Abstract
The Legionella pneumophila Dot/Icm type IV secretion system is essential for the biogenesis of a phagosome that supports bacterial multiplication, most likely via the functions of its protein substrates. Recent studies indicate that fundamental cellular processes, such as vesicle trafficking, stress response, autophagy and cell death are modulated by these effectors. However, how each translocated protein contributes to the modulation of these pathways is largely unknown. In a screen to search substrates of the Dot/Icm transporter that can cause host cell death, we identified a gene whose product is lethal to yeast and mammalian cells. We demonstrate that this protein, called SidI, is a substrate of the Dot/Icm type IV protein transporter that targets the host protein translation process. Our results indicate that SidI specifically interacts with eEF1A and eEF1Bγ, two components of the eukaryotic protein translation elongation machinery and such interactions leads to inhibition of host protein synthesis. Furthermore, we have isolated two SidI substitution mutants that retain the target binding activity but have lost toxicity to eukaryotic cells, suggesting potential biochemical effect of SidI on eEF1A and eEF1Bγ. We also show that infection by L. pneumophila leads to eEF1A-mediated activation of the heat shock regulatory protein HSF1 in a virulence-dependent manner and deletion of sidI affects such activation. Moreover, similar response occurred in cells transiently transfected to express SidI. Thus, inhibition of host protein synthesis by specific effectors contributes to the induction of stress response in L. pneumophila-infected cells.
Keywords: Bacterial pathogenesis, Virulence factor, Type IV secretion, eEF1A/HSF1
Introduction
Legionella pneumophila is a facultative intracellular bacterial pathogen capable of multiplying within fresh water amoeba and mammalian cells. Upon being phagocytosized, the bacterium initiates a unique trafficking route that bypasses the endocytic pathways to establish a Legionella-containing vacuole (LCV) with characteristics of the host rough endoplasmic reticulum (ER) compartment (Tilney et al., 2001). After 18 to 24 hours of replication, the vacuole is lysed and a large number of bacteria are released to initiate a new round of infection.
Essential to intracellular replication of L. pneumophila is the Dot/Icm type IV secretion system that translocates bacterial effector proteins into host cells during infection (Ninio and Roy, 2007). A large number of substrates of the Dot/Icm transporter have been identified but only a few of them have been assigned specific biochemical activities and roles in the biogenesis of the LCVs. It is well established that LCVs actively intercept vesicles originating from the endoplasmic reticulum to transform the bacterial vacuole into an ER-like compartment. Several bacterial effectors involved in this process have been characterized. Among them, RalF functions to recruit and activate Arf1 (Nagai et al., 2002) whereas and SidM/DrrA along with LepB, modulates the activity of Rab1 (Ingmundson et al., 2007; Machner and Isberg, 2007; Machner and Isberg, 2006; Murata et al., 2006). Another effector, SidJ, is important for efficient recruitment of ER proteins to the bacterial vacuole but its mechanism of action is unknown (Liu and Luo, 2007).
Reverse genetics and analysis of global gene expression profiles of infected cells revealed that in addition to vesicle trafficking, the Dot/Icm system modulates more cellular processes than previously anticipated (Dorer et al., 2006; Losick and Isberg, 2006). These observations may explain the large number of effectors transferred by the system. Depletion of components of host proteasome degradation machinery by siRNA leads to inhibition of L. pneumophila replication (Dorer et al., 2006). As anticipated, proteins predicted to be involved in ubiquintination pathways have been identified in L. pneumophila genomes and one such protein has been shown to possess ubiquitin ligase activity (Kubori et al., 2008; de Felipe et al., 2005).
Mammalian cells infected by L. pneumophila in its early stage exhibited strong resistance to exogenous apoptotic stimuli, this may be due to activities of cell death inhibitory effectors such as SdhA and SidF (Banga et al., 2007; Laguna et al., 2006) and by the virulence-dependent induction of many anti-apoptotic genes (Abu-Zant et al., 2007; Losick and Isberg, 2006). Mutations in certain Dot/Icm substrate genes resulted in host cell death, suggesting the existence of bacterial factors detrimental to the host (Banga et al., 2007; Laguna et al., 2006). In this study, we describe one such effector SidI, which functions as an inhibitor of host protein synthesis by targeting eEF1A and eEF1Bγ, two components of the elongation machinery of eukaryotic proteins synthesis (Browne and Proud, 2002). We also show that inhibition of protein synthesis by SidI leads to the induction of the host stress shock response, possibly by making eEF1A available to activate the heat shock response regulator HSF1.
Results
Identification of a L. pneumophila protein exhibits toxicity to eukaryotic cells
The availability of various genetic tools makes the yeast Saccharomyces cerevisiae a very useful model in the study of bacterial effectors (Siggers and Lesser, 2008). Several L. pneumophila effectors have been identified by their ability to kill yeast cells (Campodonico et al., 2005) or to interfere with its vesicle trafficking process (Heidtman et al., 2008; Shohdy et al., 2005). Furthermore, because some mammalian pro-apoptotic proteins exhibit lethality to yeast (Tao et al., 1997), the toxic phenotypes may also lead to the identification of cell-death inducing L. pneumophila effectors. The use of random libraries may not allow one to identify all such genes, as many genes require a full-length protein for their activity. Thus, we cloned full-length hypothetical L. pneumophila genes into a yeast vector and the resulting plasmids were individually introduced into a yeast strain. Transformation that yielded few colonies suggested the identification of a potentially toxic protein (Fig. S1). Several proteins exhibiting lethality to yeast were identified from the first ninety-six candidates (Table S1). Here we described one of such genes (lpg2504, ceg32 (Zusman et al., 2007)) that codes for a protein of 942 amino acids with a predicted molecular weight of approximately 110 kDa, which we referred to as sidI.
To confirm the lethality phenotype, we expressed SidI from the yeast galactose-inducible promoter and examined the inducer-dependent toxicity. Whereas all yeast strains grew normally on medium containing glucose, the strain harboring the chromosome-encoded Pgal::sidI failed to grow on medium containing galactose (Fig. 1A). These data are consistent with an earlier study showing that sidI cannot be expressed in yeast (Heidtman et al., 2008). Over-expression of foreign proteins sometimes leads to non-specific toxicity to yeast; so we performed two sets of experiments to examine the possibility that the observed SidI-mediated lethality was caused by a specific activity. First, we screened for SidI mutants that no longer were toxic to yeast. Plasmid DNA carrying sidI was mutagenized with hydroxylamine and the treated DNA was transformed into yeast. Surviving yeast strains were screened by immunoblotting for their ability to code for a full-size SidI protein (Fig. 1B and Fig. 2SA). Three such mutants were identified from about 400 candidates. In the first mutant, Glu482 was changed to lysine and the mutant was designated SidI*. In the other two mutants, Arg453 was substituted with either Leu or Pro. In this study, we will focus on the R453P mutant, which we referred to as SidI**. Consistent with their original non-toxic phenotypes, yeast strains carrying these alleles of sidI expressed from a galactose-inducible promoter gained the ability to grow on inducing medium (Fig. 1A lower panel). The loss of toxicity was not due to decreased protein stability because these mutants (SidI* and SidI**) encoded stable proteins in both yeast and E. coli (Fig. 1B and Fig. S2 A&B). Second, we transiently expressed SidI in mammalian cells of 80% confluency and found that cell survival significantly decreased in 24 hours (Fig. 1C left panel and D). In similar experiments with the mutants, although about 80% of the cells were transfected (expressing the GFP signal), the cells continued to proliferate to confluency (Fig. 1B right panel and data not shown). Furthermore, GFP fusions of these proteins can be detected in these cells (Fig. S2-C). These data indicate that these mutants have lost the cell killing activity. Taken together, our screening has identified a L. pneumophila protein toxic to eukaryotic cells, likely by targeting an essential host process(es) conserved among a wide spectrum of eukaryotes.
Fig. 1. Toxicity of SidI to eukaryotic cells.
A. SidI-mediated toxicity to yeast. Growth of yeast cells expressing sidI, sidI*, or sidI** from the galactose inducible promoter on glucose medium (upper panel) or on galactose medium (lower panel). A yeast strain harboring an empty vector was used as a control (vector). Serially diluted yeast cultures grown in glucose medium were spotted followed by incubation at 30 °C for 2 days (glucose) or 3 days (galactose). B. Untagged sidI* and sidI** was expressed in yeast. Total lysates of yeast cells grown in 2% galactose medium were probed with an anti-SidI antibody. C. Inhibition of mammalian cell proliferation by SidI. 293T cells on coverslips with about 80% confluence were transfected to express SidI (left panel) or the SidI* mutant (right panel) (transfection efficiency was about 80%); images were acquired from fixed samples 24 hours posttransfection. Bar: 50 μm. D. Relative viable cell number of samples transfected to express different alleles of sidI. 293T cells transfected to express the indicated proteins for 24 hours were lifted and viable cells were enumerated. Data were expressed in relative to samples transfected with the vector set at 100%. Similar results were obtained from at least three independent experiments each done in triplicate. **, p<0.004.
SidI is a substrate of the Dot/Icm system that is induced at exponential growth phase
sidI (ceg32) is one of genes identified to be regulated by PmrA, a protein that controls expression of many substrates of the Dot/Icm system (Zusman et al., 2007). We employed three independent methods, the interbacterial protein transfer assay (Luo and Isberg, 2004), the SidCΔC100 fusion assay (VanRheenen et al., 2006) and the Cya fusion assay (Bardill et al., 2005) to determine whether SidI is an L. pneumophila effector delivered into host cells by the Dot/Icm system. Dot/Icm-dependent transfer of SidI was readily detected by all three assays (Fig. 2A-E), indicating that this protein is a substrate of this type IV protein transfer system. We also used the saponin fractionation method (VanRheenen et al., 2006) to further examine the translocation of SidI into infected cells during infection. SidI was detected in soluble fraction from cells infected by the wild type L. pneumophila but not by the dotA mutant or the sidI deletion mutant (Fig. 2F 1st lane), indicating that SidI is delivered into host cytosol by the bacterium in a Dot/Icm-dependent manner.
Fig. 2. Dot/Icm-mediated translocation and growth phase-dependent expression of SidI.
A. Transfer of SidI by the Dot/Icm system between bacterial cells. Protein transfer was performed by using either Lp02(Dot/Icm+) or Lp03(dotA−) expressing a Cre::SidI fusion as the donor strains and a strain containing a floxed reporter as a recipient (Luo and Isberg, 2004). The transfer of Cre mediated by the effector SidF (Banga et al., 2007) was used as a positive control. Solid bars, donor strain L. pneumophila Lp02(Dot/Icm+); striped bars, donor strain L. pneumophila Lp03(dotA−). B-D. SidI-mediated translocation of the transfer-deficient SidCΔC100 into macrophages. Mouse macrophages were infected with bacteria expressing SidC, SidCΔC100 or SidCΔC100::SidI for one hour; L. pneumophila and SidC was differently labeled with distinctive antibodies followed by appropriate secondary antibodies as described (VanRheenen et al., 2006). At least 150 vacuoles in triplet samples were scored in each experiment. Representative images of vacuoles harboring sidCΔC100 (C) or sidCΔC100::sidI (D); bacteria were labeled in red and SidC was stained in green. E. Dot/Icm-mediated transfer of SidI measured by using the pertusis toxin Cya as a reporter. U937 infected with Lp02(Dot/Icm+) or Lp03 (dotA−) containing cya-sidI fusion plasmid were analyzed for cAMP production. The translocated effector SidJ (Liu and Luo, 2007) was used as positive control. Solid bars, L. pneumophila Lp02(Dot/Icm+); striped bars, L. pneumophila Lp03 (dotA−). F. SidI was translocated into host cells by the Dot/Icm system during infection. U937 macrophages were infected with relevant bacterial strains at an MOI of 5 for 6 hours and the collected cells were lysed with 0.2% saponin. Soluble proteins precipitated with methanol were separated by SDS-PAGE, transferred to nitrocellulose membranes. SidI in insoluble fraction was also probed (low panel). The cytosolic protein isocitrate dehydrogenase was probed as a control for bacterial cell lysis. Lanes: 1, wild type strain Lp02(Dot/Icm+); 2, Lp03(dotA−); 3, Lp02ΔsidI(Dot/Icm+); 4, total cell lysate of Lp02(Dot/Icm+). G-H. Expression of sidI is induced when L. pneumophila was grown to exponential phase. L. pneumophila cultures established by diluting stationary phase cells at 1:20 (starting OD600=0.2) in fresh AYE broth were grown at 37°C with vigorous shaking. The growth of the bacteria was monitored by measuring the values of OD600 at a 2-hour interval (G). At indicated time points (hours), equal amounts of cells (estimated by OD600 values) were withdrawn and lysates of the samples were separated by SDS-PAGE; proteins transferred to nitrocellulose membranes were probed with anti-SidI antibodies (H); the sidI deletion mutant grown at OD600=1.8 was loaded as a control (H last lane). The bacterial isocitrate dehydrogenase (ICDH) was detected as loading controls.
We examined the expression pattern of sidI in relation to bacterial growth phase. Interestingly, unlike most characterized substrates of the Dot/Icm transporter whose expression is induced at post-exponential phase, expression of sidI appears to be induced in cells of exponential growth phase (Fig. 2G-H), suggesting the transferring of this protein into host cell when bacteria are actively replicating (Liu et al., 2008).
In order to determine whether this gene is required for intracellular replication of L. pneumophila, we constructed an in-frame deletion mutant of sidI (Fig. 2H last lane). The mutant did not display detectable growth defect in mouse bone marrow-derived macrophages, Dictyostelium discoideum or the U937 cell line (Fig. S3 and data not shown), indicating that similar to many other substrates of the Dot/Icm transporter, sidI is not essential for intracellular multiplication of L. pneumophila in these infection models.
SidI interacts with components of the host translation elongation machinery
Since SidI shares no detectable homology to proteins in the database nor does it contain motifs suggestive of known biochemical activities, we attempted to determine its biochemical functions by identifying its targets in host cell. To this end, we incubated Affigel beads (Bio-Rad) coated with His6-SidI or GST with lysates of U937 cells. After washing with PBS buffer, proteins retained on the beads separated by SDS-PAGE were visualized by silver staining. Several proteins with molecular weights ranging from 40 kDa to 130 kDa were retained by beads coated with His6-SidI, but not by beads coated with GST (Fig. 3A lane 1 and 3). These proteins were identified by mass spectrometry analysis as components of the mammalian protein translation elongation machinery, including eEF1A, eEF1Bα, eEF1Bγ and the valinyl tRNA synthetase (Fig. 3A lane 3). Since these protein translation factors often are purified as a stable multi-subunit complex (Bec et al., 1989), we attempted to identify the direct binding targets of SidI by incubating purified GST-SidI with rabbit reticulocyte lysates (RRL) enriched in protein synthesis components and retrieved the tagged protein with glutathione beads. After washing with a more stringent buffer (Materials and Methods), two components of the protein synthesis elongation complex, eEF1A and eEF1Bγ were co-purified with GST-SidI but not with the control GST-SidF, suggesting that these two proteins interact directly with SidI (Fig. 3B).
Fig. 3. SidI interacts with components of the mammalian translation elongation machinery.
A. Components of the host protein synthesis machinery were retained by SidI. Affigel beads coated with GST (lane 1) or His6-SidI (lane 3) were incubated with mammalian cell lysates. His6-SidI coated beads incubated with cell lysis buffer (lane 2) was used as another control. After washing with lysis buffer, proteins separated by SDS-PAGE were visualized by silver staining; bands only retained by the His6-SidI coated beads were identified by MALDI /mass spectrometry analysis. Relevant protein size markers (in kDa) were indicated. B. Co-purification of eEF1A and eEF1Bγ with GST-SidI from rabbit reticulocyte lysates (RRL). Each GST-tagged protein was incubated with RRL and glutathione coated beads were used to retrieve GST-SidI or GST-SidF. After removing unbound proteins by extensive washing, the retained proteins resolved by SDS-PAGE were detected by Coomassie bright blue staining and identified by mass/spectrometry analysis. Relevant markers (in kDa) were indicated. C. Direct binding of SidI to eEF1A and eEF1Bγ. His6-SidI was incubated with GST-tagged eEF1A, eEF1Bγ or eEF1Bβ, and the protein complex was captured with glutathione beads, retained SidI was detected by immunoblotting. D. SidI* formed complexes with eEF1A or eEF1Bγ in mammalian cells. Lysates of 293T cells transfected to express GFP-SidI* or GFP-SidF were subjected to immunoprecipitation with an anti-GFP antibody, the precipitates resolved by SDS-PAGE were detected for eEF1A and eEF1Bγ using specific antibodies. 5% (50 μg) of total protein was probed as input controls (lanes 1 and 2). TCL: total cell lysates.
We further determined the direct binding of SidI to eEF1A and eEF1Bγ with individually purified proteins. We produced GST fusion proteins of eEF1A, eEF1Bα and eEF1Bγ and each of these proteins was incubated with His6-SidI. Glutathione beads were used to capture the GST fusion protein. After extensive wash with the TEN buffer (Materials and Methods), we found that SidI was retained by GST-eEF1A or by GST-eEF1Bγ (Fig. 3C). No interaction between SidI and eEF1Bα or eEF1Bβ was detected (Fig. 3C and data not shown). These data indicate that eEF1A and eEF1Bγ are the direct targets of SidI.
We also determined that the SidI* still interacts with both eEF1A and eEF1Bγ (see below and data not shown), indicating that the loss of lethality in this mutant is not due to the loss of target binding activity. Thus, we used SidI* to examine in vivo binding of SidI to its targets. Lysates of 293T cells transfected to express GFP::SidI* were subjected to immunoprecipitation with an anti-GFP antibody and co-precipitated proteins were detected with antibodies against eEF1A or eEF1Bγ. Although SidI* was expressed at a low level (Fig. 3D bottom panel 1st lane), eEF1A and eEF1Bγ was detected in the precipitates (Fig. 3D lane 3). Collectively, these data establish that SidI specifically binds eEF1A and eEF1Bγ of the translation elongation complex.
SidI inhibits mammalian protein synthesis
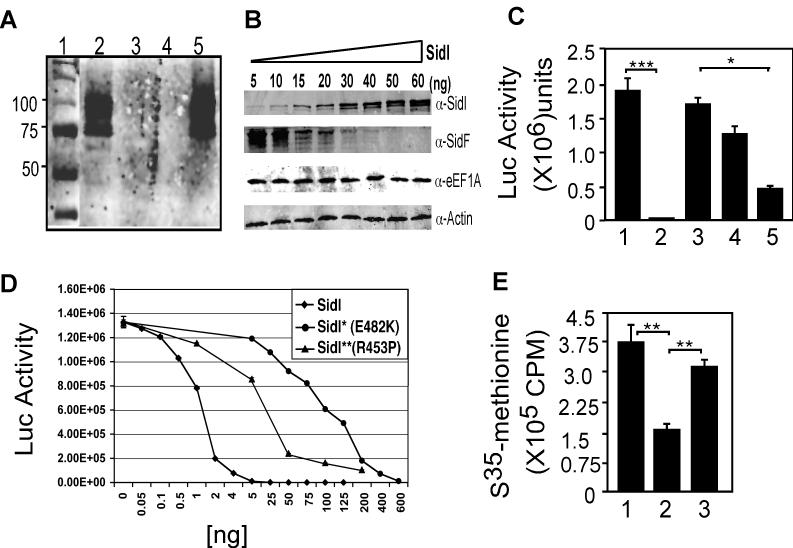
Our observations that SidI interacts with components of the eukaryotic translation elongation machinery suggest that this protein modulates host protein synthesis. Thus, we examined the effect of SidI on protein synthesis of mammalian cells using the RRL system that allows in vitro transcription/translation analysis (Promega, L1171). Adding a plasmid containing the sidF gene (Banga et al., 2007) led to robust production of SidF. However, inclusion of 180 ng of SidI completely inhibited the synthesis of SidF (Fig. 4A). The inhibitory effect was dependent upon the activity of SidI since heat-treated protein failed to block protein synthesis (Fig. 4A). Furthermore, as little as 30 ng of SidI was able to completely inhibit SidF synthesis (Fig. 4B). Such inhibitory effects were also observed when pre-synthesized mRNA encoding luciferase was used for analysis (Fig. 4C). The mRNA directed the synthesis of luciferase in the RRL system, but no light production was detected when active SidI but not when heat inactivated SidI was added. When added at 50 ng, the mutants SidI* and SidI** still exhibited partial inhibitory activity (Fig. 4C 4th and 5th bar).
Fig. 4. Inhibition of protein synthesis by SidI.
A. SidI abolished in vitro synthesis of SidF from plasmid pcDNA3-sidF. SidI or heat-inactivated SidI was added in an in vitro protein synthesis system (Promega) and the production of SidF was measured by immunoblotting. Lanes: 1, markers (kDa); 2, H2O; 3, 180 ng SidI; 4, 540 ng SidI; 5, 540 ng heat-inactivated SidI. B. Dose-dependent inhibition of SidF production. Different amounts of SidI were added to RRL containing pcDNA3-sidF and the production of SidF was detected by Western blot. The levels of relevant proteins were monitored by immunoblotting with the appropriate antibodies. Actin was detected as a loading control (lowest panel). C. Inhibition of translation of luciferase mRNA. Fifty ng of GST (1), His6-SidI (2), heat-inactivated His6-SidI (3), His6-SidI** (4) or His6-SidI* (5) was added to RRL containing luciferase mRNA. Production of light was detected after adding the luciferase assay reagent. D. Dose-dependent inhibition of protein synthesis by SidI and its mutants. His6-SidI or indicated mutants were added to RRL containing luciferase mRNA and the synthesis of luciferase was monitored by the production of light. E. Inhibition of in vivo protein synthesis by SidI. Seven hours after transfection, 35S-methionine was added to 293T cells for 3 hours and the incorporation of S35 was measured. 1, vector pEGFP-C1; 2, wild type sidI; 3, sidI*. Similar results were obtained from at least three independent experiments and the data shown are from one representative experiments done in triplicate. ***, p<0.0002; **, p<0.007; *, p<0.05.
Since both SidI* and SidI** still exert considerable inhibitory effect, we further analyzed their activity in a dose-response experiment. Under our experimental conditions, 1 ng of wild type SidI is sufficient to exert 50% inhibition (Fig. 4D). On the other hand, 100 ng of SidI* (E482K) or 25 ng of SidI**(R453P) is needed to achieve similar level of inhibition (Fig. 4D). These data indicate that these mutations severely impaired but did not abolish the protein synthesis inhibition activity of SidI.
Transfection of mammalian cells with a sidI expressing plasmid within 10 hours did not cause significant cell death (Fig. S4), we thus examined the effect of SidI on in vivo protein synthesis by the 35S-methionine incorporation assay (18). Under our experimental conditions where transfection efficiency was about 80% for the GFP vector or the plasmid carrying sidI* (data not shown), cells similarly transfected with the sidI construct incorporate 35S-methionine at about 35% of those transfected to express GFP (Fig. 4E). SidI* also detectably affects the incorporation of 35S-methionine, consistent with the partial inhibitory activity observed in in vitro experiments (Fig. 4E). Taken together, our data indicate that the direct consequence of binding of SidI to eEF1A and eEF1Bγ is the inhibition of host protein synthesis. Given the high degree of similarity among protein translation machinery from different eukaryotes, it is clear that the toxicity of SidI to yeast is due to a similar mechanism. The protein synthesis inhibition activity of SidI resembles the Lgt proteins that abolish eEF1A activity by glucosylating the G protein (Belyi et al., 2008; Belyi et al., 2006). To examine whether SidI and Lgt1 are functionally redundant, we constructed a L. pneumophila mutant lacking both genes, but no growth defect was detected for this double mutant in several hosts examined (Fig. S5).
SidI interacts with eEF1A independent of its nucleotide binding status
Despite the apparently similar effect of SidI and Lgt1 on protein synthesis, extensive mass/spectrometry analysis of eEF1A co-purified with active SidI (Fig. 3B) did not reveal any novel post-translational modification of this protein (data not shown). Like many monomeric G proteins, eEF1A transits between active and inactive forms; depending upon the nucleotides (GTP or GDP) it binds. GTPase activating proteins (GAP) or guanine nucleotide exchange factors (GEF) for classic small GTPases often exhibit higher binding affinity to the G protein associated with a specific nucleotide. To determine the biochemical activity of SidI, we first tested its interactions with eEF1A associated with different nucleotides. In a GST pull down assay, SidI exhibits higher affinity for untreated recombinant eEF1A than for its nucleotide free form (Fig. 5A). However, association with GDP or GTP does not affect the binding affinity of eEF1A to SidI (Fig. 5B). These results indicate that although SidI has a higher affinity for nucleotide-bound form of eEF1A, it does not preferentially bind eEF1A associated with a specific guanine nucleotide. This property is similar to that of eEF1Bα, the cognate GEF of eEF1A, whose binding constant to the G protein is independent of guanine nucleotides (Olarewaju et al., 2004). However, in our experiments to examine whether SidI modulates the nucleotide binding cycle of eEF1A, we did not detect any effect of SidI on the loading of GTP to eEF1A or the disassociation of GDP from the protein, nor did we detect any significant effect of SidI on the intrinsic GTPase activity of eEF1A (data not shown). SidI also does not possess detectable intrinsic GTPase activity because incubation of the protein with 32P γ-GTP did not lead to detectable release of 32PO4 (data not shown).
Fig. 5. SidI binds eEF1A independent of guanine nucleotide it associates.
A. SidI has a lower affinity for nucleotide free eEF1A. GST-eEF1A (lane 1), GST-eEF1A that had been dialyzed against 5 mM EDTA for 20 min. (lane 2) or GST (lane 3) was incubated with His6-SidI, and the protein complex was captured with glutathione beads. Retained His6-SidI was detected by immunoblotting. B. SidI binds eEF1A independent of the guanine nucleotides. Nucleotide free GST-eEF1A loaded with nonhydrolyzable GDPβS or (lane 1) or GTPγS (lane 2) was incubated with His6-SidI and the protein complexes were captured and detected as described in A. The GST protein was used as a control (lane 3). In each case, 5% of the input was detected as controls. C. Mutants SidI*(E482K) and SidI**(R453P) binds eEF1A with affinities comparable to that of wild type protein. The indicated amounts of His6-SidI or its mutants were added to wells of immulon-2 plates coated with eEF1A, SidI or the mutant proteins retained by eEF1A was detected by ELISA.
To determine whether binding to eEF1A by SidI is sufficient for the protein synthesis inhibition activity, we examined whether SidI* and SidI** exhibit any decrease in their binding affinity to eEF1A by an ELISA assay. Similar amounts of SidI and the mutants were required to saturate immobilized eEF1A (Fig. 5C), all with a Kd of about 10 nM, indicating that the mutations did not affect the binding of SidI to eEF1A. Taken together, these data suggest that physical binding is not sufficient for the inhibitory effect of SidI on eEF1A. Other activities such as subtle but effective modulation of eEF1A GTP hydrolysis cycle or yet undetected post-translational modifications of the target proteins may be responsible for its function.
SidI contributes to the induction of host stress response during L. pneumophila infection
Recent studies indicate that in addition to its canonic function in protein synthesis, eEF1A is involved in the regulation of diverse cellular processes including cell death, cytoskeleton structure and stress response (Borradaile et al., 2006; Shamovsky et al., 2006; Gross and Kinzy, 2005). Infection of wild-type L. pneumophila in both amoeba and mammalian hosts results in a robust induction of stress response genes (Farbrother et al., 2006; Losick and Isberg, 2006). Moreover, a recent study indicates that eEF1A is essential for the activation of heat shock factor 1 (HSF1), the major stress response regulatory protein in mammalian cells (Shamovsky et al., 2006). Therefore, we examined whether HSF1 is activated during L. pneumophila infection and, if so, whether the activity of SidI contributes to such activation. The activation of HSF1 was monitored by the formation of an HSF1/eEF1A protein complex detected by co-immunoprecipitation (Shamovsky et al., 2006). A large amount of the HSF1/eEF1A complex were detected in cells subjected to heat shock at 42 °C for 20 min (Fig. 6A lane 5). Basal level association between these two proteins was also detected in resting cells or cells infected with a Dot/Icm-deficient L. pneumophila strain (Fig. 6A lanes 1 & 2). The lack of HSF1 activation in infections using the dotA mutant was not due to degradation of internalized bacteria because these bacteria were viable by the time the cells were lysed (Fig. S6). Importantly, a high level of HSF1/eEF1A complex was detected in cells infected with wild-type bacteria (Fig. 6A lane 3). Furthermore, the amount of the protein complex was reproducibly decreased in cells infected with the sidI mutant but not the lgt1 mutant (Fig. 6A lanes 4 and 6). Expression of SidI from a plasmid partially restored the reduction (Fig. 6A lane 7). Interestingly, the formation of the eEF1A/HSF1 complex also occurred in cells transfected to transiently express SidI but not the SidI* mutant (Fig. 6B lanes 2 and 3).
Fig. 6. SidI contributes to the activation of HSF1 during L. pneumophila infection.
A. L. pneumophila infection-mediated activation of HSF1. Upper panel, cell lysates of U937 macrophages infected with the relevant bacterial strains at an MOI of 5 were subjected to immunoprecipitation with an anti-eEF1A antibody and the precipitated HSF1 was detected after SDS-PAGE. Lanes: 1, uninfected; 2, Lp03(dot/icm−); 3, Lp02(dot/icm+); 4, Lp02ΔsidI; 5, heat shock; 6, Lp02Δlgt1; 7, Lp02ΔsidI/psidI. Middle two panels: eEF1A and HSF1 was probed respectively to detect cellular levels of these proteins. Lower panel, quantitation of band intensity of upper panel relative to that of the heat shock treated cells set at 100%. B. Activation of HSF1 by SidI but not by the SidI* mutant. Immunoprecipitates were obtained with an eEF1A specific antibody from lysates of 293T cells transfected for 6 hours with the appropriate plasmids. HSF1 was detected as described in A. Lanes: 1, GFP; 2, SidI; 3, SidI*; 4, heat shock. C. Infection of L. pneumophila increases the DNA binding activity of HSF1. Lysates prepared from U937 macrophages infected with relevant bacterial strains were used for assay the binding of HSF1 to HSE. The probe was detected with an antibody specific for digoxigenin used to label the binding target. Lanes: 1, Uninfected; 2, Lp02(dot/icm+); 3, Lp03(dot/icm−); 4, heat shock; 5, Lp02ΔsidI; 6, Lp02ΔsidI/pSidI. Competition experiments received 5 times more unlabeled probe were performed to determine the specificity of the binding. D. Induction of hsp70. qPCR analysis for hsp70 was performed with cDNA derived from total RNA of U937 cells infected for 8 hours (Infection) or 293T cells transfected to express indicated genes for 6 hours (Transfection). The levels of expression were normalized with mRNA specific for the GAPDH gene. Relative gene expression represents the normalized values of the indicated samples against cDNA of uninfected cells or cells transfected to express GFP. Lanes: 1, uninfected; 2, Lp03(dot/icm−); 3, Lp02(dot/icm+); 3, Lp02ΔsidI; 5, heat shock; 6, Lp02Δlgt1; 7, Lp02ΔsidI/psidI; 8, vector expressing GFP; 9, GFP::SidI; 10, GFP::SidI*; 10, heat shock. Data shown are representatives of more than three independent experiments.
Under heat shock conditions, the activation of HSF1 by eEF1A and a non-coding RNA molecule enhances the binding of HSF1 to the heat shock element (HSE), thus leading to induction of target gene expression (Shamovsky et al., 2006). To determine whether infection by L. pneumophila has similar effect on HSF1, we examined the DNA binding activity of HSF1 from cells infected with relevant L. pneumophila strains by the electrophoretic mobility shift assay. No shift was detected when lysates of uninfected cells or cells infected by the Dot/Icm-deficient strain were used for binding (Fig. 6C lane 1 and 3). However, retardation of the probe was detected in reactions using lysates from cells infected by wild type bacteria or cells treated by heat shock (Fig. 6C lane 2 and 4). Importantly, infection with the sidI deletion mutant resulted in no detectable shift (Fig. 6C lane 5), a defect that can be restored partially by expressing SidI from a plasmid (Fig. 6C lane 6 arrow). The binding of HSF1 to the heat shock element was specific because the addition of 5X non-labeled probe abolished the shift (Fig. 6C). These results further point to a role of SidI in the activation of HSF1.
To determine whether the activation of HSF1 leads to induction of hsp genes, we examined mRNA levels of hsp70, one of the target genes of HSF1, by real-time PCR analysis. Consistent with the observed HSF1 activation, heat shock treatment led to about 18-fold induction of hsp70 transcription (Fig. 6D). Similarly, expression of the hsp70 gene was induced about 12-fold in cells infected with wild-type L. pneumophila for 8 hours but no induction was observed in cells infected with a dot/icm-deficient strain. Such induction detectably decreased in cells infected by the sidI deletion mutant (Fig. 6D, 4th bar). Interestingly, transient expression of wild-type SidI, but not the non-active mutant SidI* led to about 15-fold induction of hsp70 (Fig. 6D), indicating that SidI-mediated inhibition of protein synthesis led to increased expression of hsp70. Unexpectedly, we did not detect significant increase of the Hsp70 protein in cells infected by L. pneumophila throughout the infection cycle (Fig. S7). HSF1 is known to regulate the expression of a large number of genes and the gene list is still expanding (Westerheide and Morimoto, 2005), it is possible that proteins encoded by some of these genes are elevated at detectable levels in L. pneumophila infected cells. Taken together, our data suggest that HSF1 is activated by L. pneumophila infection and that SidI contributes to such activation.
Discussion
Infection by L. pneumophila leads to significant inhibition of host protein synthesis (McCusker et al., 1991). Here we show that SidI, a substrate of the Dot/Icm transporter may contribute to such inhibition in a process that results in the induction of the host stress response.
It is well established that activated eEF1A is responsible for loading charged amino acids to the A site of the ribosome during protein synthesis, whereas eEF1Bγ, along with two other components eEF1Bα and eEF1Bβ, is the GEF responsible for activating eEF1A by loading GTP to this G protein (Browne and Proud, 2002). There is no detectable homology between eEF1A and eEF1Bγ, suggesting that these proteins bind to different regions of SidI. The observation that the two non-toxic SidI substitution mutants still bind eEF1A in a manner similar to that of wild type protein indicates that simple physical association is insufficient for SidI to exert its full inhibitory activity. Whether the residual inhibitory activity of SidI* and SidI** (Fig. 4C-D) results from their interactions with eEF1Bγ remains to be studied. SidI could modulate the GTP hydrolysis cycle of eEF1A; our inability to detect such effect may result from the extremely low intrinsic GTPase activity of purified eEF1A (Crechet and Parmeggiani, 1986). Equally possible is that SidI biochemically modifies eEF1A or eEF1Bg at post-translational level. However, despite extensive effort, we did not detect novel post-translational modification on eEF1A associated with SidI (Fig. 3B). The identification of novel post-translational modifications on eEF1A is complicated by its natural extensive and dynamic modifications (Lamberti et al., 2004).
Given its toxicity to cells when overexpressed, SidI could contribute to the demise of host cells during L. pneumophila infection, though active killing of the host cells apparently would be detrimental to an intracellular pathogen. Three lines of evidence suggest that SidI plays a role in the induction of the host heat shock genes associated with L. pneumophila infection. First, deletion of sidI but not lgt1 led to a reduction in the activation of HSF1 in infected cells (Fig. 6). Second, transient expression of SidI induced HSF1 activation (Fig. 6B). Although the binding of HSF1 to HSE was not detected in cells infected by the sidI mutant (Fig. 6C), active HSF1 clearly existed in these cells (Fig. 6 A and D). These discrepancies may result from the sensitivity of the assays employed in these experiments. According to a current model, one of the most direct effect of heat shock or other stress is a drastic decrease in protein synthesis rate that leads to the release of eEF1A for the activation of HSF1 (Shamovsky et al., 2006). Interestingly, perturbations that interfere with protein synthesis often lead to stress resistance. For example, in yeast mutations in gene coding for eEF1Bγ result in constitutive resistance to oxidative stress (Olarewaju et al., 2004). The protein synthesis inhibitory activity of SidI could exert similar effect to make eEF1A available for the activation of HSF1. Finally, the expression of sidI is induced when the bacterial cells reach exponential growth phase, concomitant with the occurrence of hsp induction during infection (Fig. 2H and ref. 10), further suggesting a role of SidI in this process. Deletion of sidI does not completely abolish the induction of stress response in L. pneumophila-infected cells (Fig. 6), suggesting the presence of other effectors in this process. In agreement with this notion, two additional proteins targeting eEF1A were recently reported (Belyi et al., 2008).
Stress shock response is a fundamental mechanism necessary for cell survival under a variety of unfavorable conditions, such as cellular stress induced by physical, chemical or biological agents. Accordingly, the induction of hsp genes is very likely beneficial to L. pneumophila replication. For example, the chaperone activity of Hsps may be important for the proper folding of relevant host proteins or L. pneumophila effectors delivered by the Dot/Icm transporter. Alternatively, since Hsps also are involved in important cellular processes such as cell death and resistance to ER stress, it is possible that Hsps-mediated modulation of these pathways is important for intracellular growth of L. pneumophila (Losick and Isberg, 2006). Taken together, we postulate that protein synthesis inhibition mediated by SidI plays a role in induction of the host stress response, possibly to provide a cellular environment more conducive for bacterial replication. The induction of a host heat stress response has been documented in infections caused by a variety of pathogens and its roles in infections can be either beneficial or detrimental to the infecting agents (Singh and Aballay, 2006; Glotzer et al., 2000). Clearly, different pathogens have evolved different strategies to modulate this cellular pathway.
The common activities of Lgt proteins and SidI in inhibiting host protein synthesis indicates that L. pneumophila had evolved more than one mechanism to modulate the activity of eEF1A, which may represent one example of the functional redundancy among virulence factors of L. pneumophila (Luo and Isberg, 2004). Recent studies indicate that eEF1A is involved in several non-canonic cellular processes, including the activation of HSF1 (Shamovsky et al., 2006), the TCTP-mediated tumor proliferation (Cans et al., 2003) and the resistance to cell death induced by lipotoxin or ER stress (Borradaile et al., 2006; Talapatra et al., 2002). It is possible that regulation of eEF1A activity by SidI and other bacterial factors also alters other host cellular processes necessary for bacterial intracellular growth. That a mutant lacking both Lgt1 and SidI did not detectably affect intracellular replication of L. pneumophila suggests the existence of more L. pneumophila effectors dedicated to modulate cellular pathways regulated by this multi-functional protein.
Materials and Methods
Bacterial, yeast strains and plasmid construction
All L. pneumophila strains used in this study were derivatives of the Philadelphia 1 strain Lp02 (Berger and Isberg, 1993). Bacteria were grown and maintained on CYE medium as previously described (Conover et al., 2003). In complementation experiments, the vector used for expressing the gene of interest was introduced into the wild type strain or mutants and the resulting strains were used for infection. E. coli strains were grown and maintained on LB agar or LB broth. When necessary antibiotics were included as described (Conover et al., 2003). Following a standard procedure (Dumenil and Isberg, 2001), we generated the sidI in-frame deletion mutant ZL07 with plasmid pZLΔsidI, which was constructed by ligating into SacI/SalI digested pSR47s (Berger and Isberg, 1993) two PCR products amplified from Lp02 genomic DNA using SidIup5′SalI/SidIup3′BamHI (Table S3), SidId5′BglII/SidId3′SacI (digested with BglII/SacI). To complement the mutation, the coding region of sidI was amplified using SidICom5′SacI/SidI3′SalI and the PCR product was inserted into pJB908 (Bardill et al., 2005) as a SacI/Sal I fragment. The plasmid used for deleting Lgt1 was constructed similarly with primer pairs PL301/ PL302 and PL303/PL304. All infections were conducted with bacterial cultures grown to the post-exponential phase as judged by optical density of the cultures (OD600=3.3-3.8) as well as an increase of bacterial motility. SidI and its derivatives were cloned into pEGFPC1 (Clontech) for expression in mammalian cells. cDNA clones of used mammalian genes were amplified from a human kidney cDNA library (Clontech) or from clones purchased from ATCC. All genes were verified by sequencing analysis. The sequences of all primers used are in Table S3.
Yeast manipulation and screening of L. pneumophila proteins toxic to eukaryotic cells
Yeast strains were PJ69-4A (James et al., 1996) or W303 (Fan et al., 1996). Yeast was grown in YPD medium or in appropriate amino acid dropout minimal media. Full-length genes larger than 350 amino acids of L. pneumophila were individually cloned into pGBKT7 (BD Biosciences) as a BamHI/SalI or BglII/SalI fragment and the resulting plasmids was introduced into yeast strain PJ69-4A (James et al., 1996) by a standard transformation protocol (Gietz et al., 1995). After 3 days incubation at 30 °C, transformations that resulted in few no transformants were retained for further analysis. For the determination of expression-dependent toxicity, SidI or its derivatives SidI* and SidI** was cloned into pSB157 (Fazzio and Tsukiyama, 2003) (courtesy of Sue Biggins, Fred Hutchinson Cancer Research Center, Seattle, WA) and the resulting plasmids were linearized with an appropriate restriction enzyme and then transformed into yeast strain W303 (Fan et al., 1996). Several transformants were analyzed for galactose-dependent toxicity by spotting serially diluted cultures grown in glucose medium. To prepare cell lysates for protein analysis, cells from 5 ml overnight cultures were first lysed with a cracking buffer (40 mM Tris-Cl [pH6.8], 5% SDS, 0.1 mM EDTA, 8 M urea, bromothymol Blue 0.4 mg/ml) with glass beads. Samples were resolved by SDS-PAGE after adding Laemmli buffer.
Random mutagenesis of sidI
DNA of plasmid pSB3 (Table S2) was treated with hydroxylamine essentially as described (Luo and Farrand, 1999). Five μg of plasmid DNA was incubated in a 100 μl reaction mixture containing 0.5 M hydroxylamine, 0.5 mM Na2EDTA, and 5 mM Tris-HCl (pH 6.0) either for 10 hours at 37°C or for 1 hour at 65 °C. Treated DNA was purified with an affinity column (Zymo), dissolved in ddH2O and was introduced into the yeast strain PJ69-4A (James et al., 1996). Transformants were plated onto Trp dropout plates and surviving colonies were purified. Protein samples prepared from 5 ml culture were resolved by SDS-PAGE, transferred to nitrocellulose membranes and detected by immunoblotting with an anti-SidI antibody. Clones expressing full-size SidI were further verified by cloning the mutated alleles of sidI into new vector. The mutations were identified by double strand DNA sequencing analysis.
Protein purification
Native eEF1A was purified from rat livers as described (Shamovsky et al., 2006). To express and purify GST-SidI and His6-SidI, the open reading frame of this gene was amplified by PCR using SidIP5′BamHI/SidIF3′SalI and the resulting DNA fragment was inserted into pGEX-6P-1 (GE Healthcare) or pQE30 (Qiagen). GST-Lgt1. GST-eEF1A, GST-eEF1Bα, GST-eEF1Bβ, GST-eEF1Bγ, were expressed similarly from pGEX-6P-1 (GE Healthcare). For protein production, bacteria were grown at 37°C in LB medium (100 mg/ml Ampicillin) to an OD600 of 0.5, shifted to 22°C, and then induced with 0.2 mM isopropylthio-D-galactopyranoside (IPTG), and cultivated for an additional 15 hr at 22°C. Harvested cells were lysed in a French press at 1,500 psi, and the soluble fraction obtained by centrifugation at 6,000g for 10 min at 4°C was incubated with glutathione Sepharose resin or Ni2+ resin (Amersham Pharmacia) equilibrated with PBS in order to purify recombinant affinity-tagged proteins. We washed the resin with 20 times of the column volume with PBS; for Ni2+ resin, imidazole was included at 20 mM in the washing buffer to facilitate the removal of non-specifically bound proteins. The GST tag was removed by incubation with PreScission Protease (100 U) (Amersham Pharmacia) for 18 hours at 4°C, and recombinant proteins were eluted from the glutathione resin with PBS. Bound His6-tagged proteins were eluted with PBS buffer containing 200 mM imidazole. After dialyzing against PBS to remove imidazole, the protein was further purified by gel filtration with an FPLC system using a Superdex 200 10/300 GL column (GE Healthcare). TBST buffer (50 mM Tris-Cl, 150 mM NaCl, 0.1% Triton-X100, pH 7.4) was used as eluent and the flow rate was set at 0.4 ml/min. The single peak corresponding to the protein was collected, dialyzed in the appropriate buffer for subsequent use. Protein concentrations were determined by the Bradford assay; the purity of all proteins was more than 95% as assessed by SDS-PAGE followed by Coomassie bright blue staining (Fig. S2-B).
Cell culture and transfection
U937 cells were cultured in RPMI medium supplemented with 10% fetal bovine/calf serum (FBS) and 5 mM glutamate. 293T cells were cultured in Dulbecco’s modified minimum Eagle’s medium (DMEM) supplemented with 10% FBS. When necessary, U937 cells were differentiated into macrophages by incubating at 37 °C with 5% CO2 for 36-48 hours with 50 ng/ml phorbol myristic acid (PMA). Cells grown to about 80% confluence were transfected with Lipofectamine 2000 (Invitrogen) following manufacturer’s instructions. Mouse macrophages were prepared from bone marrow of female A/J mice of 6-10 weeks of age following published protocols (Swanson and Isberg, 1995). For infection, macrophages were plated into 24-well plates at 2×105 cell per well. Infection was carried out at the indicated MOIs as required by the particular experiments. In all cases, one hour after adding bacteria to cultured cells, infection was synchronized by washing the infected cells three times with warm PBS buffer.
Affinity chromatography from cell lysates and in vitro protein binding
We coated Affigel beads using purified His6-SidI with protocols supplied by the manufacturer (Bio-Rad). U937 cells collected from 500 ml culture were lysed in a glass homogenizer Dounce (Wheaton) with 0.5 ml PBS supplemented with 5 mM DTT and protease inhibitor cocktail (Roche). After removing unbroken cells and nuclei by centrifugation at 10,000 × g for 10 min at 4°C, the post-nuclear supernatant was added to beads and incubated for 14 hr at 4°C. Beads were washed five times with PBS and bound proteins were treated with SDS sample buffer. Proteins resolved by SDS-PAGE were visualized by silver staining (Bio-Rad). Individual protein gel bands were excised, digested with trypsin, and analyzed by matrix-assisted laser desorption/ionization /mass spectrometry (MALDI/MS) (Taplin Biological Mass Spectrometry Facility, Harvard Medical School).
For GST pull down experiments from rabbit reticulocyte lysate (RRL), 5 μg purified GST fusion protein was mixed with 200 μl RRL on a rotator for 3 hrs at 4 °C and 100 μl pre-washed glutathione beads were added to the reactions. After another 2 hours of incubation at 4 °C, the beads were washed three times with TEN buffer (100 mM Tris-Cl [pH 8.0], 10 mM EDTA, 200 mM NaCl). Proteins associated with beads were solubilized with SDS sample buffer and were separated by SDS-PAGE. Proteins were detected by coomassie bright blue staining.
To analyze protein-protein interactions with purified proteins, 2 μg purified GST-fusion protein was mixed with 2 μg His-tagged protein in PBS for 1 hour at 4 °C. After adding 50 μl of pre-washed glutathione beads, binding was allowed to proceed for 1 hour. The beads were then washed 5 times with TEN buffer containing 400 mM NaCl. Retained proteins were detected by immunoblot after SDS-PAGE.
ELISA
To compare the binding affinity between wild-type SidI protein and its point mutant forms, His-tagged proteins were purified from bacteria first using Ni-NTA affinity column, and then by passing through gel filtration column (Superdex 200 10/300 GL). For ELISA, immulon-2 96-well plate was coated overnight with purified GST-EF1A (75 ng/well) in bi-carbonate buffer (pH 9.6). The wells were subsequently blocked with 5% BSA for 1hr at 30 °C. Purified SidI proteins were then added at different concentrations ranging from 0.01-2000 nM to the wells and the plate incubated at 25 °C for 90 min to allow binding of proteins. Wells were then washed 5× with PBS containing 0.1% Tween 20 and incubated with affinity-purified SidI antibody (1:1000) at 37 °C for 1hr. The plate was again thoroughly washed 5× with above PBS wash buffer followed by addition of 1:10,000 dilution of secondary antibody conjugated with horseraddish peroxidase. After incubation at 37 °C for 1hr, the wells are washed 5× and developed with 1-Step Ultra-TMP-ELISA (Pierce) substrate. The reactions were terminated with 2 M sulfuric acid as per manufacture’s instructions and quantified by measuring absorbance at 450 nm. For the controls, wells coated with BSA protein were subjected to binding with SidI protein.
Cell Viability assay
Viability of 293T cells transfected to express SidI, SidI* or GFP was measured using the trypan blue dye according to a published protocol (Freshneyy, 2007). Briefly, 10 hours after transfection, cells were resuspended in a concentration of about 106 cells/ml and were diluted 1:1 with a 0.4% trypan blue solution. The number of stained cells and total number of cells were determined using a hemocytometer. The percentage of viable cells (unstained) was calculated.
In vitro protein synthesis
For transcription/translation analysis, we added 0.1 μg DNA of a plasmid carrying sidF (Banga et al., 2007) to 10 μl of the TNT system (Promega, L1171). For protein translation from pre-made mRNA, mRNA coding for the firefly luciferase was added to the RRL system (Promega, L4960) at a final concentration of 0.03 μg/μl. In each case, the reactions were allowed to proceed for 2 hours at 30°C. Purified SidI or its mutants were added to the reactions at indicated concentrations. The production of proteins was detected by Western blot or by measuring production of light with the luciferase assay reagent (LAR, Promega) substrate following by detection with a luminometer (Thermo Scientific).
In vivo protein synthesis
To measure the effect of SidI on de novo protein synthesis, we followed a published protocol (Belyi et al., 2006). Briefly, we transfected 293T cells seeded in 24-well plates with plasmids carrying gfp, sidI or sidI* with Lipofectamine 2000 according to instructions of the manufacturer (Invitrogen). After incubating at 37°C for 6 hours, cells were washed with DMEM-Met (Invitrogen) and were incubated for another one hour. The protein synthesis was then chase-labeled with 35S-methionine (0.5 Ci/well)(Perkin Elmer) for 3 h in DMEM-Met medium. Cells were then washed with PBS for 5 times and were lysed by 0.1% SDS supplemented with 0.2 mg/ml BSA. Proteins were precipitated by 10% trichloroacetic acid, filtered onto nitrocellulose membranes. The incorporated 35S-methionine was measured by liquid scintillation counting.
Immunoprecipitation
We seeded 293T cells on 100-mm plates at 5×105 cells per plate one day before transfection, and then transiently transfected cells with indicated plasmids. At the time points required by the experiments, we washed the cells with ice-cold PBS and lysed them with a solution containing Tris (20 mM, pH 8.0), NaCl (150 mM), 0.2 % NP-40 (w/v), 10% glycerol (w/v), NaF (100 mM), PMSF (1 mM), sodium orthovanadate (Na3VO4)(1 mM), aprotinin (20 μg/ml), and leupeptin (40 μg/ml). After incubation on ice for 30 min, cell lysates were centrifuged at 12,000g for 15 min at 4 °C. To prepare immunoprecipitates, we incubated cell lysates with an appropriate antibody and protein G agarose (Roche) for 7-12 hours at 4 °C. We then washed the agarose beads five times with lysis buffer, and solubilized adsorbed proteins by boiling the washed beads for 5 min in SDS–PAGE loading buffer. Proteins of interest were detected by Western blot after SDS–PAGE. To detect the second protein, membranes were treated with a stripping buffer (2% SDS, 100 mM β-mercaptoethanol, 62.5 mM Tris-Cl, pH 6.8) for 30 min at 70 °C and the proteins were detected by appropriate antibodies. For co-immunoprecipitation using lysates from infected cells, 5×107 U937 macrophages seeded in 100 mm Petri dish were infected at an MOI of 5 for 8 hours with indicated L. pneumophila strains. Cell lysates and precipitates were prepared as described above.
Antibodies, immunostaining and Western Blot
To prepare anti-Legionella antisera, we first fixed bacterial cells of strain Lp03 with 0.5% paraformaldehyde in PBS for 30 min at 37 °C. After extensively washing the cells with PBS (10 times with 50× volume of the pellet), the cells were used to immunize rat or rabbit following standard protocols (Pocono Rabbit Farm and Laboratory, Canadensis, PA). SidI specific antibodies were prepared by injecting rabbits with purified proteins with a similar procedure by the same service provider. When necessary, antibodies were affinity-purified against the same proteins covalently coupled to an Affigel matrix (Bio-Rad) using standard protocols (3). Monoclonal and polyclonal antibodies against GFP, eEF1A, eEF1Bγ were purchased from Sigma, Millipore and Bethyl Laboratories (Montgomery, TX), respectively. For immunostaining, anti-Legionella antisera were used at 1:10,000; purified anti-SidC was used at 1:500. Cell fixation, permeabilization and immunostaining were performed as described (Conover et al., 2003). Intracellular bacteria were distinguished from extracellular bacteria by differential immunostaining with anti-L. pneumophila antibodies.
For Western blots, samples resolved by SDS-PAGE were transferred onto nitrocellulose membranes. After blocking with 4% milk, membranes were incubated with the appropriate primary antibody: anti-SidI, 1:1,000; anti-eEF1A, 1:500; anti-GFP, 1:50,000; anti- eEF1Bγ, 1: 2000; anti-actin, 1:1000. Serum specific for Bacillus subtilis isocitrate dehydrogenase (ICDH) was generously provided by A. L. Sonenshein, Tufts University Medical School, Boston, MA and was used at 1:10,000. Horseradish peroxidase conjugated secondary antibodies and enhanced bioluminescence reagents were used to detect the signals (Pierce, Rockford, IL). Alternatively, membranes were incubated with an appropriate IRDye infrared secondary antibody (Li-Cor’s Biosciences Lincoln, Nebraska, USA) and the signals were detected, and if necessary, the intensity of the bands are quantitiated by using the Odyssey infrared imaging system.
Immunofluorescence Microscopy
Immunofluorescence microscopy studies on BMMs were performed and processed as described (Swanson and Isberg, 1995). SidC (Luo and Isberg, 2004) were labeled by using affinity-purified polyclonal antibody followed by FITC-conjugated goat anti-rabbit antibody (Jackson LAB). Rat anti-Legionella antibodies generated by immunizing a rat with fixed L. pneumophila (Pocono Rabbit Farm and Laboratory) were used to label the bacterium. Samples were inspected and scored with an Olympus IX-81 fluorescence microscope.
Guanine nucleotide-dependent protein binding assay
GST-EF1A was purified from bacterial lysate using glutathione affinity column. The purified protein was dialyzed in 5 mM EDTA for 20 min. on ice to generate nucleotide-free eEF1A. To determine the affinity of SidI for the nucleotide -free and bound form of eEF1A, the EDTA-treated or untreated GST-eEF1A (5 μg each) was incubated with 10 μg of gel filtration column (Superdex 200 10/300 GL) purified His6-SidI at 30 °C for 30 min. Washed GST-resin was then added and the tubes were rotated at 4 °C for 90 min. The GST beads were washed five times in wash buffer (PBS containing 1% Triton) and solubilized in Laemmli buffer. For control purified GST protein was incubated with purified His-SidI. To detect binding preference of SidI between GTP and GDP bound eEF1A, the dialyzed GST-eEF1A (5μg) was first loaded with 100 μM non-hydrolyzable GTPγS or GDPβS in 20 mM MgCl2 at 4 °C for 30 min. followed by 10 min. at 30 °C. The nucleotide- bound GST-EF1A was then incubated with 10 μg of gel filtration column purified His6-SidI as described above. The samples were run on 10% SDS-PAGE gel for western blot analysis.
Electrophoretic mobility shift assay
Differentiated U937 macrophages were infected with wild-type Lp02, the dotA mutant Lp03 or the sidI mutant at an MOI of 5 for 8 hours. Infected cells were collected, frozen immediately and stored at −80 °C. The cell pellet was resuspended in a buffer containing 20 mM HEPES (pH 7.9), 25% glycerol, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM PMSF and 0.2 mM DTT, kept on ice for 20 min with intermittent mixing and centrifuged at max speed for 10 min at 4 °C. The total protein content of the cell lysates was measured using Bradford assay and was adjusted to a concentration of 1.0 μg/μl, 1.0 μg protein from each treatment was mixed with 0.8 ng digoxigenin-labeled probe (obtained by annealing 5′-GATCTCGGCTGGAATATTCCCGACCTGGCAGCCGA-3′ and 5′-GATCTCGGCTGCCAGGTCGGGAATATTCCAGCCGA-3′). The volume of the binding reactions was brought to 20 μl with deionized water and binding was allowed to proceed for 15 min at 25 °C. For competition experiments, 5 times more unlabeled probe was added to the same reactions. Gel loading, electrophoresis and signal detection were performed in accordance with protocols of the DIG Gel Shift Kit (Roche). Uninfected and heat shocked cells were used as appropriate controls and treated with the same procedures.
Real-time PCR
We infected differentiated U937 with wild type, the dotA mutant and the sidI deletion mutant at an MOI of 5 for 8 hours. For transfection samples, we transfected 80% confluent 293T cells with plasmids harboring gfp, gfp-sidI or gfp-sidI* for 6 hours. In all experiments, resting cells and cells that had been heat treated at 42 °C for 15 min. were used as controls. After washing collected cells with PBS, we isolated total RNA using the RNAqueous-4PCR (Ambion) following instructions supplied by the manufacturer. One microgram of RNA was used to synthesize cDNA with the RETROscript kit following instructions of the manufacturer (Ambion). Two microliters of the products were used for qRT-PCR with the Absolute™ QPCR SYBR Green ROX mix (Thermo Scientific). To measure the hsp70 mRNA levels, primers 5′-GCCAACAAGATCACCATCAC-3′ and 5′-TTTGTACTTCTCCGCCTCCT-3′, designed to amplify a 150 bp DNA fragment were used and 70 nmol of the primers were used for each reaction. The GAPDH gene were used for internal control with primers 5′-TTGCCATCAATGACCCCTTCA-3′ and 5′-CGCCCCACTTGATTTTGGA-3′.
Data collection and statistic analyses
All experiments involved in quantitative data were performed in triplicate for at least three independent times; statistic analysis was carried out with Student’s t test.
Supplementary Material
Acknowledgements
We thank Drs. Ruben Aguilar (Purdue University, IN), Sue Biggins and Marion Dorer (Fred Hutchinson Cancer Research Center, Seattle, WA) for yeast strains, plasmids and helpful discussion for yeast work. We also thank Dr. Arthur Aronson for critical reading of the manuscript. This work was supported by Grant 0535451Z from the American Heart Association and National Institute of Health grants R01AI069344, R03AI073326 (Z.-Q. L) and R01GM069800 (E. N.).
Footnotes
Gene accession number: The gene described in this manuscript is lpg2504 with an accession number of YP_096511 in the Genebank.
Reference
- Abu-Zant A, Jones S, Asare R, Suttles J, Price C, Graham J, Kwaik YA. Anti-apoptotic signalling by the Dot/Icm secretion system of L. pneumophila. Cell Microbiol. 2007;9:246–264. doi: 10.1111/j.1462-5822.2006.00785.x. [DOI] [PubMed] [Google Scholar]
- Banga S, Gao P, Shen X, Fiscus V, Zong WX, Chen L, Luo ZQ. Legionella pneumophila inhibits macrophage apoptosis by targeting pro-death members of the Bcl2 protein family. Proc Natl Acad Sci U S A. 2007;104:5121–5126. doi: 10.1073/pnas.0611030104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardill JP, Miller JL, Vogel JP. IcmS-dependent translocation of SdeA into macrophages by the Legionella pneumophila type IV secretion system. Mol Microbiol. 2005;56:90–103. doi: 10.1111/j.1365-2958.2005.04539.x. [DOI] [PubMed] [Google Scholar]
- Bec G, Kerjan P, Zha XD, Waller JP. Valyl-tRNA synthetase from rabbit liver. I. Purification as a heterotypic complex in association with elongation factor 1. J Biol Chem. 1989;264:21131–21137. [PubMed] [Google Scholar]
- Belyi Y, Tabakova I, Stahl M, Aktories K. Lgt: a family of cytotoxic glucosyltransferases produced by Legionella pneumophila. J Bacteriol. 2008;190:3026–3035. doi: 10.1128/JB.01798-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyi Y, Niggeweg R, Opitz B, Vogelsgesang M, Hippenstiel S, Wilm M, Aktories K. Legionella pneumophila glucosyltransferase inhibits host elongation factor 1A. Proc Natl Acad Sci U S A. 2006;103:16953–16958. doi: 10.1073/pnas.0601562103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger KH, Isberg RR. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol. 1993;7:7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- Borradaile NM, Buhman KK, Listenberger LL, Magee CJ, Morimoto ET, Ory DS, Schaffer JE. A critical role for eukaryotic elongation factor 1A-1 in lipotoxic cell death. Mol Biol Cell. 2006;17:770–778. doi: 10.1091/mbc.E05-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne GJ, Proud CG. Regulation of peptide-chain elongation in mammalian cells. Eur J Biochem. 2002;269:5360–5368. doi: 10.1046/j.1432-1033.2002.03290.x. [DOI] [PubMed] [Google Scholar]
- Campodonico EM, Chesnel L, Roy CR. A yeast genetic system for the identification and characterization of substrate proteins transferred into host cells by the Legionella pneumophila Dot/Icm system. Mol Microbiol. 2005;56:918–933. doi: 10.1111/j.1365-2958.2005.04595.x. [DOI] [PubMed] [Google Scholar]
- Cans C, Passer BJ, Shalak V, Nancy-Portebois V, Crible V, Amzallag N, et al. Translationally controlled tumor protein acts as a guanine nucleotide dissociation inhibitor on the translation elongation factor eEF1A. Proc Natl Acad Sci U S A. 2003;100:13892–13897. doi: 10.1073/pnas.2335950100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conover GM, Derre I, Vogel JP, Isberg RR. The Legionella pneumophila LidA protein: a translocated substrate of the Dot/Icm system associated with maintenance of bacterial integrity. Mol Microbiol. 2003;48:305–321. doi: 10.1046/j.1365-2958.2003.03400.x. [DOI] [PubMed] [Google Scholar]
- Crechet JB, Parmeggiani A. Characterization of the elongation factors from calf brain. 3. Properties of the GTPase activity of EF-1 alpha and mode of action of kirromycin. Eur J Biochem. 1986;161:655–660. doi: 10.1111/j.1432-1033.1986.tb10490.x. [DOI] [PubMed] [Google Scholar]
- de Felipe KS, Pampou S, Jovanovic OS, Pericone CD, Ye SF, Kalachikov S, Shuman HA. Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. J Bacteriol. 2005;187:7716–7726. doi: 10.1128/JB.187.22.7716-7726.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorer MS, Kirton D, Bader JS, Isberg RR. RNA interference analysis of Legionella in Drosophila cells: exploitation of early secretory apparatus dynamics. PLoS Pathog. 2006;2:e34. doi: 10.1371/journal.ppat.0020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumenil G, Isberg RR. The Legionella pneumophila IcmR protein exhibits chaperone activity for IcmQ by preventing its participation in high-molecular-weight complexes. Mol Microbiol. 2001;40:1113–1127. doi: 10.1046/j.1365-2958.2001.02454.x. [DOI] [PubMed] [Google Scholar]
- Fan HY, Cheng KK, Klein HL. Mutations in the RNA polymerase II transcription machinery suppress the hyperrecombination mutant hpr1 delta of Saccharomyces cerevisiae. Genetics. 1996;142:749–759. doi: 10.1093/genetics/142.3.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farbrother P, Wagner C, Na J, Tunggal B, Morio T, Urushihara H, et al. Dictyostelium transcriptional host cell aresponse upon infection with Legionella. Cell Microbiol. 2006;8:438–456. doi: 10.1111/j.1462-5822.2005.00633.x. [DOI] [PubMed] [Google Scholar]
- Fazzio TG, Tsukiyama T. Chromatin remodeling in vivo: evidence for a nucleosome sliding mechanism. Mol Cell. 2003;12:1333–1340. doi: 10.1016/s1097-2765(03)00436-2. [DOI] [PubMed] [Google Scholar]
- Freshneyy RI. Culture of Animal Cells: A Manual of Basic Technique. Wiley-Liss; New York: 2007. [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- Glotzer JB, Saltik M, Chiocca S, Michou AI, Moseley P, Cotten M. Activation of heat-shock response by an adenovirus is essential for virus replication. Nature. 2000;407:207–211. doi: 10.1038/35025102. [DOI] [PubMed] [Google Scholar]
- Gross SR, Kinzy TG. Translation elongation factor 1A is essential for regulation of the actin cytoskeleton and cell morphology. Nat Struct Mol Biol. 2005;12:772–778. doi: 10.1038/nsmb979. [DOI] [PubMed] [Google Scholar]
- Heidtman M, Chen EJ, Moy MY, Isberg RR. Large-scale identification of Legionella pneumophila Dot/Icm substrates that modulate host cell vesicle trafficking pathways. Cell Microbiol. 2008 doi: 10.1111/j.1462-5822.2008.01249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingmundson A, Delprato A, Lambright DG, Roy CR. Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature. 2007;450:365–369. doi: 10.1038/nature06336. [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubori T, Hyakutake A, Nagai H. Legionella translocates an E3 ubiquitin ligase that has multiple U-boxes with distinct functions. Mol Microbiol. 2008;67:1307–1319. doi: 10.1111/j.1365-2958.2008.06124.x. [DOI] [PubMed] [Google Scholar]
- Laguna RK, Creasey EA, Li Z, Valtz N, Isberg RR. A Legionella pneumophila-translocated substrate that is required for growth within macrophages and protection from host cell death. Proc Natl Acad Sci U S A. 2006;103:18745–18750. doi: 10.1073/pnas.0609012103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamberti A, Caraglia M, Longo O, Marra M, Abbruzzese A, Arcari P. The translation elongation factor 1A in tumorigenesis, signal transduction and apoptosis: review article. Amino Acids. 2004;26:443–448. doi: 10.1007/s00726-004-0088-2. [DOI] [PubMed] [Google Scholar]
- Liu Y, Luo ZQ. The Legionella pneumophila effector SidJ is required for efficient recruitment of endoplasmic reticulum proteins to the bacterial phagosome. Infect Immun. 2007;75:592–603. doi: 10.1128/IAI.01278-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Gao P, Banga S, Luo ZQ. An in vivo gene deletion system for determining temporal requirement of bacterial virulence factors. Proc Natl Acad Sci U S A. 2008;105:9385–9390. doi: 10.1073/pnas.0801055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick VP, Isberg RR. NF-kappaB translocation prevents host cell death after low-dose challenge by Legionella pneumophila. J Exp Med. 2006;203:2177–2189. doi: 10.1084/jem.20060766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo ZQ, Farrand SK. Signal-dependent DNA binding and functional domains of the quorum-sensing activator TraR as identified by repressor activity. Proc Natl Acad Sci U S A. 1999;96:9009–9014. doi: 10.1073/pnas.96.16.9009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo ZQ, Isberg RR. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc Natl Acad Sci U S A. 2004;101:841–846. doi: 10.1073/pnas.0304916101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machner MP, Isberg RR. Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev Cell. 2006;11:47–56. doi: 10.1016/j.devcel.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Machner MP, Isberg RR. A bifunctional bacterial protein links GDI displacement to Rab1 activation. Science. 2007;318:974–977. doi: 10.1126/science.1149121. [DOI] [PubMed] [Google Scholar]
- McCusker KT, Braaten BA, Cho MW, Low DA. Legionella pneumophila inhibits protein synthesis in Chinese hamster ovary cells. Infect Immun. 1991;59:240–246. doi: 10.1128/iai.59.1.240-246.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata T, Delprato A, Ingmundson A, Toomre DK, Lambright DG, Roy CR. The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat Cell Biol. 2006;8:971–977. doi: 10.1038/ncb1463. [DOI] [PubMed] [Google Scholar]
- Nagai H, Kagan JC, Zhu X, Kahn RA, Roy CR. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science. 2002;295:679–682. doi: 10.1126/science.1067025. [DOI] [PubMed] [Google Scholar]
- Ninio S, Roy CR. Effector proteins translocated by Legionella pneumophila: strength in numbers. Trends Microbiol. 2007;15:372–380. doi: 10.1016/j.tim.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Olarewaju O, Ortiz PA, Chowdhury WQ, Chatterjee I, Kinzy TG. The translation elongation factor eEF1B plays a role in the oxidative stress response pathway. RNA Biol. 2004;1:89–94. doi: 10.4161/rna.1.2.1033. [DOI] [PubMed] [Google Scholar]
- Shamovsky I, Ivannikov M, Kandel ES, Gershon D, Nudler E. RNA-mediated response to heat shock in mammalian cells. Nature. 2006;440:556–560. doi: 10.1038/nature04518. [DOI] [PubMed] [Google Scholar]
- Shohdy N, Efe JA, Emr SD, Shuman HA. Pathogen effector protein screening in yeast identifies Legionella factors that interfere with membrane trafficking. Proc Natl Acad Sci U S A. 2005;102:4866–4871. doi: 10.1073/pnas.0501315102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siggers KA, Lesser CF. The Yeast Saccharomyces cerevisiae: a versatile model system for the identification and characterization of bacterial virulence proteins. Cell Host Microbe. 2008;4:8–15. doi: 10.1016/j.chom.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V, Aballay A. Heat shock and genetic activation of HSF-1 enhance immunity to bacteria. Cell Cycle. 2006;5:2443–2446. doi: 10.4161/cc.5.21.3434. [DOI] [PubMed] [Google Scholar]
- Swanson MS, Isberg RR. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect Immun. 1995;63:3609–3620. doi: 10.1128/iai.63.9.3609-3620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talapatra S, Wagner JD, Thompson CB. Elongation factor-1 alpha is a selective regulator of growth factor withdrawal and ER stress-induced apoptosis. Cell Death Differ. 2002;9:856–861. doi: 10.1038/sj.cdd.4401078. [DOI] [PubMed] [Google Scholar]
- Tao W, Kurschner C, Morgan JI. Modulation of cell death in yeast by the Bcl-2 family of proteins. J Biol Chem. 1997;272:15547–15552. doi: 10.1074/jbc.272.24.15547. [DOI] [PubMed] [Google Scholar]
- Tilney LG, Harb OS, Connelly PS, Robinson CG, Roy CR. How the parasitic bacterium Legionella pneumophila modifies its phagosome and transforms it into rough ER: implications for conversion of plasma membrane to the ER membrane. J Cell Sci. 2001;114:4637–4650. doi: 10.1242/jcs.114.24.4637. [DOI] [PubMed] [Google Scholar]
- VanRheenen SM, Luo ZQ, O’Connor T, Isberg RR. Members of a Legionella pneumophila family of proteins with ExoU (phospholipase A) active sites are translocated to target cells. Infect Immun. 2006;74:3597–3606. doi: 10.1128/IAI.02060-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerheide SD, Morimoto RI. Heat shock response modulators as therapeutic tools for diseases of protein conformation. J Biol Chem. 2005;280:33097–33100. doi: 10.1074/jbc.R500010200. [DOI] [PubMed] [Google Scholar]
- Zusman T, Aloni G, Halperin E, Kotzer H, Degtyar E, Feldman M, Segal G. The response regulator PmrA is a major regulator of the icm/dot type IV secretion system in Legionella pneumophila and Coxiella burnetii. Mol Microbiol. 2007;63:1508–1523. doi: 10.1111/j.1365-2958.2007.05604.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.