Abstract
Calcium modulating cyclophilin ligand (CAML) is a ubiquitously expressed cytoplasmic protein that is implicated in the EGFR and LCK signaling pathways and required for early embryonic and thymocyte development. To further define the critical biological functions of CAML at the cellular level, we generated CAML-deleted mouse embryonic fibroblasts (MEFs) using an in vitro Cre-loxP mediated conditional knockout system. We found that CAML−/− MEFs have severely impaired proliferation and a strong reduction of normal anaphases. The primary mitotic defect of CAML−/− MEFs is that duplicated chromosomes fail to segregate in anaphase, resulting in nuclear bisection by the cleavage furrow as cells decondense their DNA and exit mitosis, highly reminiscent of the “cut” phenotype in fission yeast. This phenotype is due to spindle dysfunction rather than inability to resolve physical connections between sister chromatids. Furthermore, CAML−/− MEFs display defects often seen in cells with mitotic checkpoint gene deficiencies, including lagging and misaligned chromosomes and chromatin bridges. Consistent with this, we found that CAML−/− MEFs have a modestly weakened spindle assembly checkpoint (SAC) and increased aneuploidy. Thus, our data identify CAML as a novel chromosomal instability gene and suggest that CAML protein acts as a key regulator of mitotic spindle function and a modulator of SAC maintenance.
Keywords: chromosome segregation, aneuploidy, spindle assembly checkpoint, CAML, mitosis, cut phenotype
Introduction
Mitosis is a precisely regulated process featured by nuclear division and cytokinesis, during which cells undergo profound changes, including macro-structural re-arrangement, signaling pathway activation and de-activation, protein recruitment and degradation. Multiple complex steps that are not fully defined regulate this process in order to prevent incorrect sorting of chromosomes to daughter cells. Some of the key regulators include DNA topoisomerase IIα (Topo IIα,1 securin-separase complex,2 cohesin3 and condensin,4 chromosome passenger complex (Aurora-B protein kinase, the inner centromere protein INCENP, survivin and borealin),5 spindle motors, kinetochore microtubule (de)stabilizers,6 Shugoshin1 (Sgo),7 RanGTPase,8 anaphase-promoting complex/cyclosome (APC/C) ubiquitin E3 ligase as well as its coactivators and regulators.9–11 Although the total number of genes that are directly or indirectly implicated in accurate chromosome separation is estimated to be in the several hundred, only a small fraction have been identified to date.12 In addition to the critically important biological role played by the process of chrosomome segregation, it also has a medically relevant aspect, because aneuploidy is a common trait of human neoplasms and is thought to contribute to the malignant pheno-type.13,14 To fully understand its role in cancer, it will be important to define the molecular networks that regulate chromosome segregation of mitotic chromosomes in greater detail.
Among the most intensely studied mitotic regulators are the mitotic checkpoint proteins. Together these proteins establish the spindle assembly checkpoint (SAC), also known as the mitotic checkpoint, a surveillance mechanism that ensures accurate chromosome segregation by delaying sister-chromatid separation and anaphase onset until all chromosomes are properly connected to the mitotic spindle.9 When one or more kinetochores are not properly attached to the microtubules (lacking either bi-oriented attachment or appropriate attachment tension), kinetochore-associated mitotic checkpoint proteins send out inhibitory signals.15 Sensing these signals are the mitotic checkpoint proteins MAD2, BUBR1 and BUB3, which form inhibitory complexes with the APC/C activating subunit Cdc20,16 thereby preventing APC/C-mediated degradation of securin and cyclin B, two key inhibitors of separase, an endo-peptidase that separates sister chromatids by cleaving the cohesin subunit Scc1.4 Recent studies have implicated various proteins outside the family of core mitotic checkpoint proteins in the regulation of the SAC. For instance, the nuclear transport factors Rae1 and Nup98 were shown to stabilize securin in early mitosis by inhibiting Cdh1-activated APC/C.17,18 Another nucleoporin, Tpr, has also been shown to regulate the SAC, most likely through its Mad1 and Mad2 binding ability. Furthermore, several deubiquitinating enzymes, including Usp24 and Usp44,19 and signaling proteins such as Chk1,20 the ATR family of checkpoint kinases21 and B-Raf22 have been linked to SAC activity.
CAML was previously identified as an endoplasmic reticulum associated protein.23 CAML is ubiquitously expressed in all cell lines and tissues examined, and is highly conserved throughout evolution. Genetic ablation of CAML in mice results in embryonic lethality, but it is not required for embryonic stem cell viability.24 It was also found to be critical for efficient EGF-induced proliferation of ES cell-derived epithelioid cells, and was shown to participate in EGFR recycling. In addition, CAML is essential for thymocyte development, and was implicated in mediating appropriate subcellular localization of the src-like kinase Lck.25 In this report, we identify a novel role for CAML in maintaining chromosomal stability. Using CAML conditional knockout MEFs we found that CAML is required for the proper movement of separated sister chromatids to opposite spindle poles in anaphase. In addition, we found that CAML contributes to SAC robustness, as the time of mitotic arrest in the presence of spindle depolymerization is modestly, but significantly, shortened.
Results
CAML-deleted MEFs are growth retarded
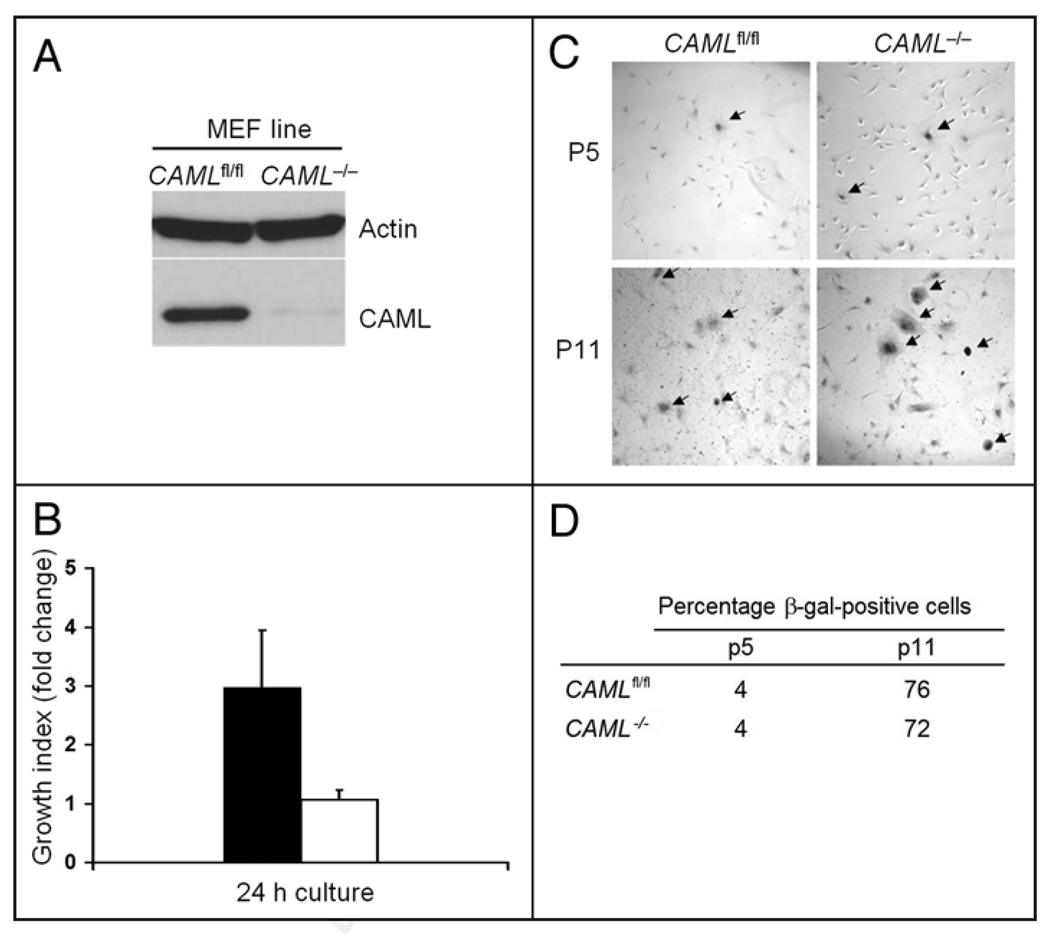
In order to explore the function of CAML in greater detail, we generated MEFs from mice bearing homozygous loxP flanked (“floxed”) alleles of the CAML gene (CAMLfl/fl).25 The CAML allele was conditionally deleted from these cells by retroviral transduction of the pMSCV-Cre/Puro vector.26 Immunoblotting performed on the whole cell lysates confirmed that CAML protein was efficiently ablated (Fig. 1A). CAML−/− MEFs were viable in culture without obvious morphological changes when observed by light microscopy in comparison to CAMLfl/fl MEFs transduced with a control retrovirus (from hereon referred to as CAMLfl/fl MEFs). However, we noticed that CAML−/− MEFs grew dramatically slower than CAMLfl/fl MEFs, which we confirmed by a direct cell count assay (Fig. 1B). Importantly, reduced proliferation was not an artifact due to Cre-toxicity,27 because wildtype MEFs (CAML+/+) transduced with pMSCV-Cre/Puro or pMSCV/Puro had normal growth rates (data not shown).
Figure 1.
Reduced growth of CAML−/− MEFs. (A) Immunoblotting of lysates from CAML−/− and CAMLfl/fl MEFs. The CAML gene was inactivated in CAMLfl/fl MEFs by retroviral transduction of Cre recombinase. (B) Characterizing the growth feature of CAML−/− MEFs by direct cell count. Passage 4 (p4) CAML−/− and CAMLfl/fl MEFs were seeded in triplicates in culture medium with 10% FBS. Cell numbers were counted after overnight culture and this number was set as the initial number. After 24 h, the final cell numbers for each cell line were counted. Data is presented as the growth index (fold change = final cell count/initial number). Error bars indicate standard deviation. (C) CAML−/− and CAMLfl/fl MEFs stained for senescence associated β-galactosidase (SA-β-Gal) and photographed at 60× magnification. Cells from both early passage (p5) and late passage (p11) are shown. (D) Quantitation of SA-β-Gal-positive CAML−/− and CAMLfl/fl MEFs at p5 and p11.
Previous studies in thymocytes suggested a possible role for CAML in preventing apoptosis.25 To explore whether increased apoptosis might explain the growth defect in CAML−/− MEFs, we examined the rate of spontaneous programmed cell death by Hoechst staining. However, in regular culture, CAML−/− MEFs were found to have similar or slightly lower rates of spontaneous apoptosis, in comparison to CAMLfl/fl MEFs (2.2% vs. 2.7%, data not shown). Furthermore, when exposed to stressful conditions, including serum starvation, etoposide or gamma-irradiation, CAML−/− and CAMLfl/fl MEFs also showed very similar rates of apoptosis (data not shown). We also tested whether CAML−/− MEFs might undergo early senescence by staining the cells for senescence-associated (SA) β-galactosidase. The percentage of cells with positive staining increased at similar rates in both CAML−/− and CAMLfl/fl MEFs from passages 5 to 11 in culture (Fig. 1C and D). From the above data, we conclude that MEFs lacking CAML are growth retarded, but that this defect does not seem to result from increased apoptosis or early senescence.
High-frequency anaphase failure in MEFs lacking CAML
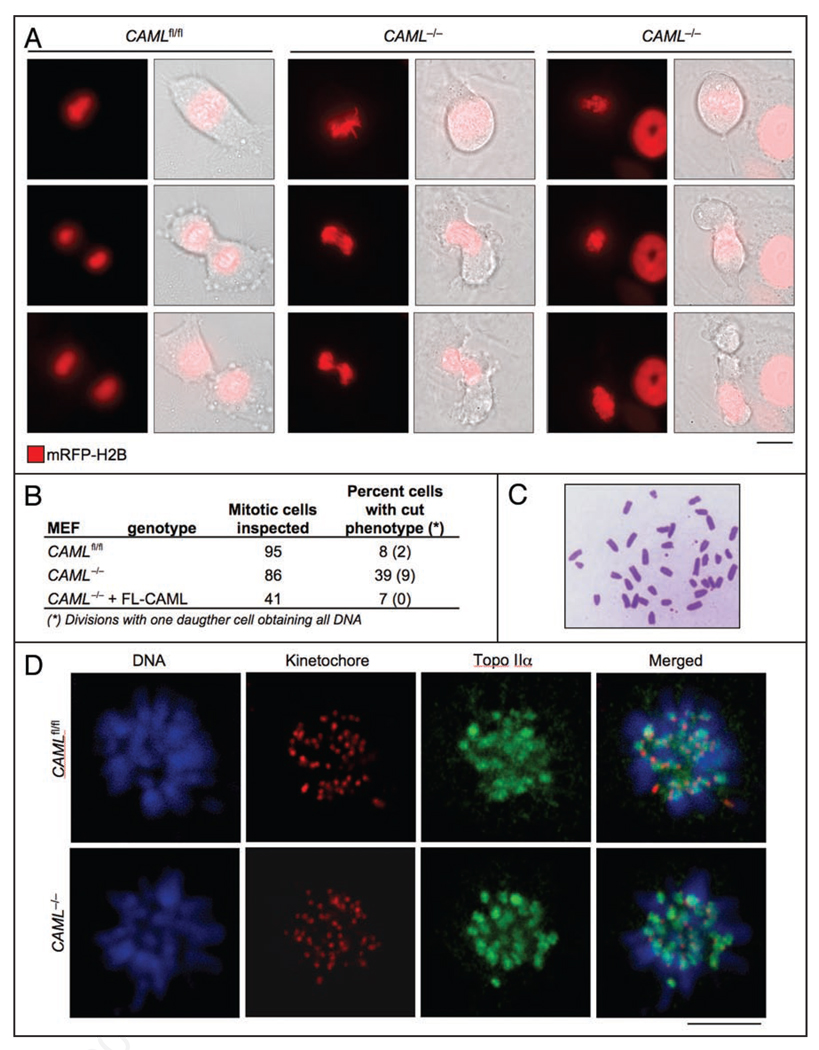
To determine whether growth retardation in the absence of CAML was due to aberrant cell division, we monitored CAML−/− and CAMLfl/fl MEFs by live-cell imaging. To visualize DNA, MEFs were transduced with a retrovirus directing expression of monomeric red fluorescent-protein (mRFP) tagged histone H2B.1 We discovered that mitosis of CAML−/− MEFs is highly abnormal. We found that 30% of CAML−/− MEFs were unable to segregate their chromosomes in anaphase, compared to only 6% of CAMLfl/fl MEFs. These CAML−/− MEFs then decondensed their chromosomes and exited mitosis as binu-cleate cells due to cleavage furrow formation and ingression (Fig. 2A and B). An additional 9% of CAML−/− MEFs showed a related defect in which the cleavage furrow asymmetrically split the DNA content into two daughter cells, with one cell obtaining all chromosomes and the other none (Fig. 2A and B). Both phenotypes are commonly observed in yeast mutants bearing one of the “cut” (cell untimely torn) mutations.28 Only 13% of CAML−/− MEFs displayed normal mitoses, while the remaining cells had minor mitotic abnormalities (see below).
Figure 2.
Loss of CAML causes a cut phenotype. (A) High incidence of anaphase failure in CAML−/− MEFs. Chromosomes were visualized by transducing MEFs with retrovirus encoding mRFP-H2B and followed by time-lapse microscopy. Time-lapse images showing a CAMLfl/fl MEF undergoing normal mitosis and two CAML−/− MEFs with distinct cut phenotypes. Scale bar = 10 µm. (B) Incidence of cut phenotypes in the indicated MEFs. (C) CAML−/− metaphase spread illustrating absence of diplochromosomes. (D) Topo IIα accumulates normally at inner centromeric regions of CAML−/− prometaphases. Monastrol-treated CAML−/− and CAMLfl/fl MEFs were immunostained for centromeres and Topo IIα, while DNA was stained with Hoechst.
Anaphase failure and the “cut” phenotype have previously been reported for separase−/− MEFs,29,30 which prompted us to test whether loss of CAML might perturb the stability of this protease by western analysis. We found that lysates from mitotic CAML−/− and CAMLfl/fl MEFs had similar amounts of separase (data not shown). A distinguishing feature of MEFs lacking separase are diplochromosomes.29,30 These abnormal chromosomes consist of four sister chromatids lying side-by-side and arise when cells go through two rounds of DNA replication without physically separating sister chromatids. Metaphase spreads were prepared from CAML−/− MEFs and screened for the presence of diplochromosomes, but none were observed (Fig. 2C). Besides separase, chromosome separation is dependent on Topo IIα, an enzyme that targets to inner centromeres during the early stages of mitosis to disentangle intertwined sister chromatids.1,31,32 Immunolocalization studies revealed that Topo IIα properly accumulates at inner centromeres of monastrol-treated CAML−/− MEFs (Fig. 2D). Thus, there is no evidence to suggest that CAML loss impairs chromosome separation by perturbing separase stability or Topo IIα targeting to inner centromeres.
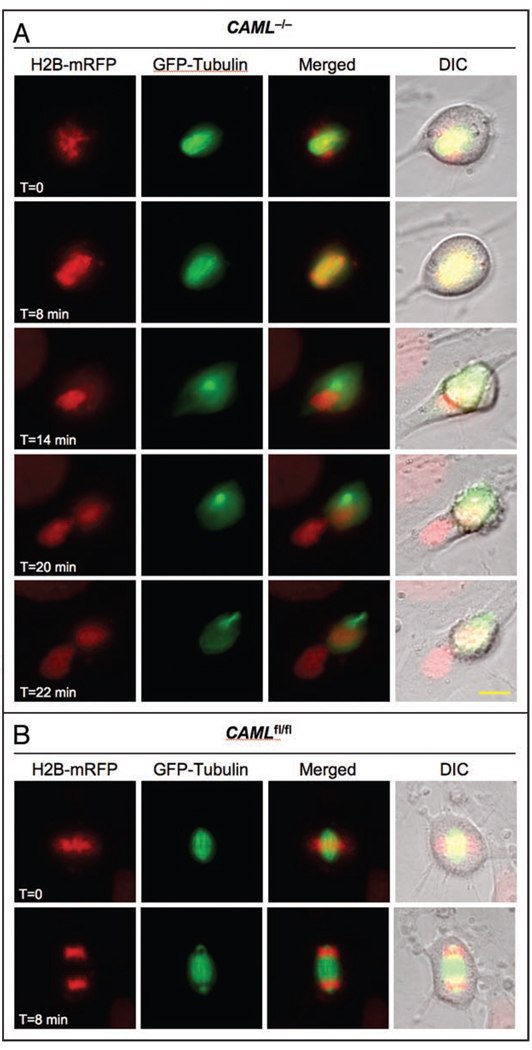
To investigate whether the cut phenotype might be due to a mitotic spindle defect, we coexpressed tubulin-GFP and mRFP-H2B in CAML−/− MEFs and monitored chromosome and spindle microtubule dynamics by live-cell imaging. CAML−/− MEFs demonstrated normal formation of the bipolar spindle and kinetochore attachment up to the beginning of anaphase. At that point, cells undergoing anaphase failure stretched the chromatin but were unsuccessful in moving duplicated chromosomes to opposite poles (Fig. 3A). These spindles then suddenly collapsed and moved to one of the two poles while the decondensing chromatin regrouped at the spindle equator. The ensuing cytokinesis bisected the undivided nucleus, producing one daughter cell containing part of the chromatin and the entire spindle apparatus, and one with only chromatin (n = 25 cells of which 6 displayed a cut phenotype). Some failed anaphases bisected into two cells with one cell lacking both chromatin and spindle microtubules and the other harboring both (data not shown). Mitotic CAMLfl/fl MEFs (n = 24 cells) that we examined in parallel all had normal spindles and successful chromosome segregations (Fig. 3B). Taken together, these data suggest that spindle dysfunction rather than persistent physical connections between sister chromosomes causes anaphase failure in the absence of CAML.
Figure 3.
CAML loss causes mitotic spindle dysfunction. (A) CAML−/− MEF expressing H2B-mRFP and tubulin-GFP followed through an unchallenged mitosis by live-cell imaging. Images were taken at the indicated timepoints. (B) Same as (A) for a CAMLfl/fl MEF. Snapshots taken at metaphase and anaphase are shown. Scale bar = 10 µm.
Decreased accuracy of chromosome segregation in CAML-deficient MEFs
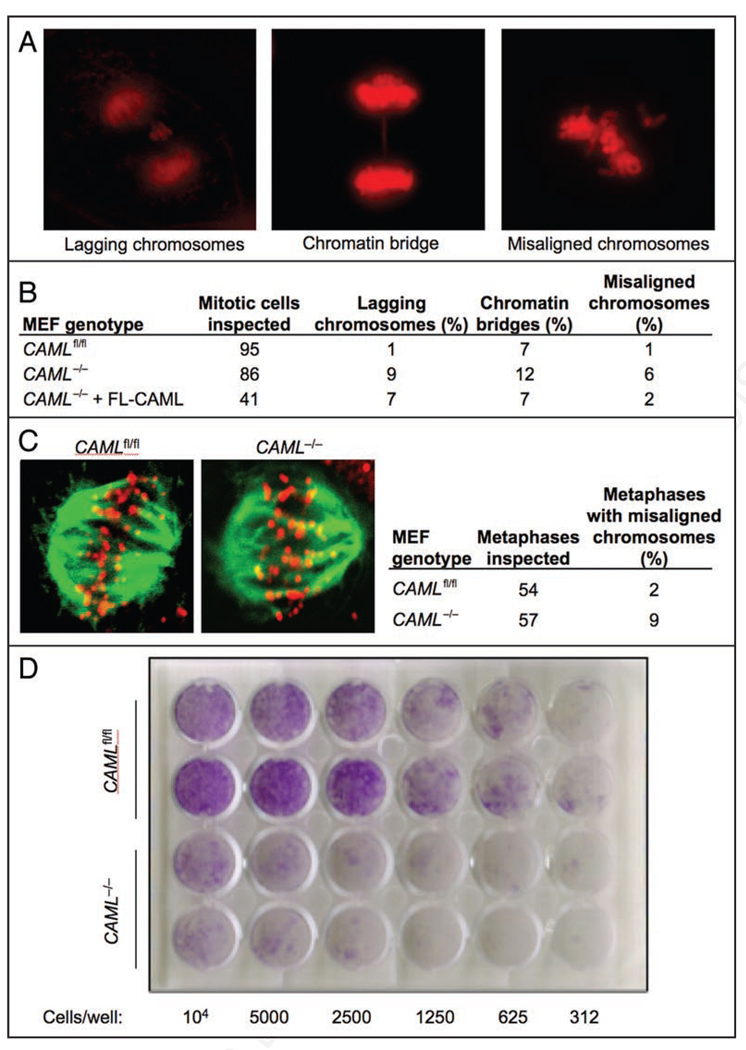
In addition to the catastrophic chromosomal segregation failure seen in MEFs lacking CAML, we also observed somewhat more minor disturbances of mitosis; 9% had lagging chromosomes and 12% had chromatin bridges, compared to 1% and 7% of CAMLfl/fl MEFs, respectively (Fig. 4A and B). Furthermore, 6% of CAML−/− metaphases showed misalignment of one or several chromosomes prior to anaphase onset, a defect that was only seen in 1% of CAMLfl/fl MEFs (Fig. 4A and B). Inefficient attachment of kinetochores to microtubules would be one way that both major and minor mitotic errors could happen in cells. In order to further explore this possibility, MEFs were fixed and co-stained with antibodies to tubulin and kinetochores, and examined by confocal microscopy for metaphase chromosome alignment. The majority of cells demonstrated appropriate co-localization of microtubultes and kinetochores, with only 9% of CAML−/− and 2% of CAMLfl/fl MEFs displaying misalignment (Fig. 4C).
Figure 4.
CAML−/− MEFs have increased chromosome missegregation. (A) Examples of mitotic CAML−/− MEFs displaying the indicated chromosome segregation defects. (B) Quantitation of chromosome segregation errors observed in the indicated MEFs by live-cell imaging. (C)CAMLfl/fl or CAML−/− MEFs were fixed and stained for tubulin and kinetochores. Examples of cells showing normal attachment of microtubules to kinetochores are shown. (D) Colony formation assay of CAML−/− and CAMLfl/fl MEFs. The number of cells seeded per 24-well is indicated. Each MEF line was tested in duplicate.
To determine whether these errors were a direct result of CAML gene inactivation, we tested whether they could be corrected by reconstitution of full-length CAML protein. Retroviral-mediated expression of CAML into CAML−/− MEFs completely reversed anaphase failure (Fig. 2B) and the formation of anaphase bridges, while chromosome misalignment and lagging were partially corrected (Fig. 4B). Together, these experiments suggested that the mitotic defects in CAML-deficient MEFs resulted from loss of CAML, and not from an effect of Cre recombinase. To further verify this, we transduced CAML+/+ MEFs with pMSCV-Cre/Puro virus and observed cells for mitotic progression by live-cell imaging. Cre-expressing CAML+/+ MEFs did not have increased incidence of major or minor mitotic errors observed in CAML−/− MEFs (data not shown). Taken together, our data demonstrate that CAML plays an important role in the proper separation of mitotic chromosomes and identify CAML is a novel chromosomal instability gene.
If the increased tendency to undergo abortive mitosis were to play a role in the defect in cell growth of CAML−/− MEFs, then the colony forming ability of these cells should be reduced. To test this, serial dilutions of CAML−/− and CAMLfl/fl MEFs were plated and colonies counted 2 weeks later. We found that CAML−/− MEFs gave 10-fold fewer numbers of colonies compared to those produced by CAMLfl/fl MEFs (Fig. 4D). This severe loss of colony formation may actually be an over-estimate of their ability to generate colonies, as there remain in Cre-transduced cultures a small percentage of cells that fail to delete the CAML gene, since prolonged growth of these cultures over two months gave rise to cells containing CAML (data not shown). Thus, the high incidence with which CAML−/− MEFs produce daughter cells unable to divide again likely explains the growth retardation phenotype of these cells.
Loss of CAML weakens the spindle assembly checkpoint
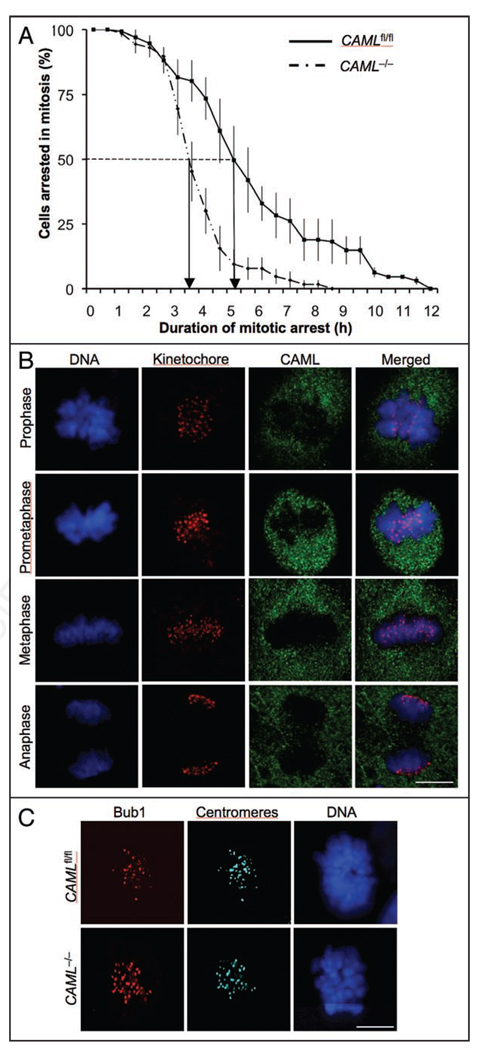
Some of the mitotic defects observed in CAML−/− MEFs, including misaligned and lagging chromosomes, and chromatin bridges, have also been reported for MEFs that are insufficient for certain mitotic checkpoint proteins.13,33–36 To examine the SAC, both CAML−/− and CAMLfl/fl MEFs were subjected to a nocodazole-challenge assay.18,34 MEFs were transduced with a retrovirus encoding mRFP-H2B to facilitate visualization of chromosomes. Next, microtubules were depolymerized by adding nocodazole to the culture medium. MEFs undergoing nuclear envelope breakdown (NEBD) were then marked and monitored by live cell imaging at 15 min intervals until the moment of chromosome decondensation. The duration of arrest in mitosis, which is defined as the interval between NEBD (onset of mitosis) and chromatin decondensation (exit from mitosis without cytokinesis), was then calculated and plotted. The time at which 50% of the cells had exited mitosis was used for comparison. When blocked by nocodazole, there was no defect in activation of the checkpoint in CAML−/− MEFs (Fig. 5A). However, CAML−/− MEFs exited mitosis significantly faster than CAMLfl/fl MEFs, with 50% of CAML−/− MEFs exiting at 3.5 h and 50% of CAMLfl/fl MEFs by 5 h (Fig. 5A). These data suggest that prolonged SAC signaling requires CAML.
Figure 5.
CAML−/− MEFs demonstrated reduced mitotic checkpoint function. (A) CAML−/− and CAMLfl/fl MEFs were examined by a nocodazole-challenge assay. Data was presented by using a survival analysis curve. CAML−/− MEFs were significantly different from CAMLfl/fl MEFs (Log rank test, p = 0.0288). Data shown represent the average values from three independent experiments. Error bars indicate standard deviation. (B) Immunolocalization of endogenously expressed CAML (green) during mitosis. Endogenous CAML protein was visualized with rabbit anti-CAML antibody and centromeres with ACA antibody. DNA was stained with Hoechst. Note that CAML- and centromere-specific signals do not overlap. (C) Immunolocalization of Bub1 in monastrol-treated CAML−/− and CAMLfl/fl MEFs. Centromeres were stained with ACA antibody and DNA with Hoechst. Note that targeting of Bub1 to kinetochores is not impaired in the absence of CAML. Scale bar = 10 µm.
The SAC consists of kinetochore-associated components that produce the “anaphase wait” signal and soluble components residing in the mitotic cytosol that bind to and inhibit the activating subunits of the APC/C, Cdc20 and Cdh1.9,13 To explore how CAML might regulate the spindle checkpoint, we determined the localization of CAML in mitosis. We found that endogenous CAML was distributed throughout the mitotic cytosol of MEF cells in the previously observed punctate pattern23,24 and did not accumulate at unattached kinetochores (Fig. 5B). Furthermore, ectopically expressed GFP-CAML fusion protein had the same distribution pattern (data not shown). The data suggest that CAML is a soluble regulator of the mitotic checkpoint. One possibility is that CAML enhances the efficiency with which kinetochore-associated mitotic checkpoint proteins target to unattached kinetochores at the onset of mitosis. To test for this possibility, we stained CAML−/− and CAMLfl/fl MEFs for a variety of kinetochore-bound mitotic checkpoint proteins, including Bub1, BubR1, Cenp-E, Mad1 and Mad2. However, none of these proteins was mislocalized in the absence of CAML (Fig. 5C, and data not shown). To test whether CAML might exert its SAC control by binding to the activating subunits of the APC/C, we performed coimmunoprecipitation experiments of mitotic MEF extracts, but neither Cdc20 nor Cdh1 coprecipitated with CAML (data not shown). The same holds true for the APC/C components APC3 and APC6 (data not shown). These experiments suggest that CAML regulates the SAC as a soluble cytoplasmic protein through a mechanism that does not involve direct inhibition of the APC/C activating subunits.
We then asked if deletion of CAML had an effect on the timing of cyclin B degradation. Normally, this key APC/C substrate is degraded in late metaphase when chromosomes are bioriented,13,37 but in MEFs in which the APC/C is not properly inhibited by BubR1, premature cyclin B degradation has been shown to occur.18 Synchronized and nocodazole blocked CAML−/− and CAMLfl/fl MEFs were collected by mitotic shake-off. Lysates were then generated and examined by western blotting. Blots were first probed with phospho-histone H3 (pH3) antibody, which confirmed that equal amounts of mitotic cells were indeed present in the CAML−/− and CAMLfl/fl lysates (data not shown). Cyclin B was also present at equal levels in these lysates, suggesting that Cdc20-activated APC/C is properly inhibited in the absence of CAML (data not shown). To further examine the degradation of cyclin B in the absence of CAML, we transiently transfected cyclin B-GFP38 into CAML−/− and CAMLfl/fl MEFs and monitored its level of expression by live-cell imaging (data not shown). In CAMLfl/fl MEFs, cyclin B-GFP localized to the cytoplasm during G2 phase. At mitosis onset, it translocated into the nucleus and diffused into the mitotic cytosol after NEBD. Cyclin B-GFP disappeared in late metaphase just prior to the onset of anaphase. A similar pattern of cyclin B-GFP translocation and degradation was observed in CAML−/− MEFs. Collectively, these data suggest that the timing of cyclin B destruction by APC/CCdc20 is not perturbed by the absence of CAML.
We also examined the degradation of securin in the absence of CAML by transiently expressing a securin-YFP fusion protein into CAML−/− and CAMLfl/fl MEFs. In MEFs, proper timing of securin destruction is regulated by the nuclear transport factors Rae1 and Nup98, which bind to and inhibit Cdh1-activated APC/C at the onset of mitosis.18 We found that securin-YFP was properly degraded at the metaphase-to-anaphase transition in CAML−/− MEFs (data not shown), indicating that CAML loss does not perturb the inhibition of APC/CCdh1 by Nup98 and Rae1.
Increased aneuploidy and micronuclei in MEFs lacking CAML
Chromosome lagging, chromosome misalignment and chromatin bridge formation are expected to promote aneuploidy. To examine whether CAML−/− MEFs are indeed more prone to aneuploidy, we prepared metaphase spreads of colcemid-treated CAML−/− and CAMLfl/fl MEFs and performed chromosome counts. As shown in Table 1, 43% of CAML−/− MEFs were aneuploid versus 13% of CAMLfl/fl MEFs. Furthermore, 25% of CAML−/− MEFs were tetraploid or near tetraploid compared with 12% of CAMLfl/fl MEFs, indicating failure to undergo cytokinesis in a prior mitosis in the absence of CAML. Increased tetraploidization is consistent with the high incidence of anaphase failure seen in CAML−/− MEFs (Table 1 and Fig. 2B).
Table 1.
CAML−/− and CAMLfl/fl MEFs were arrested in metaphase by treatment with colcemid
| Mitotic MEF genotype (n) |
Mitotic figures inspected |
Aneuploid figures |
Tetraploid & near tetrapolid figures |
Karyotypes with indicated chromosome number |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 37 | 38 | 39 | 40 | 41 | 42 | 44 | 75 | 76 | 78 | 79 | 80 | 81 | 82 | 84 | 85 | 86 | ||||
| CAMLfl/fl (2) | 100 | 13% | 12% | 2 | 4 | 79 | 3 | 2 | 8 | 1 | 1 | |||||||||
| CAML−/− (2) | 100 | 43% | 25% | 2 | 4 | 8 | 47 | 8 | 5 | 1 | 1 | 1 | 2 | 1 | 10 | 2 | 4 | 1 | 2 | 1 |
Aneuploid cells = near-diptoid cells + near-tetraploid cells
Metaphase spreads were prepared and the total number of chromosomes per cell counted.
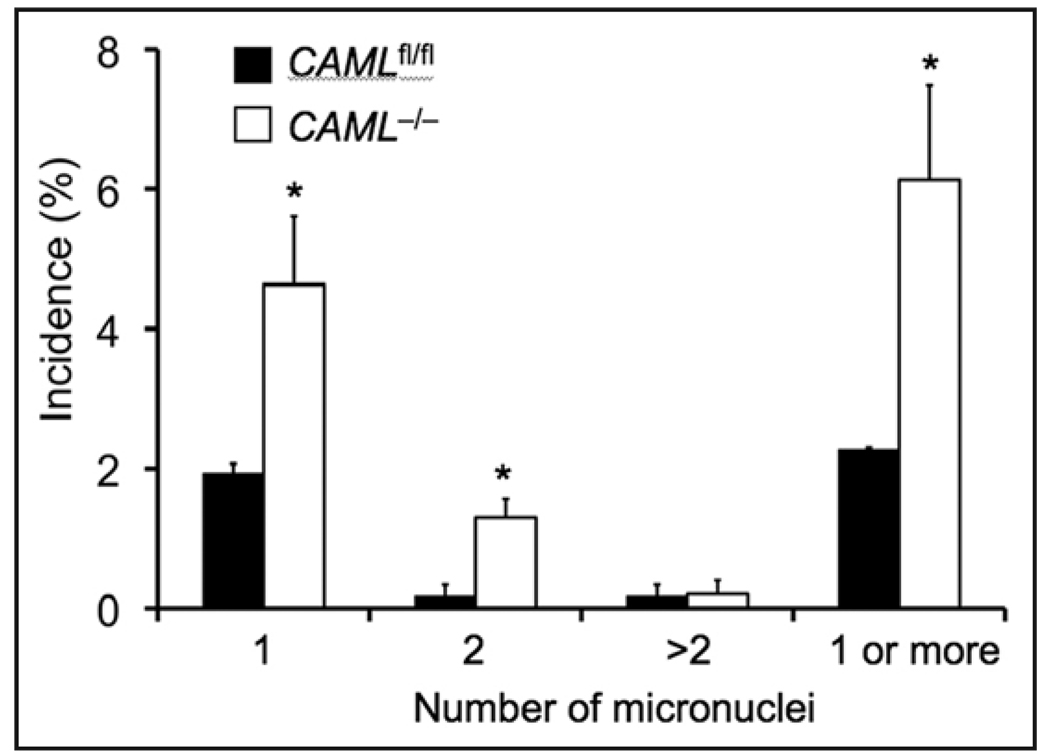
We also inspected CAML−/− and CAMLfl/fl MEFs for the incidence of micronuclei as an indicator of chromosome missgregation. Consistent with the increased aneuploidy, we found that CAML−/− MEFs formed micronuclei at significantly higher rates than CAMLfl/fl MEFs (Fig. 6). The data demonstrate that CAML is essential to maintain the appropriate cellular complement of chromosomes, and confirm that the CAML gene is indeed a chromosomal instability gene.
Figure 6.
CAML−/− MEFs have increased micronuclei. P4 CAML−/− and CAMLfl/fl MEFs were seeded on slides, fixed with 4% paraformaldehyde and processed with Hoechst staining. In each experiment approximately 200 cells from each cell line were examined for micronuclei by microscopy. The graph shows a summary of three independent experiments. Error bars indicate standard deviation. *p < 0.05 versus CAMLfl/fl MEFs (t-test).
Discussion
Here, we identify CAML as an important regulator of chromosome segregation. We demonstrate that CAML−/− MEFs displayed several types of mitotic errors with much higher incidence than CAMLfl/fl controls. The primary defects seen (“cut” appearance with nuclear DNA trapped by the constriction of the cytokinetic ring, and the “aberrant septation” defect whereby all of the chromosomal DNA ends up in just one of the two daughter cells) were highly reminiscent of yeast “cut” mutants, and were seen in up to 39% of mitoses in CAML−/− cells. These defects were rescued by reconstitution of CAML−/− MEFs with full-length CAML protein, thus excluding the possibility that these mitotic errors were induced by Cre-recombinase.27 In addition, Cre-expressing wild-type MEFs displayed normal growth rates and normal mitotic progression, further supporting this conclusion.
The initial descriptions of the “cut” phenotype in fission yeast were of gene mutants in which cytokinesis occurred in spite of unsuccessful sister-chromatid separation during anaphase.28,39,40 Some of these mutations were in genes that resolve the physical connections between duplicated chromosomes, including top2 (Topo IIα),41 cut1 (separase),42,43 cut2 (securin),44 or in genes encoding subunits of the cohesin complex, including cut3 (Smc4) and cut14 (Smc2).45 It is unlikely that the cut phenotype resulting from CAML loss is due to inability to physically separate sister chromatids because CAML−/− MEFs lack diplochromosomes. In support of this notion is the observation that two inhibitors of separase, cyclin B and securin, are properly degraded in the absence of CAML. Moreover, we observed equal levels of separase by western blots in CAML-deficient cells compared to normal controls, thus providing additional support for this notion.
CAML is not the only cut gene lacking an overt functional connection to the process of sister-chromatid separation.40 For instance, cut6 and Lsd1 are two S. pombe cut genes implicated in fatty acid biosynthesis.46 How defects in these genes produce cut pheno-types is currently unclear. Cut7 is another cut gene that is not directly involved in sister-chromatid separation.47,48 It encodes a kinesin-like motor protein that is required for spindle formation and separation of the two spindle pole bodies in early mitosis. The most likely explanation for the anaphase failure in CAML−/− cells was the striking defect in spindle morphology and function. We observed a very high frequency of cells with poorly formed spindles, starting from metaphase and lasting to the end of mitotic failure. Live microscopy demonstrated aberrant function of these atypical spindles, which ended up in only one of the daughter cells, even in cells displaying the “cut” abnormality with approximately equal amounts of chromosomal DNA divided between the two.
CAML has previously been characterized as an integral membrane protein localized to the endoplasmic reticulum and cytosolic vesicles.23 Although it does not bear significant sequence similarity to known proteins, it has been shown to physically interact with several cell surface receptors, the tyrosine kinase Lck25 and the HIV vpu protein.49 In these cases, it has been implicated in regulation of intracellular trafficking of the interacting partners. How CAML might regulate the mitotic spindle at the molecular level remains to be determined in future studies. Although we did not identify a physical interaction between CAML and known spindle proteins, others have shown that portions of the endoplasmic reticulum lie in tight apposition to the spindle body and orient along microtubules close to the spindle poles during all phases of mitosis.50 Thus, it is tempting to speculate that CAML might participate in trafficking of spindle components known to be important for microtubule motor function, such as the mitotic kinesins KLP59C, KLP10A,51 KLP38B/KIF14,52,53 or CENP-E.54
In addition to complete anaphase failure, CAML-depleted MEFs also show milder chromosome segregation defects, including chromosome misalignment, lagging and bridging. It seems reasonable that the modest SAC impairment in CAML−/− cells contributes to these segregation defects, although it is possible that aberrant spindle function could underlie this problem, as well. But how could CAML regulate the mitotic checkpoint? CAML did not colocalize with centromeres during mitosis and is therefore unlikely to have a direct role in generating anaphase “wait” signals at unattached kinetochores. Furthermore, the core mitotic checkpoint proteins that have been implicated in the production of such signals translocated with normal efficiency to unattached kinetochores in the absence of CAML. The possibility that CAML acts as a soluble inhibitor of the APC/C is highly unlikely because neither the APC/C components APC3 and APC6 nor the APC/C activating subunits Cdc20 and Cdh1 seem to interact with CAML in mitosis. The observation that timing of securin and cyclin B proteolysis in mitosis was normal in the absence of CAML supports this notion. Normal securin and cyclin B degradation is not necessarily inconsistent with reduced checkpoint activity as several mitotic checkpoint gene defects, including Bub3 and Bub1 insufficiency, do not cause premature securin and cyclin B destruction while substantially weakening the SAC (reviewed in refs. 34 and 55; and Deursen JMv and Jeganathan KB, unpublished data). Thus, based on the current evidence, it is plausible that CAML exerts its effect on the SAC in an indirect fashion. It is conceivable that, as a regulator of intracellular trafficking, CAML may participate in relaying negative signals from unattached kinetochores to the soluble APC/C inhibitors in the mitotic cytosol.
The data presented in this report identify CAML as a novel chromosomal instability gene, and reveal its requirement for normal function of the mitotic spindle in MEF cells. Although its precise mechanism in chromosome segregation remains to be determined, this work is significant because it adds to the relatively limited number of mammalian chromosomal instability and cut genes identified to date. Given the link between chromosomal instability and cancer, it will be important in future studies to examine the possibility that CAML dysfunction is causally implicated in oncogenesis.
Materials and Methods
Generation of CAML conditional knockout MEFs
CAML−/− MEFs were generated from 13.5-day CAMLfl/fl embryos. In most experiments unless noted otherwise, the cells were divided into two populations and treated under identical conditions with a retrovirus (MSCV) expressing either Puro plus Cre recombinase to delete an essential CAML exon or a puro resistance cassette alone.26 Cells were exposed to retroviruses three times per day for 2 days, and then selected by culture medium (10% FBS) containing 5 µg/ml puromycin for 24 h. The puromycin-selected MEFs (termed as CAML−/− and CAMLfl/fl) cells were used in subsequent experiments. To measure apoptosis, the entire cell population was collected and 300 cells were counted for apoptotic events by fluorescence microscopy after nuclear counter staining with Hoechst 33258.
Western blot analysis
Cells were lysed on ice in DPBS + 10% glycerol, 1% Triton X-100 and protease inhibitors. 1–2 mg of lysate was precipitated with 4 µg of antigen-specific antibodies or appropriate control antibodies (Pharmingen). Protein samples were resolved by SDS PAGE (10% gels). Membranes were blotted for CAML,20 Cdc27 (BD), cyclin B1 (Santa Cruz), Cdc20 (Santa Cruz), phospho-histone H3 (Upstate) and actin (Sigma).
Indirect immunofluorescence staining
MEF cells were fixed by 4% paraformaldehyde for 12 min and permeabilized by addition of 0.2% Triton X-100 for 10 min at room temperature. Primary antibodies applied in immunostaining included, CAML,25 Topo IIα (TopoGEN), Bub1, BubR1,26 Mad2,26 and tubulin (Sigma-Aldrich). Primary antibodies were visualized with appropriate secondary antibodies conjugated to AlexaFluor594, 488 or 647 (Invitrogen). Nuclear counterstaining was done using Hoechst 33258 (2 µM; Molecular Probes). Fluorescence was detected using the 100x objective of a Provis AX-70 confocal fluorescence microscope (Olympus).
Live cell imaging
MEFs (p3) were first transduced with a retrovirus encoding a monomeric red fluorescent protein (mRFP)-tagged H2B to allow visualization of chromosomes by fluorescence microscopy. Cells were then seeded onto 35 mm glass-bottomed culture dishes (MatTek Corporation) and cultured in DMEM/10% FBS. Approximately 24 h later, experiments were performed using a Zeiss Axio Observer Z1 system with: CO2 Module S, TempModule S, Heating Unit XL S, Pln Apo 63X/1.4 oil DICIII objective, AxioCam MRm camera and AxioVision 4.6 software. The imaging medium was DMEM/10% FBS. The temperature of the imaging medium was kept at 37°C.
For Nocodazole-challenge assays, nocodazole was added to a final concentration of 100 ng/ml. Cells undergoing nuclear envelope breakdown (NEBD) were marked and monitored at 15 min intervals to determine when they decondensed their chromosomes. The duration of arrest in mitosis, which is defined as the interval between NEBD (onset of mitosis) and chromatin decondensation (exit from mitosis without cytokinesis), was then calculated and plotted. The time at which 50% of the cells had exited mitosis was used for comparison.
Analyses of fluorescent protein-tagged cyclin B and securin levels were as follows: H2B-mRFP expressing MEFs were infected with pMSCV-Cre/Puro for 48 h and Cre expressing MEFs were selected in culture medium containing 5 µg/ml puromycin for 24 h. Cells were harvested and nucleofected with 5 µg pCMX-Cyclin B1-GFP or 2 µg pEYFP-N1-Securin plasmid DNA using an Amaxa Nucleofector II. Two x106 MEFs were used per nucleofection. Nucleofections were done in MEF2 buffer (Amaxa, VPD-1005). MEFs were immediately seeded into 35-mm dishes and analyzed 4 h later. G2 phase cells were marked and images acquired every 6 min. Exposure times were identical among experiments for each fluorochrome/filters set. For quantification of fluorescence levels, at least 10 cells were analyzed per MEF line. The mean fluorescence intensity at each cell stage was determined, after background subtraction of images transformed to 8-bit grayscale, using NIH Image J software (http://rsb.info.nih.gov/ij/). Mean fluorescence intensities were expressed in arbitrary units.
To study spindle dynamics, an NheI-BamHI fragment containing EGFP-tubulin cDNA was prepared pEGFP-Tub (Clontech, 632349). This fragment was blunted and cloned into the HpaI site of pMSCVpuro (Clontech, 634401). The resulting retroviral vector was cotransfected with pVSV-G into GP2–293 cells (Clontech) to produce to produce pantropic virus. By transducing EcoPACK cells (Clontech) with these viruses we established stable viral producer cell line. Virus-containing supernatants from this line were used to express EGFP-tubulin in MEFs.
Cell synchronizations and mitotic shake-off
MEFs were synchronized by growing highly confluent MEF cultures (at p4) in DMEM medium plus 0.1% fetal bovine serum (FBS) for 14 h. Cells were then released into DMEM plus 20% FBS. At 23 h, nocodazole was added to a final concentration of 100 ng/ml. In the mitotic shake-off procedure, loosely attached mitotic MEF cells were collected at 28 h by washing with ice cold PBS.
β-galactosidase staining
Cells were washed in PBS, fixed for 3–5 min (room temperature) in 2% formaldehyde/0.2% glutaraldehyde, washed and incubated at 37°C with freshly made β-Gal staining solution: 5-bromo-4-chloro-3-indolyl P3-D-galactoside (X-Gal) 1 mg/ml, 40 mM citric acid sodium phosphate, pH 6.0 5 mM potassium ferrocyanide, pH 6.0 5 mM potassium ferricyanid, 150 mM NaCl, 2 mM MgCl2. All reagents were from Sigma.
Chromosome counts
Karyotype analysis of CAML−/− and CAMLfl/fl MEFs was done as previously described.56 Cells were classified as diploid (40 chromosomes), tetraploid (80 or near 80 chromosomes) or aneuploid (near-diploid and near-tetraploid combined).
Acknowledgements
This work was supported by a grant from the Joseph Bloom Children’s Diseases Research Fund, as well as NIH grant 5R01CA112414.
References
- 1.Dawlaty MM, Malureanu L, Jeganathan KB, Kao E, Sustmann C, Tahk S, et al. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell. 2008;133:103–115. doi: 10.1016/j.cell.2008.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu H. Cdc20: a WD40 activator for a cell cycle degradation machine. Mol Cell. 2007;27:3–16. doi: 10.1016/j.molcel.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 3.Losada A. The regulation of sister chromatid cohesion. Biochim Biophys Acta. 2008;1786:41–48. doi: 10.1016/j.bbcan.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Nasmyth K, Haering CH. The structure and function of SMC and kleisin complexes. Annu Rev Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- 5.Maia AF, Lopes CS, Sunkel CE. BubR1 and CENP-E have antagonistic effects upon the stability of microtubule-kinetochore attachments in Drosophila S2 cell mitosis. Cell Cycle. 2007;6:1367–1378. doi: 10.4161/cc.6.11.4271. [DOI] [PubMed] [Google Scholar]
- 6.Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- 7.Pouwels J, Kukkonen AM, Lan W, Daum JR, Gorbsky GJ, Stukenberg T, et al. Shugoshin 1 plays a central role in kinetochore assembly and is required for kinetochore targeting of Plk1. Cell Cycle. 2007;6:1579–1585. doi: 10.4161/cc.6.13.4442. [DOI] [PubMed] [Google Scholar]
- 8.Li HY, Ng WP, Wong CH, Iglesias PA, Zheng Y. Coordination of chromosome alignment and mitotic progression by the chromosome-based Ran signal. Cell Cycle. 2007;6:1886–1895. doi: 10.4161/cc.6.15.4487. [DOI] [PubMed] [Google Scholar]
- 9.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nature reviews. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 10.Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- 11.Sullivan M, Morgan DO. Finishing mitosis, one step at a time. Nat Rev Mol Cell Biol. 2007;8:894–903. doi: 10.1038/nrm2276. [DOI] [PubMed] [Google Scholar]
- 12.Michor F, Iwasa Y, Vogelstein B, Lengauer C, Nowak MA. Can chromosomal instability initiate tumorigenesis? Semin Cancer Biol. 2005;15:43–49. doi: 10.1016/j.semcancer.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 13.Ricke RM, van Ree JH, van Deursen JM. Whole chromosome instability and cancer: a complex relationship. Trends Genet. 2008;24:457–466. doi: 10.1016/j.tig.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weaver BA, Cleveland DW. Aneuploidy: instigator and inhibitor of tumorigenesis. Cancer Res. 2007;67:10103–10105. doi: 10.1158/0008-5472.CAN-07-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams GL, Roberts TM, Gjoerup OV. Bub1: escapades in a cellular world. Cell Cycle. 2007;6:1699–1704. doi: 10.4161/cc.6.14.4493. [DOI] [PubMed] [Google Scholar]
- 16.Sudakin V, Chan GK, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20 and MAD2. J Cell Biol. 2001;154:925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeganathan KB, Baker DJ, van Deursen JM. Securin associates with APCCdh1 in prometaphase but its destruction is delayed by Rae1 and Nup98 until the metaphase/anaphase transition. Cell Cycle. 2006;5:366–370. doi: 10.4161/cc.5.4.2483. [DOI] [PubMed] [Google Scholar]
- 18.Jeganathan KB, Malureanu L, van Deursen JM. The Rae1-Nup98 complex prevents aneuploidy by inhibiting securin degradation. Nature. 2005;438:1036–1039. doi: 10.1038/nature04221. [DOI] [PubMed] [Google Scholar]
- 19.Stegmeier F, Rape M, Draviam VM, Nalepa G, Sowa ME, Ang XL, et al. Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature. 2007;446:876–881. doi: 10.1038/nature05694. [DOI] [PubMed] [Google Scholar]
- 20.Kim EM, Burke DJ. DNA damage activates the SAC in an ATM/ATR-dependent manner, independently of the kinetochore. PLoS Genet. 2008;4:1000015. doi: 10.1371/journal.pgen.1000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McSherry TD, Kitazono AA, Javaheri A, Kron SJ, Mueller PR. Non-catalytic function for ATR in the checkpoint response. Cell Cycle. 2007;6:2019–2030. doi: 10.4161/cc.6.16.4553. [DOI] [PubMed] [Google Scholar]
- 22.Borysova MK, Cui Y, Snyder M, Guadagno TM. Knockdown of B-Raf impairs spindle formation and the mitotic checkpoint in human somatic cells. Cell Cycle. 2008;7:2894–2901. doi: 10.4161/cc.7.18.6678. [DOI] [PubMed] [Google Scholar]
- 23.Holloway MP, Bram RJ. Co-localization of calcium-modulating cyclophilin ligand with intracellular calcium pools. J Biol Chem. 1998;273:16346–16350. doi: 10.1074/jbc.273.26.16346. [DOI] [PubMed] [Google Scholar]
- 24.Tran DD, Russell HR, Sutor SL, van Deursen J, Bram RJ. CAML is required for efficient EGF receptor recycling. Dev Cell. 2003;5:245–256. doi: 10.1016/s1534-5807(03)00207-7. [DOI] [PubMed] [Google Scholar]
- 25.Tran DD, Edgar CE, Heckman KL, Sutor SL, Huntoon CJ, van Deursen J, et al. CAML is a p56Lck-interacting protein that is required for thymocyte development. Immunity. 2005;23:139–152. doi: 10.1016/j.immuni.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 26.Malureanu L, Jeganathan KB, Hamada M, Wsilewski L, Davenport J, van Deursen J. BubR1 N terminus acts as a soluble inhibitor of cyclin B degradation by APC/C Cdc20 in interphase. Dev Cell. 2009 doi: 10.1016/j.devcel.2008.11.004. in Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loonstra A, Vooijs M, Beverloo HB, Allak BA, van Drunen E, Kanaar R, et al. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc Natl Acad Sci USA. 2001;98:9209–9214. doi: 10.1073/pnas.161269798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirano T, Funahashi SI, Uemura T, Yanagida M. Isolation and characterization of Schizosaccharomyces pombe cutmutants that block nuclear division but not cytokinesis. EMBO J. 1986;5:2973–2979. doi: 10.1002/j.1460-2075.1986.tb04594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumada K, Yao R, Kawaguchi T, Karasawa M, Hoshikawa Y, Ichikawa K, et al. The selective continued linkage of centromeres from mitosis to interphase in the absence of mammalian separase. J Cell Biol. 2006;172:835–846. doi: 10.1083/jcb.200511126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wirth KG, Wutz G, Kudo NR, Desdouets C, Zetterberg A, Taghybeeglu S, et al. Separase: a universal trigger for sister chromatid disjunction but not chromosome cycle progression. J Cell Biol. 2006;172:847–860. doi: 10.1083/jcb.200506119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhat MA, Philp AV, Glover DM, Bellen HJ. Chromatid segregation at anaphase requires the barren product, a novel chromosome-associated protein that interacts with Topoisomerase II. Cell. 1996;87:1103–1114. doi: 10.1016/s0092-8674(00)81804-8. [DOI] [PubMed] [Google Scholar]
- 32.Clarke DJ, Johnson RT, Downes CS. Topoisomerase II inhibition prevents anaphase chromatid segregation in mammalian cells independently of the generation of DNA strand breaks. J Cell Sci. 1993;105:563–569. doi: 10.1242/jcs.105.2.563. [DOI] [PubMed] [Google Scholar]
- 33.Baker DJ, Jeganathan KB, Cameron JD, Thompson M, Juneja S, Kopecka A, et al. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet. 2004;36:744–749. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- 34.Jeganathan K, Malureanu L, Baker DJ, Abraham SC, van Deursen JM. Bub1 mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis. J Cell Biol. 2007;179:255–267. doi: 10.1083/jcb.200706015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michel LS, Liberal V, Chatterjee A, Kirchwegger R, Pasche B, Gerald W, et al. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409:355–359. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- 36.Weaver BA, Bonday ZQ, Putkey FR, Kops GJ, Silk AD, Cleveland DW. Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J Cell Biol. 2003;162:551–563. doi: 10.1083/jcb.200303167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Musacchio A, Hardwick KG. The spindle checkpoint: structural insights into dynamic signalling. Nat Rev Mol Cell Biol. 2002;3:731–741. doi: 10.1038/nrm929. [DOI] [PubMed] [Google Scholar]
- 38.Hagting A, Den Elzen N, Vodermaier HC, Waizenegger IC, Peters JM, Pines J. Human securin proteolysis is controlled by the spindle checkpoint and reveals when the APC/C switches from activation by Cdc20 to Cdh1. J Cell Biol. 2002;157:1125–1137. doi: 10.1083/jcb.200111001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uemura T, Yanagida M. Isolation of type I and II DNA topoisomerase mutants from fission yeast: single and double mutants show different phenotypes in cell growth and chromatin organization. EMBO J. 1984;3:1737–1744. doi: 10.1002/j.1460-2075.1984.tb02040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yanagida M. Fission yeast cut mutations revisited: control of anaphase. Trends Cell Biol. 1998;8:144–149. doi: 10.1016/s0962-8924(98)01236-7. [DOI] [PubMed] [Google Scholar]
- 41.Uemura T, Tanagida M. Mitotic spindle pulls but fails to separate chromosomes in type II DNA topoisomerase mutants: uncoordinated mitosis. EMBO J. 1986;5:1003–1010. doi: 10.1002/j.1460-2075.1986.tb04315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baum P, Yip C, Goetsch L, Byers B. A yeast gene essential for regulation of spindle pole duplication. Mol Cell Biol. 1988;8:5386–5397. doi: 10.1128/mcb.8.12.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uzawa S, Samejima I, Hirano T, Tanaka K, Yanagida M. The fission yeast cut1+ gene regulates spindle pole body duplication and has homology to the budding yeast ESP1 gene. Cell. 1990;62:913–925. doi: 10.1016/0092-8674(90)90266-h. [DOI] [PubMed] [Google Scholar]
- 44.Funabiki H, Yamano H, Kumada K, Nagao K, Hunt T, Yanagida M. Cut2 proteolysis required for sister-chromatid seperation in fission yeast. Nature. 1996;381:438–441. doi: 10.1038/381438a0. [DOI] [PubMed] [Google Scholar]
- 45.Saka Y, Sutani T, Yamashita Y, Saitoh S, Takeuchi M, Nakaseko Y, et al. Fission yeast cut3 and cut14, members of a ubiquitous protein family, are required for chromosome condensation and segregation in mitosis. EMBO J. 1994;13:4938–4952. doi: 10.1002/j.1460-2075.1994.tb06821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saitoh S, Takahashi K, Nabeshima K, Yamashita Y, Nakaseko Y, Hirata A, et al. Aberrant mitosis in fission yeast mutants defective in fatty acid synthetase and acetyl CoA carboxylase. J Cell Biol. 1996;134:949–961. doi: 10.1083/jcb.134.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Enos AP, Morris NR. Mutation of a gene that encodes a kinesin-like protein blocks nuclear division in A. nidulans. Cell. 1990;60:1019–1027. doi: 10.1016/0092-8674(90)90350-n. [DOI] [PubMed] [Google Scholar]
- 48.Hagan I, Yanagida M. Novel potential mitotic motor protein encoded by the fission yeast cut7+ gene. Nature. 1990;347:563–566. doi: 10.1038/347563a0. [DOI] [PubMed] [Google Scholar]
- 49.Varthakavi V, Heimann-Nichols E, Smith RM, Sun Y, Bram RJ, Ali S, et al. Identification of calcium-modulating cyclophilin ligand as a human host restriction to HIV-1 release overcome by Vpu. Nat Med. 2008;14:641–647. doi: 10.1038/nm1778. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.McCullough S, Lucocq J. Endoplasmic reticulum positioning and partitioning in mitotic HeLa cells. J Anat. 2005;206:415–425. doi: 10.1111/j.1469-7580.2005.00407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rogers GC, Rogers SL, Schwimmer TA, Ems-McClung SC, Walczak CE, Vale RD, et al. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature. 2004;427:364–370. doi: 10.1038/nature02256. [DOI] [PubMed] [Google Scholar]
- 52.Carleton M, Mao M, Biery M, Warrener P, Kim S, Buser C, et al. RNA interference-mediated silencing of mitotic kinesin KIF14 disrupts cell cycle progression and induces cytokinesis failure. Mol Cell Biol. 2006;26:3853–3863. doi: 10.1128/MCB.26.10.3853-3863.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Molina I, Baars S, Brill JA, Hales KG, Fuller MT, Ripoll P. A chromatin-associated kinesin-related protein required for normal mitotic chromosome segregation in Drosophila. J Cell Biol. 1997;139:1361–1371. doi: 10.1083/jcb.139.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanudji M, Shoemaker J, L’Italien L, Russell L, Chin G, Schebye XM. Gene silencing of CENP-E by small interfering RNA in HeLa cells leads to missegregation of chromosomes after a mitotic delay. Mol Biol Cell. 2004;15:3771–3781. doi: 10.1091/mbc.E03-07-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baker DJ, Jeganathan KB, Malureanu L, Perez-Terzic C, Terzic A, van Deursen JM. Early aging-associated phenotypes in Bub3/Rae1 haploinsufficient mice. J Cell Biol. 2006;172:529–540. doi: 10.1083/jcb.200507081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Babu JR, Jeganathan KB, Baker DJ, Wu X, Kang-Decker N, van Deursen JM. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J Cell Biol. 2003;160:341–353. doi: 10.1083/jcb.200211048. [DOI] [PMC free article] [PubMed] [Google Scholar]