Abstract
Lactobacilli are probiotics that, among other health-promoting effects, have been ascribed immunostimulating and virus-preventive properties. Certain Lactobacillus spp. have been shown to possess strong interleukin-12 (IL-12) -inducing properties. As IL-12 production depends on the up-regulation of type I interferons (IFNs), we hypothesized that the strong IL-12-inducing capacity of Lactobacillus acidophilus NCFM in murine bone-marrow-derived dendritic cells (DCs) is caused by an up-regulation of IFN-β, which subsequently induces IL-12 and the double-stranded RNA binding Toll-like receptor-3 (TLR-3). The expression of the genes encoding IFN-β, TLR-3, IL-12 and IL-10 in DCs upon stimulation with L. acidophilus NCFM was determined. Lactobacillus acidophilus NCFM induced a much stronger expression of Ifn-β, Il-12 and Il-10 compared with the synthetic double-stranded RNA ligand Poly I:C, whereas the levels of expressed Tlr-3 were similar. Whole genome microarray gene expression analysis revealed that other genes related to viral defence were significantly up-regulated and among the strongest induced genes in DCs stimulated with L. acidophilus NCFM. The ability to induce IFN-β was also detected in another L. acidophilus strain (X37), but was not a property of other probiotic strains tested, i.e. Bifidobacterium bifidum Z9 and Escherichia coli Nissle 1917. The IFN-β expression was markedly reduced in TLR-2−/− DCs, dependent on endocytosis, and the major cause of the induction of Il-12 and Tlr-3 in DCs stimulated with L. acidophilus NCFM. Collectively, our results reveal that certain lactobacilli trigger the expression of viral defence genes in DCs in a TLR-2 manner dependent on IFN-β.
Keywords: dendritic cells, gene regulation, innate immune response, Lactobacillus acidophilus, Toll-like receptor-2, virus
Introduction
Lactic acid bacteria are inhabitants of the gastrointestinal tract, and some species are considered to have probiotic properties offering a number of benefits to health and well-being.1–3 Some probiotics have been shown to reduce the risk of virus infections such as the common cold and influenza.4–6 So far, the mechanisms causing the reductions in respiratory tract infections and other symptoms are unknown. It is likely that these positive effects are due to the ability of probiotics to modulate immune stimulatory responses upon interaction with dendritic cells (DCs).
Dendritic cells are central gatekeepers and regulators of the immune response, interacting with mucosally encountered antigens including the gut microbiota and viruses. The innate immune cell activation occurs predominantly through the interaction of Toll-like receptors (TLRs) and other pathogen recognition receptors on the surfaces of antigen-presenting cells.7 Exposure to micro-organisms induces up-regulation of surface markers and the production of the several cytokines that modulate the function of DCs.8 Probiotics exert differential stimulatory effects on DCs in vitro, giving rise to varying production of different cytokines and accordingly different effector functions.9,10 Members of the Lactobacillus and Bifidobacterium genera are well-recognized for their probiotic properties, but certain other bacteria, including some Escherichia coli strains, have also been shown to exert probiotic features.
Upon virus infection type I interferons (IFNs), cytokines with anti-viral and immune-regulatory functions, are produced. The TLRs of DCs have emerged as key transducers of type I IFNs during viral infections.11 Toll-like receptor-3, a receptor localized in the endosomal compartment, recognizes double-stranded (ds) RNA motifs of viruses and Poly I:C (a synthetic dsRNA) and induces the transcription of type I IFNs (IFN-α and IFN-β).12,13 Recently, TLR-2, normally associated with Gram-positive bacteria, was shown to induce type I IFN in response to viral ligands but not in response to the bacterial ligand Pam3CSK4.14 Type I IFNs exert their antiviral function by binding specifically to a unique receptor (IFNAR), thereby initiating a signalling cascade that controls the expression of hundreds of IFN-stimulated genes (ISGs) and other genes involved in an innate host response against viruses.15 Type I IFNs, although best known for their antiviral properties, are potent regulators of cell growth and can modulate both innate and adaptive immune responses. Synthesis of type I IFNs was originally associated with viral infections; however, many pathogenic bacteria are equally able to induce the up-regulation of type I IFN, leading to modulation of the innate antibacterial response. Several Gram-negative bacteria, such as Salmonella enterica Serovar Typhimurium, Shigella flexneri and Escherichia spp., stimulate type I IFN synthesis in phagocytosing cells.16 Recently, pathogenic Gram-positive bacteria, such as group A and B Streptococcus spp.,17–19Listeria monocytogenes,20,21 and the spirochaete bacterium Borrelia burgdorferi22 were likewise reported to induce the production of high quantities of type I IFN during infection. Mancuso et al.19 reported that the production of type I IFNs was critical for the clearance of infection by the host. In relation to intracellular bacteria in particular TLR-3, TLR-7 and TLR-9 have been shown to be involved in the type I IFN induction,23 whereas in connection with other bacteria TLRs and other pathogen recognition receptors on the cell surface seem of particular importance. However, no clear picture of which receptors are involved exists or of which role these receptors play in the bacterially induced IFN-β production. For Streptococcus spp. and Listeria spp., the intracellular TLR-9 was essential for the induction of IFN-β in monocytes or DCs stimulated in vitro.18,20 In Borrelia burgdorferi, the induction of IFN-β was independent of TLR-2.22 Only for the Gram-negative Pseudomonas aeruginosa a role of TLR-2 has been suggested in the induction of a pro-inflammatory response in human monocytes.24 It was demonstrated that a TLR-2 and mannose receptor synergistically were involved in the induction of the cytokines tumour necrosis factor-α (TNF-α), interleukin-6 (IL-6) and IL-1β. However, IFN-β was not included in the study. To our knowledge, neither lactic acid bacteria nor commensal bacteria have been shown to possess the capability to induce IFN-β in DCs upon stimulation.
It has been reported that TLR-mediated IL-12p70 synthesis is strongly reduced in the absence of type I IFN,25 demonstrating a critical role of type I IFN in controlling the production of the pro-inflammatory cytokine IL-12p70. We have previously reported that certain members of the Lactobacillus genus, including L. acidophilus, demonstrated remarkable IL-12-inducing properties.26 On account of these observations, we hypothesized that L. acidophilus, despite its non-pathogenic phenotype and health-promoting properties, is able to induce IFN-β production in DCs and consequently matures DCs into anti-virus phenotype cells.
The aim of this study was to investigate whether L. acidophilus has the ability to induce anti-viral defence gene expression in DC. We analysed the gene expression profile of TLR-3 and IFN-β, key players involved in viral defence, in murine bone-marrow-derived DCs stimulated in vitro with L. acidophilus NCFM. Genome-wide microarray analysis confirmed our hypothesis showing a general, significant up-regulation of anti-viral defence genes. The IFN-β-inducing property was likewise detected in another L. acidophilus strain, but not in a probiotic Bifidobacterium sp. or E. coli strain. This ability to induce IFN-β was dependent on TLR-2 recognition and required phagocytic activity in the DCs. Our results reveal that, in contrast to Poly I:C stimulation, the expression of Tlr-3 in L. acidophilus-stimulated DCs was dependent on the production of IFN-β. This study is the first to report that L. acidophilus NCFM, a widely used probiotic bacterium, is able to induce viral defence in murine bone-marrow-derived DC and that TLR-2 plays a pivotal role in IFN-β induction in DCs stimulated with this bacterium.
Materials and methods
Bacterial strains, growth conditions and preparation of UV-killed bacteria
Lactobacillus acidophilus NCFM (Danisco, Copenhagen, Denmark), L. acidophilus X37 (Copenhagen University, Department of Food Microbiology, Faculty of Life Sciences, Denmark), Bifidobacterium bifidum Z9 (Copenhagen University, Department of Food Microbiology, Faculty of Life Sciences, Denmark), which are all considered to have probiotic properties, were grown anaerobically overnight at 37° in de Man Rogosa Sharp (MRS) broth (Merck, Darmstadt, Germany) and sub-cultured twice. Cells were harvested by centrifugation at 2000 g for 15 min, washed twice in phosphate-buffered saline (PBS; Bio Whittaker, East Rutherford, NJ) and resuspended in 1/10 the growth volume of PBS. The bacteria were killed by a 20-min exposure to UV light. Escherichia coli Nissle 1917 O6:K5:H1 (Statens Serum Institut, Copenhagen, Denmark), a Gram-negative probiotic bacterium, was grown aerobically overnight at 37° in Luria–Bertani (LB) broth (Merck) and killed by a 45-min exposure to UV light. In all experimental set-ups, UV-killed bacteria were used. The bacteria were stored at − 80°, the concentration was determined as the content of dry matter per ml upon lyophilization, and the dry weight was corrected for buffer salt content. Absence of viable cells was verified by plating the UV-exposed bacteria on MRS and LB agar.
Generation of murine dendritic cells
Bone-marrow-derived DCs were prepared as previously described.9 Briefly, bone marrow from wild-type (WT) or TLR-2−/− knock out C57BL/6 mice was flushed out from the femur and tibia and washed twice in sterile PBS. Then, 3 × 105 bone marrow cells were seeded into Petri dishes in 10 ml RPMI-1640 (Sigma-Aldrich, St Louis, MO) containing 10% (volume/volume) heat-inactivated fetal calf serum supplemented with penicillin (100 U/ml), streptomycin (100 μg/ml), glutamine (4 mm), 50 μm 2-mercaptoethanol (all purchased from Cambrex Bio Whittaker, Charles City, IA) and 15 ng/ml murine granulocyte–macrophage colony-stimulating factor (GM-CSF; harvested from a GM-CSF-transfected Ag8.653 myeloma cell line). The cells were incubated for 8 days at 37° in 5% CO2 humidified atmosphere. On day 3, 10 ml of complete medium containing 15 ng/ml GM-CSF was added. On day 6, 10 ml were removed and replaced by fresh medium. Non-adherent immature DC were harvested on day 8.
Stimulation of murine dendritic cells with bacteria
Immature DCs (2 × 106 cells/ml) were resuspended in fresh medium supplemented with 10 ng/ml GM-CSF, and 500 μl/well were seeded in 48-well tissue culture plates (Nunc, Roskilde, Denmark). The stimuli were suspended in medium and added (100 μl/well) in a final concentration of 10 μg/ml (L. acidophilus NCFM, L. acidophilus X37 and E. coli Nissle 1917) and 40 μg/ml (B. bifidum Z9). Optimal bacterial concentrations were determined in a previous study.26 The TLR-2 ligands Pam2CSK4 and Pam3CSK4 (InvivoGen, San Diego, CA), which in previous experiments induced only minor amounts of IL-12 and IL-10 when added in the concentrations 0·03–3 μg/ml, were used in final concentrations of 0·1 μg/ml and 1 μg/ml, respectively. As a positive control, Poly I:C (InvivoGen), a synthetic analogue of dsRNA, was added in a final concentration of 10 μg/ml. The cell cultures were incubated at 37° in 5% CO2.
Effect of endocytic activity during stimulation
The DCs were pre-treated with cytochalasin D (0·5 μg/ml), chlorpromazine (25 μg/ml), methyl-β-cyclodextrin (1 mm) (Sigma-Aldrich) or medium alone for 1 hr at 37° in 5% CO2 before addition of L. acidophilus NCFM (10 μg/ml) or Poly I:C (10 μg/ml) as previously described.27 The cells were harvested after 3 hr of incubation at 37° in 5% CO2, and RNA was extracted.
Interferon-β inhibition assay
Mouse IFN-β polyclonal antibody (R&D Systems, Minneapolis, MN) was added in two different concentrations (10 and 50 μg/ml) immediately after addition of L. acidophilus NCFM (10 μg/ml) to the DCs. The cells were harvested after 10 hr of stimulation at 37° in 5% CO2, and RNA was extracted.
RNA extraction
Murine DCs were harvested at various stimulation time-points, homogenized by QIAshredder (Qiagen, Ballerup, Denmark), and RNA was extracted using the RNeasy Plus Mini Kit (Qiagen). RNA quality was verified by Bioanalyzer (Agilent, Santa Clara, CA), and the concentration was determined by Nanodrop (Thermo, Wilmington, DE).
Microarray analysis
Immature DCs from three C57BL/6 mice were stimulated with L. acidophilus NCFM, and DCs were harvested after 4, 10 and 18 hr. RNA was extracted, 1 μg RNA per stimulation was converted to complementary DNA (cDNA), and biotin-labelled amplified RNA (aRNA) was synthesized using the MessageAmp™ II-Biotin Enhanced kit (Ambion, Austin, TX) according to the manufacturer's instructions. The aRNA samples were hybridized to Gene Chip Mouse genome 430 2.0 Array (Affymetrix, Santa Clara, CA), comprising 45 000 probe sets representing over 34 000 mouse genes. The arrays were stained, washed and scanned according to the manufacturer's instructions. The microarray data were analysed using R and Bioconductor (Gentleman etal, 2004; Workman etal, 2002; Irizarry etal, 2003) from the three independent stimulations of DC from three individual mice (in total 12 arrays).28 Raw probe intensities were normalized using qspline and expression index calculations were performed using rma.29,30 For statistical testing, analysis of variance (anova) was performed using stimulation time as factor where all untreated samples were treated as one group. The false discovery rate (FDR) was estimated using a Monte Carlo approach, and statistical significance was set at an FDR of 0 yielding 4947 highly significant probe sets corresponding to 3319 unique genes annotated by Mouse Genome Informatics (MGI).31
Quantitative real-time polymerase chain reaction analysis
Dendritic cells were harvested after 2, 4 and 10 hr of stimulation. RNA was extracted, and 1 μg of total RNA was reverse transcribed by the TaqMan Reverse Transcription Reagent kit (Applied Biosystems, Foster City, CA) using random hexamer primers according to the manufacturer's instructions. The cDNA obtained was stored in aliquots at −80°. For the selection of primer and probe sequences, the regions coding for the genes investigated were retrieved from the GenBank EMBL databases. The following gene sequences were applied: TLR-3 (NM_126166), IFN-β (NM_010510), IL-12 p40 (NM_008352), IL-10 (NM_010548) and β-actin (NM_007393). Primers and probes were designed using the software Primer Express 3.0 (Applied Biosystems) and tested for specificity by the basic alignment search tool BLAST. HPLC-purified forward and reverse primers were manufactured by DNA Technology (Aarhus, Denmark). The probes were labelled with the 5′ reporter dye 6-carboxy-fluorescein (FAM) and the 3′ quencher dye NFQ-MGB (Applied Biosystems). Sequences of primers and probes are listed in Table 1. Primer and probe concentrations were optimized and to determine the efficiency of the amplification dilutions, standard curves were made for each set of primers and probe (data not shown). The amplifications were carried out in a total volume of 20 μl containing 1× TaqMan Universal PCR Master Mix (Applied Biosystems), forward and reverse primer (concentration 900 nm each), 200 nm TaqMan MGB probe, and purified target cDNA. The cycling parameters were initiated by 20 seconds at 95°, followed by 40 cycles of 3 seconds at 95° and 30 seconds at 60° using the ABI Prism 7500 (Applied Biosystems). Amplification reactions were performed in triplicates, and DNA contamination controls were included. The amplifications were normalized to the expression of β-actin. Relative transcript levels were calculated applying the equation described by Pfaffl.32
Table 1.
Primers and probes used for real-time polymerase chain reaction analysis
| Target | Sequence (5′–3′) | |
|---|---|---|
| Interferon-β (NM_010510) | Forward | CGGACTTCAAGATCCCTATGGA |
| Reverse | TGGCAAAGGCAGTGTAACTCTTC | |
| Probe | ATGACGGAGAAGATGC | |
| Toll-like receptor-3 (NM_126166) | Forward | GATTCTTCTGGTGTCTTCCACAAA |
| Reverse | AATGGCTGCAGTCAGCTACGT | |
| Probe | CAATGCACTGTGAGATAC | |
| Interleukin-12 p40 (NM_008352) | Forward | TGGAGCACTCCCCATTCCT |
| Reverse | TGCGCTGGATTCGAACAA | |
| Probe | CTTCTCCCTCAAGTTC | |
| Interleukin-10 (NM_010548) | Forward | GATGCCCCAGGCAGAGAA |
| Reverse | CACCCAGGGAATTCAAATGC | |
| Probe | CATGGCCCAGAAAT | |
| β-Actin (NM_007393) | Forward | CGATGCCCTGAGGCTCTTT |
| Reverse | TGGATGCCACAGGATTCCA | |
| Probe | CCAGCCTTCCTTCTT |
Cytokine quantification by enzyme-linked immunosorbent assay
After 24 hr of stimulation, culture supernatants were collected and stored at −80° for later cytokine analysis. The production of murine IL-12(p70), IL-10, IL-6, TNF-α and IFN-β was analysed using commercially available enzyme-linked immunosorbent assay (ELISA) kits (R&D Systems).
Statistical analysis
Statistical calculations were performed using the software program GraphPad Prism 5 (San Diego, CA). For each experiment, results were analysed by anova with Bonferroni as post test, and P-values of < 0·05 were considered significant.
Results
Lactobacillus acidophilus NCFM induces IFN-β and TLR-3 up-regulation in murine dendritic cells
Stimulation with L. acidophilus NCFM, Poly I:C, and the TLR-2 ligands Pam2CSK4 and Pam3CSK4 gave rise to highly distinct protein concentrations of IFN-β and IL-12 (Fig. 1a). By far the strongest production of IFN-β was detected after stimulation with L. acidophilus NCFM, as the concentration of IFN-β was more than five-fold higher in comparison to induction by Poly I:C. Pam2CSK4 and Pam3CSK4 did not induce any detectable levels of IFN-β. A similar picture was seen for IL-12, as the production induced by Poly I:C was more than seven-fold less relative to cells stimulated with L. acidophilus NCFM. The two TLR-2 ligands gave rise to low amounts of IL-12. In contrast, even though L. acidophilus NCFM triggered the highest production of TNF-α, the two TLR-2 ligands also induced higher amounts of TNF-α compared with stimulation with Poly I:C. The cytokine IL-10 was exclusively produced by DCs stimulated with L. acidophilus NCFM; stimulation with Poly I:C or the two TLR-2 ligands did not induce IL-10. Flow cytometry showed an up-regulation of CD80, CD86, CD40 and MHC class II upon stimulation with both Pam3CSK4 and L. acidophilus NCFM, indicating that the stimulation with the TLR-2 ligand and the whole bacteria induced a state of maturation (data not shown).
Figure 1.
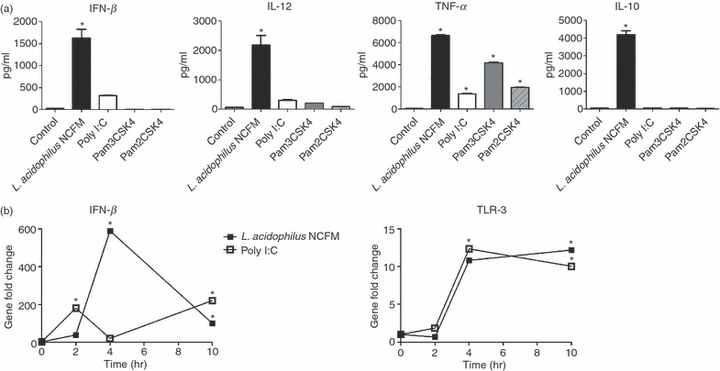
Lactobacillus acidophilus NCFM induces gene expression of interferon-β (IFN-β) and toll-like receptor 3 (TLR-3). Bone-marrow-derived dendritic cells (DCs) were stimulated with L. acidophilus NCFM (10 μg/ml), the synthetic double-stranded RNA ligand Poly I:C (10 μg/ml) and the TLR-2 ligands Pam2CSK4 (0.1 μg/ml) and Pam3CSK4 (1 μg/ml). Protein concentrations of IFN-β, IL-12, tumour necrosis factor-α (TNF-α) and IL-10 in the supernatants after 24 hr were measured by enzyme-linked immunosorbent assay (a). Two hours, 4 hr and 10 hr after stimulation RNA was extracted, and the induction of the genes encoding IFN-β and TLR-3 was determined by reverse transcription–polymerase chain reaction (b). The messenger RNA levels were normalized to the relative expression of β-actin. The error bars depict the mean value ± standard deviation of three individual measurements from one experiment. The data represent one of at least seven independent experiments, *P < 0.05 versus non-stimulated DC.
The expression of the genes encoding IFN-β and TLR-3 was determined after 2, 4 and 10 hr of stimulation with L. acidophilus NCFM or the synthetic dsRNA analogue Poly I:C (Fig. 1b). The strongest up-regulation of Ifn-β was detected after stimulation with L. acidophilus NCFM; it was only slightly up-regulated after 2 hr (38-fold) but reached a significant maximum after 4 hr (589-fold) that declined to a gene expression of 100-fold after 10 hr. In contrast to L. acidophilus NCFM, Poly I:C induced strong expression of Ifn-β after 2 hr (180-fold). However, this induction decreased to 20-fold after 4 hr and was raised to 220-fold again after 10 hr. In contrast to Ifn-β, both L. acidophilus NCFM and Poly I:C strongly induced the gene encoding TLR-3 after 4 hr. As this induction was sustained after 10 hr stimulation in both treatments, we were able to show that L. acidophilus NCFM is capable of triggering up-regulation of TLR-3 to the same extent as the synthetic TLR-3 ligand Poly I:C.
Identification of multiple virus-defence-related genes by genome-wide expression analysis in dendritic cells stimulated with L. acidophilus NCFM
Genome-wide microarray analysis was performed to further investigate the up-regulation of virus-related genes during stimulation of DCs with L. acidophilus NCFM. To generate a comprehensive view of the expression profile, samples were harvested at different time-points (4, 10 and 18 hr). Differential expression was assessed using anova resulting in 3319 significant regulated genes at an FDR of 0 (P-value < 1e−4). These findings point to a very strong response of DC upon stimulation, which is in good agreement with the phenotypic changes (e.g. production of cytokines and up-regulation of various surface markers) observed. The data generated were deposited in NCBI's Gene Expression Omnibus33 and are accessible through GEO Series accession number GSE18460.
Focusing on genes that are virus-defence-related, we used the Gene Ontology (GO) term GO:0009615 ‘Response to virus’ to test whether the distribution of their expressions was different from the entire distribution. The Wilcoxon rank sum test (Mann–Whitney test) with a P-value of 3e11 revealed a strong, significant up-regulation of these genes (Fig. 2). The induction of virus-related genes was most prominent for the gene encoding Rsad2 (700-fold). Rsad2 (radical S-adenosyl methionine domain containing 2), also known as viperin, encodes a cytoplasmic antiviral protein induced by IFNs. This protein impairs virus budding by disrupting lipid rafts at the plasma membrane, a feature which is essential for the budding process of many viruses.34 The genes encoding IFN-induced T-cell specific GTPase (TGTP2), IFN-stimulated gene 15 (ISG15), IFN-regulatory factor (IRF-7) and TLR-3, all involved in viral immune defence and induced by IFN-β, were likewise among the highest significantly up-regulated genes.
Figure 2.
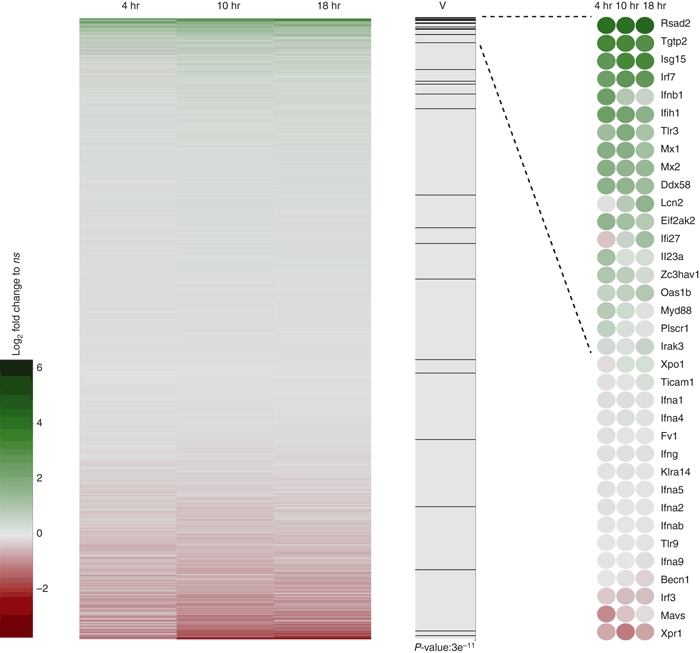
Lactobacillus acidophilus NCFM induces expression of multiple genes related to viral immune defence. Bone-marrow-derived dendritic cells (DCs) from three mice were individually stimulated with L. acidophilus NCFM (10 μg/ml) for 4 hr, 10 hr and 18 hr, RNA was extracted, and microarray analysis was performed. Heatmap of log2-fold changes versus no stimulation for all probes on the array. Probe sets are sorted according to maximal log2-fold change at any time-point. Green and red represent up-regulation and down-regulation, respectively. In column ‘V’ the position of genes in the gene ontology term GO:0009615 ‘Response to virus’ is presented as black lines together with the significance of this distribution in a two-sided Wilcoxon Rank sum test (Mann–Whitney). Detailed expression of the genes is shown rightmost.
In addition to the genes in the ‘Response to virus’ GO term, microarray data analysis revealed a significant induction of numerous genes related to virus infection (Table 2). The majority of these genes are classical IFN-sensitive genes (ISG) induced upon stimulation with IFN-β, e.g. members of the IFN-stimulated gene 56 family (ISG56), which are known to be strongly induced in response to virus infection, type I IFNs and dsRNA. In mouse, this family comprises three members (ISG56, ISG54 and ISG49) that associate with large protein complexes and block the translation pathway at different steps.35,36 Another strongly induced gene belonging to the classical family of ISGs codes for the well-studied antiviral enzyme dsRNA-dependent protein kinase (EIF2AK2, also termed PKR), which phosphorylates various substrates including the protein synthesis initiation factor eIF2α and acts by blocking the translation of viral RNA.37 The 2′,5′-oligoadenylate synthetases (OAS), a family of enzymes activated by dsRNA, were likewise strongly induced. These enzymes produce 2′,5′-linked oligoadenylates activating the latent ribonuclease RNAse L, which degrades viral messenger RNA.38 The myxovirus-resistance (Mx) proteins, IFN-inducible GTPases, were up-regulated in a similar manner. These proteins have a wide antiviral spectrum against different types of viruses and form complexes with dynamin, which disrupts intracellular transport or interferes with the activity of viral polymerases.39
Table 2.
Significant up-regulation of interferon-induced genes in murine dendritic cells stimulated with Lactobacillus acidophilus NCFM
| Gene number | Annotation | 4 hr | 10 hr | 18 hr | Name |
|---|---|---|---|---|---|
| NM_126166 | TLR3 | 3·1 | 4·2 | 2·6 | Toll-like receptor 3 |
| NM_010510 | IFNB1 | 4·1 | 1·9 | 1·2 | Interferon-β |
| NM_021384 | RSAD2 | 5·9 | 6 | 6·6 | Interferon-induced protein Viperin |
| NM_020583 | ISG20 | 3·9 | 5·5 | 5·4 | Interferon-stimulated exonuclease |
| NM_011163 | PKR | 2·7 | 2·2 | 1·5 | dsRNA-activated protein kinase |
| P56 family | |||||
| NM_008331 | ISG56 | 5·8 | 5·1 | 5·2 | Interferon-stimulated gene 56 |
| NM_008332 | ISG54 | 5·9 | 5·9 | 5·4 | Interferon-stimulated gene 54 |
| NM_010501 | ISG49 | 5·3 | 5·7 | 4·9 | Interferon-stimulated gene 49 |
| OAS family | |||||
| NM_145209 | OASL1 | 4·1 | 4·7 | 4·6 | Oligoadenylate synthetase-like 1 |
| NM_011854 | OASL2 | 3·7 | 3·3 | 3 | Oligoadenylate synthetase-like 2 |
| NM_145227 | OAS2 | 2·2 | 2·3 | 1·6 | Oligoadenylate synthetase 2 |
| NM_145226 | OAS3 | 2·4 | 2·6 | 2·4 | Oligoadenylate synthetase 3 |
| NM_011852 | OAS1G | 1·9 | 2 | 1·7 | Oligoadenylate synthetase 1G |
| NM_033541 | OAS1C | 1·1 | 1·2 | 1·2 | Oligoadenylate synthetase 1C |
| Mx proteins | |||||
| NM_013606 | MX2 | 3·3 | 3·1 | 2·6 | Myxovirus resistance 2 |
| NM_010846 | MX1 | 2·7 | 2·7 | 1·9 | Myxovirus resistance 1 |
| p200 gene family | |||||
| NM_001045481 | IFI203 | 3·4 | 2·9 | 2·5 | Interferon-activated gene 203 |
| NM_008329 | IFI204 | 2·5 | 3·4 | 2·7 | Interferon-activated gene 204 |
| NM_172648 | IFI205 | 3·1 | 3·2 | 3 | Interferon-activated gene 205 |
| LOC623121 | XM_001477431 | 4·4 | 5·1 | 4·6 | Novel interferon-β induced gene similar to IFN-inducible protein 203 |
| NM_027320 | IFI35 | 1·9 | 1·5 | 0·89 | Interferon-induced protein 35 |
| NM_133871 | IFI44 | 3·5 | 4·8 | 4·6 | Interferon-induced protein 44 |
| Interferon-induced GTPases | |||||
| NM_021792 | IIGP1 | 4·7 | 5·2 | 4·9 | Interferon-inducible GTPase 1 |
| NM_019440 | IIGP2 | 3·1 | 2·3 | 1·8 | Interferon-inducible GTPase 2 |
| NM_001039160 | GVIN1 | 1·8 | 1·4 | 1·3 | Interferon-inducible GTPase |
| Interferon-induced helicases | |||||
| NM_172689 | DDX58 | 2·9 | 2·7 | 2·3 | RNA helicase DDX58 |
| NM_030150 | DHX58 | 3·2 | 3 | 2·2 | RNA helicase DHX58 |
| NM_027835 | IFIH1 | 3·6 | 3·5 | 2·6 | Interferon-induced with helicase C domain 1 |
| Protein ubiquitination | |||||
| NM_022329 | ISG15 | 1·9 | 1·7 | 2 | Interferon-stimulated gene 15 |
| XM_001478484 | HERC5 | 3 | 3·4 | 3·3 | IFN-induced E3 protein ligase |
| NM_019949 | UBE2L6 | 1·9 | 2·7 | 2·1 | ISG-15-conjugating enzyme |
| NM_011909 | USP18 | 3·7 | 3·6 | 3·3 | Protease specifically removing ISG15 |
| NM_023738 | UBE1l | 1·6 | 2·3 | 2 | Ubiquitin-activating enzyme E1-like |
| NM_019949 | UBE2l6 | 1·9 | 2·7 | 2·1 | Ubiquitin-conjugating enzyme E2L 6 |
| LOC677168 | XR_005074 | 4·2 | 4·8 | 4·8 | Novel interferon-β induced gene similar to ISG15 ubiquitin-like modifier |
| NM_028864 | Zc3hav1 | 1·6 | 1·2 | 0·76 | Antiviral zinc and RNA binding protein |
| NM_001038587 | Adar | 1·9 | 1·7 | 1·8 | Adenosine deaminase (binds dsRNA) |
| NM_175397 | Sp110 | 1·5 | 1·1 | 0·43 | Sp110 nuclear body protein (inhibits virus replication) |
| NM_011636 | Plscr1 | 1·1 | 0·35 | 0·2 | Phospholipid scramblase 1(enhances IFN response) |
| Interferon regulatory factors (IRF) | |||||
| NM_016850 | IRF7 | 4·1 | 4·5 | 4·2 | Interferon regulatory factor 7 |
Induction of anti-viral mechanisms in dendritic cells is confined to certain probiotic strains
To elucidate whether the induction of the antiviral response is unique for L. acidophilus NCFM, universal for L. acidophilus strains, or a common property of probiotics, we further stimulated DCs with another L. acidophilus (X37), a B. bifidum strain (Z9), and the Gram-negative probiotic E. coli Nissle 1917. Gene expression analysis by reverse trasncription–polymerase chain reaction (RT-PCR) revealed that L. acidophilus X37 was similarly able to trigger expression of the genes encoding IFN-β and TLR-3 (Fig. 3a). In contrast, neither B. bifidum Z9 nor E. coli Nissle 1917 gave rise to a strong up-regulation. Both strains resulted in a small peak of Ifn-β expression after 2 hr of stimulation, followed by a rapid decrease to almost background level and a lower and less sustained up-regulation of Tlr-3 transcription compared with the L. acidophilus strains. The rapid but low up-regulation of Ifn-β transcription upon stimulation with B. bifidum Z9 and E. coli Nissle 1917 corresponded to the peak observed upon stimulation with Poly I:C. However, stimulation with Poly I:C showed a re-emerging rise in the transcription after 10 hr. The results obtained for Ifn-β were verified on a protein level by ELISA (Fig. 3b). The highest production of IFN-β was measured upon stimulation of DCs with L. acidophilus X37, which induced 18 times more IFN-β compared with E. coli Nissle 1917. Lactobacillus acidophilus NCFM induced more than 14 times the production of IFN-β compared with E. coli Nissle 1917, whereas DCs stimulated with B. bifidum Z9 did not produce detectable levels of IFN-β.
Figure 3.
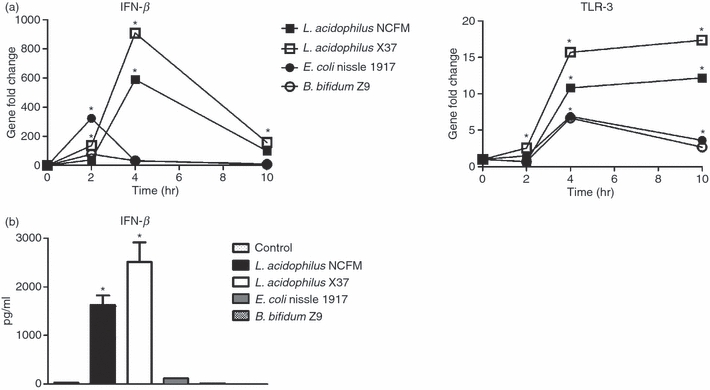
Lactobacillus acidophilus strains, but not Bifidobacterium bifidum and Escherichia coli, induce interferon-β (IFN-β) expression in dendritic cells (DCs). (a) Bone-marrow-derived DCs were stimulated with L. acidophilus NCFM (10 μg/ml), L. acidophilus X37 (10 μg/ml), E. coli Nissle 1917 (10 μg/ml) and B. bifidum Z9 (40 μg/ml) for 2 hr, 4 hr and 10 hr. RNA was extracted, and the induction of the gene encoding IFN-β and toll-like receptor 3 (TLR-3) was determined by reverse transcription–polymerase chain reaction analysis. The messenger RNA levels were normalized to the relative expression of β-actin. (b) Bone-marrow-derived DCs were stimulated with L. acidophilus NCFM (10 μg/ml), L. acidophilus X37 (10 μg/ml), E. coli Nissle 1917 (10 μg/ml) and B. bifidum Z9 (40 μg/ml) for 24 hr. The supernatant was harvested and protein concentrations were measured by enzyme-linked immunosorbent assay. The error bars depict the mean value ± standard deviation of three individual measurements from one experiment. The data represent one of at least three independent experiments,*P < 0.05 versus non-stimulated DC.
Induction of IFN-β and TLR-3 is dependent on TLR-2
The bacterial strains investigated in this study, capable of inducing strong Ifn-β and Tlr-3 expression levels, were also the strains that gave rise to a high IL-12 production. As we have previously found that the IL-12 production is to a great extent dependent on TLR-2 stimulation,26 we hypothesized that TLR-2 might likewise be involved in the stimulation of DCs with L. acidophilus, leading to the transcription of Ifn-β and Tlr-3 (along with other virus-related genes).
To investigate whether TLR-2 is required for the induction of IFN-β, we generated bone-marrow-derived DCs from WT and TLR-2−/− mice. The expression of the gene encoding IFN-β was determined in DCs upon stimulation with L. acidophilus NCFM, Poly I:C, E. coli Nissle 1917, and B. bifidum Z9 after 2, 4 and 10 hr. As depicted in Fig. 4(a), the lack of TLR-2 resulted in a dramatic decrease in the Ifn-β expression peak after 4 hr induced by L. acidophilus NCFM. The Ifn-β expression profile was only moderately affected upon Poly I:C stimulation, with a slight increase in Ifn-β expression after 2 hr and a decrease after 10 hr. In contrast, when the DCs were stimulated with either B. bifidum Z9 or E. coli Nissle 1917, the weak expression peaks observed after 2 hr in WT DCs were markedly increased in TLR-2−/− cells. As a consequence, whereas the absence of TLR-2 was central for the IFN-β production upon stimulation with L. acidophilus NCFM, TLR-2 seemingly exhibited the opposite role upon stimulation with E. coli Nissle 1917 and B. bifidum Z9, as the Ifn-β induction was higher in TLR-2−/− DCs. Our gene expression results were confirmed by the presence of IFN-β in culture supernatants measured by ELISA after 24 hr of stimulation (Fig. 4b).
Figure 4.
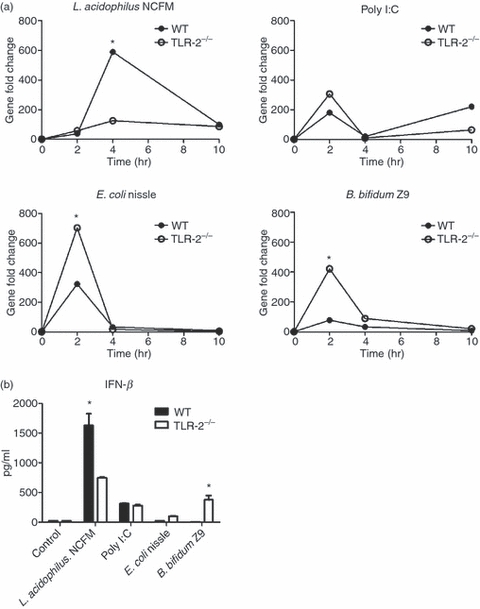
Interferon-β (IFN-β) stimulating activity of Lactobacillus acidophilus NCFM is dependent on toll-like receptor 2 (TLR-2). (a) Bone-marrow-derived dendritic cells (DCs) from both wild-type (WT) and TLR-2−/− mice were stimulated with L. acidophilus NCFM (10 μg/ml), Poly I:C (10 μg/ml), Escherichia coli Nissle 1917 (10 μg/ml), and Bifidobacterium bifidum Z9 (40 μg/ml) for 2 hr, 4 hr and 10 hr. RNA was extracted, and the induction of the gene coding for IFN-β was determined by reverse transcription–polymerase chain reaction analysis. The messenger RNA levels were normalized to the relative expression of β-actin. (b) Bone-marrow-derived DCs from both WT and TLR-2−/− were stimulated with L. acidophilus NCFM (10 μg/ml), Poly I:C (10 μg/ml), E. coli Nissle 1917 (10 μg/ml), and B. bifidum Z9 (40 μg/ml) for 24 hr. The supernatant was harvested and protein concentrations of IFN-β were measured by ELISA. The error bars depict the mean value ± standard deviation of three individual measurements from one experiment. The data represent one of at least two independent experiments. *P < 0.05 values (WT versus TLR-2−/−) are indicated.
Figure 5(a) illustrates the expression of Tlr-3 upon stimulation of WT DCs and TLR-2−/− DCs with L. acidophilus NCFM, Poly I:C, E. coli Nissle 1917 and B. bifidum Z9 for 2, 4 and 10 hr. In the case of L. acidophilus NCFM and Poly I:C, the expression of Tlr-3 was not affected by the absence of TLR-2 after 2 and 4 hr. However, after 10 hr Tlr-3 was significantly reduced in TLR-2−/− DCs compared with WT DCs. In TLR-2−/− DCs stimulated with E. coli Nissle 1917, the expression of Tlr-3 was, in contrast to WT DC, only slightly lower (one-fold after 2, 4 and 10 hr). Upon incubation of DC with B. bifidum Z9, the up-regulation of Tlr-3 was increased in TLR-2−/− DCs compared with WT DCs at all time-points.
Figure 5.
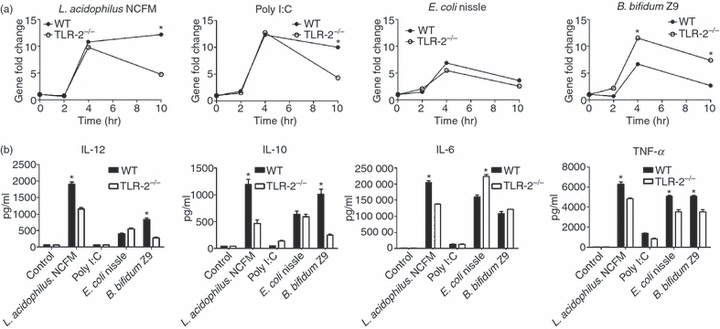
The toll-like receptor-3 (TLR-3) stimulating activity of Lactobacillus acidophilus NCFM is dependent on TLR-2. (a) Bone-marrow-derived dendritic cells (DCs) from both wild-type (WT) and TLR-2−/− were stimulated with L. acidophilus NCFM (10 μg/ml), Poly I:C (10 μg/ml), Escherichia coli Nissle 1917 (10 μg/ml), and Bifidobacterium bifidum Z9 (40 μg/ml) for 2 hr, 4 hr and 10 hr. RNA was extracted, and the induction of the gene coding for TLR-3 was determined by reverse transcription–polymerase chain reaction. The messenger RNA levels were normalized to the relative expression of β-actin. *P < 0.05 values (WT versus TLR-2−/−) are indicated. (b) Cytokine concentration [interleukin-12 (IL-12), IL-10, IL-6 and tumour necrosis factor-α (TNF-α)] measured in supernatants from DCs stimulated for 24 hr with L. acidophilus NCFM (10 μg/ml), Poly I:C (10 μg/ml), E. coli Nissle (10 μg/ml) and B. bifidum Z9 (40 μg/ml), respectively, as indicated. The data represent one of at least two independent experiments.
To further investigate the dependency of IL-12 on IFN-β, and hence indirectly on TLR-2, we measured the protein production of IL-12 and three other cytokines (IL-10, IL-6 and TNF-α) in WT and TLR-2−/− DCs upon stimulation with L. acidophilus NCFM, Poly I:C, B. bifidum Z9, and E. coli Nissle 1917 in the supernatants by ELISA after 24 hr of stimulation (Fig. 5b). The protein concentration of IL-12 corresponded largely to the concentration of IFN-β measured. Both the production of IL-12 and IL-10 was significantly reduced in TLR-2−/− DCs stimulated with L. acidophilus NCFM and B. bifidum Z9 compared with WT DCs. For all four stimulation regimes, the TNF-α protein concentration was slightly reduced in the supernatants of TLR-2−/− DCs, whereas IL-6 concentration was increased upon E. coli Nissle 1917 stimulation and decreased upon L. acidophilus NCFM stimulation.
Taken together, these results show that TLR-2 plays an important role in the strong induction of IFN-β in DCs upon stimulation with L. acidophilus NCFM. This observation is also reflected in the expression of the genes encoding IL-12 and TLR-3. In contrast, the same genes were largely unaffected when DCs were stimulated with Poly I:C. In case of E. coli Nissle 1917 and B. bifidum Z9, TLR-2 seems to hold a suppressive role.
The clathrin-mediated endocytic pathway is required for the induction of IFN-β and TLR-3 upon stimulation with L. acidophilus
Poly I:C-stimulated IFN-β induction in DC has recently been shown to depend on clathrin-mediated endocytosis.27 We have observed in previous studies that a prerequisite for a strong IL-12 response upon stimulation with L. acidophilus is that the bacterium is intact.26 As a consequence, we speculated that the IFN-β and strong IL-12-inducing mechanism could involve phagocytosis- or endocytosis-triggering events. Accordingly, we used pharmacological inhibitors to investigate whether bacterial uptake of L. acidophilus NCFM is required for the induction of IFN-β, and, in turn, IL-12 and TLR-3. The effect of cytochalasin D (phagocytosis inhibitor), methyl-β-cyclodextrin (calveolae-mediated endocytosis inhibitor) and chlorpromazine (clathrin-mediated endocytosis inhibitor) on the stimulation profile of DCs after incubation with either L. acidophilus NCFM or Poly I:C was investigated (Fig. 6). Upon stimulation with L. acidophilus NCFM, the expression of the genes encoding IFN-β, TLR-3 and IL-12 was significantly inhibited when the DCs were pre-treated with cytochalasin D and chlorpromazine. This inhibition was absent when the DCs were pre-treated with methyl-β-cyclodextrin. The pharmacological inhibitors did not have the same impact on the expression of the gene encoding IL-10, as only a slight reduction was observed. We obtained similar results when DCs were stimulated with Poly I:C. Pre-treatment with cytochalasin D and chlorpromazine of DCs had a significant inhibitory effect on the expression of the genes coding for IFN-β and TLR-3, whereas pre-treatment with methyl-β-cyclodextrin did not have an impact. Our results indicate that the clathrin-mediated endocytic pathway participates in uptake of L. acidophilus NCFM as an important step in the stimulation of the transcription of IFN-β and TLR-3 and, in turn, IL-12.
Figure 6.
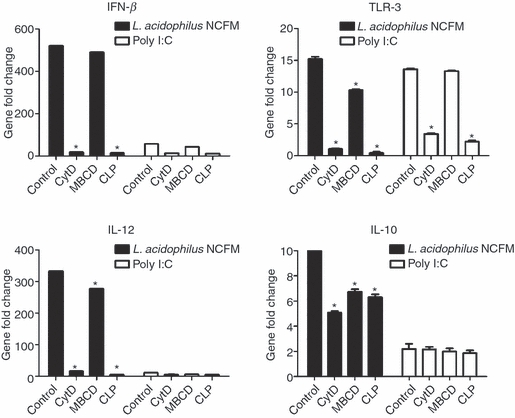
A clathrin-dependent endocytic pathway participates in Lactobacillus acidophilus NCFM-induced interferon-β (IFN-β) production. Bone-marrow-derived dendritic cells (DCs) were pre-treated with cytochalasin D (CytD, 0.5 μg/ml), chlorpromazine (CLP, 25 μg/ml), methyl-β-cyclodextrin (MBCD, 1 mm) or medium alone for 1 hr. Subsequently, the cells were stimulated with L. acidophilus NCFM (10 μg/ml) and Poly I:C, (10 μg/ml) for 3 hrs, RNA was extracted, and the induction of the gene coding for IFN-β, toll-like receptor-3 (TLR-3), interleukin-12 (IL-12) and IL-10 was determined by reverse transcription–polymerase chain reaction analysis. The messenger RNA levels were normalized to the relative expression of β-actin, *P < 0.05 values (control versus inhibitors). The data represent one of at least four independent experiments.
Induction of IL-12 and TLR-3 by L. acidophilus NCFM is dependent on IFN-β
Despite the vast difference in the Ifn-β expression profiles of DCs stimulated with L. acidophilus NCFM and Poly I:C, the Tlr-3 expression profiles obtained were highly similar (Fig. 1b). We therefore speculated that the Tlr-3 expression was caused by distinct mechanisms, i.e. that L. acidophilus NCFM Tlr-3 expression was induced through the action of IFΝ-β and that the Poly I:C-induced Tlr-3 expression was the result of another mechanism induced by direct binding of Poly I:C to TLR-3. To investigate the role of IFN-β in expressing Tlr-3, we added polyclonal anti-IFN-β antibodies to the cell cultures simultaneously with L. acidophilus NCFM, and measured the expression profiles of Tlr-3 and Il-12 (Fig. 7). The expression of Tlr-3 was almost completely inhibited upon addition of 10 μg/ml and 50 μg/ml polyclonal anti-IFN-β antibodies. By contrast, the expression of Il-12 was down-regulated by 20% and 46%, respectively. These results were confirmed by ELISA in the supernatant harvested after 24 hr (data not shown).
Figure 7.
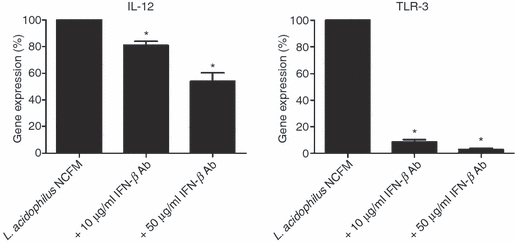
Induction of Il-12 and Tlr-3 in Lactobacillus acidophilus-stimulated dendritic cells (DCs) is dependent on interferon-β (IFN-β) Simultaneously with addition of L. acidophilus NCFM (10 μg/ml) to DCs, polyclonal IFN-β antibody was added in various concentrations (0 μg/ml, 10 μg/ml and 50 μg/ml). Cells were harvested after 10 hr, RNA was extracted and the gene expression of Il-12 and Tlr-3 was analysed by reverse transcription–polymerase chain reaction. The messenger RNA levels were normalized to the relative expression of β-actin, *P < 0.05 values (control versus L. acidophilus NCFM + IFN-β Ab). The data represent one of at least two independent experiments.
Discussion
In this study we have shown that the probiotic bacterium L. acidophilus possesses the capability to induce a viral defence phenotype in bone-marrow-derived murine DCs. Such properties have been demonstrated earlier using pathogenic bacteria,18,40 but to our knowledge this has not been demonstrated for bacteria regarded as non-pathogenic or even beneficial for the immune system. The induction of viral defence mechanisms may explain the ability of some probiotic bacteria to stimulate the immune system as demonstrated in a number of clinical trials, including their ability to protect against viral infection.4–6
The up-regulation of viral response genes seems to a great extent to be caused by a rapid and strong transient up-regulation of Ifn-β, which in turn stimulates transcription of a high number of other genes involved in viral defence. This was demonstrated in our microarray-based kinetics study, as the gene encoding IFN-β appeared to belong to a minor group of genes with a rapid transient profile. By this approach we showed that virtually all genes related to viral defence were among the most up-regulated genes and that a high number of these genes are known to be directly regulated through the action of type I IFNs.13,15 Strikingly, only the whole bacteria and not the TLR-2 ligands were able to induce detectable amounts of IFN-β and IL-12. This corresponds to our earlier findings26 that only intact bacteria are able to induce appreciable amounts of IL-12.
The up-regulation of Ifn-β in DCs was much stronger upon stimulation with L. acidophilus NCFM compared with cells stimulated with Poly I:C, E. coli Nissle 1917 and B. bifidum Z9. The up-regulation of Ifn-β correlated with an increased expression of Tlr-3 as well as Il-12, supporting the connection between IFN-β and IL-12 that was found by others.25 In contrast, the up-regulation of Tlr-3 was similar after stimulation of DCs with L. acidophilus NCFM and Poly I:C. This indicates that up-regulation of Tlr-3 does not exclusively depend on IFN-β, but may be affected by other mechanisms induced by virus recognition. Poly I:C has recently been shown to up-regulate IFN-β in a TLR-3-dependent manner in HEK293 cells and DCs,41 hence there is evidence that ligand binding to TLR-3 induces IFN-β and, conversely, that IFN-β is able to induce Tlr-3 expression. This may explain our observation that Tlr-3 is up-regulated to the same extent upon Poly I:C stimulation as upon L. acidophilus NCFM stimulation despite the considerable difference in the produced IFN-β. Conversely, the marked difference in IFN-β production by L. acidophilus NCFM and Poly I:C, respectively, may reflect two different mechanisms of induction which are likely to involve different receptors.
Not all probiotic bacteria were able to induce an up-regulation of Ifn-β and Tlr-3 in DCs, as demonstrated here with B. bifidum Z9, whereas another Lactobacillus strain, L. acidophilus X37, induced an IFN-β and TLR-3 response in a similar manner to L. acidophilus NCFM. The Gram-negative E. coli Nissle 1917, also considered probiotic, was not capable of inducing a significant expression of the genes coding for IFN-β or TLR-3. This is in accordance with the lack of capability of these bacteria to induce an extensive IL-12 production in DCs.26 To which extent other probiotic bacteria are capable of inducing IFN-β and viral defence genes is currently under investigation.
As the IL-12 response was shown to be dependent on TLR-2 in a previous study,26 we investigated the IFN-β response in DC from TLR-2−/− mice and found that TLR-2, as for IL-12 expression, is mandatory for an induction of IFN-β upon L. acidophilus NCFM stimulation. In contrast, lack of TLR-2 resulted in an increase of IFN-β upon stimulation with B. bifidum Z9 and E. coli Nissle 1917, although the IFN-β response was much lower compared with the level induced by L. acidophilus NCFM in WT mice. Hence, TLR-2 is not only playing a major role in the strong IL-12 and IFN-β response induced by L. acidophilus NCFM, it is simultaneously important for the suppression of the same response upon stimulation of DCs with other bacteria such as B. bifidum Z9 and E. coli Nissle 1917 investigated in the present study. This dualism in TLR-2's role is not well described, but confirms our previous studies on TLR-2's role for IL-12 induction.26 Whereas the response to the TLR-2 ligand Pam3CSK4 is generally reported to be weak,26,42 stimulation with whole bacteria through TLR-2 is reported to give rise to a strong pro-inflammatory response.22,26 Furthermore, Barbalat et al.14 have demonstrated that virus in contrast to Pam3CSK4 induces type I IFNs in bone-marrow-derived cells and other cells. Hence, as in our data, this points towards the importance of recognition of whole micro-organisms containing TLR-2 ligands for the induction of a strong Th1-promoting response in contrast to the effect of stimulation with TLR-2 ligands. The fact that certain Gram-negative bacteria are equally capable of inducing IFN-β15 further indicates that either other mechanisms involving other TLRs give rise to the same response or that TLR-2 is involved as well. A common mechanism involving distinct TLRs might be required for cellular uptake of the whole bacterium. Our results and the study by Barbalat et al.14 may point towards an IFN-β-inducing mechanism that is common to virus and certain bacteria.
In human DCs, IFN-β was found to be induced through a clathrin-dependent endocytotic mechanism.27 We also found that the induction of Ifn-β was dependend on phagocytosis, possibly through a clathrin-mediated mechanism, as addition of both the actin inhibitor cytochalasin and the clathrin inhibitor chlorpromazine abolished the induction of the gene coding for IFN-β in DC stimulated with L. acidophilus NCFM. We have previously shown that UV-killed, but intact, bacteria, in particular L. acidophilus, induce a response corresponding to live bacteria, which leads to much stronger IL-12 and TNF-α production compared with fragments or isolated cell walls of the bacteria.26 Taken together, this indicates that active uptake of the bacteria by endocytosis is important for the IFN-β induction. As TLR-2 was shown to be involved, our study suggests that TLR-2 plays an active role in the endocytosis-dependent IFN-β up-regulation, a phenomenon that to our knowledge has not been described before. However, whether there is a connection between the dependency of TLR-2 and endocytosis cannot be firmly established from the presented results. Maturation of DCs is generally considered to abolish endocytosis in these cells, but a number of studies reports that some degree of maturation may take place in DCs without abolishment of endocytosis. Weck et al.42 found that, in contrast to activation through TLR-3 and TLR-4, activation through TLR-2 with the synthetic TLR-2 agonist Pam3CSK4 did not abolish endocytosis. However, in contrast to Pam3CSK4, ligands like peptidoglycan and lipopeptides present in close proximity and in high numbers in an intact micro-organism may stimulate several TLRs – or other receptors – simultaneously and hence work through a distinct mechanism. Such receptor collaboration is well established for TLR-2 together with TLR-1 or TLR-6.43 Moreover, P. aeruginosa was shown to induce a strong pro-inflammatory response by a TLR-2 and mannose-receptor-dependent mechanism.24 The mannose receptor and TLR-2 form complexes on the cell surface during early phagocytosis and are found co-localized in endosomes for up to 1 hr after addition of the bacteria to the cells. From the present data, we cannot obtain further information about which receptors other than TLR-2 are important for the induction of IFN-β. Hence, we can neither exclude that TLR-3 or other TLRs play a role, nor that carbohydrate receptors, such as dectin-1 or scavenger receptors, are involved in the IFN-β induction. We did not investigate the involvement of the mannose receptor in the present study but it is conceivable that the mannose receptor, or another receptor, collaborates with TLR-2 in the activation of a pro-inflammatory response.
Charrel-Dennis et al.18 found that only live bacteria (streptococci) stimulated a strong induction of IFN-β; however, they compared with heat-killed bacteria while we stimulated with UV-killed bacteria. This indicates that some protein-containing or heat-vulnerable compound may be involved. Our previous studies showed that lipoteichoic acid, a major immunostimulatory component of Gram-positive bacteria, but not Pam2CSK4 or Pam3CSK4 was involved in the TLR-2-dependent stimulation of IL-12 production in DCs.26 Hence, other proteins or molecules important for the intact bacterium may be a prerequisite for triggering an appropriate immune response, perhaps in collaboration with a TLR-2 ligand. Salazar et al.22 stimulated monocytes with Borrelia burgdorferi, which responded through both a TLR-2-dependent and TLR-2-independent pathway, but only the TLR-2-independent response lead to an induction of IFN-β. This is in contrast to our finding, as we observed a dramatic effect in TLR-2−/− DCs. Consequently, different micro-organisms may stimulate antigen-presenting cells by distinct mechanisms giving rise to various cellular phenotypes or the specific cell may play an important role for the type of response to a given stimulus.
Taken together, these results add to the picture of TLR-2 as an important receptor for both pro-inflammatory and regulatory responses in antigen-presenting cells. Our study reveals that L. acidophilus is capable of stimulating a pro-inflammatory and antiviral response by a TLR-2-dependent mechanism, which suggests that TLR-2 acts a receptor, playing a central role in endocytosis-dependent stimulation of a pro-inflammatory response in DCs.
Acknowledgments
This study was supported by the Danish Strategic Research Council under the programme Food and Health. The skilled technical support of Marianne K. Pedersen, Anni Mehlsen and Pia Friis was highly appreciated.
Disclosures
The Authors declare that there is no conflict of interest.
References
- 1.Fuller R. Probiotics in human medicine. Gut. 1991;32:439–42. doi: 10.1136/gut.32.4.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gill HS, Guarner F. Probiotics and human health: a clinical perspective. Postgrad Med J. 2004;80:516–26. doi: 10.1136/pgmj.2003.008664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parvez S, Malik KA, Kang SA, Kim HY. Probiotics and their fermented food products are beneficial for health. J Appl Microbiol. 2006;100:1171–85. doi: 10.1111/j.1365-2672.2006.02963.x. [DOI] [PubMed] [Google Scholar]
- 4.Hatakka K, Savilahti E, Ponka A, et al. Effect of long term consumption of probiotic milk on infections in children attending day care centres: double blind, randomised trial. BMJ. 2001;322:1327. doi: 10.1136/bmj.322.7298.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rautava S, Salminen S, Isolauri E. Specific probiotics in reducing the risk of acute infections in infancy – a randomised, double-blind, placebo-controlled study. Br J Nutr. 2009;101:1722–6. doi: 10.1017/S0007114508116282. [DOI] [PubMed] [Google Scholar]
- 6.Leyer GJ, Li SG, Mubasher ME, Reifer C, Ouwehand AC. Probiotic effects on cold and influenza-like symptom incidence and duration in children. Pediatrics. 2009;124:E172–9. doi: 10.1542/peds.2008-2666. [DOI] [PubMed] [Google Scholar]
- 7.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 8.Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392:245–52. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 9.Christensen HR, Frokiaer H, Pestka JJ. Lactobacilli differentially modulate expression of cytokines and maturation surface markers in murine dendritic cells. J Immunol. 2002;168:171–8. doi: 10.4049/jimmunol.168.1.171. [DOI] [PubMed] [Google Scholar]
- 10.Zeuthen LH, Christensen HR, Frokiaer H. Lactic acid bacteria inducing a weak interleukin-12 and tumor necrosis factor alpha response in human dendritic cells inhibit strongly stimulating lactic acid bacteria but act synergistically with gram-negative bacteria. Clin Vaccine Immunol. 2006;13:365–75. doi: 10.1128/CVI.13.3.365-375.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat Immun. 2004;5:987–95. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- 12.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappa B by Toll-like receptor 3. Nature. 2001;413:732–8. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 13.Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–81. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 14.Barbalat R, Lau L, Locksley RM, Barton GM. Toll-like receptor 2 on inflammatory monocytes induces type I interferon in response to viral but not bacterial ligands. Nat Immunol. 2009;10:1200–7. doi: 10.1038/ni.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katze MG, He YP, Gale M. Viruses and interferon: a fight for supremacy. Nat Rev Immunol. 2002;2:675–87. doi: 10.1038/nri888. [DOI] [PubMed] [Google Scholar]
- 16.Bogdan C, Mattner J, Schleicher U. The role of type I interferons in non-viral infections. Immunol Rev. 2004;202:33–48. doi: 10.1111/j.0105-2896.2004.00207.x. [DOI] [PubMed] [Google Scholar]
- 17.Gratz N, Siller M, Schaljo B, et al. Group A streptococcus activates type I interferon production and MyD88-dependent signaling without involvement of TLR2, TLR4, and TLR9. J Biol Chem. 2008;283:19879–87. doi: 10.1074/jbc.M802848200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Charrel-Dennis M, Latz E, Halmen KA, et al. TLR-independent type I interferon induction in response to an extracellular bacterial pathogen via intracellular recognition of its DNA. Cell Host Microbe. 2008;4:543–54. doi: 10.1016/j.chom.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mancuso G, Midiri A, Biondo C, et al. Type I IFN signaling is crucial for host resistance against different species of pathogenic bacteria. J Immunol. 2007;178:3126–33. doi: 10.4049/jimmunol.178.5.3126. [DOI] [PubMed] [Google Scholar]
- 20.Stockinger S, Kastner R, Kernbauer E, et al. Characterization of the interferon-producing cell in mice infected with Listeria monocytogenes. PLoS Pathog. 2009;5:e1000355. doi: 10.1371/journal.ppat.1000355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Connell RM, Vaidya SA, Perry AK, Saha SK, Dempsey PW, Cheng GH. Immune activation of type IIFNs by Listeria monocytogenes occurs independently of TLR4, TLR2, and receptor interacting protein 2 but involves TNFR-associated NF-kappa B kinase-binding kinase 1. J Immunol. 2005;174:1602–7. doi: 10.4049/jimmunol.174.3.1602. [DOI] [PubMed] [Google Scholar]
- 22.Salazar JC, Duhnam-Ems S, La Vake C, et al. Activation of human monocytes by live Borrelia burgdorferi generates TLR2-dependent and -independent responses which include induction of IFN-β. PLoS Pathog. 2009;5:e1000444. doi: 10.1371/journal.ppat.1000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawai T, Akira S. Toll-like Receptor and RIG-1-like Receptor Signaling. Oxford: Blackwell Publishing; 2008. [DOI] [PubMed] [Google Scholar]
- 24.Xaplanteri P, Lagoumintzis G, Dimitracopoulos G, Paliogianni F. Synergistic regulation of Pseudomonas aeruginosa-induced cytokine production in human monocytes by mannose receptor and TLR2. Eur J Immunol. 2009;39:730–40. doi: 10.1002/eji.200838872. [DOI] [PubMed] [Google Scholar]
- 25.Gautier G, Humbert M, Deauvieau F, et al. A type I interferon autocrine–paracrine loop is involved in Toll-like receptor-induced interleukin-12p70 secretion by dendritic cells. J Exp Med. 2005;201:1435–46. doi: 10.1084/jem.20041964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeuthen LH, Fink LN, Frokiaer H. Toll-like receptor 2 and nucleotide-binding oligomerization domain-2 play divergent roles in the recognition of gut-derived lactobacilli and bifidobacteria in dendritic cells. Immunology. 2008;124:489–502. doi: 10.1111/j.1365-2567.2007.02800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Itoh K, Watanabe A, Funami K, Seya T, Matsumoto M. The clathrin-mediated endocytic pathway participates in dsRNA-induced IFN-β production. J Immunol. 2008;181:5522–9. doi: 10.4049/jimmunol.181.8.5522. [DOI] [PubMed] [Google Scholar]
- 28.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Workman C, Jensen L, Jarmer H, et al. A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-9-research0048. research0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bult CJ, Eppig JT, Kadin JA, Richardson JE, Blake JA. The Mouse Genome Database (MGD): mouse biology and model systems. Nucleic Acids Res. 2008;36:D724–8. doi: 10.1093/nar/gkm961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suh HS, Zhao ML, Rivieccio M, et al. Astrocyte indoleamine 2,3-dioxygenase is induced by the TLR3 ligand Poly(I:C): mechanism of induction and role in antiviral response. J Virol. 2007;81:9838–50. doi: 10.1128/JVI.00792-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fensterl V, White CL, Yamashita M, Sen GC. Novel characteristics of the function and induction of murine p56 family proteins. J Virol. 2008;82:11045–53. doi: 10.1128/JVI.01593-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sen GC, Lu L, Fensterl V, et al. Induction, functions and viral evasion of the ISG56 family of genes. Cytokine. 2008;43:234. [Google Scholar]
- 37.Lu JF, O’Hara EB, Trieselmann BA, Romano PR, Dever TE. The interferon-induced double-stranded RNA-activated protein kinase PKR will phosphorylate serine, threonine, or tyrosine at residue 51 in eukaryotic initiation factor 2 alpha. J Biol Chem. 1999;274:32198–203. doi: 10.1074/jbc.274.45.32198. [DOI] [PubMed] [Google Scholar]
- 38.Zhou AM, Hassel BA, Silverman RH. Expression cloning of 2-5A-dependent Rnase – a uniquely regulated mediator of interferon action. Cell. 1993;72:753–65. doi: 10.1016/0092-8674(93)90403-d. [DOI] [PubMed] [Google Scholar]
- 39.Stranden AM, Staeheli P, Pavlovic J. Function of the mouse Mx1 protein is inhibited by overexpression of the Pb2 protein of influenza-virus. Virology. 1993;197:642–51. doi: 10.1006/viro.1993.1639. [DOI] [PubMed] [Google Scholar]
- 40.Sing A, Merlin T, Knopf HP, et al. Bacterial induction of beta interferon in mice is a function of the lipopolysaccharide component. Infect Immun. 2000;68:1600–7. doi: 10.1128/iai.68.3.1600-1607.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trumpfheller C, Caskey M, Nchinda G, et al. The microbial mimic poly IC induces durable and protective CD4+ T cell immunity together with a dendritic cell targeted vaccine. Proc Natl Acad Sci USA. 2008;105:2574–9. doi: 10.1073/pnas.0711976105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weck MM, Grunebach F, Werth D, Sinzger C, Bringmann A, Brossart P. TLR ligands differentially affect uptake and presentation of cellular antigens. Blood. 2007;109:3890–4. doi: 10.1182/blood-2006-04-015719. [DOI] [PubMed] [Google Scholar]
- 43.Warshakoon HJ, Hood JD, Kimbrell MR, et al. Potential adjuvantic properties of innate immune stimuli. Hum Vaccin. 2009;5:381–94. doi: 10.4161/hv.5.6.8175. [DOI] [PubMed] [Google Scholar]


