Abstract
Objective
We investigated the role of CCAAT enhancer-binding protein-α (C/EBPα) during zebrafish embryonic blood development.
Methods
Whole-mount mRNA in situ hybridization was performed to determine the spatio-temporal expression pattern of zebrafish cebpa in developing hematopoietic progenitors. A deletion mutation of cebpa (zD420), which mimics the human dominant-negative mutations of C/EBPα, was transfected into CV1 cell line to evaluate its transcriptional activity in vitro and injected into zebrafish embryos at the one- to two-cell stage to examine its effects on primitive hematopoiesis during early zebrafish development.
Results
Zebrafish cebpa is expressed in the anterior and posterior lateral plate mesoderm at 12 hours postfertilization, along with scl, pu.1 and gata1 in developing hematopoietic progenitors. In vitro, the deletion mutation of cebpa (zD420) prevents expression of the full-length protein, allowing the expression of truncated isoforms from internal translational initiation sites. As in the human, the truncated zebrafish C/ebpα proteins did not activate the expression of known target granulocytic genes, and in fact, suppressed transactivation that was induced in vitro by the full-length protein. Forced expression of the zD420 mRNA in zebrafish embryos led to an expansion of primitive erythropoiesis, without a discernible effect on granulopoiesis.
Conclusion
Expression of the truncated isoforms of cebpa alters the developmental pattern of hematopoietic progenitor cells during embryogenesis.
Keywords: Zebrafish, C/ebpα, erythropoiesis
In the mammalian hematopoietic system, the CCAAT/enhancer-binding protein α (C/EBPα, encoded by the CEBPA gene) functions as a critical regulator of granulocytic differentiation. CEBPA is strongly expressed in granulocyte/monocyte progenitors (GMPs) of human and rodent bone marrow and is specifically upregulated during granulocytic differentiation [1-4]. Induced expression of CEBPA in GMPs can initiate granulocytic differentiation along with simultaneous inhibition of monocytic maturation [2,5]. Mice lacking the CEBPA gene also lack mature granulocytes and show an accumulation of myeloblasts in blood and bone marrow [6]. Additionally, murine hematopoietic transplantations of wild-type cells expressing the BCR-ABL oncogene results in myeloid leukemia, while the transplantation of CEBPA null cells expressing this oncogene developed erythroleukemia [7].
Heterozygous mutations in CEBPA were recently found in cases of acute myeloid leukemia (AML) [8]. These mutations affect the amino terminus and eliminate expression of the 42-kDa full-length, wild type C/EBPα protein, but do not affect a 30-kDa isoform initiated further downstream. This truncated protein inhibits DNA binding of the wild-type C/EBPα, and, thus, inhibits transactivation of key granulocytic target genes in a dominant-negative manner [8,9]. Despite major advances in understanding the biochemical properties of C/EBPα, the roles of this transcription factor and its dominant-negative isoform in early blood development and AML remain largely unknown.
Emerging evidence suggests that the zebrafish might provide a useful model for elucidating the regulatory functions of mammalian C/EBPα during primitive hematopoiesis. Zebrafish orthologues of many key transcriptional factors necessary for mammalian hematopoietic development have been identified, suggesting a conserved genetic program regulating vertebrate hematopoiesis [10,11]. The zebrafish system affords several advantages for this analysis, including external embryonic development and optical clarity, which allow direct visualization of the hematopoietic process. In the fish, hematopoiesis begins in two anatomically and functionally distinct regions: the anterior and posterior -lateral plate mesoderm (A- and P-LPM) [12-14]. Cell fate mapping experiments at the shield stage of development show that P-LPM arises from the most ventral region opposite the embryonic organizer (or shield) [15,16]. Cells in the P-LPM migrate medially in an anterior-to-posterior wave to form the intermediate cell mass (ICM, or “ventral” mesoderm) [10,17]. P-LPM/ICM is the major site of primitive erythropoiesis, with gata1 being exclusively expressed in this region during somitogenesis [10,18]. However, the expression of granulocyte-specific genes, such as pu.1 and mpo [12,19] in the posterior ICM prior to the onset of circulation (less than 24 hpf) suggests that P-LPM/ICM may also maintain a level of granulopoietic activity, during early development of the zebrafish hematopoietic system. During gastrulation, A-LPM arises in a region closer to the shield than to the ventral end destined to become P-LPM/ICM [12] and unlike P-LPM, appears to be exclusively occupied by cells that express granulocytic genes, such as pu.1[12], mpo [19], c/ebp1 [20] and l-plastin [14]. Despite these spatially-restricted gene expression patterns, recent studies in the zebrafish have determined that both the A- and P-LPM contain bipotential cells, similar to the common myeloid progenitor cells in mammalian hematopoiesis, with the ability to differentiate into either erythroid or myeloid cells [21].
The rapid development of granulopoiesis in the A-LPM and its coexistence with erythropoiesis in the P-LPM region of zebrafish embryos prompted us to study the functions of zebrafish C/ebpα and its dominant-interfering isoforms in vertebrate blood development. We show here that cebpa is coexpressed with scl, gata1 and pu.1 in the hematopoietic progenitors in the P-LPM of embryos at 12 to 16 hours postfertilization (hpf). Forced expression of dominant-interfering C/ebpα isoforms induced an expansion of primitive erythropoiesis in the P-LPM and ectopic erythropoiesis in the A-LPM, without a discernible effect on granulopoiesis in both the A- and P-LPM. These results suggest that the aberrant expression of truncated forms of C/ebpα can interfere with the development of embryonic hematopoietic progenitors and drive them to adopt an erythroid cell fate.
Materials and methods
Fish care
Zebrafish maintenance, breeding and staging were performed as described previously [22,23].
Cloning and mapping
Messenger RNA was extracted and isolated from zebrafish adult kidney, and degenerate PCR was used to amplify a 383-bp fragment of zebrafish cebpa. An adaptor-ligated SMART cDNA library was then constructed by using the kidney mRNA for 5’ and 3’ RACE (rapid amplification of cDNA ends) to obtain the cDNA of full-length cebpa. Radiation hybrid mapping was performed with the Goodfellow zebrafish T51 panel. Primer pairs were designed from the 3’UTR of zebrafish cebpa using OILGO 6 software (forward primer: 5’-GGTAAAATCATGCCCATTAGCTGC-3’; reverse primer: 5’-CGGAGCGAGCTTGACTTTTGAA-3’). Zebrafish putative orthologues were identified by the “reciprocal best hit” method, as described previously [24].
Whole-mount mRNA in situ hybridization and single-embryo RT-PCR
Antisense probes were generated from the 1.8-kb fragment of the 3’ RACE product of the cebpa gene. The synthesis of RNA probes and hybridizations were performed as described previously [19]. Double in situ hybridization assays were performed with digoxigenin- and fluorescein-labeled probes and developed with the chromagenic substrates BCIP/NBT and Fast Red, as described previously [19]. For single-embryo RT-PCR, an individual embryo injected with either zD420 or GFP mRNA was rinsed twice with PBS and transferred into an RNase-free tube containing 100 μl of Trizol (GIBCO) and 5 μl glycogen (Ambino). Extracted total RNA was diluted in 10 μl of RNase-free water. One microliter of total RNA was used as a template and one-step RT-PCR (Qiagen) was performed in a volume of 25 μl. Intron-spanning primers were designed with PRIMER 3 software. Primer sequences of zebrafish genes are: gata1 (F: ATTATTCCACCAGCGTCCAG and R: CCACTTCCACTCATGGGACT); α-hemaglobin (F: TTGTCTACCCCCAGACCAAG and R: AGAGCCAGAGCTGAGAGGAA) and β-actin (F: CCCAGACATCAGGGAGTGAT and R: CACCGATCCAGACGGAGTAT). The PCR conditions were as follows: 50°C, 30 min; 95°C, 15min; 28 cycles of 94°C, 30 s; 60°C, 30 s; 72°C, 1 min; then 72°C, 10 min; 4°C stored.
Constructs
The wild type, full-length cebpa gene (zWT), defined by Sac I/Xho I, was subcloned into either the pCS2+ or PBK vector. Site-directed mutagenesis was performed with the zWT to delete the third base of the triplet TTC encoding the conserved phenylalanine at amino acid position 65 of zebrafish C/ebpα. The resultant deletion mutant was designated zD420. zWT-GFP and zD420-GFP were generated by in-frame fusion of the gene encoding green fluorescent protein (GFP) with the 3’ end of the zWT and zD420 open reading frame.
Electrophoretic mobility shift assay
The zWT and zD420 proteins (both GFP-tagged and untagged) were translated in vitro with the reticulocyte system (Promega). To compare translation efficiencies, we performed parallel reactions in the presence of 35S-labeled methionine, and separated protein products on a 12% SDS-polyacrylamide gel. The binding reaction was performed as described previously [8]. The sequence of the human GCSFR promoter oligonucleotide (bp −57 to −38, with C/EBPα binding site underlined) was 5’-AAGGTGTTGCAATCCCCAGC-3’. GFP monoclonal antibody (0.2ul; Clontech) was used in electrophoresis supershift experiments performed on a 4% polyacrylamide gel at 10V/CM in 1×TBE buffer.
Immunoblotting
CV1 cells (5×105) were transfected with expression plasmid (1 μg) using Lipofectamine (Gibco). Twenty-four hours after transfection, the cells were lysed in RIPA buffer (1×PBS, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 100 μg/ml PMSF, 1 μg/ml aprotinin), and protein extracts were diluted 1:1 with Laemmli sample buffer (Bio-Rad), fractionated on 10% SDS-polyacrylamide gels and transferred to nitrocellulose membranes. The blots were blocked in 1×TBS, 5% nonfat milk and 0.05% Tween-20 at room temperature for 30 minutes, followed by incubation with a 1:1,000 dilution of either a mouse monoclonal anti-GFP antibody (Clontech) or a rabbit polyclonal antiserum against zebrafish C/ebpα (using a peptide fragment, ‘N’-GIFRQLPDGSFVKAMGNCA-‘C’, that represented the last 20 amino acids 268-288 of zebrafish C/ebpα as an antigen source) at room temperature for 1 hour. The blots were then washed with TBST. After incubation with an anti-mouse or -rabbit IgG-horseradish peroxidase (HRP)-conjugated secondary antibody (diluted at 1:2,000) at room temperature for 45 minutes, the blots were visualized with an ECL kit (Santa Cruz) according to the manufacturer’s protocol.
Transactivation assay
CV1 cells (2×104) were seeded to 24-well dishes and transfected using Lipofectanin (Gibco) with reporter plasmid (200 ng; tetramer of the CEBP site of human GCSFR inserted into the promoterless luciferase vector pTK81-luc), the CMV-LacZ construct (40 ng), and selected C/ebpα expression plasmids (60 ng). Luciferase assays were carried out at 24 hours post-transfection according to the manufacturer’s instruction (Dual-luciferase reporter assay, Promega). Luciferase activities were normalized for transfection efficiency using the cotransfected CMV-LacZ construct and chemiluminescent reporter assay kit (to detect β-galactosidase [TROPIX]). All transactivation experiments were repeated three times with two different versions of each plasmid (cloned in CMV-containing pCS2 and PBK vector). The expression of C/ebpα protein was detected by Western blotting using either anti-GFP or rabbit antiserum against the zebrafish C/ebpα protein.
Microinjection
Capped mRNAs were transcribed from a NotI-linearized plasmid (zWT and zD420, cloned in pCS2) using SP6 RNA polymerase (Message Machine, Ambion), purified by phenol-chloroform extraction, dissolved in DEPC-treated water and quantified with a spectrophotometer.
Histology
Embryos to be sectioned were rinsed briefly in PBS twice and fixed in 4% paraformaldehyde (PFA) overnight at 4°C prior to paraffin sectioning. Sections were stained with hematoxylin and eosin. After in situ hybridization, the embryos were rehydrated and embedded in 3% sucrose overnight at 4° C before cryostat sectioning. Sections were counterstained with Eosin (Sigma) and photographed.
Results
Structural features of the zebrafish cebpa orthologue
The full-length cDNA sequence of the zebrafish cebpa orthologue has been reported previously [25]. We obtained cebpa genomic P1-artificial clones (PACs) by screening the zebrafish genomic PAC library, and sequenced the 5-kb genomic sequence that comprised the entire cebpa open reading frame. As in other species, the mRNA of the full-length zebrafish cebpa gene was transcribed from a single exon containing a highly conserved upstream open reading frame (u-ORF), followed by a spacer of seven nucleotides in the 5’-untranslated region (Figure 1A, top). Comparison of the zebrafish C/ebpα protein with its human, mouse, bovine, chicken, frog, and xenopus counterparts revealed several conserved regions, including a C-terminal domain consisting of a DNA-binding nuclear localization signal and leucine zipper dimerization region (LZ) and two transactivation domains (AD1 and 2) at the N-terminus (Figure 1A, bottom), which reportedly possess transactivation potential [26].
Figure 1.
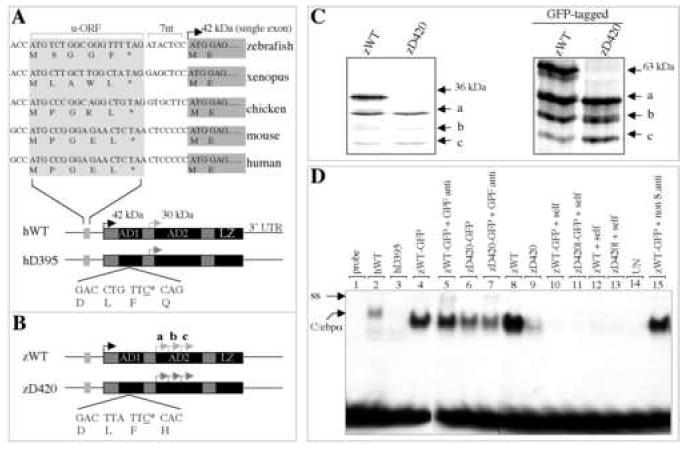
Deficiency of dominant-interfering C/ebpα isoforms in binding to human GCSFR promoter DNA. (A) Structural conservation of CEBPA gene during evolution. The deleted cytosine at position 395 of human CEBPA gene (referred to as hD395) is underlined (bottom). u-ORF, upstream of open reading frame; nt, nucleotides; AD1, transactivation domain 1; AD2, transactivation domain 2; LZ, lucine zipper; UTR, untranslated region. Black arrow denotes the translational initiation site that encodes the full-length 42-kDa C/EBPα (referred to as hWT), while gray arrow denotes the in-frame, internal initiation site that encodes a dominant-interfering, 30-kDa N-terminal truncated isoform. (B) Schematic diagram of full-length (zWT) and alternative (zD420) isoforms of zebrafish cebpa gene. The deleted cytosine homologue at position 420 of zebrafish cebpa (zD420) is underlined. Black arrow denotes the translational initiation site that encodes the full-length C/ebpα protein (36-kDa, zWT). Gray arrows mark three putative, in-frame internal initiation sites (a, b and c). (C) Proteins translated in vitro in the presence of 35S-labeled methionine from zWT and mutant zD420 (left panel), as well as GFP-tagged zWT and zD420 (right panel) expression plasmids. (D) Comparison of the ability of in vitro translated zD420 mutant proteins (lanes 6 and 9) and zWT proteins (lanes 4 and 8) to bind to the CEBP site of the human GCSFR promoter. Human wild type (hWT, lane 2) and mutant (hD395, lane 3) proteins served as positive controls. Also indicated are specific, unlabeled competitor oligonucleotides (self, lanes 10-13) used in 100-fold molar excess, unprogrammed lysates used for in vitro translation (UN, lane 14) and GFP antibody (anti, lanes 5 and 7) or nonspecific antibody (non S.anti, sheep anti-mouse IgG, lane 15) used as a negative control for supershift experiments. ss, supershift; C/ebpα, shifted band.
Radiation hybrid (RH) mapping found the cebpa gene in a chromosome fragment between z3076 and z5563 on zebrafish chromosome 7, which is syntenic with the human and mouse CEBPA loci (data not shown). These results indicate that zebrafish cebpa is a bona fide structural orthologue of mammalian CEBPA, with the exception of an important structural divergence. In mammals (human, mouse and bovine), a single internal translational initiation site (ATG) encodes a 30-kDa dominant-interfering protein isoform, residing between AD1 and AD2 (Figure 1A, bottom), whereas, in zebrafish three such sites were found in the same reading frame within the AD2 domain (a, b and c in Figure 1B).
Internal translation initiation sites direct the synthesis of truncated isoforms
In human AML cases, deletion of cytosine 395 of the human CEBPA cDNA (hD395; Figure 1A, bottom) abolishes expression of the full-length (42-kDa) protein and promotes the expression of a 30-kDa isoform. This truncated protein blocks the DNA-binding activity of wild-type C/EBPα, and the transactivation of granulocytic target genes, in a dominant-interfering manner [8]. To determine whether the three potential internal initiation sites of the zebrafish cebpa gene (proteins a, b and c; Figure 1B) direct protein synthesis, we deleted the evolutionarily conserved cytosine 420 (corresponding to the cytosine 395 in human CEBPA cDNA) of the cebpa cDNA (zD420; Figure 1B, bottom). While the in vitro translation of the wild-type cebpa expression plasmid (designated zWT) resulted in expression of four C/ebpα proteins, one full-length (36 kDa) and three isoforms, only the three shorter isoforms were obtained with the zD420 expression plasmid (Figure 1C, left panel). In vitro translation of green fluorescence protein (GFP)-tagged zWT and zD420 (zWT-GFP and zD420-GFP) expression plasmids yielded the same results (Figure 1C, right panel), indicating that fusion with GFP did not affect the translation of these protein isoforms in vitro.
Truncated zebrafish C/ebpα proteins show decreased binding to the CEBP site in the human GCSFR promoter
Binding of the C/EBPα protein to the functional region (-57 to -38) of the human granulocyte colony-stimulating factor receptor (GCSFR) gene is required for granulocytic differentiation [27]. Therefore, we performed gel-shift and supershift assays to assess the ability of truncated and full-length zebrafish C/ebpα proteins to bind to the human GCSFR promoter. Proteins obtained from in vitro translation (Figure 1C) of zWT expression plasmid displayed strong DNA binding, while those from the zD420 expression plasmid showed markedly decreased binding activity (Figure 1D, lanes 8-9). The results indicate that truncated C/ebpα isoforms lose most of their ability to bind to the human GCSFR promoter, consistent with the previous observation for human hWT and hD395 [8]. Decreased DNA binding activity was also observed for zD420-GFP proteins, although it was not as pronounced as that for zD420 proteins (Figure. 1D, lane 4 and lane 6).
Truncated proteins suppress transactivation by full-length C/ebpα
We next investigated transactivation by full-length and truncated C/ebpα proteins binding through a CCAAT site in the human GCSFR promoter in transfected CV1 cells (lack endogenous C/EBPα protein). Both GFP-tagged and untagged zWT and zD420 expression plasmids were transiently transfected into CV1 cells. Immunoblotting with either GFP antibody (for the GFP-tagged proteins) or a rabbit antiserum against C/ebpα (for untagged proteins) detected the full-length and truncated proteins (Figure 2A, top panels). The result indicated that the zD420 mutation results in expression of the three truncated isoforms initiated further downstream.
Figure 2.
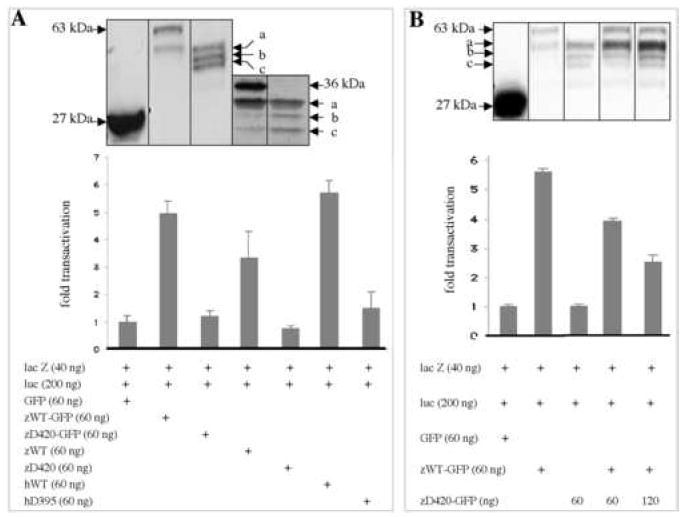
Transactivation potential of truncated C/ebpα isoforms versus wild-type protein. (A) CV1 cells were transiently cotransfected with 60 ng of each indicated expression plasmid, with CMV-lacZ plasmid (lacZ, 40 ng) and with luciferase reporter plasmid (luc, 200 ng), which contains a tetramer of the CEBP site of the human GCSFR promoter. Transactivation results (bottom) of the luciferase assay are compared with that of the pCS2+GFP expression vector alone (“GFP”=1.0) and were normalized for transfection efficiency by measuring β-galactosidase activity from the cotransfected lacZ plasmid. The bars denote mean (± SD) values for three repetitions. Western analysis of transfected CV1 cells (top) were performed in parallel to detect the expression of wide-type and mutated C/ebpα proteins using either an mouse monoclonal antibody against GFP or an rabbit antiserum against zebrafish C/ebpα. (B) CV1 cells were transiently transfected with 60 ng of GFP-tagged zWT and increasing amounts of GFP-tagged zD420 plasmids. Total amounts of transfected plasmids were 420 ng (otherwise, pCS2 plasmid was added). The proteins were detected in parallel by Western blots (top) from parts of the lysates used for luciferase assay.
The zWT and zWT-GFP proteins stimulated luciferase transactivation 3.5- and 5-fold above background, while the zD420 and zD420-GFP expressed proteins were the same as GFP controls (Figure 2A). As a control, full-length C/ebpα proteins failed to activate the mutated GCSFR promoter (data not shown). To test for dominant interfering activity, we transfected a constant amount of zWT (60 ng) plasmid with an increasing amount of zD420 plasmid. A 1:1 ratio of mutant to wild-type plasmid reduced the luciferase expression by 30%, while a 2:1 ratio reduced it by more than 50% (Figure 2B). Increasing amounts of the truncated proteins were demonstrated by Western analysis (Figure 2B, top).
Spatial and temporal expression of cebpa during embryogenesis
The zebrafish cebpa expression pattern in developing embryos was evaluated by in situ hybridization analysis. The gene was first detected in the yolk syncytial layer at 50% epiboly at 5.3 hours postfertilization (hpf) (Figure 3A, arrowheads). By 12 hpf, cebpa activity was detected in two stripes along the lateral plate mesoderm in both the anterior and posterior regions of the embryo (A- and P-LPM, Figure 3B,C). By 13 hpf, the A-LPM cebpa+ cells began to migrate medially, converging to the mid-line by 17 hpf, and subsequently dispersed over the surface of the yolk cell (Figure 3C,E, top). In the P-LPM between 12-17 hpf, cebpa+ cells converged medially in an anterior-to-posterior wave, resulting in a single stripe in the position of the ICM (Figure 3C). The cebpa expression decreased by 20 hpf (Figure 3C, bottom) and is largely gone by 24 hpf in the ICM (Figure 3E, top). The cebpa+ cells in the gut primordium, between the rostral and caudal LPM, could be detected at 13 hpf and has converged towards the midline by 20 hpf (Figure 3C, arrowheads). By 48 hpf, LPM-derived cebpa+ cells were reduced overall, with a few positive cells remaining on the yolk cells (Figure 3D, arrow). At that time, cebpa expression was observed in the developing liver and gut (Figure 3D, star and arrowhead), in accordance with previous observations [25].
Figure 3.
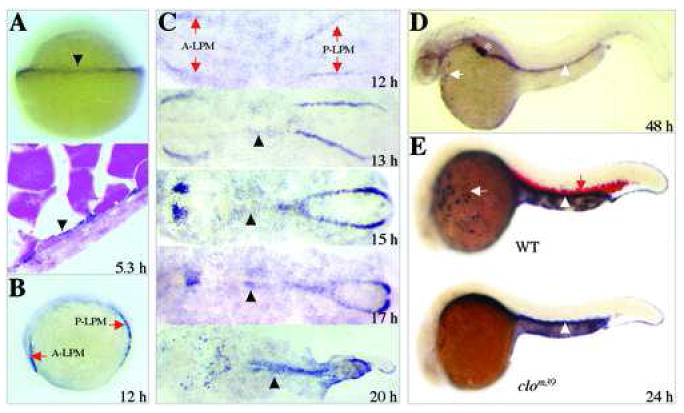
Spatio-temporal expression of cebpa during zebrafish embryogenesis. (A) At the 50%-epiboly stage (5.3 hpf), expression of cebpa is first detected in the yolk syncytial layer (YSL) at the margin of the blastoderm (top, arrowhead). Cells expressing cebpa are seen in a representative section across the YSL (bottom. arrowhead). (B) At the 6-somite stage (12 hpf), cebpa expression can be seen as distinct stripes, one is located in the anterior lateral plate mesoderm and the other in the posterior lateral plate mesoderm (A-LPM and P-LPM, arrows). (C) Dynamic expression of cebpa in cells of A- and P-LPM during the segmentation period (stages 12 to 20 hpf). (D) At 48 hpf, expression of cebpa can been observed in anterior myeloid cells (arrow), liver (star) and developing gut primordium (arrowhead). (E) At 24 hpf, detection of cebpa (white arrow for myeloid cells; white arrowhead for gut) and α-hemoglobin (red arrow) in wild-type sibling (top) as well as the cloche mutant (clom39, bottom). Note that the cloche mutant lacks both cebpa and α-hemoglobin in hematopoietic cells, but maintains the gut expression of cebpa. Panels A, B, D and E are lateral views, anterior to the left in B, D and E, with animal pole up in A. Panel C shows dorsal views of flat-mounted embryos, anterior to the left.
The cloche (clom39) mutant lacks the capacity for hematopoiesis and vasculogenesis due to an as-yet-unidentified mutation upstream of the early hematopoietic transcription factor scl [28]. Embryos homologous for the clom39 allele show loss of most hematopoietic gene expression at 24 hpf, including gata1 and α-globin [29]. Unlike the normal expression pattern of cebpa and α-globin in wild type or heterozygous cloche siblings (Figure 3E, top, red and black arrows), neither gene was expressed in the regions normally containing hematopoietic cells of the homozygous clom39 embryos (Figure 3E, bottom), although cebpa expression remained unaffected in the gut (Figure 3E, arrowheads). These results indicate that cebpa was specifically expressed in blood cells of developing embryos.
Colocalization of cebpa with blood markers in developing zebrafish hematopoietic progenitor cells
The spatial and temporal distribution of cebpa mRNA transcripts in the A- and P-LPM during zebrafish embryogenesis correlates with the described expression patterns of scl and pu.1 in the A-LPM [12,30] and scl, pu.1 and gata1 in the P-LPM [17]. Two-color in situ hybridization analysis confirmed that cebpa and scl expression are coexpressed in a lateral subset of cells of both the P- (Figure 4A) and A-LPM (data not shown) at 12 hpf. However, unlike some scl+ cells in the A-LPM, none of the cebpa+ cells contribute to the developing central nervous system (data not shown).
Figure 4.
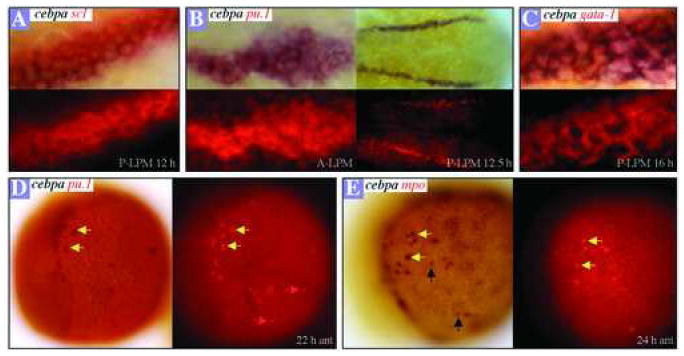
Analysis of cebpa coexpression with other blood markers. Two-color in situ hybridization gene expression analysis in wild-type embryos (flat mount, anterior to the left; dorsal views in A, B, C and lateral views in D, E). Fast Red labeling of blood markers (RITC filter set), is shown at the bottom of panels A, B, C and to the right in panels D, E. (A) Cells expressing cebpa (black) and scl (red) in the P-LPM of a 12 hpf embryo. (B) Cells expressing cebpa (black) and pu.1 (red) in the A-LPM (left) and P-LPM (right) of a 12.5-hpf embryo. (C) Expression of cebpa (black) and gata1 (red) in the P-LPM of a 16-hpf embryo. (D) Cells coexpressing cebpa and pu.1 (yellow arrows) in the anterior yolk sac of a 22-hpf embryo. Red arrows denote the cells expressing pu.1 only. (E) Colocalization of cebpa with mpo is marked with yellow arrows in the anterior yolk sac of a 24-hpf embryo. Black arrows indicate cells expressing cebpa only.
Colocalization of cebpa with pu.1 mRNA transcripts was observed at 12.5 hpf in the A-LPM (Figure 4B, left panels) and P-LPM (Figure 4B, right panels) after the onset of pu.1 expression. Both cebpa and pu.1 are coexpressed with gata1 in the same cells of the P-LPM through 16 hpf (Figure 4C, and data not shown). At 22 hpf, most of the cebpa+ cells also expressed pu.1 in cells dispersed on the yolk (Figure 4D, yellow arrows), but some pu.1+ cells did not express cebpa (Figure 4D, red arrows). Coexpression of cebpa with the granulocyte-specific gene mpo was assayed and most of the mpo-positive cells expressed cebpa (Figure 4E, yellow arrows), consistent with its required role in mammalian granulopoiesis. Because cebpa is expressed earlier in the development of myeloid cells than mpo, it is not surprising that mpo was not expressed in all cebpa+ cells (Figure 4E, black arrows).
Dominant-interfering cebpa enhances erythropoiesis in P-LPM
The colocalization of the hematopoietic transcription factors cebpa, gata1, pu.1 and scl in the P-LPM of 12-16 hpf embryos led us to test the consequence of misexpressing the dominant-interfering isoform, zD420, in living zebrafish. Injection of zD420 mRNA into one-cell stage embryos resulted in a striking expansion of gata1-expressing cell at 16 hpf in the P-LPM (Figure 5B, arrow), compared to controls (Figure 5A, Table 1). Interestingly, ectopic expression of gata1 was observed in cells located medially to the P-LPM in some of the zD420-injected embryos (Figure 5B, arrowhead), as well as in cells on the anterior yolk of nearly all zD420-injected embryos (Figure 5D, arrowheads). Such ectopic expression was absent in normal embryos (Figure 5A,C). The increased gata1 expression in the P-LPM reflects an expansion of erythropoietic progenitors, and this results in enhanced erythropoiesis as demonstrated by the increased expression of α-hemaglobin in the 22-hpf precirculation embryos (Figure 5E,F).
Figure 5.
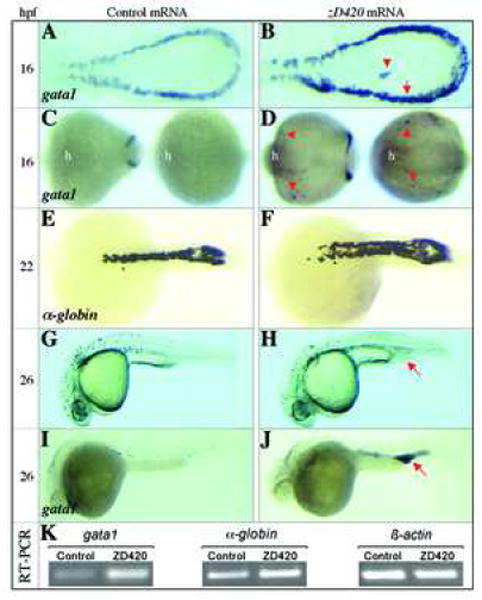
Effects of dominant-interfering cebpa on primitive erythropoiesis. (A,B) Expression of gata1 detected by whole-mount mRNA in situ in P-LPM of zD420- and control (GFP) mRNA-injected embryos (16 hpf, dorsal view with head to the left). Red arrow in B denotes gata1-positive cells in the P-LPM, while the arrowhead represents ectopic cells expressing gata1. (C,D) Expression of gata1 in the anterior part of the zD420- and control mRNA-injected embryos (16 hpf; left embryo, ventral view; right embryo, dorsal view). Note that ectopic gata1 positive cells (arrowheads) are detected only in zD420-injected embryos. The head is indicated (h) as a point of reference. (E,F) Expression of α-hemaglobin detected in the ICM of the zD420- and control mRNA-injected embryos (22 hpf, dorsal view). (G,H) Morphology of 26-hpf embryos. Arrow in H denotes an expanded posterior blood island (EPBI). (I,J) Expression of gata1 detected by whole mount mRNA in situ hybridization in 26-hpf embryos. Arrow in J denotes gata1-positive cells in the EPBI. (K) Single-embryo RT-PCR analyses of zebrafish gata1, α-hemaglobin and β-actin genes in GFP and zD420 mRNA-injected embryos at 22 hpf. Similar results were repeated three times with separate embryos injected with GFP and zD420 mRNA.
Table 1.
RNA injection studies
| mRNA | E-gata1a (16 h) | Ant.Ec-gata1b (16 h) | EPBIc (24 h) |
|---|---|---|---|
| Control (GFP) | 0/ 26 (0.0 %) | 0/ 26 (0.0 %) | 1/82 (1.2 %) |
| zD420 | 30/30 (100 %) | 30/30 (100 %) | 40/90 (44%) |
Expanded gata1 expression.
Anterior Ectopic gata1 expression.
Expanded posterior blood island.
At 26 hpf, half of the zD420-injected embryos exhibited an abnormally expanded posterior blood island (EPBI) at the posterior end of the ICM (Figure 5G,H, red arrow, Table 1). Each EPBI contained increased gata1-expressing cells compared to controls (Figure 5I,J). Consistently, analyses of the expression of gata1 and α-globin genes by semi-quantitative RT-PCR in single-embryo level showed that the transcripts of gata1and α-globin in zD420-injected embryo increased twice more than that in control GFP-injected embryo (Figure 5K). We do not believe that the EPBI was caused by abnormal development of the vasculature or a block in circulation, as both of these processes appeared to be normal at this stage of development (data not shown). Surprisingly, myeloid cell development appeared normal in the zD420-injected embryos, as indicated by the normal expression pattern of granulocyte- and monocyte-specific genes such as mpo and l-plastin, respectively, at 26 hpf (data not shown). These findings would be unexpected if the zD420-encoded proteins were functioning in vivo as a dominant negative protein in hematopoietic progenitors, because loss of Cebpa in the mouse results in a loss of granulopoiesis [6]. However, opposing data from in vitro studies indicate that the expression of a dominant negative CEBPA in murine bone marrow cells does not block granulocyte differentiation [9]. Thus our results suggest a gain-of-function activity due to the aberrant expression of truncated forms of cebpa in hematopoietic progenitors in vivo, which stimulates GATA-1 expression and results in ectopic erythropoiesis. Since pu.1 expression is not affected in zD420-injected embryos, it is likely that pu.1 acts upstream or parallel to cebpa, which is consistent with recent observations showing that pu.1 is required for normal embryonic zebrafish myelopoiesis and may regulate the myelopoietic expression of cebpa [21].
Discussion
The translationally regulated expression of critical transcription factors plays a pivotal role in determining hematopoietic cell fate [31,32]. For example, expression of the full-length human SCL protein drives uncommitted hematopoietic cells toward the megakaryocyte lineage, while truncated isoforms (generated by an alternative translation initiation mechanism) favor the erythroid lineage [32]. In patients with Down’s syndrome-related acute megakaryocytic leukemia, mutations that introduce a stop codon within the first coding exon of GATA1 abolish the 50-kDa full-length GATA1 protein, but permit an alternative 40-kDa isoform to be expressed from a downstream initiation site [33,34]. A similar mechanism generates truncated isoforms of C/EBPα that contribute to the molecular pathogenesis of AML [9].
Despite the importance of translational control mechanisms in the regulation of hematopoiesis, it is surprising that expression of truncated C/ebpα isoforms causes an expansion of primitive erythropoiesis activity during zebrafish development. Unfortunately, we are not able to assess the effects of the truncated isoform on definitive hematopoiesis, because of the technical limitation of the mRNA injections. The primitive erythroid expansion in the posterior of the embryo in the experiments using mutant cebpa mRNA encoding only truncated protein may be in part result of the inhibition of full-length C/ebpα function. In the livers of Cebpa-null mice, a fourfold increase in Epo receptor mRNA is detected [35]. Induced expression of C/EBPα in primary human CD34+ cells blocks - erythropoiesis through the downregulation of inhibitor of differentiation-1 (ID1), a transcriptional repressor known to interfere with erythrocyte differentiation [36]. Zebrafish cebpa is co-expressed with gata1 and scl in subsets of progenitors during embryonic primitive hematopoiesis (Figure 4) and recent studies in the zebrafish have demonstrated that, as in mammals, mature erythroid and myeloid cells arise from a common myelo-eyrthroid progenitor cell (MEP) [21]. Induction of human CD34+ bone marrow cells towards different hematopoietic lineages resulted in corresponding changes in gene expression; erythroid differentiation correlated with expression of SCL and GATA1 while myeloid cells expressed C/EBPα and PU.1 [37]. Thus, it is possible that dominant-interfering inhibition of wild-type C/ebpα function in these scl+ hematopoietic stem cells or MEP cells could favor erythroid progenitor proliferation and differentiation by increasing the expansion of gata1 expressing cells. Alternatively, overexpression of truncated forms of C/ebpα may act through as-yet-undefined gain of function mechanisms to promote erythropoiesis, as has been shown recently for truncated forms of GATA-1 and their actions in promoting abnormal megakaryopoiesis in genetically engineered mice [34].
In contrast to its effect on erythropoiesis, the truncated C/ebpα isoform appears to have only limited influence on the differentiation of granulocytes, as indicated by the appropriate number of cells expressing mpo in embryos injected with zD420 mRNA. It remains possible that wild-type C/ebpα is incompletely inhibited by the truncated C/ebpα isoform or that parallel pathways mediated by pu.1 compensate for the loss of cebpa function. Unfortunately, we were not able to study the effects of either overexpressing or knocking-down full-length C/ebpα in zebrafish embryos. Because forced expression of full-length cebpa mRNA, even in low concentrations (less than 50 pg/per embryo), caused embryonic death during gastrulation prior to the onset of hematopoiesis. Furthermore, four different morpholinos against different sites of C/ebpα around the ATG do not cause detectable blood phenotypes, although at higher doses leading to ventralized monsters (Dr. John Kanki, unpublished data). We are not allowed to design alternative morpholino against a splice site because of C/ebpα being a single exon.
Our results provide novel evidence for the involvement of alternatively translated C/ebpα proteins in the control of erythroid development. Given the previously observed antagonism between PU.1 and GATA1 proteins [21,38] and the loss of PU.1 function in erythroleukemia [39], our results suggest that full length C/ebpα may also be involved antagonistically in the Gata1-associated pathway of blood development. Deregulation of normal C/ebpα translation is already known to contribute to AML, and our studies in zebrafish expressing dominant-interfering C/ebpα suggest that this gene should also be examined in erythroleukemia.
Acknowledgments
We thank Drs. Yi Zhou and Leonard I. Zon for radiation hybrid mapping of cebpa; Hui Ying Piao, George Kourkoulis, Jessica Vinokur and Walter Saganic in the DFCI zebrafish facility for excellent fish care; and John R. Gilbert for editorial assistance with manuscript preparation. The work was supported by a Leukemia and Lymphoma Society Center grant, and by NIH grants CA93152 (ATL), HD-41330 (JR), and CA96785 (KH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hohaus S, Petrovick MS, Voso MT, Sun Z, Zhang DE, Tenen DG. PU.1 (Spi-1) and C/EBP alpha regulate expression of the granulocyte-macrophage colony-stimulating factor receptor alpha gene. Mol Cell Biol. 1995;15:5830. doi: 10.1128/mcb.15.10.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Radomska HS, Huettner CS, Zhang P, Cheng T, Scadden DT, Tenen DG. CCAAT/enhancer binding protein alpha is a regulatory switch sufficient for induction of granulocytic development from bipotential myeloid progenitors. Mol Cell Biol. 1998;18:4301. doi: 10.1128/mcb.18.7.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scott LM, Civin CI, Rorth P, Friedman AD. A novel temporal expression pattern of three C/EBP family members in differentiating myelomonocytic cells. Blood. 1992;80:1725. [PubMed] [Google Scholar]
- 4.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 5.Wang X, Scott E, Sawyers CL, Friedman AD. C/EBPalpha bypasses granulocyte colony-stimulating factor signals to rapidly induce PU.1 gene expression, stimulate granulocytic differentiation, and limit proliferation in 32D cl3 myeloblasts. Blood. 1999;94:560. [PubMed] [Google Scholar]
- 6.Zhang DE, Zhang P, Wang ND, Hetherington CJ, Darlington GJ, Tenen DG. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc Natl Acad Sci U S A. 1997;94:569. doi: 10.1073/pnas.94.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagner K, Zhang P, Rosenbauer F, Drescher B, Kobayashi S, Radomska HS, Kutok JL, Gilliland DG, Krauter J, Tenen DG. Absence of the transcription factor CCAAT enhancer binding protein alpha results in loss of myeloid identity in bcr/abl-induced malignancy. Proc Natl Acad Sci U S A. 2006;103:6338. doi: 10.1073/pnas.0508143103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pabst T, Mueller BU, Zhang P, Radomska HS, Narravula S, Schnittger S, Behre G, Hiddemann W, Tenen DG. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-alpha (C/EBPalpha), in acute myeloid leukemia. Nat Genet. 2001;27:263. doi: 10.1038/85820. [DOI] [PubMed] [Google Scholar]
- 9.Schwieger M, Lohler J, Fischer M, Herwig U, Tenen DG, Stocking C. A dominant-negative mutant of C/EBPalpha, associated with acute myeloid leukemias, inhibits differentiation of myeloid and erythroid progenitors of man but not mouse. Blood. 2004;103:2744. doi: 10.1182/blood-2003-07-2280. [DOI] [PubMed] [Google Scholar]
- 10.Amatruda JF, Zon LI. Dissecting hematopoiesis and disease using the zebrafish. Dev Biol. 1999;216:1. doi: 10.1006/dbio.1999.9462. [DOI] [PubMed] [Google Scholar]
- 11.Zon LI. Zebrafish: a new model for human disease. Genome Res. 1999;9:99. [PubMed] [Google Scholar]
- 12.Lieschke GJ, Oates AC, Paw BH, Thompson MA, Hall NE, Ward AC, Ho RK, Zon LI, Layton JE. Zebrafish SPI-1 (PU.1) marks a site of myeloid development independent of primitive erythropoiesis: implications for axial patterning. Dev Biol. 2002;246:274. doi: 10.1006/dbio.2002.0657. [DOI] [PubMed] [Google Scholar]
- 13.Shepard JL, Zon LI. Developmental derivation of embryonic and adult macrophages. Curr Opin Hematol. 2000;7:3. doi: 10.1097/00062752-200001000-00002. [DOI] [PubMed] [Google Scholar]
- 14.Herbomel P, Thisse B, Thisse C. Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development. 1999;126:3735. doi: 10.1242/dev.126.17.3735. [DOI] [PubMed] [Google Scholar]
- 15.Kimmel CB, Warga RM, Schilling TF. Origin and organization of the zebrafish fate map. Development. 1990;108:581. doi: 10.1242/dev.108.4.581. [DOI] [PubMed] [Google Scholar]
- 16.Warga RM, Nusslein-Volhard C. Origin and development of the zebrafish endoderm. Development. 1999;126:827. doi: 10.1242/dev.126.4.827. [DOI] [PubMed] [Google Scholar]
- 17.Detrich HW, 3rd, Kieran MW, Chan FY, Barone LM, Yee K, Rundstadler JA, Pratt S, Ransom D, Zon LI. Intraembryonic hematopoietic cell migration during vertebrate development. Proc Natl Acad Sci U S A. 1995;92:10713. doi: 10.1073/pnas.92.23.10713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson MA, Ransom DG, Pratt SJ, MacLennan H, Kieran MW, Detrich HW, 3rd, Vail B, Huber TL, Paw B, Brownlie AJ, Oates AC, Fritz A, Gates MA, Amores A, Bahary N, Talbot WS, Her H, Beier DR, Postlethwait JH, Zon LI. The cloche and spadetail genes differentially affect hematopoiesis and vasculogenesis. Dev Biol. 1998;197:248. doi: 10.1006/dbio.1998.8887. [DOI] [PubMed] [Google Scholar]
- 19.Bennett CM, Kanki JP, Rhodes J, Liu TX, Paw BH, Kieran MW, Langenau DM, Delahaye-Brown A, Zon LI, Fleming MD, Look AT. Myelopoiesis in the zebrafish, Danio rerio. Blood. 2001;98:643. doi: 10.1182/blood.v98.3.643. [DOI] [PubMed] [Google Scholar]
- 20.Lyons SE, Shue BC, Oates AC, Zon LI, Liu PP. A novel myeloid-restricted zebrafish CCAAT/enhancer-binding protein with a potent transcriptional activation domain. Blood. 2001;97:2611. doi: 10.1182/blood.v97.9.2611. [DOI] [PubMed] [Google Scholar]
- 21.Rhodes J, Hagen A, Hsu K, Deng M, Liu TX, Look AT, Kanki JP. Interplay of pu.1 and gata1 determines myelo-erythroid progenitor cell fate in zebrafish. Dev Cell. 2005;8:97. doi: 10.1016/j.devcel.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 22.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 23.Westerfield M. The zebrafish book a guide for the laboratory use of zebrafish Danio (Brachydanio) rerio. Eugene, OR: Institute of Neuroscience University of Oregon; 1993. [Google Scholar]
- 24.Liu TX, Zhou Y, Kanki JP, Deng M, Rhodes J, Yang HW, Sheng XM, Zon LI, Look AT. Evolutionary conservation of zebrafish linkage group 14 with frequently deleted regions of human chromosome 5 in myeloid malignancies. Proc Natl Acad Sci U S A. 2002;99:6136. doi: 10.1073/pnas.072560099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyons SE, Shue BC, Lei L, Oates AC, Zon LI, Liu PP. Molecular cloning, genetic mapping, and expression analysis of four zebrafish c/ebp genes. Gene. 2001;281:43. doi: 10.1016/s0378-1119(01)00774-0. [DOI] [PubMed] [Google Scholar]
- 26.Nerlov C, Ziff EB. Three levels of functional interaction determine the activity of CCAAT/enhancer binding protein-alpha on the serum albumin promoter. Genes Dev. 1994;8:350. doi: 10.1101/gad.8.3.350. [DOI] [PubMed] [Google Scholar]
- 27.Smith LT, Hohaus S, Gonzalez DA, Dziennis SE, Tenen DG. PU.1 (Spi-1) and C/EBP alpha regulate the granulocyte colony-stimulating factor receptor promoter in myeloid cells. Blood. 1996;88:1234. [PubMed] [Google Scholar]
- 28.Liao EC, Paw BH, Oates AC, Pratt SJ, Postlethwait JH, Zon LI. SCL/Tal-1 transcription factor acts downstream of cloche to specify hematopoietic and vascular progenitors in zebrafish. Genes Dev. 1998;12:621. doi: 10.1101/gad.12.5.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stainier DY, Weinstein BM, Detrich HW, 3rd, Zon LI, Fishman MC. Cloche, an early acting zebrafish gene, is required by both the endothelial and hematopoietic lineages. Development. 1995;121:3141. doi: 10.1242/dev.121.10.3141. [DOI] [PubMed] [Google Scholar]
- 30.Gering M, Rodaway AR, Gottgens B, Patient RK, Green AR. The SCL gene specifies haemangioblast development from early mesoderm. Embo J. 1998;17:4029. doi: 10.1093/emboj/17.14.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Calkhoven CF, Muller C, Leutz A. Translational control of gene expression and disease. Trends Mol Med. 2002;8:577. doi: 10.1016/s1471-4914(02)02424-3. [DOI] [PubMed] [Google Scholar]
- 32.Calkhoven CF, Muller C, Martin R, Krosl G, Pietsch H, Hoang T, Leutz A. Translational control of SCL-isoform expression in hematopoietic lineage choice. Genes Dev. 2003;17:959. doi: 10.1101/gad.251903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wechsler J, Greene M, McDevitt MA, Anastasi J, Karp JE, Le Beau MM, Crispino JD. Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat Genet. 2002;32:148. doi: 10.1038/ng955. [DOI] [PubMed] [Google Scholar]
- 34.Li Z, Godinho FJ, Klusmann JH, Garriga-Canut M, Yu C, Orkin SH. Developmental stage-selective effect of somatically mutated leukemogenic transcription factor GATA1. Nat Genet. 2005;37:613. doi: 10.1038/ng1566. [DOI] [PubMed] [Google Scholar]
- 35.Tenen DG, Hromas R, Licht JD, Zhang DE. Transcription factors, normal myeloid development, and leukemia. Blood. 1997;90:489. [PubMed] [Google Scholar]
- 36.Cammenga J, Mulloy JC, Berguido FJ, MacGrogan D, Viale A, Nimer SD. Induction of C/EBPalpha activity alters gene expression and differentiation of human CD34+ cells. Blood. 2003;101:2206. doi: 10.1182/blood-2002-05-1546. [DOI] [PubMed] [Google Scholar]
- 37.Edvardsson L, Dykes J, Olsson ML, Olofsson T. Clonogenicity, gene expression and phenotype during neutrophil versus erythroid differentiation of cytokine-stimulated CD34+ human marrow cells in vitro. Br J Haematol. 2004;127:451. doi: 10.1111/j.1365-2141.2004.05227.x. [DOI] [PubMed] [Google Scholar]
- 38.Rekhtman N, Radparvar F, Evans T, Skoultchi AI. Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: functional antagonism in erythroid cells. Genes Dev. 1999;13:1398. doi: 10.1101/gad.13.11.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moreau-Gachelin F, Tavitian A, Tambourin P. Spi-1 is a putative oncogene in virally induced murine erythroleukaemias. Nature. 1988;331:277. doi: 10.1038/331277a0. [DOI] [PubMed] [Google Scholar]


