Abstract
Background:
The BRCA2 gene is responsible for a high number of hereditary breast and ovarian cancers, and studies of the BRCA2 biological functions are limited by the lack of models that resemble the patient's tumour features. The aim of this study was to establish and characterise a new human breast carcinoma xenograft obtained from a woman carrying a germline BRCA2 mutation.
Methods:
A transplantable xenograft was obtained by grafting a breast cancer sample into nude mice. The biological and genetic profiles of the xenograft were compared with that of the patient's tumour using histology, immunohistochemistry (IHC), BRCA2 sequencing, comparative genomic hybridisation (CGH), and qRT–PCR. Tumour response to standard chemotherapies was evaluated.
Results:
Histological profile identified the tumour as a basal-like triple-negative breast cancer. Targeted BRCA2 DNA sequencing of the xenograft showed the presence of the mutation previously identified in the carrier. Comparative genomic hybridisation array profiles of the primary tumour and the xenograft revealed a high number of similar genetic alterations. The therapeutic assessment of the xenograft showed sensitivity to anthracyclin-based chemotherapy and resistance to docetaxel. The xenograft was also highly sensitive to radiotherapy and cisplatin-based treatments.
Conclusions:
This study describes a new human breast cancer xenograft obtained from a BRCA2-mutated patient. This xenograft provides a new model for the pre-clinical drug development and for the exploration of the drug response biological basis.
Keywords: BRCA2 mutation, human breast cancer xenograft, preclinical model
Germline BRCA2 mutations in female carriers confer a cumulative breast cancer risk at age 70 years of 49% (95% CI: 40–57%), an ovarian cancer risk of 18% (95% CI: 13–23%), and a moderate increased risk of pancreatic cancer (Chen and Parmigiani, 2007).
Although the available evidence is not sufficient to decisively conclude that the clinical outcome of women with BRCA1/2-associated breast cancer differs significantly from those of women with sporadic tumours, BRCA1-associated breast cancer often manifests adverse outcome features.
Establishment of pre-clinical models, which accurately reflect the genetic and phenotypic features of primary tumours, and their response to treatment, is an important step in identifying novel therapeutic targets and testing new treatment modalities. New strategies may take advantage of the specific DNA repair defects inherent in BRCA-deficient cells, such as the defect in homologous recombination. In fact, most of the insights into the functions of the BRCA2 protein have included key insights from studies of mice by the use of gene targeting and from studies of altered mouse embryonic cells (Evers and Jonkers, 2006).
BRCA2 has a key role in DNA double-strand break repair and cell-cycle control. BRCA2-related defects are associated with chromosomal abnormalities, a hallmark of the genomic instability that could foster tumourigenesis. Moreover, BRCA2 participates in the regulation of mitosis and cytokinesis that contribute to numerical chromosomal stability.
Although conventional, non-conditional, mouse mutants might be used to model familial forms of cancer, they do not mimic sporadic tumourigenesis because the initiating mutation is present in all cells of the body, including those that constitute the tumour microenvironment.
Moreover, embryonic lethality and development of non-epithelial tumours are another important limitation of genetically mutated Brca2 mice. Some murine Brca2 mutant mammary tumour models develop mammary tumours with histopathological features that are significantly different to their human counterparts. Although some studies report a strikingly similar histopathology in BRCA1 null breast tumours from mice and humans (Dennis, 1999; Xu et al, 1999), their relevance to the human situation remains to be demonstrated. The most used human BRCA2-mutated model is the CAPAN1 pancreatic cell line that is mainly used to understand drug resistance in BRCA1/2 mutation carrier, as well as in defining functionally important domains within BRCA2 (Edwards et al, 2008). To date, only one BRCA2-mutated breast cancer xenografts (MX1) is available, (Donawho et al, 2007) but its characterisation has not been described in detail.
Genetic testing to identify BRCA1/2 mutations is widely available and commonly employed. As a result, increasing numbers of women are aware that they are mutation carriers at the time of their cancer diagnosis. Unfortunately, current knowledge is not sufficient to mandate specific local or systemic treatments that are tailored to BRCA1/2 mutation carriers. In fact, the available studies examining the issue of whether BRCA1/2-associated breast cancer should be treated differently from sporadically occurring, non-familial disease are almost exclusively retrospective and limited by small size and various ascertainment biases.
Recently, inhibitors of the DNA repair proteins, poly(ADP-ribose) polymerase 1 and 2 (PARP1/2), have been shown to be selectively cytotoxic to tumour cells with BRCA1 or BRCA2 deficiency. Preclinical data, including that generated with a limited array of tumour cell line xenografts, suggest that PARP inhibitors can act as single agents to selectively kill cancers with BRCA1 or BRCA2 mutations, and phase I clinical trial results confirm that PARP inhibitors have some single-agent activity in cancers with BRCA1/2 mutations (Fong et al, 2009). However, little is known about the long-term effects of these drugs and it seems likely that some tumours may have de novo resistance or acquired resistance (Ashworth, 2008). Thus, definitive answers remain elusive, and preclinical evaluation of new targeted therapy is limited by the lack of suitable preclinical models (Robson, 2007a).
Here, we report the establishment and characterisation of a novel xenograft, human breast cancer xenograft (HBCx-17) established from a breast cancer in a woman carrying a BRCA2 germline mutation. We show that this xenograft accurately reflects the genetic and the phenotypic features of the primary tumour, thus providing a new model to test new therapies for BRCA2-mutated patients.
Materials and methods
Animals and establishment of tumour xenografts
The breast cancer specimen was obtained with informed consent from the patient undergoing surgery. Fresh tumour fragments were grafted into the interscapular fat pad of 8–12-week-old female Swiss nude mice, under avertin anaesthesia. Mice were maintained in specific pathogen-free animal housing (Institut Curie, Paris, France) and received oestrogen (8 μg ml−1) diluted in drinking water. Xenografts appeared at the graft site about 1 month after grafting. One xenograft was subsequently transplanted from mouse to mouse and stocked frozen in DMSO-fetal calf serum solution or frozen dried in nitrogen for further studies, and a fragment was fixed in phosphate-buffered saline (PBS) 10% formol for histological studies. The experimental protocol and animal housing were in accordance with institutional guidelines as put forth by the French Ethical Committee (Agreement B75-05–18, France).
Histology and IHC
The morphology of patients’ tumour tissue and of the xenograft was compared using paraffin-embedded sections and standard protocols (Vincent-Salomon et al, 2007). Tumours were removed from mice and immediately fixed in a 10% formol/PBS solution.
Determination of oestrogen receptor, progesterone receptor, ERBB2, Ki67, cytokeratin (CK) 5/6, and epidermal growth factor receptor (EGFR) status by IHC was done according to previously published protocols (Vincent-Salomon et al, 2007).
To search for spontaneous lung metastasis of the HBCx-17 xenograft, mice were killed when the tumour reached an ethical size (about 2500 mm3), and lungs were formalin-fixed in-toto for histological evaluation.
Compounds and therapeutic assays
Doxorubicin, 2 mg kg−1 (Adriamycin, Teva Pharmaceuticals, Paris, France), and cyclophosphamide, 100 mg kg−1 (Endoxan, Baxter, Maurepas, France), diluted in 0.9% NaCl, and docetaxel, 20 mg kg−1 (Taxotere, Sanofi-Aventis, Paris, France), diluted in its specific excipient, were given by intraperitoneal (i.p.) route at 3-week intervals. Ifosfamide, 90 mg kg−1 (Holoxan, Baxter) was given by i.p. route on three consecutive days every 3 weeks. Cisplatin, 1 mg kg−1 (Mylan, Hoeilaart, Belgium) was diluted in 0.9% NaCl and given weekly by i.p. route. Capecitabine (Xeloda, Roche, Basel, Switzerland), 540 mg per kg per day was diluted in glucose 5% and given per os in two administrations a day. Radiotherapy was administered locally with one dose of 8 Gy or two weekly fraction doses of 8 and 7 Gy, respectively, for a total dose of 15 Gy. Irradiation experiments were done using a caesium source having a dose rate of 2.15 Gy per min.
Therapeutic assessments were performed as described elsewhere (Marangoni et al, 2007). Briefly, tumour volume was calculated as V=axb2/2, a being the largest diameter and b the smallest. Treatment was initiated when tumours in each group achieved an average volume of ∼170–200 mm3. For each tumour, Vs were reported to the initial volume as relative tumour volume (RTV). Means (and s.e.) of RTV in the same treatment group were calculated, and growth curves were established as a function of time. Optimal tumour growth inhibition (TGI) of treated tumours vs controls was calculated as the ratio of the mean RTV in treated group to the mean RTV in the control group at the same time. Statistical significance of TGI was calculated by the paired Student's t-test, by comparing the individual RTVs in the treated and control groups. Mice were ethically killed when the tumour volume reached about 2500 mm3.
DNA sequencing
Screening for BRCA2 point and small size mutations was performed through analysis of genomic DNA from patient's tumour. In all, 100 ng of DNA was amplified using Taq ABgene 0.025 U μl−1 (ABgene UK, ref: http://www.abgene.com), 4 × 0.2 mM dNTPs, 1.5 mM MgCl2, and 0.3 μM of each primer in a final reaction volume of 50 μl. Amplification was performed with an initial denaturation step at 94°C for 5 min followed by 35 PCR cycles: denaturation at 94°C for 30 s, annealing at 54°C for 30 s, and elongation at 72°C for 30 s.
PCR products were separated by agarose gel electrophoresis, purified (Macherey Nagel, Düren, Germany), and sequenced using one of the PCR primers (usually the forward primer, except in the case of poor sequence quality). Big Dye Cycle Sequencing Reactions and an ABI3130XL automated sequencer (Applied Biosystems, Foster City, CA, USA). Seqscape (Applied Biosystems) software was used for sequence analysis.
Genotypage
Allelic loss was analysed by amplification of two microsatellite markers flanking the BRCA2 gene: D13S260 and D13S1701. Germline DNA, obtained from a blood sample, and tumour and xenograft DNA were compared. In all, 100 ng of DNA was amplified using AmpliTaq Gold 5 U μl−1 (Applied Biosystems), 4 × 0.2 mM dNTPs, 1.5 mM MgCl2, and 0.3 μM of each primer in a final reaction volume of 10 μl. Amplification was performed with an initial denaturation step at 94°C for 5 min followed by 30 PCR cycles: denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and elongation at 72°C for 30 s. A total of 1 μl of PCR products was mixed with 19 μl of Formamide Hi-Di (Applied Biosystems) and 0.5 μl of Genescan 400 ROX Size Standard (PE Applied Biosystems) and separated on ABI3130XL automated sequencer (Applied Biosystems). Genemapper (Applied Biosystems) software was used for genotype analysis.
Quantitative multiplex PCR of short fluorescent fragments
Quantitative multiplex PCR of short fluorescent fragments is a sensitive method for the detection of large gene deletions or duplications. It is based on the simultaneous amplification of short genomic fragments using dye-labelled primers under quantitative conditions (Casilli et al, 2002; Tournier et al, 2004). PCR products were analysed on a sequencing platform used in the fragment analysis mode, where both peak heights and areas are proportional to the quantity of template present for each target sequence. Nine amplicons of BRCA2 between 180 and 300 bp were amplified in the same multiplex reaction. As an internal control, we included in each reaction a fragment of different gene in which deletion was not expected (MLH1 exon 14). One primer from each primer pair was 5′-labeled with 6-FAM fluorochrome.
In all, 100 ng of two genomic DNA from the xenograft, two normal control, and one BRCA2-mutated control were amplified in a final volume of 25 μl, including 0.06–0.24 μmol l−1 for each primer and 12.5 μl of QIAGEN master mix (QIAGEN multiplex PCR Kit). After an initial denaturation step, samples underwent 23 cycles (30 s at 95°C, 40 s at 54°C, and 60 s at 72°C). After the multiplex reactions, the DNA fragments were separated on an ABI3130XL automated sequencer and analysed using Genemapper Software (Applied Biosystems).
For this analysis, we used visual sample-to-control comparison. We estimated allele dosage by superimposing the electropherogram of the tested sample onto the corresponding image for a control DNA sample, after adjusting the vertical scale of the internal control amplicon. Allelic losses of one or more amplicons are represented by a two-fold reduction in the intensity (peak heights) of an amplicon of the sample analysed. Allelic duplication of one or more amplicons is represented by a 0.5-fold rise in the intensity (peak heights) of an amplicon of the sample analysed.
Protein analysis
Whole-cell protein extracts were prepared from tumour samples by homogenising the tissue in RIPA buffer (10 mM Tris-Cl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Nonidet P40, 0.5% sodium deoxycholate, 0.1% SDS, protease inhibitors). Protein concentrations were measured with Bio-Rad Protein Assay Reagent (Bio-Rad, Marnes-la-Coquette, France). Immunoprecipitations were performed by incubating Protein G beads (Sigma, Steinheim, Germany), 1–2 mg of precleared cell lysate, and anti-BRCA2 mouse monoclonal Ab-1 antibody (dilution 1 : 200; Merck) overnight at 4°C. Beads were subsequently washed three times in cold lysis buffer, after which 2 × loading buffer was added and the samples were boiled for 2 min before SDS–PAGE. For western blotting analysis, lysates were subjected to electrophoresis on Novex precast gels (Invitrogen, Cergy Pontoise, France) and immunoblotted overnight at 4°C with the following antibodies: anti-BRCA2 and anti-β-tubulin, T4026 (Sigma). This was followed by incubation with anti-IgG-horseradish peroxidase and enhanced chemiluminescent detection (GE Healthcare Bio-Sciences AB, Uppsala, Sweden). As a control, full-length BRCA2 was detected in lysates generated from 293T cells.
Detection of ERα, Ki67, and ERBB2 by quantitative RT–PCR
Total RNA extraction and cDNA synthesis were done as previously described from 1 μg total RNA (de Cremoux et al, 2004). ERα, Ki67, and ERBB2 transcripts were quantified using real-time quantitative reverse transcription–PCR (RT–PCR) assays. The nucleotide and probe sequences, and the conditions of PCR have been previously described (de Cremoux et al, 2007). Results were expressed as N-fold differences in target gene expression relative to a reference gene defined as ‘N target’.
Array-based CGH
A genome-wide resource of 5244 fluorescence in situ hybridisation mapped, sequenced BAC and PAC clones, verified for gene and marker content were represented as immobilised DNA targets on glass slides for array-based CGH analysis, allowing a mean resolution of 0.5 Mb all along the genome. Each clone was spotted in quadruplicate on a slide prepared by Integragen (Evry, France) and developed by the Institut National de la Sante et de la Recherche Medicale Unit U830. After extraction, 1.5 μg of each test and control DNA samples were digested with DpnII enzyme (Ozyme, Saint-Quentin en Yveline, France) and purified with a QIAquick PCR purification kit (Qiagen, Courtaboeuf, France). They were then labelled by random priming using a Bioprime DNA labelling kit (Invitrogen) with the appropriate cyanine dye (Cy3 or Cy5; PerkinElmer, Waltham, MA, USA). The control and test DNA were coprecipitated with Cot-1 DNA (Invitrogen), denatured, and resuspended in hybridisation buffer (50% formamide). Competitive cohybridisation was done on CGH array slides. After 24-h hybridisation, slides were washed with SDS and saline citrate, dried, and scanned using a 4000B scan (Axon, Orleans, CA, USA). Image analysis was done with Genepix 5.1 software (Axon) and processed using a software developed at the Curie Institute (La Rosa et al, 2006). Any BAC with less than two replicates flagged for not fulfilling qualitative spot criteria was excluded. A ratio <0.8 was considered as a loss, a ratio >1.2 was considered as a gain, and a ratio >1.5 was considered as amplification (Auger et al, 2006).
Data analysis was based on the normalised ratios of Cy5/Cy3 signals observed for each BAC clone that previously passed the flag assessment procedure. For autosomal chromosomes, the loss of a given locus was defined by a ratio ⩽0.8, a gain was defined by a ratio ⩾1.2 and <2.0, and an amplicon was defined by a ratio ⩾2.0. For X chromosomes, a loss was defined by a ratio ⩽1.2, a gain was defined by a ratio ⩾1.7, and an amplicon was defined by a ratio ⩾2.5. The data analysis was done according to previously published protocols (Vincent-Salomon et al, 2007).
Clinical features of the patient
The BRCA2 mutation carrier from whom the HBCx-17 xenograft was obtained was a 37-year-old woman who was affected at 32 years of age for a first invasive ductal carcinoma (IDC) of grade III that was treated with surgery, FEC100, and radiotherapy. After 6 years, she developed a contralateral IDC and was treated by upfront surgery followed by six cycles of docetaxel and radiotherapy. The xenograft was established from the second carcinoma. The patient tumour was C-ERBB2, oestrogen-, and progesterone-receptor negative, with a high mitotic index. The patient had a strong familial history of breast and ovarian cancer. BRCA1/2 gene testing, performed with the informed consent of the patient, identified a mutation in the BRCA2 gene: c.6033_6034delTT; p.ser2012GlnfsX5.
Results
Histology of xenograft and comparison with patient tumour
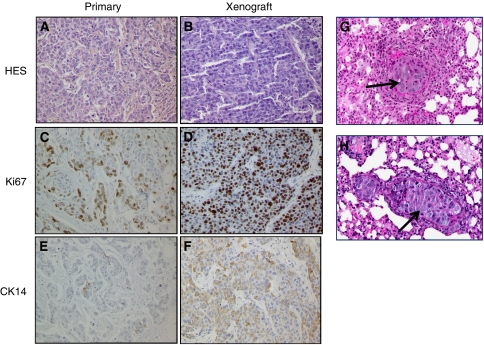
Histopathological analysis was performed with the primary tumour and the xenograft HBCx-17 at passage 6. As shown in Figure 1A and B, the histology of the original tumour was conserved in the xenograft. Indeed, HBCx-17 showed an infiltrating ductal carcinoma with typical loss of tubule formation, prominent nuclear pleomorphism, and mitotic activity. Irregular infiltration of stroma was observed in both patient and xenografts. Assessment of tumour proliferation using Ki67 staining showed a high proliferative rate in the primary tumour, which was increased in the xenograft (Figure 1C and D). Both primary and xenograft tumours were negative for CK5 and CK6 (data not shown) but positive for CK14 expression as shown in Figure 1E and F. Both tumour sample and HBCx-17 are negative for ERBB2 and oestrogen, and progesteron receptor, and the clinical sample presented a strong EGFR staining that the xenograft did not (data not shown).
Figure 1.
Representative haematoxylin-and-eosin-stained sections of patients and xenografts tumours. Haematoxylin and eosin sections Gx100 (A and B), KI67 (C and D), and CK14 (E and F). Lung metastases are shown in pictures G and H (arrows) clusters of tumour cells obstruct the lumen of a small number of pulmonary arterioles (cancerous emboli), without evident effraction of the arteriolar media.
To search for spontaneous lung metastasis of the HBCx-17 xenograft, mice lungs were analysed by histochemistry. Figure 1G and H show two examples of small metastasis: clusters of tumour cells obstructed the lumen of a small number of pulmonary arterioles (cancerous emboli), without evident effraction of the arteriolar media. Lung metastases were detected in 23% of mice (six positive animals on 26).
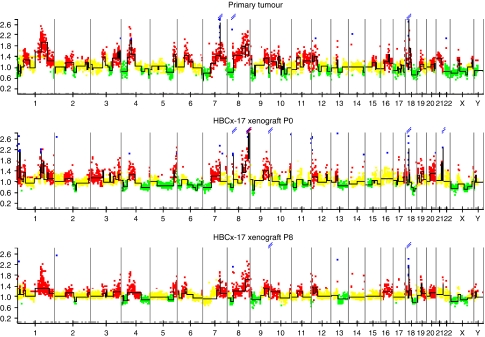
CGH array and genomic alterations
The clinical sample and xenografted tumours at passages 0, 6, and 8 were characterised for genetic parameters using CGH array technique (Figure 2). Comparative genomic hybridisation array analysis showed a very high number of alterations and quite similar gene copy number changes. The genomic profiles of the primary tumour and different passages in the xenograft were very similar, sharing large regions of DNA amplifications. Common DNA gains and deletions are described in the Tables 1 and 2. These amplifications are described in the Table 1 such as amplicons of chromosome 1q and 8p.
Figure 2.
HBCx-17 tumour array CGH profiling of patient (top) and xenograft (bottom). Loss (green points), gain (red points), or amplification (blue points) of chromosome material. Recurrence of copy number alterations (y axis) is plotted for each probe aligned along the x axis in chromosome order.
Table 1. Recurrent amplicons observed in the primary tumour and maintained in at least two passages of the xenograft.
| Cytogenetic region | Start position * | End position* | Candidate genes | Maximum xenograft ratio (passage) |
|---|---|---|---|---|
| 1q23.3–24.1 | 163.3 | 164.3 | LMX1A, RXRG, LRRC52, MGST3, ALDH9A1, TMCO1, UCK2 | 2.1 (p8) |
| 7q31.1 | 111.0 | 112.4 | DOCK4, IFRD1, NPM1P14, TMEM168 | 2.2 (p0) |
| 8p11.21 | 41.6 | 42.2 | MIRN486, ANK1, MYST3, AP3M2, PLAT, IKBKB | 2.12 (p8) |
| 13q14.11–12 | 43.0 | 44.6 | CCDC122, TSC22D1, NUFIP1 | 2.69 (p0) |
| 18q11.2–12.1 | 19.6 | 24.3 | CABYR, OSBPL1A, HRH4, RAC1P1, hCG1643695, SS18, PSMA8, TAF4B, KCTD1, hCG38400, AQP4, CHST9, CDH2 | 3.3 (p0) |
UCSC Genome Browser on Human May Assembly; positions are given in megabased.
Table 2. Recurrent losses observed in the primary tumour and maintained through passages of the xenograft.
| Cytogenetic region | Start position* | End position* | Candidate genes |
|---|---|---|---|
| 1p34.3 | 34.4 | 34.9 | MIRN552 |
| 2q22.2 | 141.5 | 142.1 | Not known |
| 4p16.1–15.33 | 8.5 | 12.3 | GPR78, CPZ, DUB4, DEFB131, DRD5, SLC2A9, MIST, MIRN572, HS3ST1 |
| 4p15.2 | 25.1 | 26.5 | SLC34A2, RBPJ, CCKAR, TBC1D19, STIM2 |
| 4q35.1 | 186.8 | 189.0 | SORB2, TLR3, CYP4V2, KLKB1, F11, MTNR1A, MRPS36P2 |
| 7p22.2–7p22.1 | 3.1 | 5.9 | SDK1, FOXK1, RADIL, PAPOLB, MMD2, RBAK, WIPI2, SLC29A4, FBXL18, MIRN589, ACTB, EGID-654231 |
| 7p15.3 | 20.2 | 22.6 | ITGB8, ABCB5, SP8, SP4, DNAH11, CDCA7L, RAPGEF5, MGC87042 |
| 7p15.1 | 28.3 | 29.9 | CREB5, CPVL, CHN2, PRR15, WIPF3 |
| 30.1 | 31.1 | MIRN550-1, NOD1, GARS, CRHR2, INMT, AQP1, GHRHR, ADCYAP1R1 | |
| 7p14.3–14.1 | 32.5 | 43.0 | MIRN550-2, KBTBD2, FKBP9, NT5C3, RP9, BBS9, BMPER, AAA1, NPSR1, TBX20, MERPUD2, hCG1642425, SEPT7, ANLN, AOAH, ELM01, GPR141, TXNDC3, SFRP4, STARD3NL, T cell receptor gamma variable genes, AMPH, RALA, CDC2L5, INHBA, GLI3, TCP1L1, PSMA2, MRPL32 |
| 7p12.3 | 46.0 | 47.1 | EPS15L2 |
| 8p21.2 | 22.9 | 24.0 | TNFRSF10C, TNFRSF10D, TNFRSF10A, CHMP7, R3HCC1, LOXL2, ENTPD4, SLC25A37, NKX3.1, NKX2.6, STC1 |
| 25.2 | 25.5 | GNRH1, KCT9, CDCA2 | |
| 25.7 | 26.3 | EBF2, PPP2R2A, SDAD1P1 | |
| 8p21.1 | 27.4 | 28.6 | GULOP, CLU, SCARA3, CCDC25, PBK, PNOC, FBXO16, EXTL3 |
| 28.8 | 30.0 | KIF13B, DUSP4, MAP2K1P1 | |
| 31.9 | 32.1 | NRG1 | |
| 9p21.3 | 20.3 | 24.7 | MIRN491, PTPLAD2, IFNB1, IFNW1, IFNxxx…, LEIF-M, MIRN31, MTAP, CDKN2A, CDKN2B, DMRTA1, ELAVL2 |
| 9p21.2–21.1 | 27.3 | 28.9 | IFNK, LING02 |
| 10q22.1 | 72.1 | 74.2 | SGPL1, PCBD1, SLC29A3, CDH23, PSAP, CHST3, SPOCK2, ASCC1, SSIT4, DNAJB12, CBARA1 |
| 10q23.31 | 89.7 | 91.4 | LIPJ, LIPF, LIPK, LIPN, LIPM, ANKRD22, STAMBPL1, ACTA2, FAS, CH25H, IFIT2, LIPA |
| 10q23.33–24.1 | 96.5 | 98.6 | CYP2C19, CYP2C9, CYP2C8, PDLIM1, SORBS1, ALDH18A1, TCTN3, ENTPD1, CC2D2B, CCNJ, BLNK, DNTT, TMEM10, TLL2, TM9SF3, PIK3AP1, MIRN606 |
| 11q25 | 131.7 | 133.7 | OPCML, SPATA19, IGSF9B, JAM, PTP4AP2, NCAPD3, THYN1, ACAD8, GLB1L3 |
| 17p11.2 | 19.9 | 21.6 | SPECC1, COTL1P2, MEIS3P2, RNASEH1P1, HSP22, DHRS7B, TMEM11, MAP2K3, KCNJ12 |
| 18q21.2–21.33 | 46.9 | 61.0 | DCC, MBD2, POLI, STARD6, C18orf54, C18orf26, RAB27B, CCDC68, TCF4, AC009271.7, TXNL1, WDR7, AC100775.3, ST8SIA3, ONECUT2, FECH, NARS, ATP8B1, AC022724.8, AC090324.7, NEDD4L, ALPK2, AC104971.5, MALT1, AC104971.5, ZNF532, SEC11L3, GRP, RAX, CPLX4, LMAN1, CCBE1, PMAIP1, AC090377.15, MC4R, AC010928.7, CDH20, RNF152, PIGN, KIAA1468, TNFRSF11A, ZCCHC2, PHLPP, AC015989.11, BCL2, FVT1, VPS4B, SERPINB5, AC036176.8, SERPINB12, SERPINB13, SERPINB4, SERPINB3, SERPINB11, SERPINB7, SERPINB2, SERPINB10, SERPINB8 |
| 18q22.3 | 66.9 | 68.6 | CBLN2 |
| 68.8 | 72.8 | FBXO15, CYB5A, FAUP1, CNDP2, CNDP1 |
UCSC Genome Browser on Human May Assembly; positions are given in megabased.
Gene expression profiles
The histological classification of the HBCx-17 as a triple-negative tumour was confirmed by quantitative RT–PCR analysis of the ER, PR, and ERBB2 receptors (Table 3). Both the clinical sample and the xenograft tumours were negative for the three receptors. By contrast, the basal-like CK5 and CK6 were not expressed, but the classification of basal-like breast cancer was done on CK14 expression as determined by IHC (Figure 1).
Table 3. Gene expression analysis of ERBB2, KI67, and ERα evaluated by quantitative RT–PCR (overexpression limit of 1000 units).
| ERBB2 | Ki67 | ERα | |
|---|---|---|---|
| Patient tumour | 8166 (−) | 6788 (++) | 34 (−) |
| Xenograft tumour | 10 533 (−) | 16 695 (+++) | 6 (−) |
The TP53 status was found to be mutated in the xenograft tumour as defined by the functional Fasay Assay (data not shown).
BRCA2 alterations
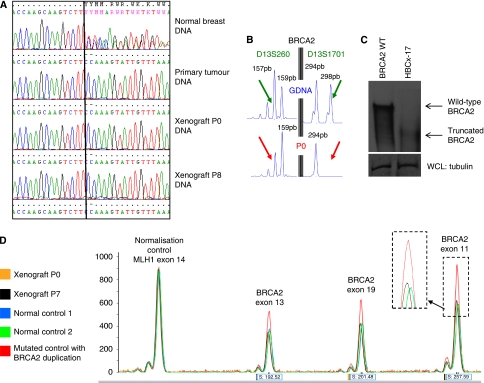
DNA sequencing of the primary tumour, and P0 and P8 from the HBCx-17 xenograft showed the presence of the germline BRCA2 mutation identified in the patient (c.6033_6034delTT; p.Ser2012GlnfsX5; Figure 3A). The informativity of the two studied markers located at the BRCA2 locus, D13S260 and D13S1701, has allowed detecting the presence of the two alleles at the germline DNA level, whereas the primary tumour and the xenograft DNA showed loss of one allele. Loss of heterozygosity (LOH) at the BRCA2 locus was confirmed in the xenograft at P0 by using two flanking microsatellite markers for BRCA2, D13S260, and D13S1701 (Figure 3B).
Figure 3.
(A) Chromatograms of tumour and normal breast tissue DNA showing BRCA2 mutation: (c.6033_6034delTT/p.Ser2012GlnfsX5) Germline DNA (GDNA) shows both mutated and wild-type alleles. Loss of the wild-type allele with retention of the mutant one is shown in the primary tumour and in passage 0 (P0) and 8 (P8) of the xenograft. (B): Amplification of two microsatellites (D13S260 and D13S1701) flanking BRCA2 shows loss of heterozygosity in P0. (C) Illustrates an anti-BRCA2 immunoprecipitation on lysates generated from tumour sample HBCX-17 and 293T cells. Western blot analysis detected full-length BRCA2 in the 293T samples and a truncated BRCA2 product of the predicted size in the tumour sample. (D) quantitative multiplex PCR of short fragment shows that mutated allele is duplicated whereas wild-type allele is lost.
To assess the LOH consequence on the BRCA2 protein, a protein analysis was performed by western blot on cell lysate obtained from a control breast tumour xenograft (having no BRCA2 mutation) and from the HBCx-17 xenograft. As shown in Figure 3C, the BRCA2-mutated xenograft lysate contains a truncated form of the BRCA2 protein and has lost the wild-type protein.
Quantitative multiplex PCR of short fluorescent fragments was used to determine BRCA2 copy number. Figure 3D shows the electrophoregrams of germline DNA, and P0 and P7 DNA (orange and black). Mutated (BRCA2 duplicated) DNAs were used as controls (green and red, respectively). Xenograft samples and control samples were perfectly superimposed after normalisation, indicating a duplication of BRCA2-mutated allele.
Tumour responses of xenograft to conventional chemotherapies and radiotherapy
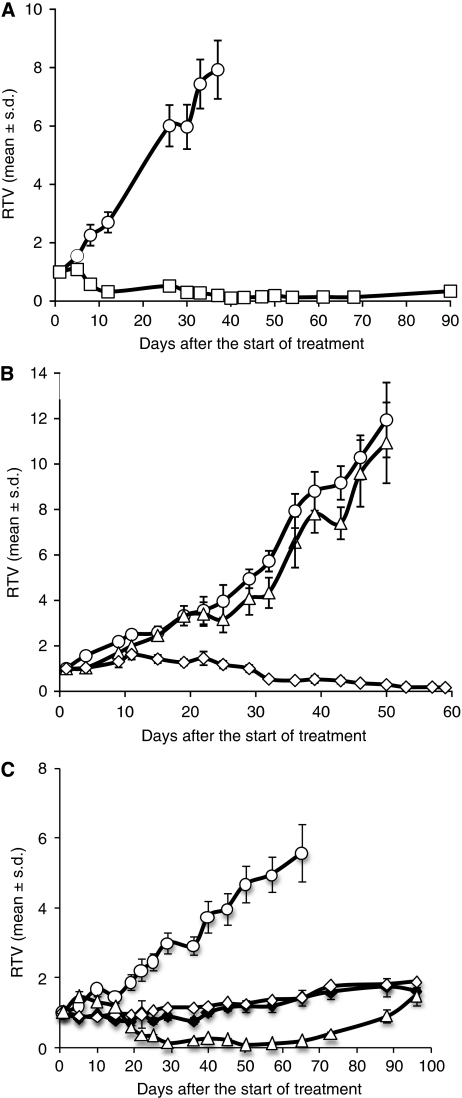
The HBCx-17 xenograft growth parameters (tumour latency and tumour take) are published (Marangoni et al, 2007). Tumour responses to standard chemotherapies used in the treatment of breast cancer and radiotherapy have been studied. As shown in Figure 4A, the HBCx-17 xenograft was a high responder to AC, with five out of eight complete regressions and a TGI of 98%. The HBCx-17 model was also sensitive to capecitabin-based treatment with a TGI of 98% 4 weeks after start of the treatment. No response was observed to docetaxel, as shown in Figure 4B. Cisplatin/ifosfamide combination gave also an important growth inhibition (98%) with 7 out of 10 complete regressions at day 50 (Figure 4C). Radiotherapy assays were done with a caesium irradiation source. The mice received either 8 or 15 Gy locally delivered in two fractions of 7 and 8 Gy. The xenograft was a high responder to irradiation with a TGI of 73 and 75% at 8 and 15 Gy, respectively (Figure 4C).
Figure 4.
Tumour growth curves of HBCx-17 xenograft as a function of time: HBCx-17 bearing mice were treated with two cycles of AC (A), a combination of doxorubicin (2 mg kg−1 i.p. every three weeks) and cyclophosphamide (100 mg kg−1 i.p. every three weeks), with docetaxel (▵) (20 mg kg−1 i.p. every three weeks) or capecitabin (◊) (540 mg kg−1 per os 5 days per week two times) (B) or with a combination of cisplatin (1 mg kg−1 i.p. once a week) and ifosfamide (90 mg kg−1, 3 consecutive days every 3 weeks) (▵) compared with 15 Gy (⧫) or 7 Gy (◊) irradiations (C). Controls (○) were not treated. Mice were treated at day 1, and tumour volume was measured twice a week. Tumour growth was evaluated by plotting the mean of the RTV±s.d. per group (each group consisted of 10 mice) over time after first treatment.
Discussion
In this work, we report the characterisation of a human breast tumour xenograft obtained from a woman carrying a BRCA2 mutation. The basal-like morphology of the patient tumour was conserved in the xenograft including the stroma component and tissular architecture. Ki67 staining was higher in the xenograft's tumour than in the primary tumour, suggesting that tumour engrafting may have selected highly proliferating cancer cells. Studies on large series of BRCA2-associated breast cancers indicate that these tumours are predominantly high-grade IDCs of no special marker subtype, and that they are more often oestrogen- and progesterone-receptor positive (Lakhani et al, 2002; Brekelmans et al, 2007; Palacios et al, 2008). As the majority of BRCA2-deficient cells, the tumour cells contain only the truncated BRCA2 protein, indicating that LOH has occurred as a consequence of an inactivating mutation in the second allele. This is what normally happens in most BRCA2-mutated tumour, where inactivation of the wild-type allele occurs by LOH, abolishing normal protein expression (Smith et al, 1992; Collins et al, 1995). In addition, multiplex PCR of short fragment showed that loss of the wild-type gene was associated with duplication of the mutated BRCA2 gene. LOH and copy number abnormalities are often associated with BRCA1, and BRCA2-associated breast or ovarian cancer (Staff et al, 2000).
Concordance between the clinical sample and xenograft was also shown by CGH array analysis. Genetic profiles were very similar not only between the patient tumour and the xenografts, but also when comparing xenografts at different passages, suggesting that although extremely altered, the HBCx-17 xenograft genetic profile was stable during subsequent passages. The clinical sample and the xenograft tumours present some of the chromosome alterations that have already been frequently described in the genomic profiles of BRCA2-mutated tumours, as gains of 8q and 20q, and loss of 13q (Palacios et al, 2008). The amplification of the MYST3 and AP3M2 genes was already described like recurrent amplicons associated with reduced survival duration in breast cancer (Chin et al, 2006).
Gains in the regions 1q32–q41, 8q22.1–24.3, and 20q12–q13, and loss in the region 8p23.3–p21.2 occur in both the primary and xenograft tumours and have also been found in primary BRCA2 tumours by different authors (Gronwald et al, 2005; Jonsson et al, 2005).
However, none of these regions have been confirmed in large number of patients and in studies of independent collections of families, and more extensive studies are necessary to find alterations specific to BRCA1/2-mutated tumours.
There is a significant body of preclinical data that supports the hypothesis BRCA1/2-deficient cells are more sensitive to certain chemotherapy agents than are the cells with intact BRCA1 and BRCA2 proteins (Foulkes, 2006). In line with this, the HBCx-17 xenograft showed a pronounced sensitivity to anthracycline-based chemotherapy with >50% of mice showing complete response (no tumour recurrence), as well as to cisplatin and ifosfamide combination. Although BRCA2 deficiency has not been extensively studied, some previous works showed that decreased BRCA2 function is associated with increased in vitro sensitivity to cisplatin, mitomycin, doxorubicin, and etoposide (Foulkes, 2006; Robson, 2007a). The HBCx-17 xenograft was highly resistant to docetaxel. Although some experiments raise the possibility that BRCA1-deficient cells may be resistant to anti-cancer agents targeting the microtubules (such as vinca alkaloids and taxanes), no preclinical in vivo studies have ever demonstrated taxane resistance in BRCA2-deficient cells (Foulkes, 2006; Robson, 2007b). In the same way, some recent clinical analyses suggest that primary resistance to docetaxel-based chemotherapy correlates with BRCA1 mutations’ high frequency. No indications exist for breast tumours occurring in BRCA2 mutation carrier. Our data suggest that, like BRCA1, wild-type BRCA2 could be required for in vivo response to mitotic spindle poisons and that the docetaxel resistance could be attributed to BRCA1/2 involvement in the taxane-induced stress response pathway.
In the clinic, the available evidence is not sufficient to conclude that BRCA1/2-associated breast cancer is differentially sensitive to specific conventional chemotherapeutic agents. In the adjuvant setting, the choice of treatment regimen is not modified based on the presence of such a predisposition. Clinical trials are in progress to further address this issue. Nevertheless, the outcome of hereditary breast cancer remains poor, addressing the questions of potential new strategies that take advantage of the specific DNA repair defects inherent in BRCA-deficient cells. The inhibition of PARP1 potentiates the activity of DNA-damaging agents, such as alkylating drugs, platinums, topoisomerase inhibitors, and radiation in in vitro and in vivo models. Tumours with DNA repair defects, such as those arising from patients with BRCA1/2 mutations, are more sensitive to PARP inhibition (Bryant et al, 2005; Farmer et al, 2005; Fong et al, 2009). In this context, the HBCx-17 model could improve preclinical assays of PARP inhibitors that are usually done in BRCA1/2 knockout mice or in pancreatic cancer cells. Different agents are undergoing phase I and II clinical trials in BRCA1/2-associated breast and ovarian cancer, and new compounds are entering in the early preclinical settings. In this perspective, the establishment of this new BRCA2 breast cancer xenograft reproducing, over successive generations, the patient's characteristics in terms of histology, genetic profile, and biological characteristics may contribute to the preclinical development of innovative therapeutics regimens.
Acknowledgments
Sophie Piperno, Olivier Delattre, Nathalie Auger (Institut Curie, Paris), Laetitia Durand, Win-Chygn Lui for their helpful contribution; Vincent Bordier (Institut Curie, Paris) and Louis-François Plassa (Hopital Saint-Louis, Paris) for their technical assistance, and also the La Ligue Nationale contre le Cancer for its support to the array-based comparative genomic hybridisation platform.
References
- Ashworth A (2008) Drug resistance caused by reversion mutation. Cancer Res 68: 10021–10023 [DOI] [PubMed] [Google Scholar]
- Auger N, Thillet J, Wanherdrick K, Idbaih A, Legrier ME, Dutrillaux B, Sanson M, Poupon MF (2006) Genetic alterations associated with acquired temozolomide resistance in SNB-19, a human glioma cell line. Mol Cancer Ther 5: 2182–2192 [DOI] [PubMed] [Google Scholar]
- Brekelmans CT, Tilanus-Linthorst MM, Seynaeve C, vd Ouweland A, Menke-Pluymers MB, Bartels CC, Kriege M, van Geel AN, Burger CW, Eggermont AM, Meijers-Heijboer H, Klijn JG (2007) Tumour characteristics, survival and prognostic factors of hereditary breast cancer from BRCA2-, BRCA1- and non-BRCA1/2 families as compared to sporadic breast cancer cases. Eur J Cancer 43: 867–876 [DOI] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T (2005) Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434: 913–917 [DOI] [PubMed] [Google Scholar]
- Casilli F, Di Rocco ZC, Gad S, Tournier I, Stoppa-Lyonnet D, Frebourg T, Tosi M (2002) Rapid detection of novel BRCA1 rearrangements in high-risk breast-ovarian cancer families using multiplex PCR of short fluorescent fragments. Hum Mutat 20: 218–226 [DOI] [PubMed] [Google Scholar]
- Chen S, Parmigiani G (2007) Meta-analysis of BRCA1 and BRCA2 penetrance. J Clin Oncol 25: 1329–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, Lapuk A, Neve RM, Qian Z, Ryder T, Chen F, Feiler H, Tokuyasu T, Kingsley C, Dairkee S, Meng Z, Chew K, Pinkel D, Jain A, Ljung BM, Esserman L, Albertson DG, Waldman FM, Gray JW (2006) Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell 10: 529–541 [DOI] [PubMed] [Google Scholar]
- Collins N, McManus R, Wooster R, Mangion J, Seal S, Lakhani SR, Ormiston W, Daly PA, Ford D, Easton DF, Stretton MR (1995) Consistent loss of the wild type allele in breast cancers from a family linked to the BRCA2 gene on chromosome 13q12-13. Oncogene 10: 1673–1675 [PubMed] [Google Scholar]
- de Cremoux P, Bieche I, Tran-Perennou C, Vignaud S, Boudou E, Asselain B, Lidereau R, Magdelenat H, Becette V, Sigal-Zafrani B, Spyratos F (2004) Inter-laboratory quality control for hormone-dependent gene expression in human breast tumors using real-time reverse transcription-polymerase chain reaction. Endocr Relat Cancer 11: 489–495 [DOI] [PubMed] [Google Scholar]
- de Cremoux P, Jourdan-Da-Silva N, Couturier J, Tran-Perennou C, Schleiermacher G, Fehlbaum P, Doz F, Mosseri V, Delattre O, Klijanienko J, Vielh P, Michon J (2007) Role of chemotherapy resistance genes in outcome of neuroblastoma. Pediatr Blood Cancer 48: 311–317 [DOI] [PubMed] [Google Scholar]
- Dennis C (1999) Branching out with BRCA1. Nat Genet 22: 10. [DOI] [PubMed] [Google Scholar]
- Donawho CK, Luo Y, Luo Y, Penning TD, Bauch JL, Bouska JJ, Bontcheva-Diaz VD, Cox BF, DeWeese TL, Dillehay LE, Ferguson DC, Ghoreishi-Haack NS, Grimm DR, Guan R, Han EK, Holley-Shanks RR, Hristov B, Idler KB, Jarvis K, Johnson EF, Kleinberg LR, Klinghofer V, Lasko LM, Liu X, Marsh KC, McGonigal TP, Meulbroek JA, Olson AM, Palma JP, Rodriguez LE, Shi Y, Stavropoulos JA, Tsurutani AC, Zhu GD, Rosenberg SH, Giranda VL, Frost DJ (2007) ABT-888, an orally active poly(ADP-ribose) polymerase inhibitor that potentiates DNA-damaging agents in preclinical tumor models. Clin Cancer Res 13: 2728–2737 [DOI] [PubMed] [Google Scholar]
- Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DA, Boyd J, Reis-Filho JS, Ashworth A (2008) Resistance to therapy caused by intragenic deletion in BRCA2. Nature 451: 1111–1115 [DOI] [PubMed] [Google Scholar]
- Evers B, Jonkers J (2006) Mouse models of BRCA1 and BRCA2 deficiency: past lessons, current understanding and future prospects. Oncogene 25: 5885–5897 [DOI] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A (2005) Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434: 917–921 [DOI] [PubMed] [Google Scholar]
- Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O’Connor MJ, Ashworth A, Carmichael J, Kaye SB, Schellens JH, de Bono JS (2009) Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med 361: 123–134 [DOI] [PubMed] [Google Scholar]
- Foulkes WD (2006) BRCA1 and BRCA2: chemosensitivity, treatment outcomes and prognosis. Fam Cancer 5: 135–142 [DOI] [PubMed] [Google Scholar]
- Gronwald J, Jauch A, Cybulski C, Schoell B, Bohm-Steuer B, Lener M, Grabowska E, Gorski B, Jakubowska A, Domagala W, Chosia M, Scott RJ, Lubinski J (2005) Comparison of genomic abnormalities between BRCAX and sporadic breast cancers studied by comparative genomic hybridization. Int J Cancer 114: 230–236 [DOI] [PubMed] [Google Scholar]
- Jonsson G, Naylor TL, Vallon-Christersson J, Staaf J, Huang J, Ward MR, Greshock JD, Luts L, Olsson H, Rahman N, Stratton M, Ringner M, Borg A, Weber BL (2005) Distinct genomic profiles in hereditary breast tumors identified by array-based comparative genomic hybridization. Cancer Res 65: 7612–7621 [DOI] [PubMed] [Google Scholar]
- La Rosa P, Viara E, Hupe P, Pierron G, Liva S, Neuvial P, Brito I, Lair S, Servant N, Robine N, Manie E, Brennetot C, Janoueix-Lerosey I, Raynal V, Gruel N, Rouveirol C, Stransky N, Stern MH, Delattre O, Aurias A, Radvanyi F, Barillot E (2006) VAMP: visualization and analysis of array-CGH, transcriptome and other molecular profiles. Bioinformatics 22: 2066–2073 [DOI] [PubMed] [Google Scholar]
- Lakhani SR, Van De Vijver MJ, Jacquemier J, Anderson TJ, Osin PP, McGuffog L, Easton DF (2002) The pathology of familial breast cancer: predictive value of immunohistochemical markers estrogen receptor, progesterone receptor, HER-2, and p53 in patients with mutations in BRCA1 and BRCA2. J Clin Oncol 20: 2310–2318 [DOI] [PubMed] [Google Scholar]
- Marangoni E, Vincent-Salomon A, Auger N, Degeorges A, Assayag F, de Cremoux P, de Plater L, Guyader C, De Pinieux G, Judde JG, Rebucci M, Tran-Perennou C, Sastre-Garau X, Sigal-Zafrani B, Delattre O, Dieras V, Poupon MF (2007) A new model of patient tumor-derived breast cancer xenografts for preclinical assays. Clin Cancer Res 13: 3989–3998 [DOI] [PubMed] [Google Scholar]
- Palacios J, Robles-Frias MJ, Castilla MA, Lopez-Garcia MA, Benitez J (2008) The molecular pathology of hereditary breast cancer. Pathobiology 75: 85–94 [DOI] [PubMed] [Google Scholar]
- Robson M (2007a) Is breast conservation a reasonable option for women with BRCA-associated breast cancer? Nat Clin Pract Oncol 4: 10–11 [DOI] [PubMed] [Google Scholar]
- Robson M (2007b) Treatment of hereditary breast cancer. Semin Oncol 34: 384–391 [DOI] [PubMed] [Google Scholar]
- Smith SA, Easton DF, Evans DG, Ponder BA (1992) Allele losses in the region 17q12-21 in familial breast and ovarian cancer involve the wild-type chromosome. Nat Genet 2: 128–131 [DOI] [PubMed] [Google Scholar]
- Staff S, Nupponen NN, Borg A, Isola JJ, Tanner MM (2000) Multiple copies of mutant BRCA1 and BRCA2 alleles in breast tumours from germ-line mutation carriers. Genes Chromosomes Cancer 28: 432–442 [DOI] [PubMed] [Google Scholar]
- Tournier I, Raux G, Di Fiore F, Marechal I, Leclerc C, Martin C, Wang Q, Buisine MP, Stoppa-Lyonnet D, Olschwang S, Frebourg T, Tosi M (2004) Analysis of the allele-specific expression of the mismatch repair gene MLH1 using a simple DHPLC-based method. Hum Mutat 23: 379–384 [DOI] [PubMed] [Google Scholar]
- Vincent-Salomon A, Gruel N, Lucchesi C, MacGrogan G, Dendale R, Sigal-Zafrani B, Longy M, Raynal V, Pierron G, de Mascarel I, Taris C, Stoppa-Lyonnet D, Pierga JY, Salmon R, Sastre-Garau X, Fourquet A, Delattre O, de Cremoux P, Aurias A (2007) Identification of typical medullary breast carcinoma as a genomic sub-group of basal-like carcinomas, a heterogeneous new molecular entity. Breast Cancer Res 9: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Wagner KU, Larson D, Weaver Z, Li C, Ried T, Hennighausen L, Wynshaw-Boris A, Deng CX (1999) Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nat Genet 22: 37–43 [DOI] [PubMed] [Google Scholar]