Abstract
Endonuclease III from Escherichia coli, yeast (yNtg1p and yNtg2p) and human and E.coli endonuclease VIII have a wide substrate specificity, and recognize oxidation products of both thymine and cytosine. DNA containing single dihydrouracil (DHU) and tandem DHU lesions were used as substrates for these repair enzymes. It was found that yNtg1p prefers DHU/G and exhibits much weaker enzymatic activity towards DNA containing a DHU/A pair. However, yNtg2p, E.coli and human endonuclease III and E.coli endonuclease VIII activities were much less sensitive to the base opposite the lesion. Although these enzymes efficiently recognize single DHU lesions, they have limited capacity for completely removing this damaged base when DHU is present on duplex DNA as a tandem pair. Both E.coli endonuclease III and yeast yNtg1p are able to remove only one DHU in DNA containing tandem lesions, leaving behind a single DHU at either the 3′- or 5′-terminus of the cleaved fragment. On the other hand, yeast yNtg2p can remove DHU remaining on the 5′-terminus of the 3′ cleaved fragment, but is unable to remove DHU remaining on the 3′-terminus of the cleaved 5′ fragment. In contrast, both human endonuclease III and E.coli endonuclease VIII can remove DHU remaining on the 3′-terminus of a cleaved 5′ fragment, but are unable to remove DHU remaining on the 5′-terminus of a cleaved 3′ fragment. Tandem lesions are known to be generated by ionizing radiation and agents that generate reactive oxygen species. The fact that these repair glycosylases have only a limited ability to remove the DHU remaining at the terminus suggests that participation of other repair enzymes is required for the complete removal of tandem lesions before repair synthesis can be efficiently performed by DNA polymerase.
INTRODUCTION
Ionizing radiation produces a wide spectrum of DNA damage including AP sites, base damage, single and double strand breaks (DSB), multiply damaged sites (MDS) and DNA–protein and DNA–DNA crosslinks (1,2). MDS are small regions in which multiple lesions are clustered, usually within a single helical turn of the DNA (3,4). DNA containing tandem base modification, closely opposed lesions and crosslinks between bases or the sugar–phosphate backbone are other examples of DNA containing MDS. It is important to point out that DNA DSB are a form of MDS that are comprised of closely opposed single strand breaks and have been estimated to contribute to ~10% of the total MDS generated per lethal dose of ionizing radiation (5). DSB are thought to be repaired primarily by the homologous recombination pathway in yeast (6–9) and bacteria (9–11), whereas in mammalian cells, DSB are repaired primarily by a non-homologous end-joining process, mediated by the Ku70/80/DNA-PK complex (9,12–15).
DNA lesions such as abasic (AP) sites, single strand breaks and base damages are repaired via the base excision repair (BER) pathway (9,16–18). It is most likely that individual lesions within MDS, in particular the base lesions and single strand breaks, are recognized by repair enzymes of the BER pathway (9). However, whether the presence of multiple lesions in close proximity will affect the efficiency of repair by these enzymes has not been well addressed. For example, inappropriate processing of closely opposed base lesions could lead to the production of DSB and thus an increase in cellular lethality and/or mutagenesis. Recently, the in vitro processing of closely opposed base lesions by bacterial repair enzymes was examined. BER enzymes such as endonucleases III (19–21), IV (22) and VIII (20), exonuclease III (22) and formamidopyrimidine N-glycosylase (21,23) are inhibited in vitro by strand breaks formed by nicking at oppositely placed base lesions. In contrast, prolonged incubation or the addition of excess amounts of repair enzymes to DNA containing closely opposed lesions leads to the formation of DSB (19,20).
MDS such as tandem base lesions have been shown to be readily formed by ionizing radiation (24–27) as well as by photosensitization reactions (28,29). Tandem base lesions consisting of N-(2′-deoxy-β-erythro-pentofuranosyl)-formamide (dβF) and 8-oxo-deoxyguanosine (8-oxoG) have been identified in irradiated dinucleotides of d(TpG), d(CpG), d(GpT) and d(GpC) as well as in calf thymus DNA (24,25). Fpg was shown to recognize DNA containing tandem dβF–8-oxoG, and its ability to fully remove both lesions was found to be dependent on the arrangement of these two base damage within MDS (23). Since 8-oxoG is not a substrate for endonuclease III, the enzyme can only remove dβF in tandem dβF–8-oxoG lesions, leaving behind 8-oxoG at the nicked terminus. In this manuscript we describe the in vitro recognition of tandem dihydrouracil (DHU) lesions by Escherichia coli, yeast and human endonuclease III as well as E.coli endonuclease VIII. We demonstrate that these enzymes can efficiently remove only one of the two DHU lesions. Once DNA is nicked either 5′ or 3′ to a DHU site, subsequent recognition of the remaining DHU by these enzymes was inhibited. These results suggested additional repair enzymes are needed to complete the repair of these tandem lesions initiated by DNA N-glycosylases before repair synthesis can be efficiently carried out by DNA polymerase. Alternatively, these lesions may not be efficiently repaired at all by cells, thus leading to increased cellular lethality.
MATERIALS AND METHODS
Enzymes
Escherichia coli endonuclease III gene nth was cloned into the NdeI restriction site of pET12(b)+ plasmid and transformed into E.coli strain BL21(DE3). Cultures of E.coli strain BL21(DE3) harboring the overproducing plasmid pET22b-nth were grown to an absorbance of 0.6 at 600 nm, then induced with 1 mM IPTG and harvested after 3 h. The collected cell paste was then resuspended in 50 mM HEPES–KOH, pH 7.6, 250 mM KCl, 0.1% Triton X-100, 1 mg/ml lysozyme and subjected to three freeze–thawing cycles. The DNA in the crude extract was then removed by the addition of PEG8000 and E.coli endonuclease III (eNTH) was then purified using MonoS, MonoQ and then Phenyl Sepharose chromatography as described previously (30). Endonuclease VIII (eNEI) was prepared from an E.coli strain BL21(DE3) host harboring the expression plasmid pET22b-nei (Dr Zafer Hetahet, University of Texas at Tyler). Similarly, logarithmic growing cultures (OD600 = 0.5) were induced with 1 mM IPTG, and the overexpressed endonuclease VIII was purified according to the procedure described previously (31). Human endonuclease III (hNTH) was purified by Dr Robindra Roy (32) and yeast endonuclease III homologs, yNtg1p and yNtg2p were purified by Dr Ho Jin You (33).
DNA substrates
All oligonucleotides were obtained from Operon and purified by polyacrylamide gel (15%) electrophoresis as described previously (34). Oligonucleotides containing DHU were 5′-end-labeled with [γ-32P]ATP (Amersham Corp.) using T4 polynucleotide kinase or 3′-end-labeled with [α-32P]cordycepin 5′-triphosphate (Du Pont NEN) using deoxynucleotidyl terminal transferase following instructions from the enzyme supplier (US Biochemical Corp.). Labeled oligonucleotides containing DHU were annealed to the appropriate complementary strands at a 1:1.5 ratio in 10 mM Tris–HCl, pH 7.5, 0.1 M NaCl and 2 mM 2-mercaptoethanol by heating the mixture to 90°C and cooling gradually to room temperature. The following oligonucleotide duplexes were used in this study (Q is DHU):
5′- TTCCAGACTGTCCTTCGTQCCTTTCCTCTCAA DHU-19/A
AAGGTCTGACAGGAAGCAAGGAAAGGAGAGTT-5′;
5′- TTCCAGACTGTCCTTCGTCQCTTTCCTCTCAA DHU-20/A
AAGGTCTGACAGGAAGCAAGGAAAGGAGAGTT-5′;
5′- TTCCAGACTGTCCTTCGTQTCTTTCCTCTCAA DHU-19/G
AAGGTCTGACAGGAAGCAAGGAAAGGAGAGTT-5′;
5′- TTCCAGACTGTCCTTCGTTQCTTTCCTCTCAA DHU-20/G
AAGGTCTGACAGGAAGCAAGGAAAGGAGAGTT-5′;
5′- TTCCAGACTGTCCTTCGTQQCTTTCCTCTCAA DHU/AG
AAGGTCTGACAGGAAGCAAGGAAAGGAGAGTT-5′;
5′- TTCCAGACTGTCCTTCGTQQCTTTCCTCTCAA DHU/GA
AAGGTCTGACAGGAAGCAAGGAAAGGAGAGTT-5′.
DHU is a good model for thymine damage with ring saturation at 5,6 double bond. Therefore, oligonucleotides containing DHU opposite an A were used as a model for DNA substrate containing thymine radiolysis damage, since thymine base lesions such as thymine glycol and dihydrothymine are lesions that paired with A. Similarly, oligonucleotides containing DHU opposite G were used as a model for a DNA substrate containing a cytosine radiolysis product. DHU is a stable radiolysis product of cytosine under hypoxic conditions, resulting from ring saturation followed by deamination.
Endonuclease assays
Endonuclease III from E.coli, human and Saccharomyces cerevisiae (yNtg1p and yNtg2p proteins) and E.coli endonuclease VIII were assayed in a standard reaction buffer (20 µl) containing 100 mM KCl, 10 mM Tris–HCl, pH 7.5, 1 mM EDTA, 10 nM labeled substrate and 40 ng of protein. Reactions were performed at 37°C for 30 min and then stopped by the addition of 10 µl of loading buffer (90% formamide, 1 mM EDTA, 0.1% xylene cyanol and 0.1% bromophenol blue) followed by heating at 90°C for 5 min. Reaction products were then analyzed by denaturing PAGE (15%). Electrophoresis was performed at a constant voltage of 2000 V for 4 h. The polyacrylamide gel was then dried under vacuum and exposed to X-ray film or analyzed with a Fuji Bio-Imaging analyzer.
RESULTS AND DISCUSSION
Double-stranded oligonucleotides containing DHU as substrates for yNtg1p and yNtg2p
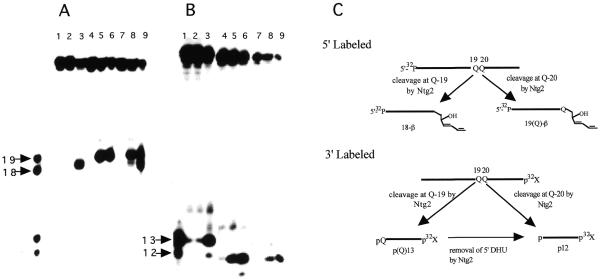
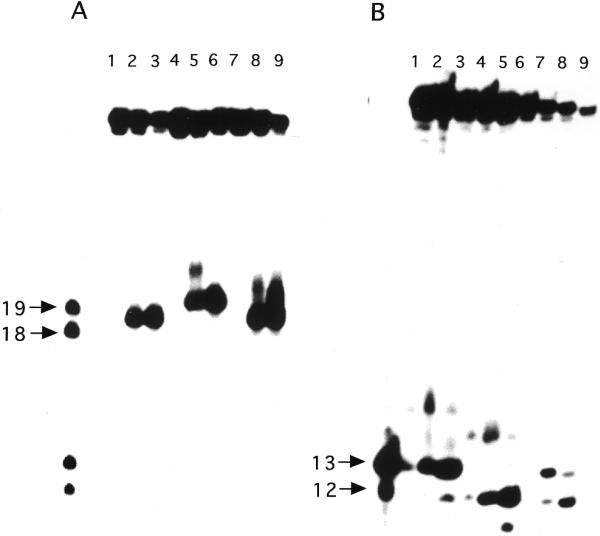
Saccharomyces cerevisiae possesses two endonuclease III activities, namely the yNtg1p and yNtg2p proteins (33,35). In order to examine whether both enzymes can efficiently remove DHU when two of these lesions are present in a tandem context on the same DNA strand, double-stranded oligonucleotides DHU/AG and DHU/GA containing tandem DHU opposite AG and GA pairs, respectively, were used as DNA substrates. Due to the fact that there might also be flanking sequence context effects on the processing of each individual lesion by repair enzymes, duplexes containing a single DHU within the same flanking sequence context were also utilized. These duplexes were constructed in such a way that DHU was placed either opposite an A or G at position 19 (DHU-19/A and DHU-19/G) or at position 20 (DHU-20/A and DHU-20/G) from the 5′-terminus. It is known that both yNtg1p and yNtg2p produce β-elimination products that contain 4-hydroxy-2-pentenal moieties attached to 3′-termini of cleavage products (9,16). The β-elimination product derived from a 5′-end-labeled DHU-A/19 is expected to migrate slower than a 18mer but faster than a 19mer. Similarly, the β-elimination product derived from a 5′-end-labeled DHU-G/20 is expected to migrate slower than a 19mer. Figure 1 shows that both yNtg1p and yNtg2p generate the expected β-elimination products from 5′-end-labeled DHU-19/A, DHU-20/G and DHU/AG. Using 5′-labeled substrates, yNtg1p recognized DHU quite poorly when the lesion was placed opposite A at position 19 (Fig. 1A, lane 2). In contrast, DHU-20/G, which contained a DHU opposite G at position 20, was a good substrate for yNtg1p (Fig. 1A, lane 5). In contrast, both DHU-19/A and DHU-20/G were good substrates for yNtg2p (Fig. 1A, lanes 3 and 6). The apparent sequence context effect on yNtg1p activity was also observed when 5′-labeled DHU/AG was used as a DNA substrate. yNtg1p produced predominantly a cleavage product that migrated to the same gel position as the cleavage product derived from DHU-20/G (Fig. 1A, lane 8), and yNtg2p produced two cleavage products corresponding to cleavages occurring at positions 19 and 20 (Fig. 1A, lane 9).
Figure 1.
Substrate specificity of yNtg1p and yNtg2p on oligonucleotides containing a DHU lesion. (A) 100 fmol of 5′-32P-labeled duplex DHU-19/A (lanes 1–3), DHU-20/G (lanes 4–6) and DHU/AG (lanes 7–9) were incubated with 40 ng of yeast yNtg1p or yNtg2p at 37°C for 30 min in a standard reaction mix. Lane 1, control, DHU-19/A; lane 2, DHU-19/A + yNtg1p; lane 3, DHU-19/A + yNtg2p; lane 4, control, DHU-20/G; lane 5, DHU-20/G + yNtg1p; lane 6, DHU-20/G + yNtg2p; lane 7, control, DHU/AG; lane 8, DHU/AG + yNtg1p; lane 9, DHU/AG + yNtg2p. Arrows indicate the positions of 5′-32P-labeled 18mer and 19mer standards. (B) 100 fmol of 3′-32P-labeled duplex DHU-19/A (lanes 1–3), DHU-20/G (lanes 4–6) and DHU/AG (lanes 7–9) were incubated with 40 ng of yeast yNtg1p or yNtg2p at 37°C for 30 min in a standard reaction mix. Lane 1, control, DHU-19/A; lane 2, DHU-19/A + yNtg1p; lane 3, DHU-19/A + yNtg2p; lane 4, control, DHU-20/G; lane 5, DHU-20/G + yNtg1p; lane 6, DHU-20/G + yNtg2p; lane 7, control, DHU/AG; lane 8, DHU/AG + yNtg1p; lane 9, DHU/AG + yNtg2p. Arrows indicate the positions of 3′-32P-labeled 12mer and 13mer standards. (C) A scheme describing the ability of yNtg2p to further remove the DHU lesion remaining on the 5′-terminus.
Using 3′-end-labeled DNA substrates, corroborating results were observed for both yNtg1p and yNtg2p on DNA containing DHU. The 3′ cleavage products generated by both yNtg1p and yNtg2p on DHU-19/A and DHU-20/G were 13mers and 12mers containing a 5′-phosphoryl group, respectively. Using DNA containing a single DHU as a substrate, yNtg1p poorly recognized DHU at position 19 when it was opposite to an A (Fig. 1B, lane 2). Conversely, when DHU was placed at position 20 opposite a G, yNtg1p recognized DHU efficiently (Fig. 1B, lane 5). yNtg2p recognized DHU equally well at both positions 19 and 20 (Fig. 1B, lanes 3 and 6). These data thus confirmed the results obtained with the 5′-end-labeled substrate. However, when 3′-labeled substrate containing tandem DHU lesions was used, both yNtg1p and yNtg2p generated a cleavage product with an electrophoretic mobility similar to the product derived from cleavage occurring at position 20 only (Fig. 1B, lanes 8 and 9). Since yNtg1p poorly recognized DHU-19/A compared to DHU-20/G (Fig. 1B, lanes 2 and 5), it was therefore expected that yNtg1p would produce predominantly a single cleavage product that migrated to the same distance as the product derived from cleavage of DHU at position 20 (Fig. 1B, lane 8). However, yNtg2p produced primarily a single cleavage product with 3′-labeled DHU/AG (Fig. 1B, lane 8) but generated two cleavage products with 5′-labeled DHU/AG (Fig. 1A, lane 8), suggesting equal recognition of DHU at either position 19 or 20. The observation that yNtg2p generated primarily a single cleavage product with 3′-labeled DHU/AG suggests that DHU left at the 5′-terminus of the 3′ fragment derived from cleavage at position 19 was further processed by yNtg2p, generating a product with an electrophoretic mobility identical to the product derived from cleavage occurring at position 20 (Fig. 1C).
The apparent sequence context effect on yNtg1p and yNtg2p activity was further examined by switching the positions of the DHU/A and DHU/G pairs in the substrate. 5′-End-labeled substrates containing a DHU/G pair at position 19 (DHU-19/G) and DHU/A pair at position 20 (DHU-20/A) were incubated with yNtg1p and yNtg2p. In this case, DHU was recognized efficiently by both yNtg1p and yNtg2p at positions 19 and 20 (Fig. 2A, lanes 2, 3, 5 and 6). As expected, with 5′-labeled DNA containing tandem DHU (DHU/GA), yNtg2 produced two cleavage products in approximately equal amounts (Fig. 2A, lane 9). However, yNtg1p generates one predominant cleavage product that corresponded to cleavage occurring at DHU-19 (DHU at this position was opposite G) and a minor amount of cleavage product that corresponded to a product derived from cleavage occurring at position 20 (DHU at this position was opposite A) (Fig. 2A, lane 8). Taken together these data suggested that when tandem DHU lesions are present, yNtg1p exhibited a higher affinity for a DHU/G pair than a DHU/A pair. When 3′-end-labeled duplexes containing a single DHU were used as substrates, both yNtg1p and yNtg2p efficiently recognized DHU at positions 19 (Fig. 2B, lanes 2 and 3) and 20 (Fig. 2B, lanes 5 and 6). However, when 3′-end-labeled DHU/GA was used as a DNA substrate, yNtg1p produced predominantly a 3′ fragment that corresponded to a cleavage product derived from cleavage occurring at DHU-19. A small amount of product that corresponded to cleavage occurring at DHU-20 was also observed. The data obtained for yNtg1p using 3′-end-labeled DHU/GA were consistent with those obtained with 5′-end-labeled DHU/GA, further supporting the idea that yNtg1p has a preference for a DHU/G pair over a DHU/A pair. Interestingly, yNtg2p generated predominantly a 3′ fragment that corresponded to the cleavage product derived from cleavage at DHU-20 (Fig. 2B, lane 9). In addition, a small amount of product that corresponded to cleavage at DHU-19 was also observed (Fig. 2B, lane 9). Taken together, these results suggested that yNtg2p was capable of removing the DHU that remained at the 5′-terminus of the cleavage product (Fig. 1C).
Figure 2.
Substrate specificity of yNtg1p and yNtg2p on oligonucleotides containing a DHU lesion. (A) 100 fmol of 5′-32P-labeled duplex DHU-19/G (lanes 1–3), DHU-20/A (lanes 4–6) and DHU/GA (lanes 7–9) were incubated with 40 ng of yeast yNtg1p or yNtg2p at 37°C for 30 min in a standard reaction mix. Lane 1, control, DHU-19/G; lane 2, DHU-19/G + yNtg1p; lane 3, DHU-19/G + yNtg2p; lane 4, control, DHU-20/A; lane 5, DHU-20/A + yNtg1p; lane 6, DHU-20/A + yNtg2p; lane 7, control, DHU/GA; lane 8, DHU/GA + yNtg1p; lane 9, DHU/GA + yNtg2p. Arrows indicate the positions of 5′-32P-labeled 18mer and 19mer standards. (B) 100 fmol of 3′-32P-labeled duplex DHU-19/G (lanes 1–3), DHU-20/A (lanes 4–6) and DHU/GA (lanes 7–9) were incubated with 40 ng of yeast yNtg1p or yNtg2p at 37°C for 30 min in a standard reaction mix. Lane 1, control, DHU-19/G; lane 2, DHU-19/G + yNtg1p; lane 3, DHU-19/G + yNtg2p; lane 4, control, DHU-20/A; lane 5, DHU-20/A + yNtg1p; lane 6, DHU-20/A + yNtg2p; lane 7, control, DHU/GA; lane 8, DHU/GA + yNtg1p; lane 9, DHU/GA + yNtg2p. Arrows indicate the positions of 3′-32P-labeled 12mer and 13mer standards.
Double-stranded oligonucleotides containing DHU as substrates for hNTH, eNTH and eNEI
The ability of both eNTH and hNTH as well as eNEI in processing DNA containing tandem DHU lesions was also examined in a similar manner as described above for yNtg1p and yNtg2p. Similar to the yeast homologs, eNTH and hNTH are AP lyases that catalyze β-elimination reactions, whereas the AP lyase activity of eNEI catalyzes a β,δ-elimination reaction (9,16). The β-elimination products generated by both human and E.coli endonuclease III have a slower electrophorectic mobility than the β,δ-elimination product generated by endonuclease VIII (Fig. 3A, compare lanes 2 and 4 with lane 3, and lanes 6 and 7 with lane 8). This is due to the fact that the 3′-terminus of a β-elimination product contains a 4-hydroxy-2-pentenal moiety, while the 3′-terminus of a β,δ-elimination product contains a phosphate group.
Figure 3.
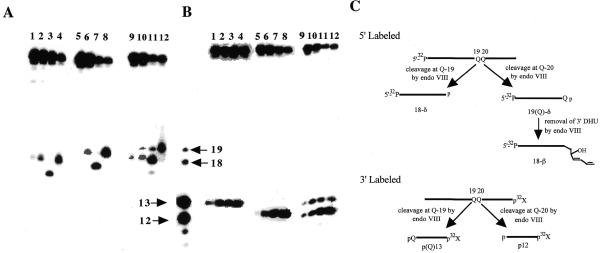
Substrate specificity of eNTH, hNTH and eNEI on oligonucleotides containing a DHU lesion. (A) 100 fmol of 5′-32P-labeled duplex DHU-19/A (lanes 1–4), DHU-20/G (lanes 5–8) and DHU/AG (lanes 9–12) were incubated with 40 ng of eNTH, hNTH or eNEI at 37°C for 30 min in a standard reaction mix. Lane 1, control, DHU-19/A; lane 2, DHU-19/A + eNTH; lane 3, DHU-19/A + eNEI; lane 4, DHU-19/A + hNTH; lane 5, control, DHU-20/G; lane 6, DHU-20/G + eNTH; lane 7, DHU-20/G + eNEI; lane 8, DHU-20/G + hNTH lane 9, control, DHU/AG; lane 10, DHU/AG + eNTH; lane 11, DHU/AG + eNEI; lane 12, DHU/AG + hNTH. (B) 100 fmol of 3′-32P-labeled duplex DHU-19/A (lanes 1–4), DHU-20/G (lanes 5–8) and DHU/AG (lanes 9–12) were incubated with 40 ng of eNTH, hNTH or eNEI at 37°C for 30 min in a standard reaction mix. Lane 1, control, DHU-19/A; lane 2, DHU-19/A + eNTH; lane 3, DHU-19/A + eNEI; lane 4, DHU-19/A + hNTH; lane 5, control, DHU-20/G; lane 6, DHU-20/G + eNTH; lane 7, DHU-20/G + eNEI; lane 8, DHU-20/G + hNTH; lane 9, control, DHU/AG; lane 10, DHU/AG + eNTH; lane 11, DHU/AG + eNEI; lane 12, DHU/AG + hNTH. (C) A scheme describing the ability of eNEI to further process the DHU remaining at the 3′-terminus, generating a β-elimination product, 18-β. Reactions described in (A) and (B) were analyzed in the same sequencing gel, separated with the four DNA standards as indicated by arrows. Arrows indicate the positions of 5′-32P-labeled 18mer and 19mer standards and 3′-32P-labeled 12mer and 13mer standards.
Figure 3 shows that DHU-19/A and DHU-20/G are good substrates for eNTH, hNTH and eNEI (Fig. 3A, lanes 2–4 and 6–8; Fig. 3B, lanes 2–4 and 6–8). When the 5′-end-labeled DHU/AG was used as a substrate, eNTH generated two cleavage products with electrophoretic mobilities identical to the expected β-elimination products generated from cleavages at positions 19 and 20 (Fig. 3A, lane 10). Similar results were obtained for eNTH using 3′-end-labeled substrate (Fig. 3B, lane 2).
hNTH showed a similar activity towards DHU-19/A and DHU-20/G (Fig. 3A, lanes 4 and 8; Fig. 3B, lanes 4 and 8) It was therefore expected that when 5′-end-labeled DHU/AG was used as a substrate, hNTH would generate approximately equal amounts of cleavage products corresponding to incisions occurring at positions 19 and 20. Instead, the enzyme generated a major product with an electrophoretic mobility similar to the β-elimination product derived from cleavage at position 20, and a minor β-elimination product corresponded to cleavage at position 19 (Fig. 3A, lane 12). The 5′ cleavage pattern observed for hNTH agreed with that obtained using 3′-end-labeled DHU/AG as the DNA substrate (Fig. 3B, lane 4). These data suggested that when the DHU lesion was present in tandem, hNTH preferentially recognized the DHU/G pair, leading to a preferential nicking at position 20. Alternatively, the appearance of a higher amount of 3′-labeled fragment p12 (cleavage at position 20) could be the result of a sequential cleavage at positions 19 and 20, thus leading to an apparent preference of cleavage at position 20.
eNEI is an AP lyase that catalyzes a β,δ-elimination reaction. eNEI showed similar activity for DHU lesions at positions 19 and 20, generating the expected β,δ-elimination products (Fig. 3A, lanes 3 and 5). Interestingly, using 5′-end-labeled DHU/AG that contained a tandem DHU, eNEI generated three cleavage products. A major product with an electrophoretic mobility similar to the β-elimination product derived from cleavage at DHU at position 19 and minor amounts of β- and β,δ-elimination products derived from cleavages occurred at positions 20 and 19, respectively (Fig. 3A, lane 12). However, using 3′-end-labeled DHU/AG as a DNA substrate, eNEI generated a major cleavage product that corresponded to cleavage occurring at DHU at position 20 (Fig. 3B, indicated by 12 and an arrow) and a smaller amount of a product that corresponds to cleavage at DHU at position 19 (Fig. 3B, indicated by 13 and an arrow). In order to explain the differences in the cleavage pattern obtained with 5′- and 3′-end-labeled substrates as well as the apparent lack of β,δ-elimination products, we suggest that after the initial removal of DHU by eNEI at position 20, the 5′ fragment that contains the DHU at position 19 is further processed by eNEI. eNEI is unable to complete the β,δ-elimination reaction, however, it is able to catalyze a β-elimination at the DHU moiety located at the 3′-terminus, generating β-elimination products at position 19 (Fig. 3C). It is important to point out that eNEI is a β,δ-AP lyase and does not normally generate a β-elimination product; thus the formation of the β-elimination product observed under such a reaction condition is interesting. The ability of a β,δ-AP lyase to generate a β-elimination product has also been observed previously for E.coli formamidopyrimidine N-glycosylase (9,16).
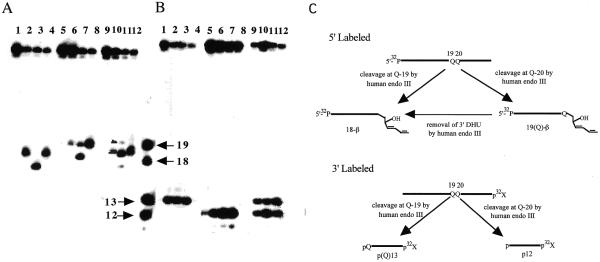
The possibility of a sequence context effect on these repair enzymes was also examined by exchanging the positions of the DHU/G and DHU/A pairs. Figure 4 shows that eNTH, hNTH and eNEI efficiently recognize both DHU-19/G and DHU-20/A (Fig. 4A, lanes 2–4 and 6–8; Fig. 4B, lanes 2–4 and 6–8). This result is comparable to those obtained with DHU-19/A and DHU-20/G. Using 5′-end-labeled DHU/GA as a substrate, eNTH generated two β-elimination products, with the major product corresponding to cleavage occurring at position 19 (Fig. 4A, lane 10). Similar results were obtained for eNTH using 3′-end-labeled DHU/GA (Fig. 4B, lane 10).
Figure 4.
Substrate specificity of eNTH, hNTH and eNEI on oligonucleotides containing a DHU lesion. (A) 100 fmol of 5′-32P-labeled duplex DHU-19/G (lanes 1–4), DHU-20/A (lanes 5–8) and DHU/GA (lanes 9–12) were incubated with 40 ng of eNTH, hNTH or eNEI at 37°C for 30 min in a standard reaction mix. Lane 1, control, DHU-19/G; lane 2, DHU-19/G + eNTH; lane 3, DHU-19/G + eNEI; lane 4, DHU-19/G + hNTH; lane 5, control, DHU-20/A; lane 6, DHU-20/A + eNTH; lane 7, DHU-20/A + eNEI; lane 8, DHU-20/A + hNTH; lane 9, control, DHU/GA; lane 10, DHU/GA + eNTH; lane 11, DHU/GA + eNEI; lane 12, DHU/GA + hNTH. (B) 100 fmol of 3′-32P-labeled duplex DHU-19/G (lanes 1–4), DHU-20/A (lanes 5–8) and DHU/GA (lanes 9–12) were incubated with 40 ng of eNTH, hNTH or eNEI at 37°C for 30 min in a standard reaction mix. Lane 1, control, DHU-19/G; lane 2, DHU-19/G + eNTH; lane 3, DHU-19/G + eNEI;. lane 4, DHU-19/G + hNTH; lane 5, control, DHU-20/A; lane 6, DHU-20/A + eNTH; lane 7, DHU-20/A + eNEI; lane 8, DHU-20/A + hNTH; lane 9, control, DHU/GA; lane 10, DHU/GA + eNTH; lane 11, DHU/GA + eNEI; lane 12, DHU/GA + hNTH. (C) A scheme describing the ability of hNTH to further process the DHU remaining at the 3′-terminus, generating a β-elimination product, 18-β. Reactions described in (A) and (B) were analyzed in the same sequencing gel, separated with the four DNA standards as indicated by arrows. Arrows indicate the positions of 5′-32P-labeled 18mer and 19mer standards and 3′-32P-labeled 12mer and 13mer standards.
The results obtained for hNTH using DHU/GA as a substrate were similar to those obtained with DHU/AG. When 5′-end-labeled DHU/GA was utilized as a substrate, hNTH generated a major product that possessed an electrophoretic mobility equivalent to the β-elimination product derived from cleavage at position 19 (Fig. 4A, lane 12). However, when 3′-end-labeled DHU/GA was used as a DNA substrate, hNTH generated the expected cleavage products in approximately equal amounts (Fig. 4B, lane 12). The hNTH endonuclease activity was found to be similar for DHU-19/G and DHU-20/A (Fig. 4A, lanes 4 and 8; Fig. 4B, lanes 4 and 8). In order to account for the difference in the 5′ and 3′ cleavage patterns, we suggest that hNTH removed the DHU remaining at the 3′-terminus of the β-elimination product derived from cleavage at position 20. Such an event would lead predominantly to the generation of a product with mobility identical to the β-elimination product produced from cleavage occurring at position 19 (Fig. 4C).
eNEI showed a similar activity for DHU-19/G and DHU-20/A, generating the expected β,δ-elimination products that migrated faster than the β-elimination product generated by endonuclease III (Fig. 4A, lanes 3 and 5; Fig. 4B, lanes 3 and 8). Similar to DHU/AG, eNEI generated three cleavage products when 5′-end-labeled DHU/GA was used as a substrate; a major product with an electrophoretic mobility similar to the β-elimination product derived from cleavage at position 19 and a minor β- and β,δ-elimination products derived from cleavages occurring at positions 20 and 19, respectively (Fig. 4A, lane 12). Similar to DHU/AG, eNEI generated two expected cleavage products that corresponded to cleavage occurring at DHU at positions 19 and 20 (Fig. 4B, lane 11). The cleavage pattern can be explained by the same rationale used for the eNEI-induced cleavage pattern for DHU/AG (Fig. 3C).
Based on the data presented above, it appears that eNTH is unable to further process DHU when left at a cleaved terminus. However, both hNTH and eNEI were able to further process the DHU remaining at the 3′-terminus. In order to further analyze the ability of these two repair enzymes to process DHU remaining at the 3′-terminus, a time course experiment was performed for hNTH and eNEI, using duplexes containing tandem DHU. Using 5′-end-labeled DHU/GA, hNTH generated a major cleavage product corresponding to the product derived from cleavage at DHU-19 (18-β). However, using 3′-end-labeled substrates, the cleavage product that corresponded to cleavage at DHU-20 (12mer) was generated at a similar rate to the product resulting from cleavage at DHU-19 (13mer) (data not shown). For eNEI, the rate of formation of 18-β was higher compared to 18-δ and 19-β when 5′-end-labeled DHU/GA was used as a substrate (data not shown). In contrast, when 3′-end-labeled DHU/GA was utilized as a substrate, both the 12mer and 13mer were formed at an approximately equal rate (data not shown).
CONCLUSION
The energy from ionizing radiation is not deposited uniformly into the absorbing medium but along the tracks of the charged particles (36). The energy is thus deposited in the form of ‘spurs’, which consist of three to four ion pairs within a diameter of 4 nm, or as a ‘blob’, which consists of as many as 12 ion pairs within a diameter of 7 nm (36). For X-rays, 95% of the energy deposition events are spurs (36). When tracks of ionization overlap the DNA helix, multiple radical reactions occur with the DNA, leading to the generation of various lesions clustered within a small region of DNA defined by the manner of energy deposition. The clustered lesion, or MDS as it is commonly referred to, consists of a variety of radiation-induced damage including base and sugar damage, DNA strand breaks as well as different types of crosslinks.
In this study, we have shown that in vitro repair of a form of MDS which consists of tandem DHU lesions located on the same DNA strand. Even though DHU is a good substrate for all endonuclease III functional homologs used in this study, DNA containing tandem DHU lesions are not efficiently removed by these N-glycosylases. eNTH and yNtg1p can only recognize one of the two DHU lesions, and once a DNA incision was made, the lesion that remained at the nick site was not further processed by these enzymes. It is possible that when the first DHU is removed by eNTH and yNtg1p, these enzymes dissociate rapidly from the nicked substrate, leading to the accumulation of nicked DNA with one of the unprocessed DHUs. For yNtg2p, hNTH and eNEI, these enzymes show limited potential in removing lesions remaining at the nick site after the first incision. However, yNtg2p is able to remove the lesion left at the 5′-terminus, but is unable to remove lesions left at the 3′-terminus. In contrast, both hNTH and eNEI can remove lesions remaining at 3′-termini but are unable to remove lesions remaining at 5′-termini. Despite the apparent overlapping substrate specificities for eNTH, eNEI, yeast Ntg1 and Ntg2, these data suggest that the apparent redundancy in these BER enzymes serves not only to increase the repair capacity for individual lesions, but also to function in the removal of lesions embedded in various sequence contexts. The presence of multiple enzymes possessing similar substrate specificities thus provides cells with a comprehensive repair repertoire for reducing the genetic damage load of cells, preventing mutations and maintaining a high fidelity in the passage of genetic material to progeny cells.
Furthermore, these data suggest that tandem DHU lesions are inefficiently removed by these DNA glycosylases. Based on the above discussion, it is clear that BER enzymes are unable to completely remove both lesions in vitro. However, it is becoming clear that repair activities of DNA glycosylases can be dramatically enhanced in the presence of other accessory proteins. Recent studies showed that interaction of N-methylpurine DNA glycosyalse (MPG) with hHR23 led to a dramatic increase in the glycosylase activity of MPG (37). Similarly, interaction of the human XPG protein with hNTH also led to a 3-fold increase in the activity of hNTH (38). It is therefore possible that accessory proteins can help to overcome the shortcomings of these DNA glycosylase, thus helping to process tandem lesions more efficiently. Otherwise, additional repair enzymes might be needed before repair synthesis and ligation could occur to complete the repair. The participation of repair enzymes that can help to remove lesions remaining at either the 3′- or 5′-terminus are essential for the repair of tandem lesions. These enzymes could be either exonucleases or novel repair glycosylases/endonucleases that have high affinity for terminal lesions are potential partners for the BER enzymes for the repair of tandem lesions. Failure to remove lesions remaining at the termini could lead to persistence of strand breaks which could result in increases in cell death or mutations.
Acknowledgments
ACKNOWLEDGEMENTS
This research was supported by grants from the National Institutes of Health (grants GM 37216 and GM 54163 to Y.W.K., grant CA78622 to P.W.D. and grant CA80917 to R.R.). H.J.Y. was also supported by Chosun University (Republic of Korea) and KOSEF (1999-1-208-001-5).
References
- 1.Huttermann J., Kuhnlein,W. and Teoule,R. (1978) Effects of Ionizing Radiation on DNA. Physical, Chemical, and Biological Aspects. Springer-Verlag, Berlin. [PubMed]
- 2.Von Sonntag C. (1987) The Chemical Basis of Radiation Biology. Taylor and Francis, London, UK.
- 3.Ward J.F. (1981) Some biochemical consequences of the spatial distribution of ionizing radiation-produced free radicals. Radiat. Res., 86, 185–195. [PubMed] [Google Scholar]
- 4.Ward J.F. (1988) DNA damage produced by ionizing radiation in mammalian cells: identities, mechanisms of formation, and repairability. Prog. Nucleic Acids Res. Mol. Biol., 35, 95–125. [DOI] [PubMed] [Google Scholar]
- 5.Ward J.F., Limoli,C.L., Calabrp-Jones,P. and Evans,J.W. (1988) In Nygaard,O.F., Simic,M. and Cerutti,P. (eds), Anticarcinogenesis and Radiation Protection. Plenum, New York, NY.
- 6.Namsaraev E.A. and Berg,P. (1998) Branch migration during Rad51-promoted strand exchange proceeds in either direction. Proc. Natl Acad. Sci. USA, 95, 10477–10488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clever B., Interthal,H., Schmuckli-Maurer,J., King,J., Sigrist,M. and Heyer,W.D. (1997) Recombinational repair in yeast: functional interactions between Rad51 and Rad54 proteins. EMBO J., 16, 2535–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wood R.D. (1996) DNA repair in eukaryotes. Annu. Rev. Biochem., 65, 135–167. [DOI] [PubMed] [Google Scholar]
- 9.Freidberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. American Society of Microbiology Press, Washington, DC.
- 10.Bien M., Steffen,H. and Schulte-Frohlinde,D. (1988) Repair of the plasmid pBR322 damaged by gamma-irradiation or by restriction endonucleases using different recombination-proficient E. coli strains. Mutat. Res., 194, 193–205. [DOI] [PubMed] [Google Scholar]
- 11.Morel P., Cherny,D., Ehrlich,S.D. and Cassuto,E. (1997) Recombination-dependent repair of DNA double-strand breaks with purified proteins from Escherichia coli. J. Biol. Chem., 272, 17091–17096. [DOI] [PubMed] [Google Scholar]
- 12.Ramsden D.A. and Gellert,M. (1998) Ku protein stimulates DNA end joining by mammalian DNA ligases: a direct role for Ku in repair of DNA double-strand breaks. EMBO J., 17, 609–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen F., Peterson,S.R., Story,M.D. and Chen,D.J. (1996) Disruption of DNA-PK in Ku80 mutant xrs-6 and the implications in DNA double-strand break repair. Mutat. Res., 362, 9–19. [DOI] [PubMed] [Google Scholar]
- 14.Rathmell W.K. and Chu,G. (1994) Involvement of the Ku autoantigen in the cellular response to DNA double-strand breaks. Proc. Natl Acad. Sci. USA, 91, 7623–7627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blunt T., Gell,D., Fox,M., Taccioli,G.E., Lehmann,A.R., Jackson,S.P. and Jeggo,P.A. (1996) Identification of a nonsense mutation in the carboxyl-terminal region of DNA-dependent protein kinase catalytic subunit in the scid mouse. Proc. Natl Acad. Sci. USA, 93, 10285–10290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doetsch P.D. and Cunningham,R.P. (1990) The enzymology of apurinic/apyrimidinic endonucleases. Mutat. Res., 236, 173–201. [DOI] [PubMed] [Google Scholar]
- 17.Ramotar D. and Demple,B. (1993) In Halliwell,B. and Aruoma,O.I. (eds), DNA and Free Radicals. Ellis Horwood Ltd, London, UK, pp. 165–191.
- 18.Wilson D.M.,III, Engelward,B.P. and Samson,L. (1998) In Nickoloff,J.A. and Hoekstra,M.F. (eds), DNA Repair in Prokaryotes and Lower Eukaryotes. Humana Press, Totawa, NJ, Vol. I, pp. 29–64.
- 19.Chaudhry M.A. and Weinfeld,M. (1995) The action of Escherichia coli endonuclease III on multiply damaged sites in DNA. J. Mol. Biol., 249, 914–922. [DOI] [PubMed] [Google Scholar]
- 20.Harrison L., Hatahet,Z., Purmal,A.A. and Wallace,S.S. (1998) Multiply damaged sites in DNA: interactions with Escherichia coli endonucleases III and VIII. Nucleic Acids Res., 26, 932–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.David-Cordonnier M.-H., Laval,J. and O’Neill,P. (2000) Clustered DNA damage, influence on damage excision by XRS5 nuclear extracts and Escherichia coli Nth and Fpg proteins. J. Biol. Chem., 275, 11865–11873. [DOI] [PubMed] [Google Scholar]
- 22.Takeshita M., Chang,C.N., Johnson,F., Will,S. and Grollman,A.P. (1987) Oligodeoxynucleotides containing synthetic abasic sites: Model substrates for DNA polymerases and apurinic/apyrimidinic endonucleases. J. Biol. Chem., 262, 10171–10179. [PubMed] [Google Scholar]
- 23.Bourdat A.-G., Gasparutto,D. and Cadet,J. (1999) Synthesis and enzymatic processing of oligodeoxynucleotides containing tandem base damage. Nucleic Acids Res., 27, 1016–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Box H.C., Freund,H.G., Budzinski,E.E., Wallace,J.C. and Maccaubbin,A.E. (1995) Free radical-induced double base lesions. Radiat. Res., 141, 91–94. [PubMed] [Google Scholar]
- 25.Box H.C., Budzinski,E.E., Freund,H.G., Evens,M.S., Patrzyc,H.B., Wallace,J.G. and Maccubbin,A.E. (1993) Vicinal lesions in X-irradiated DNA? Int. J. Radiat. Biol., 64, 261–263. [DOI] [PubMed] [Google Scholar]
- 26.Maccubbin A.E., Iijima,H., Ersing,N., Dawidzik,J.B., Patrzyc,H.B., Wallace,J.C., Budzinski,E.E., Freund,H.G. and Box,H.C. (2000) Double-base lesions are produced in DNA by free radicals. Arch. Biochem. Biophys., 375, 119–123. [DOI] [PubMed] [Google Scholar]
- 27.Box H.C., Budzinski,E.E., Dawidzik,J.B., Wallace,J.C. and Iijima,H. (1998) Tandem lesions and other products in X-irradiated DNA oligomers. Radiat. Res., 149, 433–439. [PubMed] [Google Scholar]
- 28.Delatour Y., Douki,T., Gasparutto,D., Brochier,M.C. and Cadet,J. (1998) A novel vicinal lesion obtained from the oxidative photosensitization of TpdG: characterization and mechanistic aspects. Chem. Res. Toxicol., 11, 433–439. [DOI] [PubMed] [Google Scholar]
- 29.Mariaggi N., Cadet,J. and Teoule,R. (1976) Tetrahedron, 32, 2385–2387. [Google Scholar]
- 30.Asahara H., Wistort,P.M., Bank,J.F., Bakerian,R.H. and Cunningham,R.P. (1989) Purification and characterization of Escherichia coli endonuclease III from the cloned nth gene. Biochemistry, 10, 4444–4449. [DOI] [PubMed] [Google Scholar]
- 31.Jiang D., Hatahet,Z., Melamede,R.J., Kow,Y.W. and Wallace,S.S. (1997) Characterization of Escherichia coli endonuclease VIII. J. Biol. Chem., 272, 32230–32239. [DOI] [PubMed] [Google Scholar]
- 32.Ikeda S., Biswas,T., Roy,R., Izumi,T., Boldogh,I., Kurosky,A., Sarker,A.H., Seki,S. and Mitra,S. (1998) Purification and characterization of human NTH1, a homolog of Escherichia coli endonuclease III. Direct identification of Lys-212 as the active nucleophilic residue. J. Biol. Chem., 273, 21585–21593. [DOI] [PubMed] [Google Scholar]
- 33.You H.J., Swanson,R.L., Harrington,C., Corbett,A.H., Jinks-Robertson,S., Senturker,S., Wallace,S.S., Boiteux,S., Dizdaroglu,M. and Doetsch,P.W. (1999) Saccharomyces cerevisiae Ntg1p and Ntg2p: broad specificity N-glycosylases for the repair of oxidative DNA damage in the nucleus and mitochondria. Biochemistry, 38, 11298–11306. [DOI] [PubMed] [Google Scholar]
- 34.Rabow L.E. and Kow,Y.W. (1997) Mechanism of action of base release by Fpg protein: role of lysine 155 in catalysis. Biochemistry, 16, 5084–5096. [DOI] [PubMed] [Google Scholar]
- 35.Alseth I., Eide,L., Pirovano,M., Rognes,T., Seeberg,E. and Bjoras,M. (1999) The Saccharomyces cerevisiae homologues of endonuclease III from Escherichia coli, Ntg1 and Ntg2, are both required for efficient repair of spontaneous and induced oxidative DNA damage in yeast. Mol. Cell. Biol., 19, 3779–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hall E.J. (1994) Radiobiology for the Radiobiologist. J.B. Lippincott Company, Philadelphia, PA.
- 37.Miao F., Bouziane,M., Dammann,R., Masutani,C., Hanaoka,F., Pfeifer,G.P. and O’Connor,T.R. (2000) 3-Methyladenine-DNA glycosylase (MPG protein) interacts with human RAD23 proteins. J. Biol. Chem., 275, 28433–28438. [DOI] [PubMed] [Google Scholar]
- 38.Bessho T. (1999) Nucleotide excision repair 3′ endonuclease XPG stimulates the activity of base excision repair enzyme thymine glycol DNA glycosylase. Nucleic Acids Res., 27, 979–983. [DOI] [PMC free article] [PubMed] [Google Scholar]