although the DNA damage response was dampened by the loss of H2A.X mRNA and protein, it was sufficient to activate the tumor suppressor p53 and induce apoptosis in a subset of cells and may contribute to G2/M check-point activation (see model in Figure 7G). Other phenotypes observed in Set8bΔPIP2-expressing cells, such as the induction of DNA re-replication and the repression of histone and E2F1 -driven transcripts are unlikely to be dependent on DNA damage per se. For example, DNA damage frequently leads to the phosphorylation and activation, rather than repression, of E2F1 with the subsequent activation of E2F1-dependent apoptosis (Stevens and La Thangue, 2004). Histone genes on the other hand, are repressed by ionizing radiation as part of a G1 checkpoint arrest (Su et al., 2004), but this repression is dependent on p53, while the repression of histone genes seen with Set8ΔPIP2 expression was also seen in p53-deficient cells such as H1299 (data not shown) and occurs in cells in G2. Furthermore, the dramatic reduction of all histone proteins or the accumulation of H4K20me3 on the promoter of histone genes, have not been reported after DNA damage or checkpoint activation. Additionally, the bulk of genes that are deregulated by stable Set8 are not altered in expression in cells exposed to DNA damaging agents. Because the mechanism by which stable Set8 induces DNA damage is unclear, the DNA damage cannot be experimentally prevented, and so we cannot conclusively rule out an indirect effect of DNA damage on gene expression. Based on the arguments above however, we believe that significant parts of the phenotypes produced by stable Set8 are independent of DNA damage.
Because, the PCNA binding-deficient mutant of Set8 can still mono-methylate H4K20 in vivo, the growth inhibitory effects of Set8ΔPIP2 cannot be attributed to a failure of Set8 to mono-methylate H4K20. Our attempts at separating PCNA-binding of Set8 from degradation have been unsuccessful. A single point mutation of Set8 at Thr182 to Alanine (Set8bT182A) inhibited Set8-PCNA binding in vivo and had similar phenotypes as the ΔPIP2 mutant described here. On the other hand, mutation of the +4 basic Arg189 residue of Set8b (Set8bR189A) did not disrupt PCNA-binding and did not significantly stabilize the protein and was therefore tolerated well (data not shown). Several lines of evidence however, support our conclusion that it is the increased stability and activity of Set8ΔPIP2 and not the loss of PCNA-binding that caused the deregulation of gene expression as well as the proliferation defect. First, overexpression of wt Set8 from a high-titer virus down-regulated transcription from all the tested histone genes (Fig. S7C, D), similar to that induced by Set8bΔPIP2. Second, only a catalytically active Set8bΔPIP2 suppressed proliferation, demonstrating that it is the increased activity (due to increased stability) of Set8 that is required to cause the phenotypes described. Third, a careful time course examination of the effect of wt Set8b and Set8bΔPIP2 expression on H4K20me1 demonstrates that Set8bΔPIP2 expression initially increases H4K20me1, which is rapidly converted to H4K20me2 and H4K20me3 by SUV4-20h1/2. Finally, stabilization of endogenous Set8 by depletion of Cdt2 or Cul4A had a similar effect on transcription of histone and p53-regulated genes. Thus, the preponderance of evidence supports the hypothesis that the toxic effects from the non-degradable Set8 stems from the repression of inappropriate genes due to the trimethylation of H4K20 at the promoters of specific genes such as certain histone genes. Formally however, we cannot rule out the possibility that at least some of the phenotypes might be due to the methylation of non-histone substrates of the non-degradable Set8 that may or may not be methylated by wt Set8. Furthermore, our findings do not rule out the possibility that in addition to its role in promoting the destabilization and degradation of Set8, the Set8-PCNA interaction may serve another unforeseen function, the loss of which may contribute to some of the phenotypic effects of Set8bΔPIP2.
Set8 Represses Histone Gene Transcription and Regulates Chromatin Compaction
The most striking effect of the failure to degrade Set8 in S phase via the CRL4-Cdt2-PCNA pathway is the catastrophic loss of five canonical histone mRNAs and proteins (Figure 7). In metazoans, canonical histone mRNAs are among the most highly cell-cycle-regulated with its expression restricted to S-phase, but the bulk of this regulation is due to post-transcriptional processing (reviewed in Marzluff et al., 2008). Towards the completion of S-phase and during G2, canonical histone mRNAs are rapidly degraded. Thus, the activation of G2/M checkpoint of Set8ΔPIP2-expressing cells may aggravate the reduction of histone mRNAs and proteins in these cells. However, we find that Set8ΔPIP2-specifically represses histone gene transcription through the accumulation of H4K20me3 at various histone promoters prior to the dramatic loss of these proteins. Additionally, the repression of histone mRNAs was not limited to the canonical and replication-dependent histone genes and included those that are not cell-cycle-regulated such as the histone variants H2A.X and H2A.Z. Finally, the early repression of H4 was not accompanied by de-stabilization of the mRNA, as would have been expected if it was secondary to a G2 cell-cycle arrest.
One consequence of the loss of histone proteins is chromatin decondensation and the resultant deregulation of gene expression. In fact, reduction of the linker histone H1 alone to 50% of its normal level affects chromatin structure and deregulates several genes (Fan et al., 2005). Thus, it is likely that most of the gene deregulation seen in our microarray analysis is secondary to the reduction of histone mRNA and protein and chromatin decondensation. Future experiments with cells constitutively expressing the five canonical histone genes from heterologous promoters will be needed to determine whether histone repression is the sole mechanism underlying all the toxicity seen with Set8ΔPIP2.
One of the unsolved puzzles in epigenetics is how chromatin modifications are related to time of DNA replication in S phase. Many studies have shown that late replicating genes are endowed with repressive chromatin marks that suppress gene expression (Birney et al., 2007). The repressed chromatin, in turn predicts late replication in subsequent cell cycles. Conversely, early replicating parts of chromosomes are spared these repressive marks. We propose that at least part of the mechanism is the selective degradation in early S phase of Set8, a writer of one of the repressive marks. Set8 is normally absent when the early replicating genes replicate, and so H4K20me1 and the subsequent H4K20me3 marks cannot be laid down to repress these genes. Histone genes are, of course, the paradigmatic early replicating, highly expressing genes lacking repressive marks. The stabilization of Set8 in S-phase through the disruption of the CRL4-Cdt2-PCNA-dependent degradation axis, closes the window of protection from repression in early S phase and causes several of these genes to acquire repressive marks. Not all early replicating genes however, acquire repressive marks (data not shown), so that there has to be some mechanism that target the stabilized Set8 to specific histone genes to initiate such profound repression. This profound repression of histone genes highlightes the importance of Set8 degradation for the clockwork like functioning of the cell-cycle.
EXPERIMENTAL PROCEDURES
Cell Culture, Antibodies, and Reagents
U2OS, H1299, HeLa and 293T cells were obtained from the American Type Culture Collection (ATCC), and cultured in DMEM (Invitrogen) supplemented with 10% FBS and antibiotics. The antibodies purchased were p21 (C-19), PCNA (PC10), Cul1 (D-5), Cul4 (C-19), and HA (Y-11) (Santa Cruz Biotechnologies); against tubulin, flag, and actin (Sigma); against Set8, H4, H4K20me1, H4K20me2, H2A, H2AX, Cdc2-phospho-Tyr15, p53-phospho-Ser15 and H2AX-phospho-Ser139 (Cell Signaling); against H4K20me3 (Abcam), against histone H3 (active motif); against Cul4A and DDB2 (Rockland Immunochemicals); and against DDB1 (Invitrogen). The anti-Cdt2 antibody was described before (Abbas et al., 2008). Cells were lysed in a Triton X-100 lysis buffer (50 mM Tris at pH 7.5, 250 mM NaCl, 0.1% Triton X-100, 1 mM EDTA, 50 mM NaF, protease inhibitor cocktail (Sigma).
In vivo and in vitro ubiquitylation of Set8
Details of the in vivo and in vitro ubiquitylation reactions were described before (Abbas et al., 2008). Briefly, for in vivo ubiquitin labeling, 293T cells were transiently transfected with either myc-Set8 ± HA-ubiquitin ±flag-Cdt2 or flag-DDB2-expressing plasmids. Forty-eight hours after transfection, the cells were treated with MG132 (20 µg/mL) for two hours prior to lysis. Immunoprecipitated ubiquitylated proteins (anti-HA) or the Set8 protein (anti-Myc) were fractionated on SDS-PAGE, transferred, and immunoblotted with anti-Set8 or anti-ubiquitin lys 48-specific antibody (Millipore), respectively. The procedure for Set8 ubiquitin labeling in vitro was essentially as described before (Abbas et al., 2008), with minor modifications (supplementary information).
Supplementary Material
ACKNOWLEDGMENT
We are grateful to members of the Dutta Lab for helpful suggestions and discussions throughout this work. Recombinant Ubch5c and Ubch3 were generous gift from Michele Pagano, NYU. This work was supported by NIH grant (R01 CA60499) to A.D. T.A. was supported by cancer training grant (T32CA009109) and by NCI grant (KCA140774A).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abbas T, Sivaprasad U, Terai K, Amador V, Pagano M, Dutta A. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 2008;22:2496–2506. doi: 10.1101/gad.1676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, Stein A, Woodcock CL, Skoultchi AI. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- Fang J, Feng Q, Ketel CS, Wang H, Cao R, Xia L, Erdjument-Bromage H, Tempst P, Simon JA, Zhang Y. Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr Biol. 2002;12:1086–1099. doi: 10.1016/s0960-9822(02)00924-7. [DOI] [PubMed] [Google Scholar]
- Havens CG, Walter JC. Docking of a specialized PIP Box onto chromatin-bound PCNA creates a degron for the ubiquitin ligase CRL4Cdt2. Mol Cell. 2009;35:93–104. doi: 10.1016/j.molcel.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, McCall CM, Ohta T, Xiong Y. Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat Cell Biol. 2004;6:1003–1009. doi: 10.1038/ncb1172. [DOI] [PubMed] [Google Scholar]
- Huen MS, Sy SM, van Deursen JM, Chen J. Direct interaction between SET8 and proliferating cell nuclear antigen couples H4-K20 methylation with DNA replication. J Biol Chem. 2008;283:11073–11077. doi: 10.1074/jbc.C700242200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S, Xiong Y. CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem Sci. 2009;34:562–570. doi: 10.1016/j.tibs.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen S, Elvers I, Trelle MB, Menzel T, Eskildsen M, Jensen ON, Helleday T, Helin K, Sorensen CS. The histone methyltransferase SET8 is required for S-phase progression. J Cell Biol. 2007;179:1337–1345. doi: 10.1083/jcb.200706150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien E, Herr W. A switch in mitotic histone H4 lysine 20 methylation status is linked to M phase defects upon loss of HCF-1. Mol Cell. 2004;14:713–725. doi: 10.1016/j.molcel.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Karachentsev D, Sarma K, Reinberg D, Steward R. PR-Set7-dependent methylation of histone H4 Lys 20 functions in repression of gene expression and is essential for mitosis. Genes Dev. 2005;19:431–435. doi: 10.1101/gad.1263005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Starostina NG, Kipreos ET. The CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to control replication licensing. Genes Dev. 2008;22:2507–2519. doi: 10.1101/gad.1703708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Poitelea M, Watson A, Yoshida SH, Shimoda C, Holmberg C, Nielsen O, Carr AM. Transactivation of Schizosaccharomyces pombe cdt2+ stimulates a Pcu4-Ddb1-CSN ubiquitin ligase. EMBO J. 2005;24:3940–3951. doi: 10.1038/sj.emboj.7600854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Nishioka K, Rice JC, Sarma K, Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, Valenzuela P, Tempst P, Steward R, et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell. 2002;9:1201–1213. doi: 10.1016/s1097-2765(02)00548-8. [DOI] [PubMed] [Google Scholar]
- Nishitani H, Shiomi Y, Iida H, Michishita M, Takami T, Tsurimoto T. CDK inhibitor p21 is degraded by a proliferating cell nuclear antigen-coupled Cul4-DDB1Cdt2 pathway during S phase and after UV irradiation. J Biol Chem. 2008;283:29045–29052. doi: 10.1074/jbc.M806045200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph E, Boye E, Kearsey SE. DNA damage induces Cdt1 proteolysis in fission yeast through a pathway dependent on Cdt2 and Ddb1. EMBO Rep. 2006;7:1134–1139. doi: 10.1038/sj.embor.7400827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SL, Portoso M, Mata J, Bahler J, Allshire RC, Kouzarides T. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 2004;119:603–614. doi: 10.1016/j.cell.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Sautel CF, Cannella D, Bastien O, Kieffer S, Aldebert D, Garin J, Tardieux I, Belrhali H, Hakimi MA. SET8-mediated methylations of histone H4 lysine 20 mark silent heterochromatic domains in apicomplexan genomes. Mol Cell Biol. 2007;27:5711–5724. doi: 10.1128/MCB.00482-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senga T, Sivaprasad U, Zhu W, Park JH, Arias EE, Walter JC, Dutta A. PCNA is a cofactor for Cdt1 degradation by CUL4/DDB1-mediated N-terminal ubiquitination. J Biol Chem. 2006;281:6246–6252. doi: 10.1074/jbc.M512705200. [DOI] [PubMed] [Google Scholar]
- Shi X, Kachirskaia I, Yamaguchi H, West LE, Wen H, Wang EW, Dutta S, Appella E, Gozani O. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol Cell. 2007;27:636–646. doi: 10.1016/j.molcel.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens C, La Thangue NB. The emerging role of E2F-1 in the DNA damage response and checkpoint control. DNA Repair (Amst) 2004;3:1071–1079. doi: 10.1016/j.dnarep.2004.03.034. [DOI] [PubMed] [Google Scholar]
- Su C, Gao G, Schneider S, Helt C, Weiss C, O'Reilly MA, Bohmann D, Zhao J. DNA damage induces downregulation of histone gene expression through the G1 checkpoint pathway. EMBO J. 2004;23:1133–1143. doi: 10.1038/sj.emboj.7600120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardat M, Murr R, Herceg Z, Sardet C, Julien E. PR-Set7-dependent lysine methylation ensures genome replication and stability through S phase. J Cell Biol. 2007;179:1413–1426. doi: 10.1083/jcb.200706179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G, Lange HW, Hempel K. Kinetics of histone methylation in vivo and its relation to the cell cycle in Ehrlich ascites tumor cells. Eur J Biochem. 1975;51:609–615. doi: 10.1111/j.1432-1033.1975.tb03963.x. [DOI] [PubMed] [Google Scholar]
- Xiao B, Jing C, Kelly G, Walker PA, Muskett FW, Frenkiel TA, Martin SR, Sarma K, Reinberg D, Gamblin SJ, et al. Specificity and mechanism of the histone methyltransferase Pr-Set7. Genes Dev. 2005;19:1444–1454. doi: 10.1101/gad.1315905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Mizzen CA. The multiple facets of histone H4-lysine 20 methylation. Biochem Cell Biol. 2009;87:151–161. doi: 10.1139/O08-131. [DOI] [PubMed] [Google Scholar]
- Yang H, Pesavento JJ, Starnes TW, Cryderman DE, Wallrath LL, Kelleher NL, Mizzen CA. Preferential dimethylation of histone H4 lysine 20 by Suv4-20. J Biol Chem. 2008;283:12085–12092. doi: 10.1074/jbc.M707974200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Chen Y, Dutta A. Rereplication by depletion of geminin is seen regardless of p53 status and activates a G2/M checkpoint. Mol Cell Biol. 2004;24:7140–7150. doi: 10.1128/MCB.24.16.7140-7150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.