Stem-cell-based regenerative medicine, especially in the last decade, has offered great opportunities for investigating fundamental biological processes and treating devastating diseases and injuries. In particular, human embryonic stem cells (hESCs) have two distinctive abilities: to self-renew for an unlimited number of generations and to differentiate into all cell types found in the three layers of embryo (endoderm, mesoderm, and ectoderm). Two major obstacles associated with hESCs, however, still need to be overcome: 1) ethical issues concerning the use of human embryos, and 2) immune rejection following transplantation.[1]
The advent of induced pluripotent stem cells (iPSCs) can overcome some of the current limitations of hESC-based cell therapy. For instance, using genetic engineering, the Yamanaka group manipulated murine somatic cells through the expression of four transcription factors (TFs) Oct4, Klf4, Sox2, and c-Myc, and reprogrammed the cells to a pluripotent state.[2] Similar genetic manipulations subsequently resulted in the generation of human iPSCs.[3] Efforts in many labs are now concentrated on applying the iPSC technology to tissue-replacement therapies as well as on modeling diseases in vitro.[4] Other advances include generating iPSCs from patients with different diseases in an effort to cause the in vitro differentiation of these iPSCs into the cell types affected by the disease.[5] The use of iPSCs also helps in gaining significant insights into understanding the mechanisms underlying pluripotency and differentiation.
However, before iPSCs can be clinically relevant, several limitations, including the use of viral vectors and the slow kinetics and low efficiency of induction, need to be addressed.[6] One of the most critical issues is the presence of transgenes in the iPSCs. Typically, iPSCs are generated by transducing somatic cells with transgenes, which are integrated within the cell's genome, by using retroviruses or lentiviruses. The integrated transgenes are silenced, and the endogenous genes encoding the TFs are activated. However, transgene reactivation (especially c-Myc) poses a significant threat as it can lead to tumorigenesis.[7] Furthermore, the erroneous expression of these transgenes can lead to inhibition of complete iPSC differentiation and maturation, thereby increasing the chances of forming immature teratomas.
To address the aforementioned problems associated with conventional viral vectors, many groups have tried to generate iPSCs by using nonviral methods. In a recent study,[8] Yamanaka's group successfully generated iPSCs by repeatedly transfecting mouse embryonic fibroblasts with a reprogramming plasmid containing self-cleavage sequences for expressing Oct4, Sox2, and Klf4 and another plasmid carrying the cDNA for c-Myc. Other attractive recent advances for reprogramming somatic cells include the use of Cre-excisable viruses,[9] a piggybac transposition system,[10] and an oriP/EBNA1-based expression system.[11] Yet, all these methods make use of genetic materials, and this could lead to unexpected genetic modifications by the exogenous genes present within target cells. Thus, many labs are now developing ways of reprogramming somatic cells to generate iPSCs that avoid genetic modifications. For instance, in a recent study,[12] recombinant reprogramming proteins from E. coli were used to efficiently generate iPSCs from mouse embryonic fibroblasts (mEFs). In order to generate recombinant proteins that could penetrate the plasma membrane of the somatic cells, a poly-arginine (R11) domain was fused to the C terminus of the four reprogramming factors to obtain efficient recombinant reprogramming proteins (i.e., Oct4-11R, Sox2-11R, Klf4-11R, and c-Myc-11R). This method of using recombinant proteins offers a safer mode of generating iPSCs than the previously mentioned methods as it eliminates any risk of modifying the target cell genome with exogenous genetic material (Figure 1).
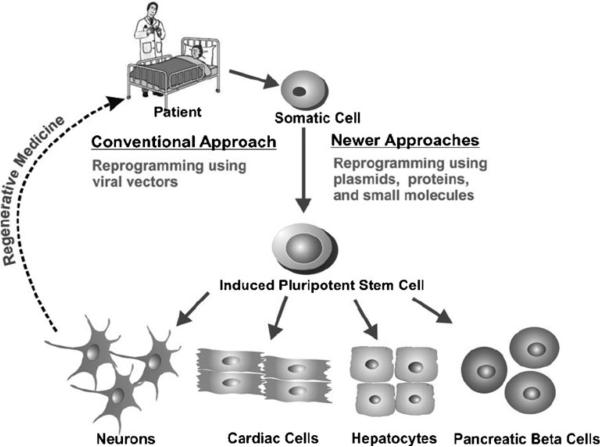
Figure 1.
The figure illustrates the manner in which a patient's somatic cells can be reprogrammed to iPSCs and then differentiated into the desired cell type for regenerative medicine. The reprogramming of somatic cells into iPSCs can be carried out by using conventional or newer approaches.
Another effort to avoid viral vector-based genetic manipulation is to develop a chemical platform for generating iPSCs. Several groups have already identified certain small molecules or a combination of small molecules that can replace one or more of the reprogramming factors (TFs) involved in generating iPSCs.[13] These small molecules not only reduce the risk of oncogene transduction, but also significantly improve the reprogramming efficiency.[6] In a recent study,[14] neural progenitor cells (NPCs), retrovirally transduced with only two TFs, Oct4 and Klf4, were reprogrammed by using the small molecule BIX, which is an inhibitor of G9a methyltransferase. The reprogramming efficiency was comparable to transduction with all four factors. As NPCs endogenously express significant levels of Sox2, which might have caused the cells to be more susceptible to reprogramming with BIX, the group went on to identify small molecules that reprogrammed mEFs, which do not express any of the TFs necessary for reprogramming.[15] They identified the small molecule BayK, a specific L-type calcium channel agonist, which, in the presence of BIX, successfully replaced Sox2 and c-Myc by reprogramming mEFs transduced with only two TFs, Oct4 and Klf4. Although small molecules seem safer than transgene integrations, most of the small molecules involved in the replacement of one or more TFs tend to impact the cells directly at the epigenetic level, and this could potentially lead to chemically induced mutations. In the previous study, BayK itself did not affect the cells at the epigenetic level, but caused mEF reprogramming only in the presence of BIX, which caused epigenetic modifications. There is a clear need to discover small molecules that can replace the TFs in reprogramming and, at the same time, do not impact the cells at the epigenetic level. To this end, research by Ichida et al., has led to the discovery of a small molecule, RepSox, that successfully replaces Sox2 in reprogramming (hence the name RepSox) by inhibiting transforming growth factor-β (Tgf-β) signaling, which in turn induces Nanog expression.[16]
The researchers discovered RepSox by first screening a library of 800 distinct compounds, with known pharmacological targets, that could potentially replace Sox2. Oct4-GFP reporter mouse embryonic and adult fibroblasts, virally transduced with Oct4, Klf4, and c-Myc, were treated with these compounds. This was the preferred approach as it was unbiased with respect to the mechanism of action of the different compounds. In the hope of distinguishing the compounds that required chromatin remodeling from those that did not, the wells containing the compounds were treated with the HDAC inhibitor, valproic acid (VPA). Of the 800 compounds in the initial screen, only three compounds successfully generated iPSCs. Of those three, RepSox was the only compound found to successfully replace Sox2 in the absence of VPA. Interestingly, RepSox was identified as a Tgf-β1 kinase inhibitor and did not require chromatin remodeling to induce reprogramming.
Another important concern when generating iPSCs has always been the presence of the transgene c-Myc, which increases the reprogramming efficiency, although its reactivation can lead to tumorigenesis.[7] Hence, the elimination of c-Myc is an important step towards significantly reducing the risk of forming tumors. It was observed that RepSox generated iPSCs in cells transduced with only two TFs, Oct4 and Klf4, thus successfully replacing both Sox2 and c-Myc. Furthermore, RepSox induced the appearance of iPSCs with an efficiency similar to viral Sox2, thus suggesting that the reprogramming efficiency was not compromised when the transgene, Sox2, was replaced with the small molecule. In addition, the RepSox-generated iPSCs successfully responded to directed differentiation signals in vitro; one example being their robust differentiation into motor neurons. The pluripotency of the cells reprogrammed by using RepSox was further confirmed by their ability to contribute to forming chimeric embryos in vivo when injected into blastocysts.
RepSox was found to be most efficient at generating iPSCs when treating cultures containing stable intermediates that were trapped in a partially reprogrammed state (RepSox-responsive cell lines), characterized by the overexpression of Oct4, Klf4, and c-Myc. These cellular intermediates were trapped in an unproductive state in the absence of RepSox treatment. The authors confirmed that RepSox did not replace Sox2 by directly activating endogenous Sox2, or a Sox family member, as no significant increase in the endogenous expression of Sox1, Sox2, Sox3, or the remaining Sox family transcription factors was observed within the first two days of RepSox treatment. Moreover, depleting Sox1, the most potent Sox family member after Sox2, by using shRNA did not affect the reprogramming rate in presence of RepSox. On further investigation, Nanog was found to be among the most increased transcription factors after RepSox treatment. Nanog expression levels, as compared to untreated controls, increased fourfold within 24 h and tenfold within 48 h. The researchers thus hypothesized that RepSox replaced Sox2 in reprogramming through the induction of Nanog transcription in the absence of Sox2.
To test if the inhibition of the Tgf-β signaling pathway induced Nanog expression, RepSox-responsive cell lines were treated with alternative inhibitors of the Tgf-β signaling pathway such as SB43152- and Tgf-β-neutralizing antibodies. In all cases (including treatment with RepSox), an increase in the expression of Nanog was observed, thus confirming that RepSox replaced Sox2 by inhibiting the Tgf-β signaling pathway, which in turn led to the sustained transcription of Nanog. Furthermore, a RepSox-responsive cell line, transduced with a lentivirus encoding the shRNA specific for Nanog, was treated with RepSox. It was observed that the Nanog knockdown cells reprogrammed at an efficiency that was 50 times lower than that of the empty vector control, thus confirming that RepSox replaces Sox2 by inducing Nanog expression. The fact that Nanog compensated for the absence of Sox2 led the researchers to reprogram mEFs retrovirally, transducing them with Oct4, Klf4, c-Myc, and Nanog (instead of Sox2). These cells generated Oct4-GFP+ colonies as efficiently as those reprogrammed with Oct4, Klf4, c-Myc, and Sox2. Nevertheless, a definite proof of pluripotency of these cells is still required to confirm the replacement of Sox2 with Nanog as transcription factor in reprogramming.
According to findings in the paper, a one-day treatment with RepSox is sufficient to replace transgenic Sox2. This is in stark contrast to the reprogramming achieved by transgenic Oct4, Klf4, and Sox2, in which each transgene must be expressed for several days. The researchers believe that small molecules such as RepSox can act as switches that induce gene-expression changes that lead to the completion of reprogramming. Results such as these demonstrate the feasibility and advantages of replacing the central reprogramming trans-genes with small molecules. In addition, molecules such as RepSox modulate discrete cellular pathways or processes rather than modifying the chromatin structure; this makes reprogramming significantly safer. The mechanism by which RepSox replaces Sox2 without compromising the efficiency of reprogramming clearly shows that small molecules can contribute to reprogramming in a very distinct manner when compared to the mechanisms of the factors that they replace.
The application of the chemical approach in stem-cell biology has advanced significantly over the last decade. Small molecules, in combination with known proteins have been used to induce the robust differentiation of embryonic and adult stem cells into specific cell types. For instance, neuropathiazol was identified as a small molecule that could differentiate multipotent neural progenitor cells into mature neurons even under gliogenic conditions.[17] Chemical libraries of pharmacologically active compounds are regularly being screened to identify small molecules that could potentially contribute to promoting the differentiation or survival of stem cells in vitro.[13] Furthermore, advances in stem-cell biology have led to the reprogramming of somatic cells into pluripotent stem cells (iPSCs). The small-molecule approach in this regard might be one of the most efficient, safe, and cost-effective ways of generating iPSCs. In the past couple of years, many research labs have identified novel small molecules that can either replace certain transgenes in reprogramming or improve the efficiency of reprogramming. It is only a matter of time before a cocktail of such small molecules will replace all reprogramming genes, opening avenues to purely chemical reprogramming.
Acknowledgements
We would like thank Shreyas Shah and Birju Shah for their helpful discussions and comments during the process of writing the highlight. In addition, we are grateful for the NIH Director's Innovator Award (1DP20D006 462-01) and the NJ Commission on Spinal Cord Research Grant (09-3085-SCR-E-0).
References
- 1.Yamanaka S. Cell. 2009;137:13. doi: 10.1016/j.cell.2009.03.034. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Yamanaka S. Cell. 2006;126:663. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Cell. 2007;131:861. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 4.Hanna J, Wernig M, Markoulaki S, Sun CW, Meissner A, Cassady JP, Beard C, Brambrink T, Wu LC, Townes TM, Jaenisch R. Science. 2007;318:1920. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- 5.a Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, Lensch MW, Cowan C, Hochedlinger K, Daley GQ. Cell. 2008;134:877. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, Croft GF, Saphier G, Leibel R, Goland R, Wichterle H, Henderson CE, Eggan K. Science. 2008;321:1218. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]; c Ebert AD, Yu JY, Rose FF, Mattis VB, Lorson CL, Thomson JA, Svendsen CN. Nature. 2009;457:277. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, Hargus G, Blak A, Cooper O, Mitalipova M, Isacson O, Jaenisch R. Cell. 2009;137:1356. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin TX, Ambasudhan R, Yuan X, Li WL, Hilcove S, Abujarour R, Lin XY, Hahm HS, Hao E, Hayek A, Ding S. Nat. Methods. 2009;6:805. doi: 10.1038/nmeth.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okita K, Ichisaka T, Yamanaka S. Nature. 2007;448:313. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 8.Okita K, Nakagawa M, Hong HJ, Ichisaka T, Yamanaka S. Science. 2008;322:949. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 9.Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, Hargus G, Blak A, Cooper O, Mitalipova M, Isacson O, Jaenisch R. Cell. 2009;136:964. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hamalainen R, Cowling R, Wang W, Liu PT, Gertsenstein M, Kaji K, Sung HK, Nagy A. Nature. 2009;458:766. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA. Science. 2009;324:797. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou HY, Wu SL, Joo JY, Zhu SY, Han DW, Lin TX, Trauger S, Bien G, Yao S, Zhu Y, Siuzdak G, Scholer HR, Duan LX, Ding S. Cell Stem Cell. 2009;4:381. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Y, Shi Y, Ding S. Nature. 2008;453:338. doi: 10.1038/nature07042. [DOI] [PubMed] [Google Scholar]
- 14.Shi Y, Do JT, Desponts C, Hahm HS, Scholer HR, Ding S. Cell Stem Cell. 2008;2:525. doi: 10.1016/j.stem.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 15.Shi Y, Desponts C, Do JT, Hahm HS, Scholer HR, Ding S. Cell Stem Cell. 2008;3:568. doi: 10.1016/j.stem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 16.Ichida JK, Blanchard J, Lam K, Son EY, Chung JE, Egli D, Loh KM, Carter AC, Di Giorgio FP, Koszka K, Huangfu D, Akutsu H, Liu DR, Rubin LL, Eggan K. Cell Stem Cell. 2009;5:491. doi: 10.1016/j.stem.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warashina M, Min KH, Kuwabara T, Huynh A, Gage FH, Schultz PG, Ding S. Angew. Chem. 2006;118:605. doi: 10.1002/anie.200503089. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2006. p. 591.