Abstract
Innate immunity mechanisms play a critical role in the primary response to invading pathogenic microorganisms and other insulting agents. The innate lung immune system includes lung surfactant, a lipoprotein complex that carries out a function essential for life, that is, reduction of the surface tension at the air–liquid interphase of the alveolar space. By means of this function, pulmonary surfactant prevents lung collapse, therefore ensuring normal lung function and lung health. Pulmonary surfactant contains a number of host-defense molecules that are involved in the elimination of pathogens, viruses, particles, allergens, and other insults, as well as in the control of inflammation. This review is concerned with one of the surfactant proteins, the human (h) surfactant protein A (hSP-A), which, in addition to its role in surfactant-related functions, plays an important role in the modulation of lung host defense. The hSP-A locus has been identified with extensive complexity that may have an impact on its function, structure, and regulation. In humans, two genes—SP-A1 (SFTPA1) and SP-A2 (SFTPA2)—encode SP-A, with SP-A2 gene products being more biologically active than SP-A1 in most of the in vitro assays investigated. Although the two hSP-A genes share a high level of sequence similarity, differences in the structure and function between SP-A1 and SP-A2 have been observed in recent studies. In this review, we discuss the human SP-A complexity and how this may affect SP-A function.
Keywords: innate immunity, phagocytosis, genetic variants, post-translational modifications, structure–function correlations
I. INTRODUCTION
Pulmonary surfactant is essential for life. It lowers surface tension at the air–liquid interphase at the distal airspaces (or alveoli) of the lung and thus prevents alveolar (or lung) collapse at low lung volumes during respiration. This surfactant-mediated function, which ensures patency of distal airspaces, enables the lung to carry out its key function, the exchange of O2 and CO2. Details of the structure and function of pulmonary surfactant have been reviewed elsewhere.1 In brief, the alveolar epithelial surface of the lung is lined with a thin layer of liquid called hypophase. Lung surfactant is synthesized by alveolar epithelial type II cells, and it can be found in several structural forms. Structural forms relevant to this review are the lamellar bodies found intracellularly within the type II cells as well as extracellularly in the hypophase, where they may unravel to form another structural form of surfactant, the tubular myelin (TM). The function of TM is not entirely understood. Surfactant from the hypophase adsorbs at the air–liquid interphase to form a monolayer (and perhaps a multilayer), and by means of this structural form, surfactant exerts its key function (i.e., lowers surface tension and prevents alveolar collapse).
Pulmonary surfactant is biochemically complex consisting of lipids (~90%) and proteins (~10%)by weight (reviewed in Ref. 2). The major lipid component of surfactant is dipalmitoylphosphatidylcholine (DPPC), and the surfactant proteins (SP-) include SP-A (~5.3%), SP-B (~0.7%), SP-C (~0.4%), and SP-D (~0.6 %). The pulmonary surfactant proteins are involved in two broad groups of functions: the surfactant-related functions and the innate host defense functions of the lung. SP-B and SP-C are hydrophobic proteins and are primarily involved in the surface-tension-lowering function of surfactant.3-5 SP-A and SP-D are hydrophilic proteins primarily responsible for host defense–related functions of surfactant (reviewed in Refs. 6-11).
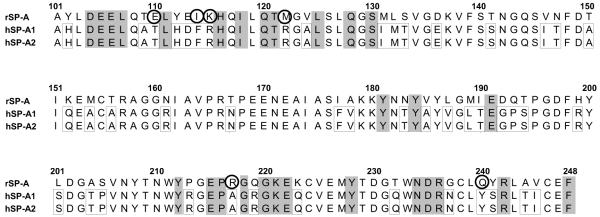
SP-A (the focus in this review) can bind DPPC12 and is involved in both host defense– and surfactant-related functions.6,8,13 In addition, SP-A has been shown to serve as a hormone in parturition through its ability to modulate proinflammatory cytokine production,14 as well as in several other functions (reviewed in Refs. 2, 6, 7, 9, 11, 13, 15-18), as depicted in Figure 1. SP-A as an innate immune molecule plays an important role in preserving lung health in the face of a considerable threat of invading or insulting agents, and perhaps in the everyday exposure to low levels of invading or insulting agents. The first line of lung innate immune defense consists of macrophages that eliminate harmful agents and produce a number of inflammatory mediators. The interactions of SP-A with the macrophage or other immune cells and its role in these and other processes, as well as the mechanisms involved, have been reviewed elsewhere.6,7,9,13,15-17,19
FIGURE 1.
Diagrammatic presentation of some of the major functions of SP-A (reviewed in Refs. 2, 6, 7, 9, 11, 13, 15-18). Examples of SP-A involvement in innate immunity, including its function in uptake and clearance of pathogens (bacteria, viruses), production of oxygen radicals, killing of bacteria, and inhibition of IAV infection; in inflammation, including its ability to modulate proinflammatory cytokine production and other anti-inflammatory processes; in adaptive immunity, including its ability to modulate dendritic cell maturation and T-cell proliferation; in surfactant structure and homeostasis, including its role in tubular myelin formation, and its ability to inhibit surfactant secretion; and in other functions, including its ability to clear apoptotic cells, modulate allergic reactions, and serve as a hormone in parturition.
Of interest, in general, as one ascends the mammalian evolutionary ladder, the genetic complexity of SP-A increases. For example, in rodents there is a single SP-A gene locus, and in primates a gene duplication event occurred generating two genes, SP-A1 and SP-A2. In humans, a more pronounced level of complexity evolved in the form of 5′ untranslated region (5′ UTR) splice variants and sequence variability. It has been postulated (reviewed in Ref. 20) that the increase in genetic complexity reflects the necessity to combat a wider variety of insulting or injurious agents as, perhaps, more physical distance is covered by mammals up the evolutionary ladder. Although the functional importance of this increased complexity of SP-A during evolution remains unknown, the question whether SP-A served initially as a host-defense molecule and subsequently was co-opted by the surfactant system to serve additional functions or vice versa remains an interesting one. Furthermore, the questions of whether the two human genes exhibit divergent functions and/or whether these are subject to differential regulation continue to be under active investigation. Below we provide an update of the genetic complexity of the human SP-A locus and review the functional differences between the two human genes.
II. GENETIC COMPLEXITY OF HUMAN SP-A
A. Genomic Locus
The genomic locus of human SP-A consists of two functional genes, SFTPA1 (or SP-A1) and SFTPA2 (or SP-A2), as well as a pseudogene.21 This locus is located on the long arm of chromosome 1022 and the two functional genes are in opposite transcriptional orientation.21 The NCBI GenBank database, last reviewed May 14, 2008, shows that there are two genetic loci for each SP-A1 (SFTPA1, SFTPA1B) (http://ncbi.nlm.nih.gov/projects/mapview/map_search.cgi?taxid=9606&build=current&advsrch=off&query=sftpa1) and SP-A2 (SFTPA2, SFTPA2B) (http://ncbi.nlm.nih.gov/projects/mapview/map_search.cgi?taxid=9606&build=current&advsrch=off&query=sftpa2) gene. However, the private genome database assembled by Celera indicates that each SP-A1 and SP-A2 is a single copy gene, and each is located between 74,655 kb and 74,600 kb (SP-A1) and between 74,600 kb and 74,606 kb (SP-A2) of chromosome 10 (http://www.ncbi.nlm.nih.gov/projects/mapview/map_search.cgi?taxid=9606&query=sftpa1&qchr=10&strain=Celera) (http://www.ncbi.nlm.nih.gov/projects/mapview/map_search.cgi?taxid=9606&query=sftpa2&qchr=10&strain=Celera). The Celera data of a single gene copy are consistent with previously published information.21-24 Moreover, analysis of the NCBI GenBank SP-A data indicates that both copies of each gene are nearly identical. For SP-A2, with the exception of a single nucleotide difference within intron I, no nucleotide differences are observed between the two copies of SP-A2 in a 4.5-kb genomic sequence. For SP-A1, with the exception of a single base difference in 3′UTR, no nucleotide differences are observed between the two SP-A1 copies. Also, the sequence of an about 3.8-kb region located between SP-A1 and the pseudogene is identical between the two corresponding sets of copies. This sequence identity between the two copies of each SP-A gene is difficult to explain (if correct), because it stands in contrast to all available relevant information indicating an extensive genetic variability among intragenic haplotypes (or variants) of each human SP-A gene (discussed below). Furthermore, the fact that the Celera genome database indicates that the human SP-A1 and SP-A2 genes are each a single copy (consistent with previous information),21-24 the possibility of an erroneous duplication in the NCBI database cannot be excluded at this point. The SP-A1 and SP-A2 intragenic haplotypes or variants (see below) of the NCBI database are 6A3 (SP-A1) and 1A0 (SP-A2), and those of the Celera database are 6A3 (SP-A1) and 1A2 (SP-A2), indicating that different genomic samples were used in the NCBI and Celera databases. The 6A3/1A0 intergenic haplotype has been observed previously,25 and both intergenic haplotypes 6A3/1A0 and 6A3/1A2 have been found in higher frequency in African Americans and Nigerians.26
Although all mammalian species studied to date have been shown to contain a single copy gene, except from primates onward where a gene duplication occurred giving rise to SP-A1 and SP-A2 genes, a recent report23 revealed additional SP-A sequences in some species. A phylogenetic analysis in which amino acid alignment of the surfactant proteins in birds and mammals was carried out, showed, in addition to providing strong support for the SP-A gene duplication in primates, the opossum genome to have three genes (SP-A1, SP-A2, SP-A3) and the chicken genome to have an SP-A and an SP-A-like gene.
B. Genetic and Splice Variants
As described above, the human SP-A locus consists of two functional genes, SP-A1 and SP-A2, and a pseudogene. On the basis of coding sequence differences, more than 30 variants or intragenic haplotypes27-29 have been collectively characterized, in part or fully, for the two human SP-A genes. Of these (Fig. 2A), four SP-A1 (6A, 6A2, 6A3, 6A4) and six SP-A2 variants (1A, 1A0, 1A1, 1A2, 1A3, 1A5) have been observed in higher frequency of > 0.01 in the general population.27,30 In addition to these coding sequence variants, nucleotide29,31,32 and/or splice28,33 differences within regulatory 3′ and 5′ untranslated regions (3′ UTR and 5′ UTR) between the SP-A1 and SP-A2 genes and/or variants have been identified and studied. Regarding splice variants, a number of untranslated exons (A, B, B’, C, C’, D, D’) have been characterized that splice in different configurations to give rise to several 5′ UTR splice variants (reviewed in Ref. 20). For SP-A1, the most frequently observed variant is the AD’, and for SP-A2, the ABD’ and ABD. SP-A1 and SP-A2 5′ UTR splice variants were recently shown to differentially affect mRNA stability and translation.33
FIGURE 2.
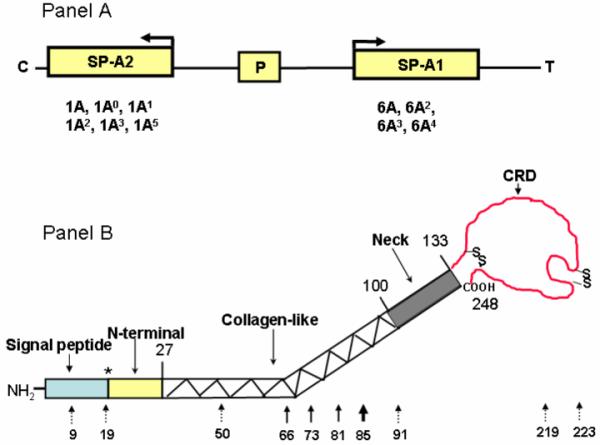
Schematic presentation of the human SP-A locus and the protein domains and genetic variations of SP-A. Panel A depicts the organization of the human SP-A locus (SP-A2, pseudogene [P], and SP-A1), the transcriptional orientation (arrow) of each gene, and the corresponding, more frequently observed coding variants below each gene. Panel B depicts the various protein domains of SP-A and the position of each of the 10 collective amino acid differences among the human SP-A variants (reviewed in Ref. 20). The number above the structure shows the numbers of the last amino acid of the SP-A precursor where the preceding domain ends. * denotes that the signal peptide could be 18, 19, or 20 amino acids. The number below the structure indicates the amino acid number of the SP-A precursor. Solid-line arrows denote the gene-specific differences that distinguish all SP-A1 variants from the SP-A2 variants. The dotted-line arrows show differences among variants of either gene.
C. Transcription Start Site
Differences between the transcription start sites of SP-A1 and SP-A2 have been described. Two groups have independently discovered alternative transcription start sites in human SP-A1 and SP-A2, and the results from both reports were obtained using primer extension.28,34 For hSP-A2 transcripts, a common transcript start site with an 5′-AACTTGGAGG -3′ terminal sequence was described.28,34 For SP-A1 transcripts, three start sites28 and two start sites34 were described. One of these transcript species was found in greater frequency in one of the studies34 and with no significant differences observed in the frequency of the 3 start sites in the other study.28 One study analyzed 100 positive clones,34 and the other analyzed 130 positive clones (87 SP-A2 and 43 SP-A1).28 The apparent discrepancy between the two reports about the SP-A1 transcription start sites may be due to variations in the individual samples from which the total lung RNA was prepared, the limited number of tested clones that may have precluded detection of less frequently observed transcripts in the particular sample, or due in part to both of these possibilities.
III. SP-A PROTEIN CHARACTERISTICS
Prior to 1988,35 SP-A was known by a variety of names, including glycoprotein, alveolyn, surfactant apoprotein, pulmonary surfactant-associated protein or PSAP, pulmonary surfactant protein A or PSP-A,36-43 as well as other variations of the above. SP-A is assembled as an octodecamer, consisting of six trimer subunits. Although it has been proposed that each trimer in humans is a heterotrimer consisting of two SP-A1 and one SP-A2 molecules,44 recent evidence of in vitro–45-52 and in vivo53–expressed SP-A variants indicate that homotrimers or homo-oligomers exist and/or that these are functional.
A. Structural Domains
SP-A, a member of the C-type collectin family, in addition to containing a signal peptide, consists of four domains, the N-terminal sequence, the collagen-like (G-X-Y) domain, the neck region, and the carbohydrate recognition domain (CRD) (reviewed in Refs. 20, 54) (Fig. 2B). The numbering of the amino acid positions in human SP-A in the present review is that of the precursor protein molecule (i.e., it includes the signal peptide sequence). For convenience, in most cases, the amino acid number of the precursor and that of the mature SP-A are shown. This cautionary note is made because the signal peptide in human (unlike in other species) can be 18, 19, or 20 amino acids long,49 which could make numbering amino acid positions rather complicated, if based on the sequence of the mature human SP-A. Therefore, in reviewing the literature caution should be exercised because there may be a difference in amino acid position by a factor of 20. For example, a key amino acid at position 85 (see below) may also be described as being located at position 65.
The most conservative of the four domains of SP-A is the neck and N-terminal regions. Amino acid differences that distinguish between SP-A1 and SP-A2 variants28 are located in the collagenlike domain (reviewed in Ref. 20). However, nucleotide differences that do or do not change the encoded amino acid among SP-A1 or SP-A2 variants are located within the sequence for the signal peptide, collagen-like region, and CRD.20 The collagen-like domain contains 23 GXY triplets and a sequence irregularity (PCPP) between triplets 13 and 14.52,55 A difference in triplet 18 (GEC for SP-A1 and GER for SP-A2) is postulated to contribute to differences in local structural stability between the two gene products.52 The human SP-A1 gene, as well as that in chimp and baboon, has an extra cysteine at position 85 (which is part of triplet 18 (GEC)) of the SP-A protein precursor. This extra Cys-85 may generate additional intermolecular disulfide bonds, and it can therefore influence the biochemical properties and/or biological function of SP-A1.55 Moreover, as noted above, the signal peptide can be cleaved at different amino acid locations, thereby generating three different N-termini in humans.49 Some of these N-terminal variants contain a cysteine. It is postulated that this cysteine plays a role in disulfide bond formation, and that by means of this, it may contribute to the oligomer structure of SP-A.49 The mechanism involved in this differential signal peptide cleavage, or the factors that may influence this process, are currently unknown. Moreover, Cys-26 (precursor or Cys-6 in mature protein) in the N-terminal sequence has been shown to be involved in the supratrimeric assembly of SP-A.56 When this cysteine was mutated to a serine, only SP-A trimers were detected. This mutated SP-A was more susceptible to trypsin degradation and exhibited reduced thermal stability in the collagen domain, but no changes were observed in its ability to modulate TNF-α production. The CRD consists of 115 amino acids, including four cysteines that are conserved from birds to mammals. These four cysteines within the CRD can form two pairs of disulfide bonds, which play a critical role in formation and stability of the SP-A conformation. Important amino acids in this domain that may contribute to its structure are discussed below.
B. Crystal Structure Analysis
Information from the crystal structure study of a recombinant rat SP-A fragment containing the neck and CRD57 revealed that the two domains are oriented virtually perpendicular to one another (“T”) and that this is primarily due to major intermolecular contacts between the two domains. In one case, Phe-228 (or phe-228+20 precursor) from one monomer makes contact with His-96 (or His-96+20 precursor) and Gln-100 (or Gln-100+20 precursor) of another monomer via the formation of a salt bridge and H-bond, respectively. These 3 residues are conserved in mammalian SP-A sequences, including all SP-A1 and SP-A2 variants, indicating that no differences in structure among various SP-As are expected to occur by means of these contacts.
A number of other amino acids were identified as important by analysis of the crystal structure, including those that form Ca2+-binding sites (Glu-195, Arg-197, Asn-214, and Asp-215 mature SP-A; the position of these amino acids in the SP-A precursor are the numbers indicated plus 20). These amino acids are the same in human SP-A1 and SP-A2, except for Arg-197 (+20 precursor), which is Ala in both human sequences. This change is not expected to alter the Ca2+-binding ability of the molecule.57 The rat SP-A amino acids shown by crystal structure analysis to play a role in SP-A structure are shown, along with the human SP-A1 and SP-A2 sequences, in Figure 3. Although out of the 34 amino acids identified as being important, only 6 differ between the rat and human neck and CRD, and the amino acids at these 6 locations do not differ between SP-A1 and SP-A2.
FIGURE 3.
Sequence alignment of neck and CRD of SP-A. The rat sequence of the neck and CRD region was expressed and used for crystal growth and analysis.57 The alignment of the 3 neck and CRD sequences shown consists of the rSP-A (top sequence), the hSP-A1 (middle), and the SP-A2 (bottom). The numbers on top indicate the amino acid residues of the SP-A precursor. The crystal structure analysis identified 34 residues as being important for SP-A structure. The shaded amino acid residues (n = 28) identify those amino acids out of the 34 significant ones that are identical among the 3 sequences. The 6 circled residues indicate important amino acids (as assessed by crystal structure analysis) that differ between the rat sequence and the 2 human sequences. In all these 6 amino acids, hSP-A1 and hSP-A2 are identical. Other amino acid differences between rat and human sequences (not identified as important by crystal structure analysis) are shown with a box around the human sequences.
In summary, the available crystal structure information of the neck and CRD does not reveal any differences between SP-A1 and SP-A2 that may explain their observed functional differences (see below).
C. SP-A1 and SP-A2 Protein Content in Lung Health and Disease
Although differences in SP-A1 and SP-A2 basal mRNA levels and in response to various hormones and to dexamethasone, in particular, have been observed in several studies,32,58-62 it has been challenging to determine if these mRNA differences reflect differences in the protein content of SP-A1, SP-A2, or both. Changes in total SP-A protein content, without distinction between SP-A1 and SP-A2 content or SP-A genotype among individuals, have been observed in several lung diseases18,63-65 or in physiological changes of lung activity.66 Determining gene-specific protein content in lung health and disease is very important, given the functional differences observed between the two human SP-A genes (see below). However, it has been difficult to do this due to the lack of necessary reagents. The two gene products are not only very similar in their amino acid sequence but the amino acid sequences that distinguish them are located within the collagen-like domain of the molecule. The difficulty in generating gene-specific antibodies (our unpublished observations) raises the possibility that the gene-specific residues within the triple helical collagen-like domain are not very accessible for antibody generation. However, with some recent success in generating an SP-A1-specific antibody (Ab),67 some interesting observations began to surface. Differences in the ratio of SP-A1 to total SP-A content in bronchoalveolar lavage fluids were observed as a function of age among healthy individuals as well as among individuals with various lung diseases. Although this SP-A1 Ab is a valuable reagent,68 the affinity of the SP-A1 Ab differs considerably from that of the SP-A Ab that recognizes both gene products,67 and it should therefore be used only as a tool to gain insight into ratios of SP-A1 to total SP-A under various conditions and not as an indicator of the SP-A1 protein concentration.
Whether and how the genetic and structural differences observed between SP-A1 and SP-A2 affect the function of SP-A1 and SP-A2 is not well understood. Below we discuss the available information on functional differences between SP-A1 and SP-A2 variants.
IV. QUALITATIVE DIFFERENCES BETWEEN SP-A1 AND SP-A2 AND/OR AMONG THEIR RESPECTIVE VARIANTS
A. Functional Differences
Qualitative differences between SP-A1 and SP-A2 in vitro–expressed variants include differences (see Table 1) in their ability to enhance phagocytosis by alveolar macrophages,47,69 enhance production of proinflammatory cytokines (TNF-α and IL-8) by macrophage-like THP-1 cells,45,46,70 and inhibit surfactant secretion by epithelial type II cells.49 Differences in their aggregation, structural stability, and oligomerization properties49,52,71,72; their sugar-binding characteristics73; and in their ability to form phospholipids monolayers48 have also been observed. In most of these assays, SP-A2 (regardless of the specific SP-A2 variant used in the assay) was shown to be more active than SP-A1 indicating that some fundamental differences may exist between SP-A1 and SP-A2. Moreover, the ability of SP-A2 to bind with higher affinity to a wider variety of sugars than SP-A173 indicates that the structural pattern or other attributes of SP-A2 make it more favorable for more diverse carbohydrate binding. Similar observations with regard to oligomerization patterns have been made with in vivo–expressed SP-A1 and SP-A2 variants in humanized transgenic mice.53 These in vivo experiments also showed that both SP-A1 and SP-A2 gene products are necessary for the formation of the extracellular structural form of surfactant, the tubular myelin (TM). The inability of either SP-A1 or SP-A2 alone to form TM and the need of both for formation is perhaps the first example of complete functional divergence of the two SP-A genes. The importance of TM has not yet been deciphered. TM has been detected in all species studied, regardless of whether these have a single gene copy (i.e., rodents) or two copies (i.e., humans) and has been shown in vitro to require, the presence of SP-A, SP-B, and simple lipid mixtures (dipalmitoylphosphatidylcholine and phosphatidylglycerol).74,75 Moreover, ozone-induced oxidation reduced the activity of SP-A1 and SP-A2.46,49,51,70 An inverse correlation was observed between ozone-induced oxidation or in vivo oxidation of SP-A and its ability to enhance phagocytosis by alveolar macrophages.51 SP-A2, albeit more active, may be more susceptible to oxidation.49,51 In summary, although it was noted initially that the trimeric form of the mature SP-A protein consists of two SP-A1 molecules and one SP-A2, the available data indicate that in vitro– or in vivo–expressed single gene products are functional, although with some functional and structural differences between the two gene products.
TABLE 1.
Differences Between SP-A1 and SP-A2
| Differences in | Activity | Refs. |
|---|---|---|
| †Cytokine (TNF-α) | Higher for SP-A2 (1A, 1A0, 1A1, 1A2) than SP-A1 (6A, 6A2, 6A4)* | 45 |
| †Cytokine (TNF-α, IL-8) | Higher for SP-A2 (1A, 1A0, 1A1) than SP-A1 (6A, 6A2, 6A4)** | 70 |
| Structure within the collagen domain | SP-A2 (1A2) more stable than SP-A1 (6A2, 6A4)* | 52 |
| Self-aggregation and LPS and phospholipid aggregation | Greater for SP-A2 (1A2, 1A1) than SP-A1 (6A2, 6A4)* | 52 |
| Oligomerization patterns | SP-A2 (1A2) and SP-A1 (6A2) differ in their oligomer pattern* | 52 |
| Carbohydrate-binding characteristics | SP-A2 (1A1) exhibits higher affinity to a wider variety of sugars than SP-A1 (6A2) | 73 |
| †Cytokine (TNF-α, IL-8) | Higher for SP-A2 (1A, 1A0) than SP-A1 (6A2, 6A4)* | 46 |
| Oligomerization patterns | SP-A2 (1A, 1A0) and SP-A1 (6A2, 6A4) differ in their oligomer pattern* | 46 |
| ‡Inhibition of ATP-stimulated phosphatidlycholine | Higher for SP-A2 (1A0) than SP-A2 (6A2)* or ** | 49 |
| Oligomerization patterns | Higher order oligomers for SP-A1 (6A, 6A2) than SP-A2 (1A, 1A0, 1A1)** | 49 |
| $Phagocytosis of P. aeruginosa | Higher for SP-A2 (1A, 1A0) than SP-A1 (6A2, 6A4)* | 47 |
| $Phagocytosis of P. aeruginosa | Higher for SP-A2 (1A0, 1A1) than SP-A2 (6A2, 6A4)** | 69 |
| #Phagocytosis of P. aeruginosa | Higher for SP-A2 (1A0) than SP-A2 (6A2)** | 69 |
| Re-LPS aggregation and bacterial P. aeruginosa aggregation | Higher for SP-A2 (1A0) than SP-A1 (6A2)** | 55 |
| $Phagocytosis of P. aeruginosa | Higher for SP-A2 (1A0) than SP-A1 (6A2)** | 55 |
| Oligomerization patterns | Higher size oligomers for SP-A1 (6A2) than SP-A2 (1A0)** | 55 |
| The characteristics of phospholipid monolayers containing SP-B | SP-A2 (1A0) and SP-A2 (6A2) differ in monolayer characteristics** | 48 |
| Susceptibility to trypsin degradation | SP-A more resistant than SP-A2* | 72 |
| Self-aggregation and LPS and phospholipid aggregation | Higher for SP-A2 than SP-A1* | 72 |
| Oligomerization patterns | SP-A1 and SP-A2 differ in their oligomer pattern* | 72 |
| Inhibition of hemaglutination activity of Influenza A | No differences between SP-A2 (1A0) and SP-A1 (6A2)** | 50 |
| Inhibition of hemaglutination activity of Influenza A | No activity for SP-A2 (1A0) and SP-A1 (6A2)* | 50 |
| $Phagocytosis of P. aeruginosa | Higher activity for SP-A2 (1A0) than SP-A1 (6A4)**Higher activity for SP-A2 (1A) than SP-A1 (6A4)* |
51 |
| ΔStructure of surfactant | Both SP-A1 (6A4) and SP-A2 (1A3) gene products are needed for the formation of tubular myelin |
53 |
| ΔOligomerization patterns | SP-A1 (6A4) AND SP-A2 (1A3) differ in their oligomer pattern | 53 |
Note:
Production by a macrophage-like cell line THP-1
production by alveolar type II cells
insect-expressed variants
mammalian-expressed variants
by alveolar rat macrophages
by human alveolar macrophages.
This is an in vivo study of humanized transgenic mice generated in the SP-A −/− mouse background.
B. Post-translational Modifications and Function
Although post-translational modifications positively affect SP-A function (i.e., phagocytosis, inhibition of surfactant secretion, cytokine production), they are not responsible for the observed differences between SP-A1 and SP-A2 because these differences are maintained regardless of the presence49,69 or absence45-47,49,52 of post-translational modifications. For example, both insect cell–derived47 and mammalian (CHO) cell–derived69 SP-A2 variants enhanced bacterial phagocytosis by either rat47,69 or human69 alveolar macrophages at higher levels than SP-A1, although a considerably higher protein concentration was required for the insect cell–expressed proteins. On the basis of the above observations, the “core” gene-specific amino acid differences between the two genes located within the collagen-like domain are likely to be responsible for such differences. These may differentially affect function either directly or indirectly (i.e., by affecting structure). But, if post-translational modifications are the binding sites of ligands, as is the case in the inhibition of the hemagglutination activity of influenza virus,50 insect cell–expressed SP-As exhibited no activity, and mammalian-expressed SP-As, although active, showed no gene-specific differences.50 The functional activity of SP-A1 and SP-A2 with respect to IAV appears to depend principally on the type of sialic acid linkage and not on sequence or oligomerization patterns of SP-A1 and SP-A2.
C. Amino Acid Differences and Structure/Function
Amino acids identified as being important in SP-A structure through the crystal structure analysis of an SP-A fragment consisting of the neck and CRD are similar between SP-A1 and SP-A2, and, therefore, the residues in neck and CRD are not expected to contribute to structural or functional differences between the two hSP-A genes. Therefore, the observed functional differences between SP-A1 and SP-A2 are likely due to direct and/or indirect influences of the core amino acid differences within the collagen-like region that distinguish the two genes and their respective variants. One of the key differences between SP-A1 and SP-A2 is amino acid 85 (precursor numbering), where SP-A1 has a cysteine (Cys-85) and SP-A2 has an arginine (Arg-85). The additional Cys-85 in SP-A1 may be involved in the formation of an SP-A intertrimeric or intratrimeric disulfide bond and account, in part, for the observed oligomerization pattern differences between SP-A1 and SP-A2 variants.49 These, in turn, may account for the observed SP-A1 and SP-A2 functional differences. In fact, a substitution of Arg-85 to Cys-85 in SP-A2 resulted in a functional activity similar to that observed in SP-A1, and a substitution of Cys-85 to Arg-85 in SP-A1 changed its activity to one similar to that of SP-A2.55 Alternatively, the Cys-85/Arg-85 residue may alter the stability of SP-A1 and SP-A252; structural differences, in turn, may have an impact on the functional capabilities not only of the collagen-like region where amino acid 85 resides but also of the CRD region. In summary, the available information supports the notion that differences within the collagen-like domain play a fundamental role in the observed functional differences between SP-A1 and SP-A2 most likely by affecting the structure and subsequently the function of the two SP-A gene products.
D. Differences among Gene-Specific Variants
Differences among SP-A1 or SP-A2 variants have been observed. These include differences (1) in the ability of insect cell–expressed45 SP-A2 variants (1A2 vs 1A or 1A0) or mammalian cell–expressed70 SP-A1 variants (6A4 vs 6A or 6A2) to enhance TNF-α production by a macrophage-like cell line; (2) in the ability of insect cell–expressed47 SP-A1 (6A2, 6A4) and SP-A2 (1A, 1A0), or mammalian cell–expressed69 SP-A1 (6A2, 6A4) and SP-A2 (1A0, 1A1) to enhance phagocytosis by alveolar macrophages; (3) in the ability of insect cell–expressed52 SP-A1 (6A2, 6A4) and SP-A2 (1A0, 1A2) to aggregate LPS. The differences among SP-A1 or SP-A2 variants may be due entirely or in part to differences other than the “core” amino acids that distinguish the two gene products. For example, a single amino acid difference (Arg-219/Trp-219) in truncated SP-A1 variants (6A2 and 6A4) altered the behavior of these two molecules.71
However, if post-translational modifications are partial, the function between two gene-specific variants may differ compared to that of the same variants but with complete post-translational modification. For example, for the insect cell–expressed SP-A1 variants, the ability to enhance phagocytosis was higher for the 6A4 (6A4 > 6A2) variant,47 whereas for the mammalian cell–expressed SP-A1, the activity was higher for the 6A2 variant (6A2 > 6A4).69 This may indicate that post-translational modifications are critical for the function of the 6A2 SP-A variant, and that in addition to sequence-dependent differences between variants, post-translational modifications may contribute to variant functional differences.
Of interest, although differences may exist among SP-A1 variants, SP-A2 gene-specific variants as noted above, were uniformly more active than SP-A1 in most of the activities investigated. However, in spite of recent advances, the nature or mechanisms responsible for the functional differences among variants of a given SP-A gene (1 or 2) are not well understood.
V. COMMENTS AND FUTURE DIRECTIONS
Although there is evidence that the human SP-A complexity is associated with differences in function and structure of SP-A1 and SP-A2, there is much to be learned about the mechanisms by which these occur. Humanized transgenic mice may provide good in vivo models to study functional differences between SP-A1 and SP-A2 under normal and perturbed conditions. Each of these mice contains a different human SP-A variant53 (the mouse SP-A gene is ablated) and, as such, may also provide the opportunity to gain insight into the basis of individual variability to disease susceptibility in response to a number of injurious agents. Mice carrying different SP-A variants could be studied by means of discovery proteomics, genomics, or other approaches to identify, for example, differential responses to environmental pollutants and/or infectious agents of one mouse type from another. Moreover, attempts to understand the structure of hSP-A1 and hSP-A2 from an evolutionary point of view could be another future endeavor.
We speculate that, given the differences in function between SP-A1 and SP-A2 and/or among gene-specific variants, the overall SP-A functional activity in the lung depends on the relative levels or the ratio of SP-A1 to SP-A2 rather than the total SP-A content (i.e., without regard to the SP-A1 and SP-A2 proportions), the status of each SP-A molecule (i.e., whether this is devoid of harmful modifications such as oxidation), and/or the genotype. We further speculate that a derangement in the regulation of SP-A1 or SP-A2 gene expression could result in an “inadequate” (or “unfavorable”) SP-A1 to SP-A2 ratio for normal host defense, and that this putative functional compromise contributes to lung disease severity.
ACKNOWLEDGMENTS
The authors thank Julie Graham for typing. This work was supported by the National Institutes of Health (NIH) grant no. HL 34788 and National Institutes of Environmental Health Science (NIEHS) grant no. ES009882.
REFERENCES
- 1.Floros J, Phelps DS. Pulmonary surfactant. In: Biebuck J, Lynch C III, Maze M, Saidman LJ, Yaksh TL, Zapol WM, editors. Anesthesia: biologic foundations. Lippincott-Raven; Philadelphia: 1997. pp. 1259–79. [Google Scholar]
- 2.Hawgood S, Poulain FR. The pulmonary collectins and surfactant metabolism. Annu Rev Physiol. 2001;63:495–519. doi: 10.1146/annurev.physiol.63.1.495. [DOI] [PubMed] [Google Scholar]
- 3.Clark JC, Weaver TE, Iwamoto HS, Ikegami M, Jobe AH, Hull WM, Whitsett JA. Decreased lung compliance and air trapping in heterozygous SP-B-deficient mice. Am J Respir Cell Mol Biol. 1997;16(1):46–52. doi: 10.1165/ajrcmb.16.1.8998078. [DOI] [PubMed] [Google Scholar]
- 4.Clark JC, Wert SE, Bachurski CJ, Stahlman MT, Stripp BR, Weaver TE, Whitsett JA. Targeted disruption of the surfactant protein B gene disrupts surfactant homeostasis, causing respiratory failure in newborn mice. Proc Natl Acad Sci U S A. 1995;92(17):7794–8. doi: 10.1073/pnas.92.17.7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glasser SW, Burhans MS, Korfhagen TR, Na CL, Sly PD, Ross GF, Ikegami M, Whitsett JA. Altered stability of pulmonary surfactant in SP-C-deficient mice. Proc Natl Acad Sci U S A. 2001;98(11):6366–71. doi: 10.1073/pnas.101500298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Floros J, Phelps DS. Pulmonary surfactant protein A; structure, expression, and its role in innate host defense. In: Nakos G, editor. Surfactant-update of intensive care medicine. University of Ioannina; Greece: 2002. pp. 87–102. [Google Scholar]
- 7.Kishore U, Greenhough TJ, Waters P, Shrive AK, Ghai R, Kamran MF, Bernal AL, Reid KB, Madan T, Chakraborty T. Surfactant proteins SP-A and SP-D: structure, function and receptors. Mol Immunol. 2006;43(9):1293–315. doi: 10.1016/j.molimm.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 8.Pastva AM, Wright JR, Williams KL. Immunomodulatory roles of surfactant proteins A and D: implications in lung disease. Proc Am Thorac Soc. 2007;4(3):252–7. doi: 10.1513/pats.200701-018AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sano H, Kuroki Y. The lung collectins, SP-A and SP-D, modulate pulmonary innate immunity. Mol Immunol. 2005;42(3):279–87. doi: 10.1016/j.molimm.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 10.van de Wetering JK, van Golde LM, Batenburg JJ. Collectins: players of the innate immune system. Eur J Biochem. 2004;271(7):1229–49. doi: 10.1111/j.1432-1033.2004.04040.x. [DOI] [PubMed] [Google Scholar]
- 11.Wright JR. Immunoregulatory functions of surfactant proteins. Nat Rev Immunol. 2005;5(1):58–68. doi: 10.1038/nri1528. [DOI] [PubMed] [Google Scholar]
- 12.Kuroki Y, Akino T. Pulmonary surfactant protein A (SP-A) specifically binds dipalmitoylphosphatidylcholine. J Biol Chem. 1991;266(5):3068–73. [PubMed] [Google Scholar]
- 13.Phelps DS. Surfactant regulation of host defense function in the lung: a question of balance. Pediatr Pathol Mol Med. 2001;20(4):269–92. [PubMed] [Google Scholar]
- 14.Condon JC, Jeyasuria P, Faust JM, Mendelson CR. Surfactant protein secreted by the maturing mouse fetal lung acts as a hormone that signals the initiation of parturition. Proc Natl Acad Sci U S A. 2004;101(14):4978–83. doi: 10.1073/pnas.0401124101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crouch E, Wright JR. Surfactant proteins A and D and pulmonary host defense. Annu Rev Physiol. 2001;63:521–54. doi: 10.1146/annurev.physiol.63.1.521. [DOI] [PubMed] [Google Scholar]
- 16.Lawson PR, Reid KB. The roles of surfactant proteins A and D in innate immunity. Immunol Rev. 2000;173:66–78. doi: 10.1034/j.1600-065x.2000.917308.x. [DOI] [PubMed] [Google Scholar]
- 17.Kingma PS, Whitsett JA. In defense of the lung: surfactant protein A and surfactant protein D. Curr Opin Pharmacol. 2006;6(3):277–83. doi: 10.1016/j.coph.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Griese M, Essl R, Schmidt R, Rietschel E, Ratjen F, Ballmann M, Paul K. Pulmonary surfactant, lung function, and endobronchial inflammation in cystic fibrosis. Am J Respir Crit Care Med. 2004;170(9):1000–5. doi: 10.1164/rccm.200405-575OC. [DOI] [PubMed] [Google Scholar]
- 19.Takahashi H, Sano H, Chiba H, Kuroki Y. Pulmonary surfactant proteins A and D: innate immune functions and biomarkers for lung diseases. Curr Pharm Des. 2006;12(5):589–98. doi: 10.2174/138161206775474387. [DOI] [PubMed] [Google Scholar]
- 20.Floros J, Wang G, Lin Z. Genetic diversity of human SP-A, a molecule with innate host defence and surfactant-related functions; characteristics, primary function, and significance. Curr Pharmacogenomics. 2005;3:87–95. [Google Scholar]
- 21.Hoover RR, Floros J. Organization of the human SP-A and SP-D loci at 10q22-q23. Physical and radiation hybrid mapping reveal gene order and orientation. Am J Respir Cell Mol Biol. 1998;18(3):353–62. doi: 10.1165/ajrcmb.18.3.3035. [DOI] [PubMed] [Google Scholar]
- 22.Bruns G, Stroh H, Veldman GM, Latt SA, Floros J. The 35 kd pulmonary surfactant-associated protein is encoded on chromosome 10. Hum Genet. 1987;76(1):58–62. doi: 10.1007/BF00283051. [DOI] [PubMed] [Google Scholar]
- 23.Hughes AL. Evolution of the lung surfactant proteins in birds and mammals. Immunogenetics. 2007;59(7):565–72. doi: 10.1007/s00251-007-0218-6. [DOI] [PubMed] [Google Scholar]
- 24.Kolble K, Lu J, Mole SE, Kaluz S, Reid KB. Assignment of the human pulmonary surfactant protein D gene (SFTP4) to 10q22-q23 close to the surfactant protein A gene cluster. Genomics. 1993;17(2):294–8. doi: 10.1006/geno.1993.1324. [DOI] [PubMed] [Google Scholar]
- 25.Floros J, DiAngelo S, Koptides M, Karinch AM, Rogan PK, Nielsen H, Spragg RG, Watterberg K, Deiter G. Human SP-A locus: allele frequencies and linkage disequilibrium between the two surfactant protein A genes. Am J Respir Cell Mol Biol. 1996;15(4):489–98. doi: 10.1165/ajrcmb.15.4.8879183. [DOI] [PubMed] [Google Scholar]
- 26.Liu W, Bentley CM, Floros J. Study of human SP-A, SP-B and SP-D loci: allele frequencies, linkage disequilibrium and heterozygosity in different races and ethnic groups. BMC Genet. 2003;4(1):13. doi: 10.1186/1471-2156-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DiAngelo S, Lin Z, Wang G, Phillips S, Ramet M, Luo J, Floros J. Novel, non-radioactive, simple and multiplex PCR-cRFLP methods for genotyping human SP-A and SP-D marker alleles. Dis Markers. 1999;15(4):269–81. doi: 10.1155/1999/961430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karinch AM, Floros J. 5′ splicing and allelic variants of the human pulmonary surfactant protein A genes. Am J Respir Cell Mol Biol. 1995;12(1):77–88. doi: 10.1165/ajrcmb.12.1.7811473. [DOI] [PubMed] [Google Scholar]
- 29.Rishi A, Hatzis D, McAlmon K, Floros J. An allelic variant of the 6A gene for human surfactant protein A. Am J Physiol Lung Cell Mol Physiol. 1992;262(5 Pt 1):L566–73. doi: 10.1152/ajplung.1992.262.5.L566. [DOI] [PubMed] [Google Scholar]
- 30.Floros J, Hoover RR. Genetics of the hydrophilic surfactant proteins A and D. Biochim Biophys Acta. 1998;1408(2–3):312–22. doi: 10.1016/s0925-4439(98)00077-5. [DOI] [PubMed] [Google Scholar]
- 31.Krizkova L, Sakthivel R, Olowe SA, Rogan PK, Floros J. Human SP-A: genotype and single-strand conformation polymorphism analysis. Am J Physiol Lung Cell Mol Physiol. 1994;266(5 Pt 1):L519–27. doi: 10.1152/ajplung.1994.266.5.L519. [DOI] [PubMed] [Google Scholar]
- 32.Wang G, Guo X, Floros J. Human SP-A 3′-UTR variants mediate differential gene expression in basal levels and in response to dexamethasone. Am J Physiol Lung Cell Mol Physiol. 2003;284(5):L738–48. doi: 10.1152/ajplung.00375.2002. [DOI] [PubMed] [Google Scholar]
- 33.Wang G, Guo X, Floros J. Differences in the translation efficiency and mRNA stability mediated by 5′-UTR splice variants of human SP-A1 and SP-A2 genes. Am J Physiol Lung Cell Mol Physiol. 2005;289(3):L497–508. doi: 10.1152/ajplung.00100.2005. [DOI] [PubMed] [Google Scholar]
- 34.McCormick SM, Boggaram V, Mendelson CR. Characterization of mRNA transcripts and organization of human SP-A1 and SP-A2 genes. Am J Physiol Lung Cell Mol Physiol. 1994;266(4 Pt 1):L354–66. doi: 10.1152/ajplung.1994.266.4.L354. [DOI] [PubMed] [Google Scholar]
- 35.Possmayer F. A proposed nomenclature for pulmonary surfactant-associated proteins. Am Rev Respir Dis. 1988;138(4):990–8. doi: 10.1164/ajrccm/138.4.990. [DOI] [PubMed] [Google Scholar]
- 36.Passero MA, Tye RW, Kilburn KH, Lynn WS. Isolation and characterization of two glycoproteins from patients with alveolar proteinosis. Proc Natl Acad Sci U S A. 1973;70(4):973–6. doi: 10.1073/pnas.70.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhattacharyya SN, Sahu S, Lynn WS. Structural studies on a glycoprotein isolated from alveoli of patients with alveolar proteinosis. Biochim Biophys Acta. 1976;427(1):91–106. doi: 10.1016/0005-2795(76)90288-9. [DOI] [PubMed] [Google Scholar]
- 38.King RJ, Ruch J, Gikas EG, Platzker AC, Creasy RK. Appearance of apoproteins of pulmonary surfactant in human amniotic fluid. J Appl Physiol. 1975;39(5):735–41. doi: 10.1152/jappl.1975.39.5.735. [DOI] [PubMed] [Google Scholar]
- 39.Metcalfe IL, Enhorning G, Possmayer F. Pulmonary surfactant-associated proteins: their role in the expression of surface activity. J Appl Physiol. 1980;49(1):34–41. doi: 10.1152/jappl.1980.49.1.34. [DOI] [PubMed] [Google Scholar]
- 40.Whitsett JA, Ross G, Weaver T, Rice W, Dion C, Hull W. Glycosylation and secretion of surfactant-associated glycoprotein A. J Biol Chem. 1985;260(28):15273–9. [PubMed] [Google Scholar]
- 41.Lynn WS. Alveolyn—structure and source: a review. Exp Lung Res. 1984;6(3–4):191–6. doi: 10.3109/01902148409109247. [DOI] [PubMed] [Google Scholar]
- 42.Floros J, Steinbrink R, Jacobs K, Phelps D, Kriz R, Recny M, Sultzman L, Jones S, Taeusch HW, Frank HA, Fritsch EF. Isolation and characterization of cDNA clones for the 35-kDa pulmonary surfactant-associated protein. J Biol Chem. 1986;261(19):9029–33. [PubMed] [Google Scholar]
- 43.Floros J, Phelps DS, Kourembanas S, Taeusch HW. Primary translation products, biosynthesis, and tissue specificity of the major surfactant protein in rat. J Biol Chem. 1986;261(2):828–31. [PubMed] [Google Scholar]
- 44.Voss T, Melchers K, Scheirle G, Schafer KP. Structural comparison of recombinant pulmonary surfactant protein SP-A derived from two human coding sequences: implications for the chain composition of natural human SP-A. Am J Respir Cell Mol Biol. 1991;4(1):88–94. doi: 10.1165/ajrcmb/4.1.88. [DOI] [PubMed] [Google Scholar]
- 45.Wang G, Phelps DS, Umstead TM, Floros J. Human SP-A protein variants derived from one or both genes stimulate TNF-alpha production in the THP-1 cell line. Am J Physiol Lung Cell Mol Physiol. 2000;278(5):L946–54. doi: 10.1152/ajplung.2000.278.5.L946. [DOI] [PubMed] [Google Scholar]
- 46.Huang W, Wang G, Phelps DS, Al-Mondhiry H, Floros J. Human SP-A genetic variants and bleomycin-induced cytokine production by THP-1 cells: effect of ozone-induced SP-A oxidation. Am J Physiol Lung Cell Mol Physiol. 2004;286(3):L546–53. doi: 10.1152/ajplung.00267.2003. [DOI] [PubMed] [Google Scholar]
- 47.Mikerov AN, Umstead TM, Huang W, Liu W, Phelps DS, Floros J. SP-A1 and SP-A2 variants differentially enhance association of Pseudomonas aeruginosa with rat alveolar macrophages. Am J Physiol Lung Cell Mol Physiol. 2005;288(1):L150–8. doi: 10.1152/ajplung.00135.2004. [DOI] [PubMed] [Google Scholar]
- 48.Wang G, Taneva S, Keough KM, Floros J. Differential effects of human SP-A1 and SP-A2 variants on phospholipid monolayers containing surfactant protein B. Biochim Biophys Acta. 2007;1768(9):2060–9. doi: 10.1016/j.bbamem.2007.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang G, Bates-Kenney SR, Tao JQ, Phelps DS, Floros J. Differences in biochemical properties and in biological function between human SP-A1 and SP-A2 variants, and the impact of ozone-induced oxidation. Biochemistry. 2004;43(14):4227–39. doi: 10.1021/bi036023i. [DOI] [PubMed] [Google Scholar]
- 50.Mikerov AN, White M, Hartshorn K, Wang G, Floros J. Inhibition of hemagglutination activity of influenza A viruses by SP-A1 and SP-A2 variants expressed in CHO cells. Med Microbiol Immunol. 2008;197(1):9–12. doi: 10.1007/s00430-007-0051-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mikerov AN, Umstead TM, Gan X, Huang W, Guo X, Wang G, Phelps DS, Floros J. Impact of ozone exposure on the phagocytic activity of human surfactant protein A (SP-A) and SP-A variants. Am J Physiol Lung Cell Mol Physiol. 2008;294(1):L121–30. doi: 10.1152/ajplung.00288.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garcia-Verdugo I, Wang G, Floros J, Casals C. Structural analysis and lipid-binding properties of recombinant human surfactant protein a derived from one or both genes. Biochemistry. 2002;41(47):14041–53. doi: 10.1021/bi026540l. [DOI] [PubMed] [Google Scholar]
- 53.Wang G, Guo X, DiAngelo S, Floros J. Characterization of humanized transgenic hSP-A mice: formation of tubular myelin in vivo requires both human SP-A1 and SP-A2 gene products. Am J Respir Crit Care Med. 2008;177:A318. [Google Scholar]
- 54.Floros J, Wang G. A point of view: quantitative and qualitative imbalance in disease pathogenesis; pulmonary surfactant protein A genetic variants as a model. Comp Biochem Physiol A Mol Integr Physiol. 2001;129(1):295–303. doi: 10.1016/s1095-6433(01)00325-7. [DOI] [PubMed] [Google Scholar]
- 55.Wang G, Myers C, Mikerov A, Floros J. Effect of cysteine 85 on biochemical properties and biological function of human surfactant protein A variants. Biochemistry. 2007;46(28):8425–35. doi: 10.1021/bi7004569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanchez-Barbero F, Strassner J, Garcia-Canero R, Steinhilber W, Casals C. Role of the degree of oligomerization in the structure and function of human surfactant protein A. J Biol Chem. 2005;280(9):7659–70. doi: 10.1074/jbc.M410266200. [DOI] [PubMed] [Google Scholar]
- 57.Head JF, Mealy TR, McCormack FX, Seaton BA. Crystal structure of trimeric carbohydrate recognition and neck domains of surfactant protein A. J Biol Chem. 2003;278(44):43254–60. doi: 10.1074/jbc.M305628200. [DOI] [PubMed] [Google Scholar]
- 58.McCormick SM, Mendelson CR. Human SP-A1 and SP-A2 genes are differentially regulated during development and by cAMP and glucocorticoids. Am J Physiol Lung Cell Mol Physiol. 1994;266(4 Pt 1):L367–74. doi: 10.1152/ajplung.1994.266.4.L367. [DOI] [PubMed] [Google Scholar]
- 59.Hoover RR, Floros J. SP-A 3′-UTR is involved in the glucocorticoid inhibition of human SP-A gene expression. Am J Physiol Lung Cell Mol Physiol. 1999;276(6 Pt 1):L917–24. doi: 10.1152/ajplung.1999.276.6.L917. [DOI] [PubMed] [Google Scholar]
- 60.Karinch AM, Deiter G, Ballard PL, Floros J. Regulation of expression of human SP-A1 and SP-A2 genes in fetal lung explant culture. Biochim Biophys Acta. 1998;1398(2):192–202. doi: 10.1016/s0167-4781(98)00047-5. [DOI] [PubMed] [Google Scholar]
- 61.Kumar AR, Snyder JM. Differential regulation of SP-A1 and SP-A2 genes by cAMP, glucocorticoids, and insulin. Am J Physiol. 1998;274(2 Pt 1):L177–85. doi: 10.1152/ajplung.1998.274.2.L177. [DOI] [PubMed] [Google Scholar]
- 62.Scavo LM, Ertsey R, Gao BQ. Human surfactant proteins A1 and A2 are differentially regulated during development and by soluble factors. Am J Physiol Lung Cell Mol Physiol. 1998;275(4 Pt 1):L653–69. doi: 10.1152/ajplung.1998.275.4.L653. [DOI] [PubMed] [Google Scholar]
- 63.Takahashi H, Honda Y, Kuroki Y, Imai K, Abe S. Pulmonary surfactant protein A: a serum marker of pulmonary fibrosis in patients with collagen vascular diseases [letter] Clin Chim Acta. 1995;239(2):213–5. doi: 10.1016/0009-8981(95)06118-w. [DOI] [PubMed] [Google Scholar]
- 64.Honda Y, Takahashi H, Kuroki Y, Akino T, Abe S. Decreased contents of surfactant proteins A and D in BAL fluids of healthy smokers. Chest. 1996;109(4):1006–9. doi: 10.1378/chest.109.4.1006. [DOI] [PubMed] [Google Scholar]
- 65.Postle AD, Mander A, Reid KB, Wang JY, Wright SM, Moustaki M, Warner JO. Deficient hydrophilic lung surfactant proteins A and D with normal surfactant phospholipid molecular species in cystic fibrosis. Am J Respir Cell Mol Biol. 1999;20(1):90–8. doi: 10.1165/ajrcmb.20.1.3253. [DOI] [PubMed] [Google Scholar]
- 66.Doyle IR, Jones ME, Barr HA, Orgeig S, Crockett AJ, McDonald CF, Nicholas TE. Composition of human pulmonary surfactant varies with exercise and level of fitness. Am J Respir Crit Care Med. 1994;149(6):1619–27. doi: 10.1164/ajrccm.149.6.8004321. [DOI] [PubMed] [Google Scholar]
- 67.Tagaram HR, Wang G, Umstead TM, Mikerov AN, Thomas NJ, Graff GR, Hess JC, Thomassen MJ, Kavuru MS, Phelps DS, Floros J. Characterization of a human surfactant protein A1 (SP-A1) gene-specific antibody; SP-A1 content variation among individuals of varying age and pulmonary health. Am J Physiol Lung Cell Mol Physiol. 2007;292(5):L1052–63. doi: 10.1152/ajplung.00249.2006. [DOI] [PubMed] [Google Scholar]
- 68.Oberley RE, George CL, Snyder JM. A new tool to investigate differences between human SP-A1 and SP-A2. Am J Physiol Lung Cell Mol Physiol. 2007;292(5):L1050–1. doi: 10.1152/ajplung.00039.2007. [DOI] [PubMed] [Google Scholar]
- 69.Mikerov AN, Wang G, Umstead TM, Zacharatos M, Thomas NJ, Phelps DS, Floros J. Surfactant protein A2 (SP-A2) variants expressed in CHO cells stimulate phagocytosis of Pseudomonas aeruginosa more than do SP-A1 variants. Infect Immun. 2007;75(3):1403–12. doi: 10.1128/IAI.01341-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang G, Umstead TM, Phelps DS, Al-Mondhiry H, Floros J. The effect of ozone exposure on the ability of human surfactant protein A variants to stimulate cytokine production. Environ Health Perspect. 2002;110(1):79–84. doi: 10.1289/ehp.0211079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Selman M, Lin HM, Montano M, Jenkins AL, Estrada A, Lin Z, Wang G, DiAngelo S, Guo X, Umstead TM, Lang CM, Pardo A, Phelps DS, Floros J. Surfactant protein A and B genetic variants predispose to idiopathic pulmonary fibrosis. Hum Genet. 2003;113:542–50. doi: 10.1007/s00439-003-1015-4. [DOI] [PubMed] [Google Scholar]
- 72.Sanchez-Barbero F, Rivas G, Steinhilber W, Casals C. Structural and functional differences among human surfactant proteins SP-A1, SP-A2 and co-expressed SP-A1/SP-A2: role of supratrimeric oligomerization. Biochem J. 2007;406(3):479–89. doi: 10.1042/BJ20070275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oberley RE, Snyder JM. Recombinant human SP-A1 and SP-A2 proteins have different carbohydrate-binding characteristics. Am J Physiol Lung Cell Mol Physiol. 2003;284(5):L871–81. doi: 10.1152/ajplung.00241.2002. [DOI] [PubMed] [Google Scholar]
- 74.Hattori A, Kuroki Y, Katoh T, Takahashi H, Shen HQ, Suzuki Y, Akino T. Surfactant protein A accumulating in the alveoli of patients with pulmonary alveolar proteinosis: oligomeric structure and interaction with lipids. Am J Respir Cell Mol Biol. 1996;14(6):608–19. doi: 10.1165/ajrcmb.14.6.8652189. [DOI] [PubMed] [Google Scholar]
- 75.Williams MC, Hawgood S, Hamilton RL. Changes in lipid structure produced by surfactant proteins SP-A, SP-B, and SP-C. Am J Respir Cell Mol Biol. 1991;5(1):41–50. doi: 10.1165/ajrcmb/5.1.41. [DOI] [PubMed] [Google Scholar]