Abstract
Microfluidics has the potential to revolutionize the way we approach cell biology research. The dimensions of microfluidic channels are well suited to the physical scale of biological cells, and the many advantages of microfluidics make it an attractive platform for new techniques in biology. One of the key benefits of microfluidics for basic biology is the ability to control parameters of the cell microenvironment at relevant length and time scales. Considerable progress has been made in the design and use of novel microfluidic devices for culturing cells and for subsequent treatment and analysis. With the recent pace of scientific discovery, it is becoming increasingly important to evaluate existing tools and techniques, and to synthesize fundamental concepts that would further improve the efficiency of biological research at the microscale. This tutorial review integrates fundamental principles from cell biology and local microenvironments with cell culture techniques and concepts in microfluidics. Culturing cells in microscale environments requires knowledge of multiple disciplines including physics, biochemistry, and engineering. We discuss basic concepts related to the physical and biochemical microenvironments of the cell, physicochemical properties of that microenvironment, cell culture techniques, and practical knowledge of microfluidic device design and operation. We also discuss the most recent advances in microfluidic cell culture and their implications on the future of the field. The goal is to guide new and interested researchers to the important areas and challenges facing the scientific community as we strive toward full integration of microfluidics with biology.
1. Introduction
Over the past decade, microfluidics has emerged as a technology with the potential to make significant impact on cell biology research. The ability to manipulate small volumes of fluid in micron-sized channels, capillaries, and other geometries has led to new methods of designing and performing biological experiments, and is paving the way for innovative approaches to understanding fundamental biology. In the process, microfluidic devices are becoming increasingly high-throughput and integrated, and closing in on realizing the potential of lab-on-a-chip systems that were promised at the beginning of the microfluidics revolution.
Although research in microfluidics was initially dominated by studies in chemistry, and by analyses of physics at the microscale,1 the integration of cell biology with microfluidics has recently become a major focus within the scientific community. The initial motivation to study chemical and physical phenomena in microfluidics was borne out of an inherent need to first understand the fundamental aspects at the microscale before embarking on research work that involved complex biological systems, such as a living cell. But once knowledge was made available on how to exploit the chemical and physical aspects of microfluidics, it was natural for microfluidics and biology to intersect and establish its own area of interdisciplinary research. Cells and their internal structures have physical dimensions on the order of microns, and thus can be suitably manipulated, tested and probed in microfluidic environments using tools developed with microscale technology.
While the often-cited advantages of microfluidics, including faster response times, lower reagent volumes, and potential for integration, are major considerations in the area of chemistry as well as in biology, the most influential benefit of using microfluidics for biology is the ability to tailor the cellular microenvironment. In moving from macro- to microscale, there is unprecedented control over spatial and temporal gradients and patterns that cannot be captured in conventional Petri dishes and well plates. To make significant strides in biology and microfluidics, therefore, it is necessary to understand the intricacies of the cell microenvironment, how it differs across physical scales in vitro, and how best to control it using benefits of the microscale.
So far, progress in the area of biology-related microfluidic systems has been mostly in proof-of-principle demonstrations, with large research efforts toward testing the behavior of various cell types in different geometries and on different platforms. However, general progress has been somewhat hindered by the lack of a complete understanding of why living cells behave differently when moved from macroscale culture to confined microscale geometries.2 Cell culture techniques cannot be directly transferred to microfluidic environments without consideration of the physics of the microscale.3 Some recent reports have also begun to reveal challenges with existing microfluidic methods, and have in some cases provided possible solutions that may lead to new directions in the field. Thus, there is an apparent need to synthesize the ideas of the past decade and the current developments in microfluidics to facilitate the advancement of microscale cell culture toward truly integrative biological experimentation and outcomes.
The main purpose of this tutorial review is to summarize the key elements of microscale cell culture and the major recent advances in the area. In the process, we define the cell microenvironment, discuss conventional cell culture techniques, and summarize the major techniques for microfluidic control as they pertain to microfluidics. We then integrate these fundamental ideas and provide an updated guide to the important factors that influence cell culture at the microscale. The hope is for this guide to direct the interested reader to the important areas that need to be addressed to further advance the field of microfluidics, and to realize the full integration of microfluidics and biology. While other useful reviews have discussed the broad area of lab-on-a-chip cellomics that encompass all cell-based microfluidics topics including cell sampling, cell manipulation and sorting, as well as cell treatment and analysis,4,5 this tutorial review focuses on the aspects of treatment and analysis, with emphasis on how to assess the microenvironment. These areas are more likely to interest the biologist and the bio- or analytical chemist, who are concerned with the effects of treatment and the sensitivity and accessibility of detection methods.
2. Fundamentals of microscale cell culture
Culturing cells in microfluidic devices requires an understanding of certain fundamental principles that span multiple disciplines, including biology, biochemistry, physics and engineering. First, it is necessary to have knowledge of key elements of the cellular microenvironment in order to help develop in vitro models that more closely mimic the conditions that exist in vivo. Second, it is imperative to have command of the techniques in cell culture to aid the translation of methods from macro- to microscale. Third, it is important to have a strong background in microfluidics, and an awareness of the state of the art in microscale technology so that tools can be designed appropriately for their intended applications. The ability to advance the area of microscale cell culture will depend on understanding these fundamental areas, which we review here in succession. We then integrate these basic ideas, and discuss how to control the microenvironment in vitro using microfluidics.
2.1. Cell microenvironment
Cells reside in a milieu composed of soluble factors, cell–matrix interactions, and cell–cell contacts, and do so while living within an environment with specific physicochemical properties (pH, oxygen tension, temperature, and osmolality) (Fig. 1). These elements give the environment a distinct physiological character, and provide a set of extracellular cues that work in concert to regulate cell structure, function, and behavior, and ultimately influence the growth, development, and repair of neighboring tissue. The combination of these biochemical, physical, and physicochemical factors constitutes the cell microenvironment, (although the term tissue microenvironment is also used depending on the context of the work). For stem cells, the local microenvironment, or stem cell niche, holds the key to regulating stem cell survival, self-renewal, and differentiation.6 In cancer biology, tumor and organ microenvironments can give rise to cancer cells that are conditioned for metastasis at ectopic locations.7 In the context of microfluidic cell culture, we focus on microenvironments at the cell and local tissue level, which have physical scales amenable to microchannel dimensions. It is reasonable to assume that examining cell and tissue microenvironments will also help elucidate aspects of the microenvironment at the larger organ level.
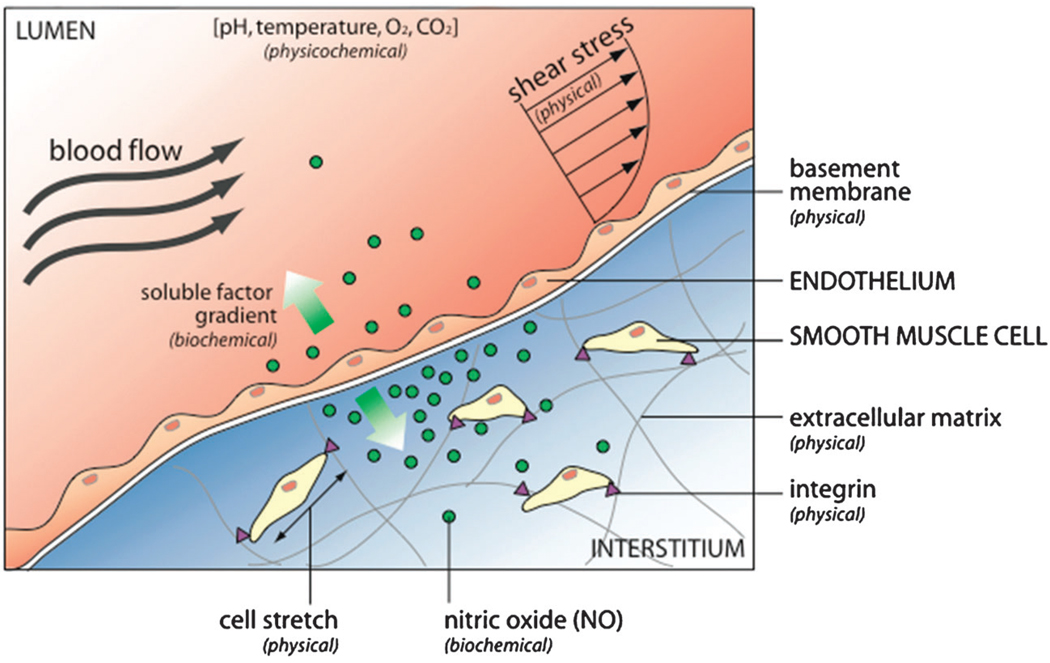
Fig. 1.
The cell microenvironment consists of physical, biochemical, and physicochemical factors. For example, the endothelium that lines blood vessels is exposed to hemodynamic shear stress (external physical force) that stimulates a biochemical response, releasing nitric oxide (NO). NO diffuses to neighboring smooth muscle cells (SMCs), where it regulates cell contraction and relaxation. The gradient of diffused NO affects nearby SMCs more than distant SMCs. Endothelial cells are anchored to the basement membrane, while SMCs are anchored to the extracellular matrix of the interstitium, both via integrins that act as sensors and transducers of physical force. Local physicochemical properties ensure proper regulation of both physical and biochemical mechanisms.
Most cells in the body are non-circulating, and therefore depend on attachment to the surrounding extracellular matrix (ECM) for survival. Cells are anchored to the ECM via cell-surface integrins that are responsible not only for the physical attachment of cells to the matrix, but also for sensing and transducing mechanical signals from focal adhesion sites to the cytoskeletal machinery within the cell.8 These signals are known to drive various cellular processes that include migration, proliferation, differentiation, and apoptosis. Some cell types such as endothelial cells also rely on cell–cell contacts via cadherins for additional physical support, allowing the endothelium to sense, transduce, and resist hemodynamic shear forces as a larger cellular monolayer.9 Together, the forces exerted on the cell through mechanical attachments and external stimuli form a dynamic three-dimensional (3D) physical microenvironment that must be carefully considered when modeling cells and tissues in vitro.
The biochemical microenvironment consists of cytokines, growth factors, hormones and other biomolecules, which combine to form complex signaling pathways that contribute to deciding the fate of the cell.10 Soluble factor signaling occurs mainly via autocrine and paracrine processes, which rely heavily on diffusion of molecules to neighboring cells either of the same or of a different type. Endocrine signaling also plays a role, but relies more on convective transport of hormonal signals from distant locations in the body to the local microenvironment. The effects of soluble factors on cell regulation depend on the concentration, half life, and receptor binding affinities of the ligand of interest. The majority of biological experiments revolve around determining these biochemical effects, and then proposing the mechanisms by which certain soluble factors regulate specific cell processes. For example, the success of certain drug candidates in drug screening tests depends on detailed pharmacokinetic analyses of turnover rates and dose-response curves that shed light on how the drug behaves in the biochemical microenvironment of the cell.
Whether the microenvironment is physical or biochemical in nature, an important aspect of the environment is the presence of gradients that can persist in the vicinity of the cell, often acting as signals themselves to regulate cell function and behavior. Chemical gradients exist naturally due to diffusion, whereas gradients in matrix stiffness or ECM composition are intrinsically built into the heterogeneity of the tissue structure. Since gradients have such an important role in many processes, including migration, angiogenesis, and tumorigenesis, more and more studies are incorporating gradients into their assays, resulting in improved understanding of chemotactic (cell motility in presence of chemical gradient), durotactic (cell motility in presence of substrate stiffness gradient), and haptotactic (cell motility in presence of gradient of surface-bound ligands) effects on cells. More importantly, microfluidics is expected to play a significant role in the design and implementation of such gradient assays because of the ability for microfluidic geometries to establish stable gradients of various forms.11
Because of the obvious coupling between biochemical and physical cues, studies are beginning to surface that examine the combinatorial effects of soluble factor signaling and cell–matrix interactions on cell behavior.12 These types of studies are especially important for drug screening tests and for optimization of microenvironments in tissue engineering applications where long-term performance of tissue constructs are dependent on many factors that act synergistically. It is not surprising that these studies are appearing at the same time that microscale technologies are beginning to integrate into the biological research community, and it is likely that microfluidics will also contribute in a significant way to how cellular microenvironments are designed and studied in vitro.
While the physical and biochemical aspects of the microenvironment receive the bulk of the attention in experimental biology, the effects of physicochemical properties on cell behavior have not been studied as extensively. This is understandable given that the physical and biochemical cues are considered the main factors that determine cell processes in normal and pathological development, while properties such as pH, temperature, and osmolality of the surrounding milieu are considered inherent to the in vivo environment, and are expected to remain unperturbed throughout the normal development and life of an organism. However, given reports that abnormal levels of pH and oxygen tension are associated with the development of various pathologies, such as cancer within tumor microenvironments,13 it is important to ensure these properties are not overlooked when designing and using in vitro microenvironments. In particular, physicochemical properties are critical to the maintenance of cell cultures (see section 2.2 below), and this issue is magnified for cultures at the microscale.
2.2. Cell culture
It is rather remarkable that only a century ago the idea of cultivating a living cell outside of a living organism was met with enormous skepticism and resistance.14 Today, cell culture is part of a huge biotechnology industry that relies on it for mass production of proteins and vaccines, and preparation of cell-based assays for drug screening applications. Moreover, cell culture techniques are an integral part of fundamental and applied cell biology research. Much of our current understanding of biology stems from in vitro experimentation with cells in Petri dishes and well plates, and biology laboratories spend significant amounts of effort and resources on designing and performing experiments based on the in vitro methods that are available to them. Proponents of in vivo methodology often cite as a major weakness of in vitro techniques the inability of a Petri dish to fully capture nuances of the in vivo cellular microenvironment. Yet, there is little doubt that cell culture has had a major impact on modern biology, and will continue to do so as pressures rise to reduce animal testing (particularly in Europe15) and we continue to make further advances in microfluidic cell culture.
Cell culture allows the researcher to isolate specific factors for experimentation outside the complex in vivo microenvironment. By doing so, scientists can make logical hypotheses of the effects of those factors, and through controlled experimentation elucidate the mechanisms that regulate cell function. The goal in cell culture is twofold: to recapitulate as closely as possible the cellular microenvironment while also maintaining enough simplicity so that experimental replicates can be performed to achieve statistically significant results in a reasonable amount of time. Often, there is a tradeoff between these two aspects, and model accuracy is sacrificed for higher throughput, or vice versa. This is where microfluidics is likely to have its largest impact: it has the potential to improve both model accuracy and throughput simultaneously, by giving scientists the freedom to tailor the microenvironment while also reducing the scale of the experimental platform. Here, we briefly summarize the main considerations when performing cell culture in hopes that it will provide insight on the needs of microscale cell culture platforms, and ultimately facilitate the integration between cell culture and microfluidics. Readers interested in a more thorough treatment of basic cell culture techniques are referred to the popular manual by Freshney.16
The basic elements of cell culture have changed little in the past fifty years. The culture medium serves as the biochemical microenvironment of the culture, and consists of essential amino acids, vitamins, salts, carbohydrates, and other components in aqueous solution. The composition of essential components were discovered in a rigorous set of experiments by Eagle more than fifty years ago (see Freshney16), and is still used today as the main source for the components of some basal media. For cells to proliferate in culture, basal media must be supplemented with factors that promote cell growth and division, and this is typically achieved by adding fetal bovine or calf serum. While sera contain the necessary growth factors and hormones for cells to proliferate, the composition of sera can also vary considerably from batch to batch, leading to variations in results from one experiment to another. Considerable progress has been made in the development of serum-free media where all components including growth factors and hormones are well defined so that experimental variations can be reduced.
For the most part, the physicochemical properties of the culture system are expected to remain unchanged throughout an experiment (as well as between separate experiments), unless the properties themselves are being tested. The pH and osmolality of the media can be measured with standard laboratory equipment (pH meter and osmometer) prior to use in culture experiments to ensure media has not deteriorated. Temperature and CO2 levels are usually monitored directly from the incubator controls, while the ambient air within the incubator provides normal oxygen tension levels for the cultures. To maintain a relatively constant and physiological pH of between 7.2 and 7.4, the media can be buffered with sodium bicarbonate, and in certain situations where CO2 cannot be supplied, with additional buffering agents such as HEPES.
In terms of cultureware, the majority of cell cultures are performed on two-dimensional flat surfaces in commercial plasticware such as polystyrene Petri dishes, flasks and well plates. Hydrophobic polystyrene surfaces are typically plasma-treated to render it hydrophilic, which facilitates cell adhesion. For certain cell types, it may be necessary to provide a coating of matrix proteins on the surface to further promote adhesion, growth, and proliferation. The key factors to healthy, viable cells during regular maintenance of the cultures include appropriate cell seeding density, regular changes of culture media, monitoring of cell growth rates, and timely subculturing of the cells. These aspects are critical to maintaining proper cell phenotype and function, and apply to both macroscale and microscale culture. Each cell type is unique, and it is still routine procedure for the biologist to fine-tune their culturing protocols to suit the needs of their cell type.
While most biologists continue to use conventional two-dimensional cultures as their main format for in vitro experimentation, the past quarter century has also seen the major development of 3D cultures that provide a more realistic model of the physical microenvironment that exists in vivo. Bissell at the Lawrence Berkeley National Laboratories has been a pioneer in the area of 3D culture systems where cells are cultured and tested in in vivo-like tissue architectures. Using techniques developed in her lab, Bissell and colleagues have made novel discoveries in breast cancer development and the mammary gland physical microenvironment,17 and have also begun to incorporate microscale technology in the form of combinatorial microenvironment microarrays to tease out the synergistic effects of combined soluble and ECM factors on stem cell fate.18
Although culture techniques have remained unchanged for the bulk of its history, recent progress in microfluidics and other microscale technologies, as evidenced in the work of Bissell described above, suggests that cell culture practices are beginning to evolve. Research laboratories will likely adopt microscale techniques at an ever-increasing pace because of their ability to create physiological microenvironments, as well as their promise to provide high-throughput solutions for intensive biological studies. Although tissue culture in industrial settings will likely follow that trend given enough time, it will perhaps require significant progress both in high-throughput capabilities and in the marketability of microfluidic systems before a massive industry such as biotechnology will change its current course.
2.3. Microfluidics: tools and techniques
Microfluidic devices designed specifically for cell culture have certain requirements that distinguish them from microscale systems used for other applications in chemistry or physics. Design considerations of particular importance to microfluidic cell culture include: (1) the choice of material for device fabrication, (2) the geometry and dimensions of the culture region, and (3) the method of pumping and controlling fluid flow. The latter consideration ultimately dictates how the microfluidic device is connected to external components of the overall system. While it is clear that microfluidics offers the engineer—and the biologist—ultimate flexibility over system and experimental design given the plethora of options, it also implies that those involved with the experiment must be aware of all the available choices so that designs can be optimized according to the application. We highlight here the types of materials and options for control that have been largely accepted as the major classes within the microfluidics community, and discuss some of the new directions being pursued.
2.3.1. Device materials and fabrication
Similar to other niche areas within microfluidics, cell-based studies made the largest strides after the introduction of soft lithography. Soft lithography was popularized by Whitesides and his group at Harvard in the late 1990s, and comprises a set of fabrication techniques similar in concept to photolithography, but with significant benefits for biochemistry and biology.19 The most popular material used in soft lithography is poly(dimethylsiloxane), or PDMS, a silicon-based elastomeric material with a number of physical and practical properties that make it desirable for experimentation. PDMS is fairly cheap and easy to mold, making it ideal for rapid prototyping of microfluidic designs and for transferring micropatterns with high fidelity via stamping techniques. PDMS is suitable for cell experiments because it is non-toxic to cells, is gas permeable, and has excellent optical properties, including low autofluorescence and optical transparency for imaging applications. Furthermore, the elastomeric properties of PDMS allow it to readily deform when subjected to local displacements, allowing the integration of built-in valves and pumps via multilayer soft lithography.20 To make enclosed microchannels, PDMS can be bonded to different materials (e.g., PDMS, glass, polystyrene) quite easily using various methods such as oxygen plasma treatment and additional curing.
While the many advantages of PDMS have established its well-known popularity among microfluidics researchers, a growing number of reports are beginning to reveal some unfavorable characteristics of PDMS that may limit its future use for microscale cell culture. Kim and co-workers alluded to these challenges in a previous review on microfluidic perfusion systems.21 More recently, however, work from our group has revealed that PDMS further confounds cell culture results by sequestering small hydrophobic molecules such as estrogen, and by leaching out uncrosslinked oligomers from the PDMS bulk during culture, which then bind to cell membranes.22 Also, as cells are exposed to PDMS for longer durations, cell metabolism and proliferation are affected, possibly as a result of the presence of PDMS.23 This growing awareness of the potential artifacts and biases associated with PDMS is providing an impetus for the microfluidics community to consider other options for materials.
The most logical choice for an alternative material for devices is polystyrene because it is the most common plastic used for traditional cell cultureware. Polystyrene microfluidic devices for cell culture applications have recently surfaced in the commercial market (e.g., Bellbrook Labs, Integrated BioDiagnostics), illustrating that the industry has already recognized a need to conform to the needs of biologists who are accustomed to certain materials. In academic research laboratories, micromolding techniques for fabrication of thermoplastic microfluidic devices has also been well-documented.24 A recent report from the Takayama group has employed a hot embossing technique using epoxy molds to fabricate polystyrene-based microfluidic devices for cell culture, with potential to incorporate soft substrates such as polyurethane or PDMS as a bonded surface.25 In yet another example, rapid prototyping of polystyrene microfluidic devices has been achieved by the use of “Shrinky-Dinks” thermoplastic sheets.26 Shrinky Dinks plastics are an arts and crafts toy for children with the property that drawn artwork on the plastic surface can be shrunk in size after heating the material. This property was recently exploited by Khine and co-workers to produce positive relief masters for microfluidic devices. Together, these developments are revealing a trend toward microfluidic devices made with more common bioware materials, as well as a trend against further investments into materials with undesirable and unknown effects on cultured cells. For the foreseeable future, however, PDMS will continue to provide an affordable rapid prototyping option for most research laboratories, and will work in concert rather than in competition with other materials such as polystyrene.
2.3.2. Geometries
Lithographic techniques allow for infinite possibilities when it comes to geometry, but for cell culture applications, some important considerations must be recognized. First, microchannel dimensions for cell culture are typically at the larger end of the spectrum for channel sizes, ranging from 100 to 1000 microns. Smaller microchannel cross-sections, on the order of tens of microns, are common in chemical applications such as electrophoretic separations, and are more suitable for single cell analyses or for chips designed for cell sorting and cell manipulation. These applications have been reviewed elsewhere.4,5,27 For biological experiments, unless single cell analysis is coupled with high throughput methods for measuring endpoints, enough cells will need to be cultured in the microchannels to permit population-based analyses, and this implies a need for larger channel culture regions.
The flexibility in geometry permits the generation of stable gradients in both soluble and surface-bound factors.11 This is particularly useful for controlling the cell microenvironment in chemotaxis studies (as well as durotaxis and haptotaxis studies) where spatial and temporal concentration gradients can induce cell responses such as migration.28
An important geometric consideration in microfluidic channels is the height-to-width aspect ratio, especially for PDMS-based devices. While the deformability of PDMS was beneficial for fabricating multilayer devices with built-in valves and pumps, the same property leads to undesirable sagging and bulging of microchannel walls when (1) the PDMS layer is thin, (2) pressure is substantial, and (3) maintaining channel cross-sectional shape is important for analysis, such as in shear flow experiments.29 This issue would be less important for devices made of stiffer materials, such as glass or polystyrene.
2.3.3. Pumps and valves: going tubeless
Within microfluidic cell culture devices, fluid volumes must be transported and displaced from region to region, using valves and pumps that are either externally connected to the device, or directly built into the system. Inlet and outlet ports of the system serve as points of interaction between the culture region and the external world. The majority of systems employ external pumps (e.g., syringe pumps for non-recirculatory flow, and peristaltic roller pumps for recirculatory flow) that can be hooked up to the access ports via tubing. This is the method of choice for perfusion systems that rely on constant fluid flow to replenish nutrients and remove waste products in a timely manner. Using multilayer soft lithography, it is possible to incorporate pneumatic valves into the system to produce fully automated, high-throughput culture systems (Fig. 2).30 Such a system provides a concrete example of the many benefits associated with microfluidic cell culture. The major concerns with this type of system are the large number of connections required, the potential for leakages at those connections, the technical expertise necessary for proper operation, and the need for elastic materials that possess properties with some confounding issues (see section 2.3.1). These concerns will likely prohibit the widespread use of such complex integrated systems in the research community at large.
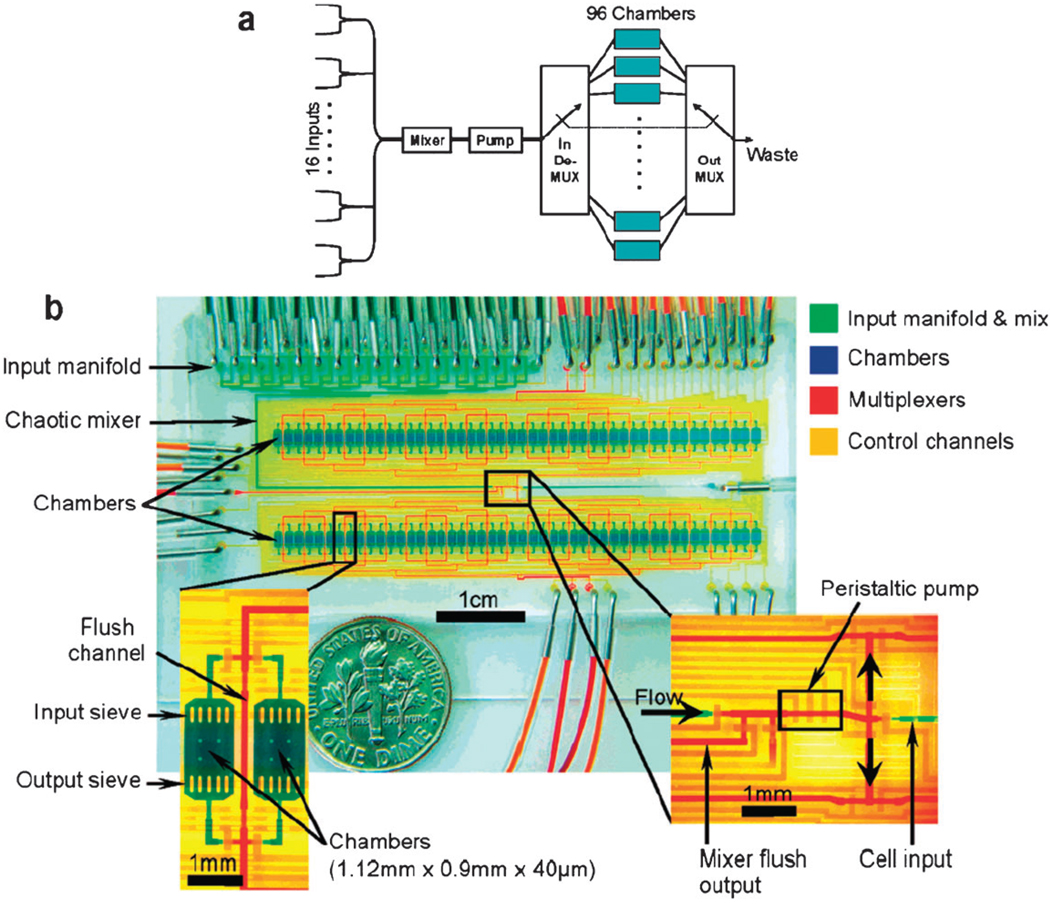
Fig. 2.
A fully automated PDMS-based microfluidic cell culture system consisting of 96 individually addressable cell culture chambers (blue dye), on-chip peristaltic pumping (lower right inset), and multiplexing capabilities that allow different mixtures of reagents to be formulated. Reprinted with permission from Gomez-Sjoberg et al30 Copyright 2007 American Chemical Society.
A particularly interesting method for pumping and valving fluids in microfluidic channels was developed by the Takayama group, and employs Braille pin displays to deform thin PDMS layers into microchannels in specific sequences to generate peristaltic flow.31 While the design and concept are both novel, it also has not been widely used in research circles, likely stemming from the challenges associated with the complexity of the equipment, and from the unconventional nature of the platform.
An alternative method of fluid replacement, which has potential for widespread acceptance because of its simplicity and compatibility with existing techniques in biology, is passive pumping. First developed by the Beebe group in the early 2000s, passive pumping relies on the surface tension of different-sized droplets placed at the inlet and outlet ports to drive fluid from one port to the other (Fig. 3). The difference in droplet volumes induces a differential pressure between ports that generates flow in the microchannel.32 The major advantage of passive pumping is that it can be performed without connecting to an external pump, eliminating the need for tubing and interconnections at the ports. Passive pumping can be performed simply by pipetting the appropriate volumes of droplets at the ports, and is therefore amenable to automated liquid handling systems that are commonly used in major biology research laboratories.33 The ability to do experiments with “tubeless” microfluidics is likely to attract an increasing number of biologists to the microscale techniques that are currently being developed. In fact, other researchers have recently begun to study the physical phenomena related to passive pumping in their own microfluidic systems,34,35 demonstrating that the technique is receiving support. While passive pumping is an effective method for many applications, it is limited to low volume flows and low pressures. Steady continuous perfusion of microchannels is better achieved with external pumps, even though droplets can theoretically be pipetted continuously in a passive pump cycle that simultaneously adds droplets at the inlet while removing droplets at the outlet.
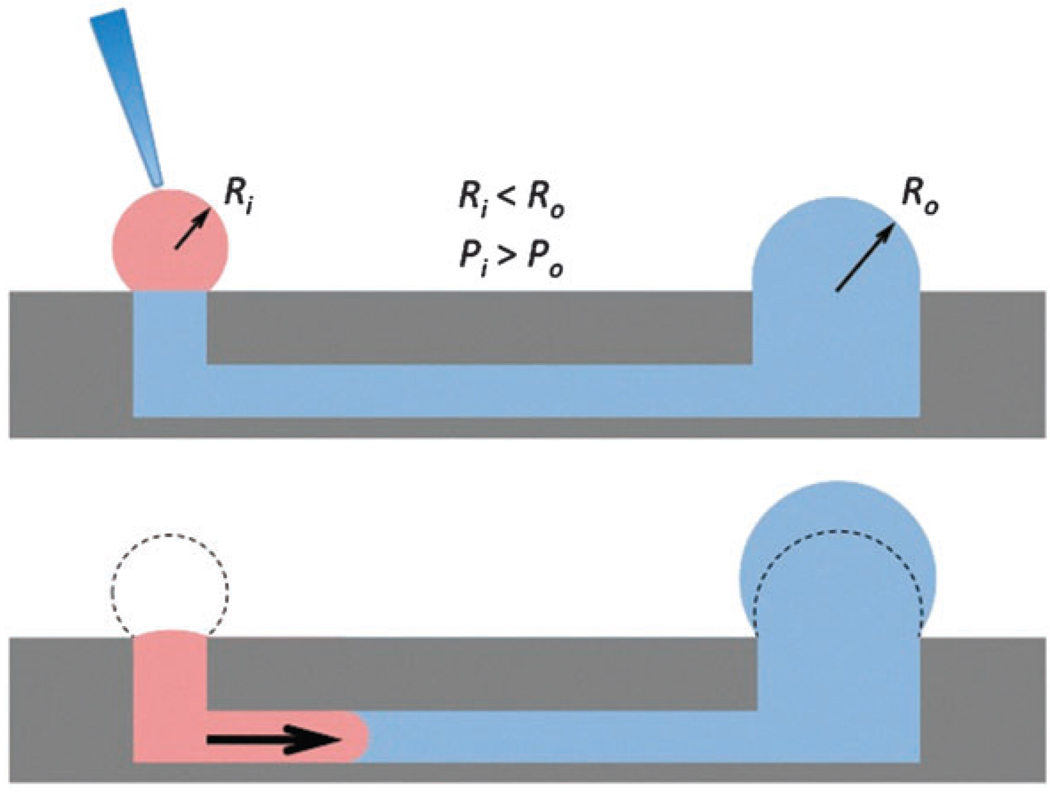
Fig. 3.
Passive pumping relies on surface tension of small droplets to pump fluid from inlet to outlet. A smaller drop of radius Ri has an internal pressure Pi greater than the pressure in a larger drop of radius Ro because of Laplace’s law (ΔP = 2γ/R), where ΔP is the pressure difference across the liquid–air interface of the droplet, and γ is the surface tension at the interface.
2.3.4. Membranes
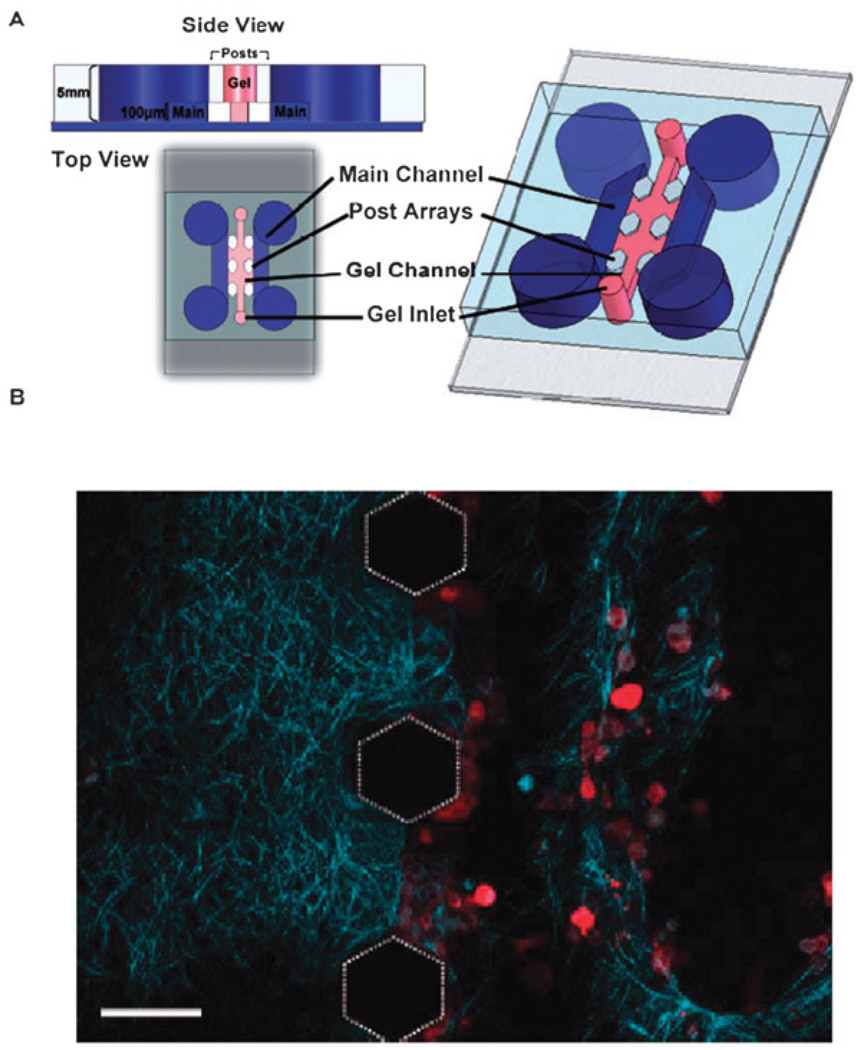
Certain on-chip accessories can be incorporated into microfluidic devices to add extra functionality to the cell culture systems. Three-dimensional networks are achievable by incorporating commercially available track-etched membranes (polycarbonate or polyethylene terephthalate) into the device during fabrication. The membrane serves as a semi-permeable barrier that separates microchannels on different horizontal planes, allowing communication only at locations where microchannels intersect. Several groups have begun using this geometry for cell-based studies.36–38 The most notable is the Takayama group, who have employed membrane-PDMS devices to study lung epithelial cell rupture, and more recently, the adhesion of cancer cells on a microfluidic endothelium (Fig. 4).36,39 The usefulness of this arrangement lies in the in vivo-like organization of the endothelium into luminal and abluminal compartments separated by a membrane that mimics the basal lamina. Such compartmentalization is potentially useful for coculture studies that involve communication between endothelial cells and neighboring cells from the stromal or smooth muscle layers.
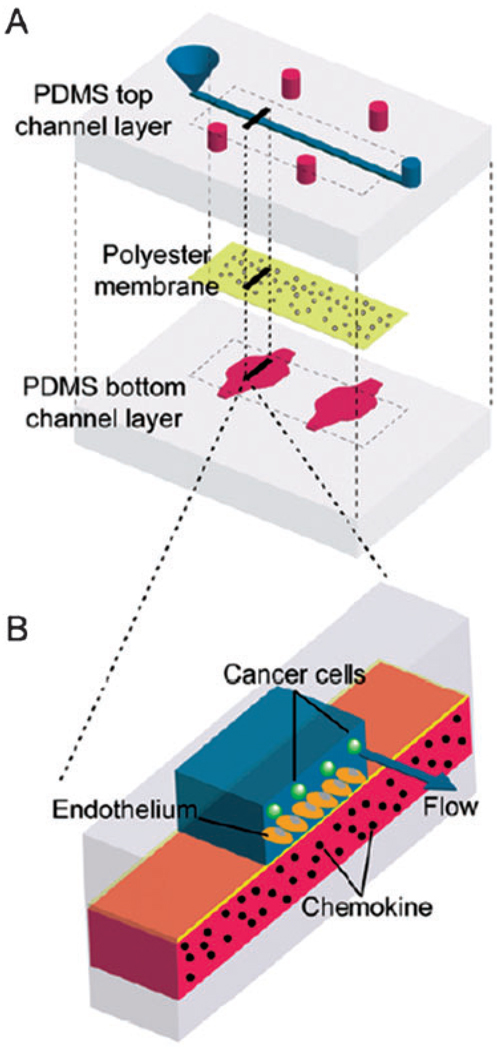
Fig. 4.
PDMS-based microfluidic device containing a porous polyester membrane for supporting growth of endothelial cells. Device was used to study adhesion of cancer cells on the endothelium in the presence of chemokines released on the basolateral side of the endothelium. (Open access: Song et al., PLoS One, 2009, 4(6); 5756.)
3. Controlling the microenvironment in vitro
The fundamental elements of microscale cell culture discussed above serve as a basis for understanding how to control the microenvironment in vitro. To be able to properly tailor the microenvironment in microfluidic devices, it is necessary to integrate the ideas developed from these elements. Previous reviews have begun examining aspects of cell culture and cellular microenvironments as they relate to microfluidics. Kim and co-workers previously outlined important practical considerations related to operating microfluidic perfusion culture systems, including cell seeding, management of physicochemical properties, mass transport, shear stress, and bubble formation.21 Walker and Beebe introduced ideas on effective culture volume and surface area-to-volume ratios as they pertain to microscale cell culture.3 Here, we extend some of the ideas developed from those reviews, and summarize new developments that have addressed previous concerns.
3.1. Cell seeding
The first crucial step to obtaining viable cultures in microfluidic devices is cell seeding. Procedures for proper cell seeding can vary considerably from one experiment to another, depending on microchannel design and geometry. While the majority of researchers employ simple syringe-based injection methods for seeding, with varied success, others have also used gravity-driven flow to achieve more uniform seeding throughout the channel. Gravity-driven flow can, for example, be used in conjunction with an open-air funnel-shaped inlet reservoir to reduce cell clogging and improve uniformity of cell distribution.39 Besides clogging, another key concern when seeding is the potential for cells to settle, accumulate and attach in the reservoir at higher density than the original suspension, even when clogging is absent. The high density of cells leads to faster nutrient depletion and waste accumulation, and causes soluble factors from the reservoir to diffuse into the channel over time. Furthermore, the physicochemical properties including pH and gas concentrations can also significantly change as a result. These factors can have adverse effects on the culture, which can sometimes be seen as a loss in cell viability propagating from the inlet port into the channel. Passive pumping systems can circumvent this issue because precise cell suspension volumes can be dispensed at the inlet port via pipetting so that little or no excess volume is introduced into the ports. Recently, a method to compartmentalize cell culture regions using a fabricated “curtain” has been shown to allow seeding of cells in specific regions of the culture surface.40 This may prove useful for applications where certain regions need to be initially cell-free, such as free migration assays. In terms of seeding cells into 3D microenvironments, Yu and co-workers in Singapore have developed a series of techniques that include the use of micropillars, 3D matrices, and more recently, a gel-free method where cells can be free to synthesize their own matrix.41 In addition, the Beebe group has begun to incorporate controlled soluble gradients into microfluidic channels with cells embedded in 3D collagen gels, demonstrating how both physical and biochemical microenvironments can be simultaneously controlled with microfluidics.42
3.2. Bubbles
Air bubbles are well known to be detrimental to cell cultures because they can rupture cell membranes when they burst. This is especially problematic in microchannels where bubbles can block an entire channel, or impede fluid flow sufficiently to alter flow patterns and disturb the cell culture environment. Due to evaporation through the PDMS (discussed below) bubbles can also grow in size if not removed, potentially drying out sections of the cell culture region. Certainly, there are unique instances where air bubbles can be deliberately applied in a two-phase flow system to model physiological phenomena that involve cell damage, such as the effects of liquid plugs on lung epithelial cells in the alveolar space.36 However, in the majority of culture studies, bubbles are undesirable, and measures should be put in place during the design phase to eliminate them. Despite various methods developed to avoid introducing bubbles into the system, as well as to passively trap those that end up entering the device anyway,21 bubbles are sometimes unavoidable. Thus, the best way to circumvent the issue is to actively remove bubbles when they appear, before they have a chance to enter the culture region.43
3.3. Evaporation
An important issue that has recently surfaced due to increasing use of open-air microscale platforms is evaporation.2 Because the majority of microfluidic cell culture systems are made from PDMS, which is permeable to gas, the media in microfluidic chambers can evaporate if the environment is not properly humidified. Evaporation can be a major problem even in tubing-connected microchannels because of the permeability of PDMS to gas, as evidenced by the shifts in osmolality that have been shown to adversely affect embryo development.44 Evaporation is an even greater concern in open microwells, and in tubeless microchannel systems where passive pumping droplets are exposed to air and prone to evaporative effects. In such systems, evaporation of droplets at the ports not only reduces total fluid volume in the system, but also concentrates soluble factors in the media and causes evaporation-induced flow, which may ruin stable gradients that were established, or otherwise transport fluid undesirably. Evaporation can be mitigated by understanding the physical principles of the evaporation process, and then designing proper humidification for the specific device under study.45
3.4. Effective culture time (ECT)
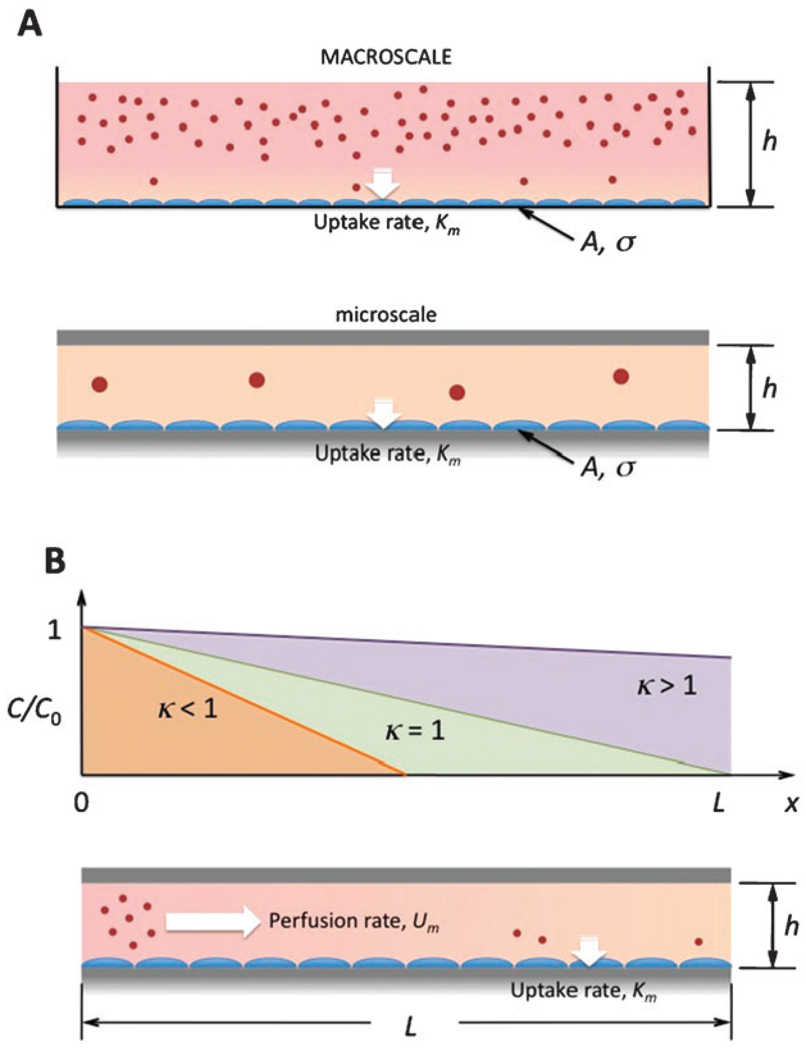
The effective culture volume (ECV) introduced by Walker and Beebe served as a necessary first step in understanding and characterizing microenvironments for microscale cell culture.3 However, the ECV as a parameter is unable to explicitly provide practical guidance on how to properly culture cells at the microscale based on our experiences at the macroscale. Specifically, the maintenance of cell cultures relies on media changes at regular time intervals, and failure to do so usually leads to cultures of low viability and poor quality because of nutrient depletion and waste accumulation. A typical concern when culturing cells in microchannels is knowing how much to adjust the time interval between media changes due to the much higher culture area-to-volume ratios. Furthermore, some microfluidic applications prefer to use perfusion to continually replenish media components, in which case the question becomes how to determine an appropriate perfusion rate such that replenishment occurs fast enough. We attempt here to solve these two problems by introducing an effective culture time (ECT) and a critical perfusion rate (CPR) based on simple geometric arguments and comparisons of time scales.
In a static culture vessel (at any scale), cells are grown on a culture surface of area A, and bathed in medium with volume V. The medium forms a column of fluid of height h = V/A above the cell layer, which corresponds to the channel height in confined microchannels, and to the height of the air–liquid interface above the cells in flasks and dishes. Height h is considered the characteristic length for microchannels because length and width are considerably larger than h, and h becomes the limiting dimension for diffusion. The key biochemical factors that are being depleted and consumed during the culture period are the growth factors contained in the serum, as well as glucose and other energy components responsible for metabolic processes. When these factors are exhausted, the medium must be replenished for cells to continue growing and dividing (Fig. 5A). The time interval between medium changes, or the effective culture time (ECT), is dependent on: (1) initial concentration of the particular substrate C0, (2) substrate uptake rate by the cells Km, (3) diffusivity of the substrate D, cell density σ, and the culture area A and medium volume V. As a first approximation, substrate uptake rate can be taken as the maximum reaction rate of the substrate with its enzyme based on Michaelis–Menten kinetics. These parameters are then related via the Damkohler number Da,46 where
| (1) |
Da is a dimensionless parameter that measures the ratio between reaction and diffusion time scales. In particular, Da can be rewritten as
| (2) |
where the numerator represents the diffusion time scale τd = h2/D, and the denominator represents the reaction time scale τr. For microscale cell culture, h is typically 5 to 10 times smaller than in macroscale culture, implying that Da is up to an order of magnitude lower in microchannels. Thus, reaction times dictate the speed of the process because diffusion to the top of the microchannel happens much more quickly than the reaction of all the substrate at the bottom. (Macroscale cultures would be heavily dependent on diffusion times to drive the speed of substrate consumption because the medium is deeper. However, at this scale, cultures are aided by free convection in the system, which mitigates the dependence on diffusion.) Since τr is equivalent to ECT in diffusion-dominant systems, and τr is linearly proportional to h, the ECT can be scaled according to the height of the microchannel, i.e., ECT ∝ h. For example, a typical macroscale culture may contain ~10 mL of medium in a Petri dish of ~80 cm2, resulting in a medium height of h ≈ 1.2 mm. For many cell types, medium is replaced every 48 h. For a microchannel of h = 200 µm, the ECT is expected to be reduced by a factor of ~ 6, which suggests that medium should be replenished every 8 h to maintain similar culture conditions. This approximation is in good agreement with previous experiments conducted by the author, who optimized feeding intervals for microscale culture of endothelial cells by trial and error, and concluded that the ECT was 8 to 12 h (maximum). The derivation of Da is also consistent with past experiments regarding the effects of channel height on microfluidic cell culture.47
Fig. 5.
Effective culture time (ECT) and critical perfusion rate (CPR). (A) Macroscale static cultures have larger h and therefore a larger Damkohler number compared to static microscale cultures. Because substrate uptake time scales dominate at the microscale, media must be replenished sooner based on the change in media height. (B) To ensure all cells in a microfluidic culture are being replenished sufficiently with perfused media, perfusion rate Um must be large enough to displace exhausted media (κ > 1). CPR is defined as the perfusion rate where κ = 1. This can be determined by dividing channel length with ECT.
3.5. Critical perfusion rate (CPR)
Similar scaling arguments can be used to obtain a critical perfusion rate (CPR), which can provide a guide for designing proper perfusion systems in microfluidic devices. In a straight microchannel of height h and length L, perfusion of the channel at a constant flow velocity Um means that it would take τc ≈ L/Um for substrates carried by the fluid to travel from inlet to outlet (the convection time scale, or media residence time21). As a first approximation, we assume that the substrate uptake rate Km is constant and uniform across the cell culture area A, and is independent of substrate concentration. We also assume that because substrate is continually being consumed along the length of the channel that substrate concentration varies linearly with the length of the channel such that C = C(x), and C = C0 at the inlet x = 0 (Fig. 5B). A dimensionless parameter, similar to Da, can be derived for the ratio of convection and reaction time scales:
| (3) |
If we define the critical perfusion rate (CPR) as the velocity at which the substrate concentration just reaches zero at the outlet, i.e., C = 0 at x = L, then CPR coincides with κ = 1 since the time scale needed for total consumption of the substrate would be on the order of the time scale for convection from inlet to outlet. For perfusion to be sufficient for replenishing media, κ > 1 such that convective time scale dominates and substrates travel through the microchannel before being completely consumed. If κ < 1, perfusion is too slow and substrate is entirely consumed before the end of the microchannel, leaving cells near the outlet end devoid of necessary media components. The CPR can be calculated then for κ = 1,
| (4) |
Since τr = ECT, eqn (4) states that the CPR can be estimated by dividing channel length by the effective culture time.
The goal of these concepts was to extend the idea of the ECV to arrive at quantitative measures that can aid in the design process of microfluidic geometries. These approximations are ballpark estimates designed to provide the practitioner with guidelines for culturing at the microscale based on experiences at the macroscale. If the system is static, and the ECT at the macroscale is known, the microscale ECT can be estimated from the change in h; and if the system is to be perfused, the CPR can be estimated from eqn (4). As a final note, the dimensionless parameter κ can also be derived by combining Da, the Peclet number, Pe = Umh/D, and the height-to-length ratio, α = h/L.
4. Recent developments & emergent challenges
The ultimate goal in microfluidic cell culture research is to simultaneously improve model accuracy and experimental efficiency in hopes of benefitting biological research. The success of this endeavor is most often measured by progress in three main areas: (1) improving the ability to control the cell microenvironment, (2) realizing the promise of high-throughput capabilities, and (3) successfully integrating additional functionality. While the past five years have seen significant strides in each of these key areas, the overall impact of microfluidics on modern biology continues to be marginal, and much of the potential of microfluidics that was promised continues to be largely unfulfilled. This is apparent from the lack of widespread acceptance within the biology community to adopt microfluidic methodologies into their laboratories. To advance the field toward more mainstream acceptance, it is beneficial to discuss the current state of the art, evaluate the recent progress within these main areas of development, and identify remaining challenges that need to be addressed in order for microfluidics to become truly useful to biologists.
First, in terms of novel techniques for creating improved cell microenvironments, recent developments have been quite significant. Membrane devices have been developed to separate microchannels into upper and lower compartments that can be used to model the apical-basolateral polarization of endothelial cells.39 More recent advances in modeling of the cell microenvironment have focused on the generation of in vivo-like ECM constructs for supporting 3D cell growth and examining cell migration.48 Certain microfluidic models have also been coupled with high-resolution optical microscopy to provide important insights about ECM remodeling in the presence of cells (Fig. 6).49 All these reports collectively represent significant advances in the area of microfluidics, especially in comparison to the early work in the field that addressed more prosaic issues (such as proper maintenance of cultures at the microscale and improving cell viability) that were no less important at the time. However, the unfortunate reality is that these novel developments may not necessarily lead to significant impact in biology because they may be too complex to implement on a larger and broader scale. Questions remain regarding the ability to expand such sophisticated systems to high throughput platforms, and the challenge will be to achieve that sophistication while also maintaining user-friendliness to biologists.
Fig. 6.
(A) Microfluidic coculture device for three-dimensional (3D) microenvironments. (B) The microfluidic coculture device allows imaging of collagen fibers (blue) via Second Harmonic Generation. Matrix remodeling is observed as cells cluster at the boundary of the gels. Scale bar = 100 µm. (Huang et al.49—reproduced by permission of The Royal Society of Chemistry.)
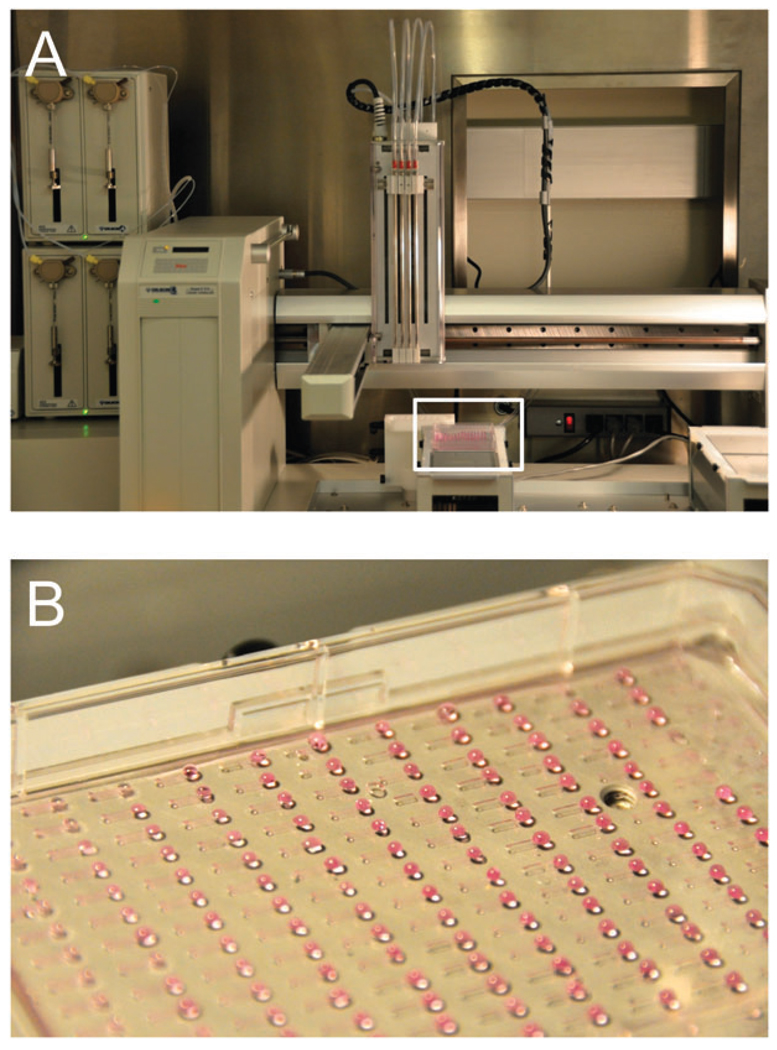
Secondly, regarding the progress of high-throughput biological platforms, a number of systems have surfaced in the literature describing organized arrays of microchannels for cell biology studies. For example, a fully automated tube-based microfluidic system has been reported that can individually address 96 separate culture chambers, culture cells for up to a week, and track cell proliferation and differentiation.30 While this is perhaps the most impressive demonstration to date of the full capabilities that can be achieved by microfluidics, it has also confirmed the various issues associated with tube-based systems regarding complexity and the lack of accessibility to biologists, who would prefer that experimental procedures remain simple and manageable even when microfluidics is expected to add more sophistication to experimental design. Ideally, traditional procedures such as cell seeding, fixation, immunostaining, western blotting, and real-time polymerase chain reactions (RT-PCR) can be performed at high throughput on microfluidic devices, with equal or better sensitivity, using existing infrastructure and equipment that are familiar to biologists. Several studies have adopted this philosophy, and have described tubeless microfluidic devices that use common laboratory equipment such as automated liquid handling systems and plate readers to carry out pipetting operations and fluorescence intensity measurements on arrays of microchannels (Fig. 7).33,50 The contrast between the two pumping strategies is evident: while scaling up a tube-based system inherently means that the number of tubes and connections must also be scaled accordingly, scaling up tubeless systems merely requires increasing the number of pipetting operations. This advantage has prompted ongoing developments in this area that will see the use of an automated liquid handling system to handle a complete experiment, from microchannel priming steps to cell seeding, media replenishment, fixation, and immunostaining, that involve over 60 000 pipetting operations over ten microchannel plates, each containing 192 microchannels.51
Fig. 7.
Interfacing microchannel arrays with automated liquid handling systems for high-throughput biological studies. (A) Four-channel automated liquid handling system (Gilson Quad-Z 215 Liquid Handler, Middleton, WI, USA) interfaced with microchannel plate (white box). (B) Array of 192 microchannels in standard microtiter plate format. Straight microchannels contain access ports to allow passive pumping.
Finally, in terms of integrating functionality, there is significant effort in the microfluidics community to develop systems with integrated on-chip components. To date, integrated components have included: on-chip optical waveguides for laser-induced fluorescence detection,52 channel-embedded carbon microelectrodes for amperometric detection of cell-secreted analytes,53 integrated micropumps and valves for manipulating fluid flow,20 electromagnetic coils for magnetic-based cell separation,54 and electrode arrays for digital microfluidic platforms interfaced with channel-based microfluidics,55 just to highlight a few of the many novel contributions. A review of on-chip analysis for biological cells is available for much of the work in this area.56 While many of these studies are excellent proof-of-principle demonstrations of what can be achieved through the integration of additional on-chip components, the overall impact of these tools on biological research at large is uncertain. The main issue appears to be how biologists perceive the practicality of on-chip components, especially when they still require external equipment, such as off-chip light sources, power supplies, and data acquisition systems, for operation. A common misconception is that adding functionality to microfluidic systems is synonymous with adding more microfluidic components on chip. This emphasis (on adding components rather than adding functionality without more components) is perhaps another reason microfluidics has yet to reach its full potential. From a biologist’s perspective, additional functionality can be achieved without adding more parts, as long as microfluidic channels are designed for compatibility with existing techniques. For example, in-cell westerns (ICW) for measuring protein expression on cells can be substituted for traditional western blotting when using microfluidic devices, provided that equipment such as fluorescence or infrared scanners are available for detection.23 Thus, as long as endpoints are measured with higher efficiency and equal or better sensitivity, the need for excessive components on-chip may become less important as the field advances.
While the three key areas of development (microenvironment control, high throughput advancement, and integrated functionality) have all experienced noticeable growth and progress the past five years, the above discussion has revealed that divisions exist between the individual topics within the field of microfluidic cell culture. Often, researchers concerned with one area of development, such as the integration of functionality, are not fully aware of the developments in another area, such as microenvironmental control. This is likely the main reason why microfluidics has yet to fulfill its promise. In fact, the subsets of microfluidic cell culture research need not be mutually exclusive. For microfluidics to have a lasting impact on biology, it will likely require a more concerted effort amongst microfluidics researchers and biologists to communicate ideas between the fields more openly, integrate the ideas between these fields, and advance microfluidic cell culture as a whole. Doing so will likely lead to a greater impact than that achieved by advancing the field as individual parts.
5. Conclusions and outlook
Culturing cells in microfluidic environments requires fundamental knowledge of multiple topics, including cell biology, cell culture techniques, microfluidics, and the physics of the microscale. In order to develop significant progress in the field, it is important to integrate ideas from these basic topics, and use them to devise new tools and concepts that have the potential to aid future design and implementation. Concepts such as ECT and CPR arise out of a need to design devices with a more systematic approach. As we continue to make progress, it would be ideal for there to be a framework on which more theoretical concepts of microfluidic cell culture can be based.
As we strive towards a full integration of biology with microfluidics, it is apparent that we must also take advantage of the flexibility in microfluidic design to conform to the needs of biologists. To this end, simple translatable technologies will likely prevail over complex systems, and control of more in vivo-like microenvironments will steer biologists to the new advances that the microscale has to offer. Trends toward the use of existing infrastructure and equipment such as automated liquid handling systems, plate readers and pipettes, and toward the use of common bioware materials like polystyrene will also help to attract biologists to microfluidics.
The field of microfluidic cell culture is growing at a remarkable pace. Myriad research areas including stem cell differentiation, neural regeneration, cell-based and point-of-care diagnostics, gene transfer, and high-throughput genetic screening are emerging and advancing quickly with the aid of microfluidics technology. These niche areas will likely continue to rely on technological breakthroughs at the microscale before major progress can be realized in their respective fields. While a critical discussion of these topics is beyond the scope of this review, we need to acknowledge that the potential for microfluidics to impact modern biology is apparent, and that the only limit to this potential is the imagination of scientists and engineers. However, the key to the further advancement in microfluidic cell culture will be our ability to seamlessly integrate the fundamental concepts developed in cell microenvironments, cell culture techniques, and microfluidic tools so that biology experiments in the future can be designed to satisfy the principles of all areas.
Acknowledgements
We acknowledge the financial support from the Natural Sciences and Engineering Research Council of Canada (NSERC) to EY in the form of a postdoctoral fellowship.
Abbreviations
- CPR
critical perfusion rate
- Da
Damkohler number
- ECM
extracellular matrix
- ECT
effective culture time
- ECV
effective culture volume
- ICW
in-cell western
- PDMS
poly(dimethylsiloxane)
- Pe
Peclet number
- RT-PCR
real-time polymerase chain reaction
Biographies

Edmond W. K. Young
Edmond W. K. Young is a postdoctoral fellow working with Dr David Beebe at the Wisconsin Institutes for Medical Research, University of Wisconsin-Madison. He received his PhD in mechanical and biomedical engineering at the University of Toronto in 2009 while studying with Drs Craig Simmons (Dept. Mechanical Engineering) and Aaron Wheeler (Dept. Chemistry). His research interests include designing and integrating microfluidic tools for cardiovascular and cancer biology, with focus on endothelial cells and multiple myeloma. He is the recipient of an NSERC postdoctoral fellowship, and was awarded the Governor General’s Gold Medal of Canada and the Norman F. Moody Award at the University of Toronto in 2009.

David J. Beebe
David J. Beebe is a Professor in the Department of Biomedical Engineering and the Wisconsin Institutes for Medical Research at the University of Wisconsin-Madison. He is the recipient of the IEEE EMBS Early Career Achievement Award, the Lab on a Chip, Royal Society of Chemistry/Corning, Pioneers of Miniaturization Prize, the Romnes Award at UW-Madison and is a Fellow of the American Institute for Medical and Biological Engineering. He has also served as an Associate Editor for Journal of MicroElectro-Mechanical Systems, the Journal of Biomechanical Engineering, Lab on a Chip, and is currently Scientific Editor of Integrative Biology. Past research topics have included development of nontraditional autonomous microfluidic devices and systems, and the study of cell and embryo development in microenvironments. David’s current interests center around the creation and use of microfluidic tools to understand cancer biology and improve cancer diagnosis and monitoring. His migration to more biological focused research was facilitated by a 5 year NIH “retraining” award in cancer biology.
Footnotes
Part of the themed issue: From microfluidic application to nanofluidic phenomena.
References
- 1.Beebe DJ, Mensing GA, Walker GM. Annu. Rev. Biomed. Eng. 2002;4:261–286. doi: 10.1146/annurev.bioeng.4.112601.125916. [DOI] [PubMed] [Google Scholar]
- 2.Paguirigan AL, Beebe DJ. BioEssays. 2008;30:811–821. doi: 10.1002/bies.20804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walker GM, Zeringue HC, Beebe DJ. Lab Chip. 2004;4:91–97. doi: 10.1039/b311214d. [DOI] [PubMed] [Google Scholar]
- 4.Andersson H, van den Berg A. Sens. Actuators, B. 2003;92:315–325. [Google Scholar]
- 5.El-Ali J, Sorger PK, Jensen KF. Nature. 2006;442:403–411. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 6.Discher DE, Mooney DJ, Zandstra PW. Science. 2009;324:1673–1677. doi: 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fidler IJ. Differentiation. 2002;70:498–505. doi: 10.1046/j.1432-0436.2002.700904.x. [DOI] [PubMed] [Google Scholar]
- 8.Janmey PA, McCulloch CA. Annu. Rev. Biomed. Eng. 2007;9:1–34. doi: 10.1146/annurev.bioeng.9.060906.151927. [DOI] [PubMed] [Google Scholar]
- 9.Davies PF. Physiol. Rev. 1995;75:519–560. doi: 10.1152/physrev.1995.75.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. New York: Garland Science; 2002. [Google Scholar]
- 11.Keenan TM, Folch A. Lab Chip. 2008;8:34–57. doi: 10.1039/b711887b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flaim CJ, Teng D, Chien S, Bhatia SN. Stem Cells Dev. 2008;17:29–39. doi: 10.1089/scd.2007.0085. [DOI] [PubMed] [Google Scholar]
- 13.Rofstad EK. Int. J. Radiat. Biol. 2000;76:589–605. doi: 10.1080/095530000138259. [DOI] [PubMed] [Google Scholar]
- 14.Landecker H. Culturing Life: How Cells Became Technologies. Harvard University Press; 2007. [Google Scholar]
- 15.Cressey D. Nat. Med. 2008;14:1292–1292. [Google Scholar]
- 16.Freshney RI. Culture of Animal Cells: A Manual of Basic Technique. Wiley-Liss; 2005. [Google Scholar]
- 17.Bissell MJ, Radisky DC, Rizki A, Weaver VM, Petersen OW. Differentiation. 2002;70:537–546. doi: 10.1046/j.1432-0436.2002.700907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LaBarge MA, Nelson CM, Villadsen R, Fridriksdottir A, Ruth JR, Stampfer MR, Petersen OW, Bissell MJ. Integr. Biol. 2009;1:70–79. doi: 10.1039/b816472j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitesides GM, Ostuni E, Takayama S, Jiang XY, Ingber DE. Annu. Rev. Biomed. Eng. 2001;3:335–373. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- 20.Unger MA, Chou HP, Thorsen T, Scherer A, Quake SR. Science. 2000;288:113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- 21.Kim L, Toh YC, Voldman J, Yu H. Lab Chip. 2007;7:681–694. doi: 10.1039/b704602b. [DOI] [PubMed] [Google Scholar]
- 22.Regehr KJ, Domenech M, Koepsel JT, Carver KC, Ellison-Zelski SJ, Murphy WL, Schuler LA, Alarid ET, Beebe DJ. Lab Chip. 2009;9:2132–2139. doi: 10.1039/b903043c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paguirigan A, Beebe DJ. Integr. Biol. 2009;1:182–195. doi: 10.1039/b814565b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heckele M, Schomburg WK. J. Micromech. Microeng. 2004;14:R1–R14. [Google Scholar]
- 25.Mehta G, Lee J, Cha W, Tung YC, Linderman JJ, Takayama S. Anal. Chem. 2009;81:3714–3722. doi: 10.1021/ac802178u. [DOI] [PubMed] [Google Scholar]
- 26.Chen CS, Breslauer DN, Luna JI, Grimes A, Chin WC, Leeb LP, Khine M. Lab Chip. 2008;8:622–624. doi: 10.1039/b719029h. [DOI] [PubMed] [Google Scholar]
- 27.Sims CE, Allbritton NL. Lab Chip. 2007;7:423–440. doi: 10.1039/b615235j. [DOI] [PubMed] [Google Scholar]
- 28.Kim D, Lokuta MA, Huttenlocher A, Beebe DJ. Lab Chip. 2009;9:1797–1800. doi: 10.1039/b901613a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gervais T, El-Ali J, Gunther A, Jensen KF. Lab Chip. 2006;6:500–507. doi: 10.1039/b513524a. [DOI] [PubMed] [Google Scholar]
- 30.Gomez-Sjoberg R, Leyrat AA, Pirone DM, Chen CS, Quake SR. Anal. Chem. 2007;79:8557–8563. doi: 10.1021/ac071311w. [DOI] [PubMed] [Google Scholar]
- 31.Gu W, Zhu XY, Futai N, Cho BS, Takayama S. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15861–15866. doi: 10.1073/pnas.0404353101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berthier E, Beebe DJ. Lab Chip. 2007;7:1475–1478. doi: 10.1039/b707637a. [DOI] [PubMed] [Google Scholar]
- 33.Meyvantsson I, Warrick JW, Hayes S, Skoien A, Beebe DJ. Lab Chip. 2008;8:717–724. doi: 10.1039/b715375a. [DOI] [PubMed] [Google Scholar]
- 34.Chen IJ, Eckstein EC, Lindner E. Lab Chip. 2009;9:107–114. doi: 10.1039/b808660e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du Y, Shim J, Vidula M, Hancock MJ, Lo E, Chung BG, Borenstein JT, Khabiry M, Cropek DM, Khademhosseini A. Lab Chip. 2009;9:761–767. doi: 10.1039/b815990d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huh D, Fujioka H, Tung YC, Futai N, Paine R, Grotberg JB, Takayama S. Proc. Natl. Acad. Sci. U. S. A. 2007;104:18886–18891. doi: 10.1073/pnas.0610868104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimura H, Yamamoto T, Sakai H, Sakai Y, Fujii T. Lab Chip. 2008;8:741–746. doi: 10.1039/b717091b. [DOI] [PubMed] [Google Scholar]
- 38.Neeves KB, Diamond SL. Lab Chip. 2008;8:701–709. doi: 10.1039/b717824g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song JW, Cavnar SP, Walker AC, Luker KE, Gupta M, Tung Y-C, Luker GD, Takayama S. PLoS One. 2009;4:e5756. doi: 10.1371/journal.pone.0005756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Neill AT, Monteiro-Riviere NA, Walker GM. Lab Chip. 2009;9:1756–1762. doi: 10.1039/b819622b. [DOI] [PubMed] [Google Scholar]
- 41.Ong SM, Zhang C, Toh YC, Kim SH, Foo HL, Tan CH, van Noort D, Park S, Yu H. Biomaterials. 2008;29:3237–3244. doi: 10.1016/j.biomaterials.2008.04.022. [DOI] [PubMed] [Google Scholar]
- 42.Abhyankar VV, Toepke MW, Cortesio CL, Lokuta MA, Huttenlocher A, Beebe DJ. Lab Chip. 2008;8:1507–1515. doi: 10.1039/b803533d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skelley AM, Voldman J. Lab Chip. 2008;8:1733–1737. doi: 10.1039/b807037g. [DOI] [PubMed] [Google Scholar]
- 44.Heo YS, Cabrera LM, Song JW, Futai N, Tung YC, Smith GD, Takayama S. Anal. Chem. 2007;79:1126–1134. doi: 10.1021/ac061990v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berthier E, Warrick J, Yu H, Beebe DJ. Lab Chip. 2008;8:852–859. doi: 10.1039/b717422e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeng Y, Lee TS, Yu P, Roy P, Low HT. J. Biomech. Eng. 2006;128:185–193. doi: 10.1115/1.2170118. [DOI] [PubMed] [Google Scholar]
- 47.Yu HM, Meyvantsson I, Shkel IA, Beebe DJ. Lab Chip. 2005;5:1089–1095. doi: 10.1039/b504403k. [DOI] [PubMed] [Google Scholar]
- 48.Chung S, Sudo R, Mack PJ, Wan CR, Vickerman V, Kamm RD. Lab Chip. 2009;9:269–275. doi: 10.1039/b807585a. [DOI] [PubMed] [Google Scholar]
- 49.Huang CP, Lu J, Seon H, Lee AP, Flanagan LA, Kim HY, Putnam AJ, Jeon NL. Lab Chip. 2009;9:1740–1748. doi: 10.1039/b818401a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu HM, Alexander CM, Beebe DJ. Lab Chip. 2007;7:388–391. doi: 10.1039/b612358a. [DOI] [PubMed] [Google Scholar]
- 51.Puccinelli JP, Su XJ, Beebe DJ. J. Assoc. Lab. Autom. 2010;15:25–32. doi: 10.1016/j.jala.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bliss CL, McMullin JN, Backhouse CJ. Lab Chip. 2008;8:143–151. doi: 10.1039/b711601b. [DOI] [PubMed] [Google Scholar]
- 53.Spence DM, Torrence NJ, Kovarik ML, Martin RS. Analyst. 2004;129:995–1000. doi: 10.1039/b410547h. [DOI] [PubMed] [Google Scholar]
- 54.Pamme N. Lab Chip. 2006;6:24–38. doi: 10.1039/b513005k. [DOI] [PubMed] [Google Scholar]
- 55.Abdelgawad M, Watson MWL, Wheeler AR. Lab Chip. 2009;9:1046–1051. doi: 10.1039/b820682a. [DOI] [PubMed] [Google Scholar]
- 56.Yi CQ, Li CW, Ji SL, Yang MS. Anal. Chim. Acta. 2006;560:1–23. [Google Scholar]