Abstract
Systems biology modeling of signal transduction pathways traditionally employs ordinary differential equations, deterministic models based on assumptions of spatial homogeneity. However, this can be a poor approximation for certain aspects of signal transduction, especially its initial steps: the cell membrane exhibits significant spatial organization, with diffusion rates approximately two orders of magnitude slower than those in the cytosol. Thus, to unravel the complexities of signaling pathways, quantitative models must consider spatial organization as an important feature of cell signaling. Furthermore, spatial separation limits the number of molecules that can physically interact, requiring stochastic simulation methods that account for individual molecules. Herein, we discuss the need for mathematical models and experiments that appreciate the importance of spatial organization in the membrane.
Keywords: Mathematical modeling, signal transduction, plasma membrane, ODE, PDE, spatial modeling, Spatial Monte Carlo
Introduction
Cell signaling is an essential, ubiquitous process that living systems use to respond to the environment. Cell signaling underlies critical cellular decisions such as development, cell growth and division, differentiation, migration, apoptosis, and it essentially provides the coordination required for the functionality of multicellular organisms. Understanding cell signaling is critical, due to its importance in cellular fate decisions and because malfunctions in cellular signaling are at the heart of many diseases such as cancer, autoimmune disease and others. To further the understanding of cellular signaling, traditional biological reductionist research is now complemented with a systems biology approach. It is the focus of this opinion article to discuss the importance of an often neglected aspect of cell signaling—the spatial organization of the cell membrane.
Signal propagation is controlled in part by spatial and temporal organization of the proteins involved in the subsequent protein-protein and protein-lipid interactions. The challenge is to understand the mechanisms that regulate the efficiency, specificity and duration of cell signaling, and how interactions among proteins in the signaling network alter signal strength and the nature of the physiological response. These are all not simply functions of the biochemical properties of the proteins involved. For example, if two components of a signaling pathway occupy separate and distinct regions of the cell membrane, there will essentially be a block in the signaling pathway. In contrast, if two proteins in the same signaling pathway exist at very low concentrations, the signal can still be transmitted effectively, if the proteins are co-clustered in the same microdomain on the cell membrane. It is likely that the cell uses spatial organization to control and regulate signaling. Therefore, the spatial and temporal complexities of cell membranes must be fully resolved, in order to properly understand cell signaling and its regulation. Furthermore, additional features in the membrane such as signaling microdomains, lipid rafts, cytoskeletal corrals and lipid shells, must be addressed [1–7].
Systems biology for signal transduction
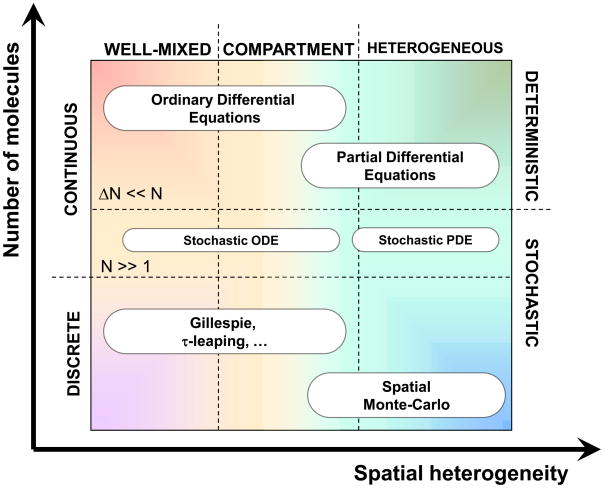
Mathematical modeling of signaling pathways has traditionally been divided into two types, deterministic and stochastic (Figure 1). In reality, biochemical reacting systems are stochastic; however, when the numbers of molecules are large, the stochastic fluctuations are insignificant relative to the absolute molecule number. This is the justification for the deterministic approach used in the vast majority of systems biology models. Another simplification that is inherent to many systems biology studies is the well-mixed assumption. In other words, these models do not consider spatial organization of the cells, tissues, organs, etc. Nonetheless, deterministic models continue to provide useful insight [8,9]. On the other hand, given the heterogeneous organization of biological systems, we view the spatial organization as an important aspect of cell biology to be included. Therefore, over the past decade we have been developing systems biology simulation tools that include realistic spatial organizations [10–14]. We have started with the plasma membrane, given its central role in signal transduction, and are extending our tools and approaches inward to the cytosol [14] and outward to the tissue level. This manuscript focuses on the systems biology tools for studying spatially non-homogeneous systems and the needs for future developments.
Figure 1.
Classes of mathematical models for biochemical processes in cells and their applicability and assumptions. Well-mixed systems: The underlying biochemical processes with cells are stochastic transformations. However, a deterministic mathematical description may be applicable depending on the number of molecules N of each molecular species in the volume (or area) of interest. This number must be large (N≫1) for a deterministic approach to provide an accurate representation. Basically, the expected stochastic fluctuations of the molecule number (ΔN, the magnitude of intrinsic fluctuations is on the order of N1/2) must be small relative to the absolute number for a deterministic description to be an acceptable assumption. Spatially heterogeneous systems: The well-mixed assumption implies that there is no significant spatial heterogeneity in the system. If this is not true but there are well-defined spatial regions that are homogeneous, then a compartment-based model may be used instead of a fully spatial model.
Spatial simulations
The majority of discrete, stochastic simulations have been limited to well-mixed (also called spatially uniform or homogeneous) systems [15–24]. The assumption of a well-mixed condition may be justified in some biological systems, whereas for other conditions spatial modeling may be necessary. The introduction of powerful microscopy methods (Figure 2)[25] has enabled the construction of spatially distributed (nonuniform) models that can impact significantly our understanding and control of biological systems at the molecular level.
Figure 2.
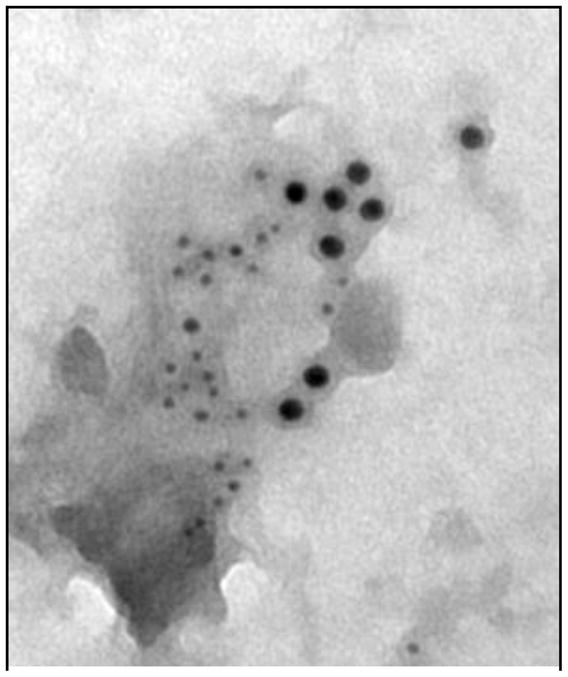
Electron micrograph of a membrane sheet as prepared by Wilson et al [75]. The figure shows colocalization of the formyl peptide receptor (FPR)(5 nm) and FcεRI (10 nm) within 1 min of simultaneous addition of their ligands.
Recent studies have emphasized the necessity of stochastic spatial, as opposed to well-mixed deterministic or partial differential equation (PDE), models for accurate quantitative analysis of biological systems; see [17,26–35] and reviews [27,28,36]. The inclusion of a spatial description is of great importance in biology [27], and spatial modeling has conventionally been performed with PDE methods. PDEs have been used to study receptor-ligand dynamics [37–46], intracellular processes [47,48], signaling processes in the plasma membrane [49,50] and other biological problems [51]. Furthermore, Fickian diffusion and Smoluchowski models have been used to study diffusion aspects of synaptic transmission [52,53]. An excellent software tool, VCell, has been developed for PDE-based spatial simulations [54].
PDE-based approaches usually have difficulty capturing the transients of bimolecular reactions, for example, receptor-receptor interactions. Two excellent examples underscoring the limitations of PDEs are found in [26,55]. These limitations arise because tracking the transient evolution of bimolecular reactions with reactants is a many-body problem [56–59], whereas most of the PDE-based efforts are based on the Smoluchowski [60,61] and Collins-Kimball [62] models which only include two-particle interactions, and are valid in the diffusion-limited regime [11]. Our intent here is not to review these efforts; rather, to point out that an exact relation between the effective reaction rate constant and diffusivity is difficult to obtain for a general two-dimensional bimolecular reaction, especially if one is interested in the transient behavior of the system.
Stochastic PDEs, such as the Langevin equation, are also used to capture the effect of noise in spatiotemporal dynamics [63]. However, these approaches rely on the idealized notion of white noise, with fluctuations on all scales. This is unrealistic and may lead to difficulties when the number of copies of proteins is small, as is often the case with plasma membrane proteins. A brief overview of the types of continuum equations that have been used for this purpose see [63] and references therein.
Kinetic or dynamic Monte Carlo (MC) based spatial modeling involves first-principles stochastic simulations of the movement, collisions and chemical transformations of individual molecules in a finite sized volume or area. It is an attractive alternative to PDE-based approaches for modeling cell surface receptor dynamics, because its computational implementation can explicitly consider (1) the creation of a spatially non-random distribution of proteins due to bimolecular reactions [26], (2) the spatial heterogeneity such as microdomains in the plasma membrane [7,64–67] and (3) the noise and correlations resulting from a small number of copies of activated receptors. The major limitation of Kinetic MC based approaches is that they are computationally demanding. Attempts at acceleration of spatial algorithms have primarily focused on algorithmic aspects: fast update and search methods. For example, the work of Bortz et al. on the n-fold or continuous time MC (CTMC) method [68] is a significant achievement in computational speedup. However, improvements are needed for stiff problems, to overcome the one event per iteration issue and to reach large length scales.
Rationale for spatial stochastic modeling: Application to the FcεRI and Formyl Peptide Receptor (FPR) systems
The rapidly growing body of simulation studies has clearly demonstrated that stochastic modeling is essential in spatially well-mixed systems when the population size of one or more key intermediates is small [22,24]. Under such conditions, large noise and significant departure of average rates from their deterministic counterparts are encountered. However, as the population size increases the noise is reduced, and for large systems deterministic behavior is recovered. In the FcεRI and FPR systems it is estimated that there are 50–200 receptors and 1–5 coated pits (diameter of 60–80 nm) in each 1 μm2 of plasma membrane [25]. Such small population sizes require a stochastic method [11].
Many microscopic-scale biological systems, including several spatial features in the FcεRI and FPR system, have yet to be evaluated by spatial stochastic models. Important spatial features of membrane systems include: (1) the necessary proximity of two receptors for dimerization to occur (nonlinear chemical events can cause significant spatial correlations and lead to pattern formation such as clustering, even in the absence of direct attractive intermolecular forces [29,69]) (Figure 3); (2) the possible attractive interactions between receptors and membrane microdomains that render Fickian diffusion structurally incorrect [70]; (3) the directional‘ or uphill‘ diffusion of receptors toward pits; and (4) the observed hop diffusion of membrane proteins [67,71–74]. With such spatial effects, the stochastic solution differs from the deterministic solution even in the infinite, macroscopic size limit. Thus, this situation is very different from spatially homogeneous systems. Consequently, we propose that the spatial multi-resolution MC framework will provide a protein-level model that can provide much insight into signal transduction.
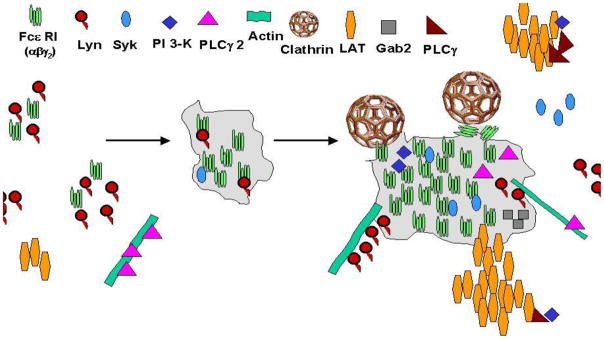
Figure 3.
Membrane organization of a subset of signaling proteins in the FcεRI (High-affinity IgE receptor) cascade, based on immunogold labeling experiments. This figure features new added complications that must be addressed by systems biology. Abbreviations used: PLCγ2 (Phospholipase C-γ2), PLCγ (Phospholipase C-γ), LAT (linker for activation of T cells), Syk (Spleen Tyrosine Kinase), Lyn (Yamaguchi sarcoma viral related oncogene homolog – a Src family tyrosine kinase), Gab2 (Grb2 associated binding protein 2), PI3K (Phosphoinositide 3-Kinase).
Need for more detailed spatial models focusing on the membrane
The fluid mosaic model portrays protein movement as Brownian motion in a sea of lipids. Although this model is the foundation of membrane biology, it has been beset by two inconsistencies: (1) the diffusion coefficients for both proteins and lipids are 5 to 50 times smaller in the plasma membrane than in artificial membranes and (2) oligomers or molecular complexes exhibit a much lower diffusion coefficient (by a factor of 40) in the plasma membrane than in artificial membranes. Also, direct observation has revealed that proteins are not randomly distributed. These discrepancies between the fluid mosaic model and experimental findings are indicative of the necessity for an improved model, and have lead researchers on an almost 40-year journey to uncover the true nature of the plasma membrane.
Associated with this lipid-based architecture arises a landscape of complexity, ranging from the coalescence of phospholipids into “lipid rafts” to the hindering interactions caused by the cytoskeleton. Thus this almost 40-year journey has enlightened the membrane biology community, and provides a new direction for systems biology as a whole. What has been established is that the plasma membrane is a highly compartmentalized surface, which affects the diffusion of signaling proteins in the membrane, and hence the initiation and activation of signal transduction pathways. These developments highlight a need for computational algorithms that take into account the observed biological complexity occurring within the cell membrane.
Acknowledgments
This work was supported by NIH grant P50GM085273 (the New Mexico Spatiotemporal Modeling Center). ÁMH is supported by NIH grant K25CA131558. We thank Dr. J Oliver, University of New Mexico, and Dr. P. Parsons-Wingerter, NASA Glenn Research Center, for helpful discussions of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jacobson K, Dietrich C. Looking at lipid rafts? Trends Cell Biol. 1999;9:87–91. doi: 10.1016/s0962-8924(98)01495-0. [DOI] [PubMed] [Google Scholar]
- 2.Brown DA, London E. Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J Biol Chem. 2000;275:17221–17224. doi: 10.1074/jbc.R000005200. [DOI] [PubMed] [Google Scholar]
- 3.Edidin M. Shrinking patches and slippery rafts: scales of domains in the plasma membrane. Trends Cell Biol. 2001;11:492–496. doi: 10.1016/s0962-8924(01)02139-0. [DOI] [PubMed] [Google Scholar]
- 4.Anderson RG, Jacobson K. A role for lipid shells in targeting proteins to caveolae, rafts, and other lipid domains. Science. 2002;296:1821–1825. doi: 10.1126/science.1068886. [DOI] [PubMed] [Google Scholar]
- 5.Dietrich C, Yang B, Fujiwara T, Kusumi A, Jacobson K. Relationship of lipid rafts to transient confinement zones detected by single particle tracking. Biophys J. 2002;82:274–284. doi: 10.1016/S0006-3495(02)75393-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang Q, Edidin M. Lowering the barriers to random walks on the cell surface. Biophys J. 2003;84:400–407. doi: 10.1016/S0006-3495(03)74860-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *7.Wilson BS, Steinberg SL, Liederman K, Pfeiffer JR, Surviladze Z, Zhang J, Samelson LE, Yang LH, Kotula PG, Oliver JM. Markers for detergent-resistant lipid rafts occupy distinct and dynamic domains in native membranes. Mol Biol Cell. 2004;15:2580–2592. doi: 10.1091/mbc.E03-08-0574. Experimental methods and results for mapping spatial distributions of membrane proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *8.Radhakrishnan K, Edwards JS, Lidke DS, Jovin TM, Wilson BS, Oliver JM. Sensitivity analysis predicts that the ERK–pMEK interaction regulates ERK nuclear translocation. IET Syst Biol. 2009;3:329–341. doi: 10.1049/iet-syb.2009.0010. Illustrates the importance of ODE simulations and sensitivity analysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brightman FA, Fell DA. Differential feedback regulation of the MAPK cascade underlies the quantitative differences in EGF and NGF signalling in PC12 cells. FEBS Lett. 2000;482:169–174. doi: 10.1016/s0014-5793(00)02037-8. [DOI] [PubMed] [Google Scholar]
- 10.Mayawala K, Vlachos DG, Edwards JS. Heterogeneities in EGF receptor density at the cell surface can lead to concave up scatchard plot of EGF binding. FEBS Lett. 2005;579:3043–3047. doi: 10.1016/j.febslet.2005.04.059. [DOI] [PubMed] [Google Scholar]
- **11.Mayawala K, Vlachos DG, Edwards JS. Spatial modeling of dimerization reaction dynamics in the plasma membrane: Monte Carlo vs. continuum differential equations. Biophys Chem. 2006 doi: 10.1016/j.bpc.2006.01.008. Illustrates the importance of stochastic Monte Carlo modeling of spatial distributed systems in biology. [DOI] [PubMed] [Google Scholar]
- *12.Niehaus AM, Vlachos DG, Edwards JS, Plechac P, Tribe R. Microscopic simulation of membrane molecule diffusion on corralled membrane surfaces. Biophys J. 2008;94:1551–1564. doi: 10.1529/biophysj.107.106484. Simulations of cell membrane with the inclusion of actin corals. Illustrates Monte Carlo simulations are efficient at capturing the non-trivial aspects of the cell membrane. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsieh MY, Yang S, Raymond-Stinz MA, Steinberg S, Vlachos DG, Shu W, Wilson B, Edwards JS. Stochastic simulations of ErbB homo and heterodimerisation: potential impacts of receptor conformational state and spatial segregation. IET Syst Biol. 2008;2:256–272. doi: 10.1049/iet-syb:20070073. [DOI] [PubMed] [Google Scholar]
- **14.Costa MN, Radhakrishnan K, Wilson BS, Vlachos DG, Edwards JS. Coupled stochastic spatial and non-spatial simulations of ErbB1 signaling pathways demonstrate the importance of spatial organization in signal transduction. PLoS One. 2009;4:e6316. doi: 10.1371/journal.pone.0006316. Hybrid Monte Carlo multiscale simulation that bridges the spatial simulation at the cell membrane with a well-mixed cytosol stochastic model. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bailey JE. Mathematical modeling and analysis in biochemical engineering: Past accomplishments and future opportunities. Biotechnology Progress. 1998;14:8–20. doi: 10.1021/bp9701269. [DOI] [PubMed] [Google Scholar]
- 16.Chatterjee A, Mayawala K, Edwards JS, Vlachos DG. Time accelerated Monte Carlo simulations of biological networks using the binomial {tau}-leap method. Bioinformatics. 2005 doi: 10.1093/bioinformatics/bti308. [DOI] [PubMed] [Google Scholar]
- 17.Vasudeva K, Bhalla US. Adaptive stochastic-deterministic chemical kinetic simulations. Bioinformatics. 2004;20:78–84. doi: 10.1093/bioinformatics/btg376. [DOI] [PubMed] [Google Scholar]
- 18.Resat H, Ewald JA, Dixon DA, Wiley HS. An Integrated Model of Epidermal Growth Factor Receptor Trafficking and Signal Transduction. Biophys J. 2003;85:730–743. doi: 10.1016/s0006-3495(03)74516-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salwinski L, Eisenberg D. In silico simulation of biological network dynamics. Nature Biotechnology. 2004;22:1017–1019. doi: 10.1038/nbt991. [DOI] [PubMed] [Google Scholar]
- 20.Ozbudak EM, Thattai M, Kurtser I, Grossman AD, Oudenaarden Av. Regulation of noise in the expression of a single gene. Nature Genetics. 2002;31:69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- 21.Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H, Luben R, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007 doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *22.Rao CV, Wolf DM, Arkin AP. Control, exploitation and tolerance of intracellular noise. Nature. 2002;420:231–237. doi: 10.1038/nature01258. Review of stochastic phenomena in the well-mixed regime. [DOI] [PubMed] [Google Scholar]
- 23.Arkin A, Shen PD, Ross J. A test case of correlation metric construction of a reaction pathway from measurements. Science. 1997;277:1275–1279. [Google Scholar]
- 24.McAdams HH, Arkin A. It’s a noisy business! Genetic regulation at the nanomolar scale. Trends Genet. 1999;15:65–69. doi: 10.1016/s0168-9525(98)01659-x. [DOI] [PubMed] [Google Scholar]
- 25.Oliver JM, Pfeiffer JR, Surviladze Z, Steinberg SL, Leiderman K, Sanders ML, Wofsy C, Zhang J, Fan H, Andrews N, et al. Membrane receptor mapping: the membrane topography of FcεRI signaling. Subcell Biochem. 2004;37:3–34. [PubMed] [Google Scholar]
- *26.Shea LD, Omann GM, Linderman JJ. Calculation of diffusion-limited kinetics for the reactions in collision coupling and receptor cross-linking. Biophys J. 1997;73:2949–2959. doi: 10.1016/S0006-3495(97)78323-1. Monte Carlo methods are used to simulate the reaction and diffusion of molecules (and determine rate constants) in the plasma membrane. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **27.Lemerle C, Ventura BD, Serrano L. Space as the final frontier in stochastic simulations of biological systems. FEBS Letters. 2005;579:1789–1794. doi: 10.1016/j.febslet.2005.02.009. Review of stochastic spatial simulations. [DOI] [PubMed] [Google Scholar]
- **28.Slepchenko BM, Schaff JC, Carson JH, Loew LM. Computational cell biology: Spatiotemporal simulation of cellular events. Annu Rev Biophys Biomol Struct. 2002;31:423–441. doi: 10.1146/annurev.biophys.31.101101.140930. Review of spatial simulations. [DOI] [PubMed] [Google Scholar]
- 29.Woolf PJ, Linderman JJ. Self organization of membrane proteins via dimerization. Biophys Chem. 2003;104:217–227. doi: 10.1016/s0301-4622(02)00369-1. [DOI] [PubMed] [Google Scholar]
- 30.Woolf PJ, Linderman JJ. Untangling ligand induced activation and desensitization of G-protein-coupled receptors. Biophys J. 2003;84:3–13. doi: 10.1016/S0006-3495(03)74828-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woolf PJ, Linderman JJ. An algebra of dimerization and its implications for G-protein coupled receptor signaling. J Theor Biol. 2004;229:157–168. doi: 10.1016/j.jtbi.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 32.Shimizu TS, Aksenov SV, Bray D. A spatially extended stochastic model of the bacterial chemotaxis signalling pathway. J Mol Biol. 2003;329:291–309. doi: 10.1016/s0022-2836(03)00437-6. [DOI] [PubMed] [Google Scholar]
- 33.Goldman J, Andrews S, Bray D. Size and composition of membrane protein clusters predicted by Monte Carlo analysis. Eur Biophys J. 2004;33:506–512. doi: 10.1007/s00249-004-0391-6. [DOI] [PubMed] [Google Scholar]
- 34.Wei W, Krone SM. Spatial invasion by a mutant pathogen. Journal of Theoretical Biology. 2005;236:335–348. doi: 10.1016/j.jtbi.2005.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mayawala K, Vlachos DG, Edwards JS. Computational modeling reveals molecular details of epidermal growth factor binding. BMC Cell Biol. 2005;6:41. doi: 10.1186/1471-2121-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi K, Arjunan SNV, Tomita M. Space in systems biology of signaling pathways – towards intracellular molecular crowding in silico. FEBS Letters. 2005;579:1783–1788. doi: 10.1016/j.febslet.2005.01.072. [DOI] [PubMed] [Google Scholar]
- *37.Lauffenburger DA, Linderman JJ. Receptors: Models for Binding, Trafficking, and Signaling. New York: Oxford University Press; 1993. Classic textbook review of mathematical model and cell signaling. [Google Scholar]
- 38.Shvartsman SY, Wiley HS, Deen WM, Lauffenburger DA. Spatial range of autocrine signaling: modeling and computational analysis. Biophys J. 2001;81:1854–1867. doi: 10.1016/S0006-3495(01)75837-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Batsilas L, Berezhkovskii AM, Shvartsman SY. Stochastic model of autocrine and paracrine signals in cell culture assays. Biophys J. 2003;85:3659–3665. doi: 10.1016/S0006-3495(03)74783-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monine MI, Berezhkovskii AM, Joslin EJ, Wiley HS, Lauffenburger DA, Shvartsman SY. Ligand Accumulation in Autocrine Cell Cultures. Biophys J. 2005;88:2384–2390. doi: 10.1529/biophysj.104.051425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pribyl M, Muratov CB, Shvartsman SY. Long-Range Signal Transmission in Autocrine Relays. Biophys J. 2003;84:883–896. doi: 10.1016/S0006-3495(03)74906-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shoup D, Szabo A. Role of diffusion in ligand binding to macromolecules and cell-bound receptors. Biophys J. 1982;40:33–39. doi: 10.1016/S0006-3495(82)84455-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goldstein B, Dembo M. Approximating the effects of diffusion on reversible reactions at the cell surface: ligand-receptor kinetics. Biophys J. 1995;68:1222–1230. doi: 10.1016/S0006-3495(95)80298-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Erickson J, Goldstein B, Holowka D, Baird B. The effect of receptor density on the forward rate constant for binding of ligands to cell surface receptors. Biophys J. 1987;52:657–662. doi: 10.1016/S0006-3495(87)83258-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang D, Gou S-Y, Axelrod D. Reaction rate enhancement by surface diffusion of adsorbates. Biophysical Chemistry. 1992;43:117–137. doi: 10.1016/0301-4622(92)80027-3. [DOI] [PubMed] [Google Scholar]
- 46.Zwanzig R, Szabo A. Time dependent rate of diffusion-influenced ligand binding to receptors on cell surfaces. Biophys J. 1991;60:671–678. doi: 10.1016/S0006-3495(91)82096-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *47.Brown GC, Kholodenko BN. Spatial gradients of cellular phospho-proteins. FEBS Letters. 1999;457:452–454. doi: 10.1016/s0014-5793(99)01058-3. The authors explore the effect of a cytosolic protein being phosphorylated and dephosphorylated at different locations within the cell and how a spatial gradient of can develop. [DOI] [PubMed] [Google Scholar]
- 48.Haugh J, Lauffenburger D. Physical modulation of intracellular signaling processes by locational regulation. Biophys J. 1997;72:2014–2031. doi: 10.1016/S0006-3495(97)78846-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *49.Haugh JM. A unified model for signal transduction reactions in cellular membranes. Biophysical Journal. 2002;82:591–604. doi: 10.1016/S0006-3495(02)75424-6. The author has developed an analytical solution for the rate of an intracellular enzyme recruitment to the plasma membrane. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goldstein B, Wofsy C, Echavarria-Heras H. Effect of membrane flow on the capture of receptors by coated pits. Biophysical Journal. 1988;53:405–414. doi: 10.1016/S0006-3495(88)83117-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rubinstein B, Jacobson K, Mogilner A. Multiscale two-dimensional modeling of a motile simple-shaped cell. Multiscale Modeling & Simulation. 2005;3:413–439. doi: 10.1137/04060370X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mariadason JM, Nicholas C, L’Italien KE, Zhuang M, Smartt HJ, Heerdt BG, Yang W, Corner GA, Wilson AJ, Klampfer L, et al. Gene expression profiling of intestinal epithelial cell maturation along the crypt-villus axis. Gastroenterology. 2005;128:1081–1088. doi: 10.1053/j.gastro.2005.01.054. [DOI] [PubMed] [Google Scholar]
- 53.Song Y, Zhang Y, Shen T, Bajaj CL, McCammon JA, Baker NA. Finite Element Solution of the Steady-State Smoluchowski Equation for Rate Constant Calculations. Biophys J. 2004;86:2017–2029. doi: 10.1016/S0006-3495(04)74263-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **54.Slepchenko BM, Schaff JC, Macara I, Loew LM. Quantitative cell biology with the Virtual Cell. Trends Cell Biol. 2003;13:570–576. doi: 10.1016/j.tcb.2003.09.002. An excellent software tool developed for spatial simulations of biological systems. [DOI] [PubMed] [Google Scholar]
- **55.Sokolov IM, Schnorer H, Blumen A. Diffusion-Controlled Reaction A + B --> 0 in One Dimension - the Role of Particle Mobilities and the Diffusion-Equation Approach. Physical Review A. 1991;44:2388–2393. doi: 10.1103/physreva.44.2388. An example underscoring the limitations of PDE simulations. [DOI] [PubMed] [Google Scholar]
- 56.Torney DC, McConnell HM. Diffusion-Limited Reactions in One Dimension. Journal of Physical Chemistry. 1983;87:1941–1951. [Google Scholar]
- 57.Lindenberg K, Argyrakis P, Kopelman R. Reaction-Diffusion Model for A+A Reaction. Journal of Physical Chemistry. 1995;99:7542–7556. [Google Scholar]
- 58.Barzykin AV, Seki K, Tachiya M. Kinetics of diffusion-assisted reactions in microheterogeneous systems. Advances in Colloid and Interface Science. 2001;89:47–140. doi: 10.1016/s0001-8686(00)00053-1. [DOI] [PubMed] [Google Scholar]
- 59.Keizer J. Diffusion Effects on Rapid Bimolecular Chemical-Reactions. Chemical Reviews. 1987;87:167–180. [Google Scholar]
- 60.Smoluchowski MV. Versuch einer mathematischen Theorie der Koagulationskinetik kolloider Lösungen. Z Phys Chem. 1917;92:129–168. [Google Scholar]
- 61.Szabo A, Zwanzig R, Agmon N. Diffusion-Controlled Reactions with Mobile Traps. Physical Review Letters. 1988;61:2496–2499. doi: 10.1103/PhysRevLett.61.2496. [DOI] [PubMed] [Google Scholar]
- 62.Collins FC, Kimball GE. Diffusion-Controlled Reaction Rates. Journal of Colloid Science. 1949;4:425–437. [Google Scholar]
- **63.Chatterjee A, Snyder MA, Vlachos DG. Mesoscopic modeling of chemical reactivity (invited) Chemical Engineering Science. 2004;59:5559–5567. An excellent review of spatial and stochastic simulations and methods. [Google Scholar]
- 64.Mayor S, Rothberg KG, Maxfield FR. Sequestration of GPI-anchored proteins in caveolae triggered by cross-linking. Science. 1994;264:1948–1951. doi: 10.1126/science.7516582. [DOI] [PubMed] [Google Scholar]
- *65.Lommerse PHM, Spaink HP, Schmidt T. In vivo plasma membrane organization: results of biophysical approaches. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2004;1664:119–131. doi: 10.1016/j.bbamem.2004.05.005. A review of experimental techniques for probing spatial organization of biological membranes. [DOI] [PubMed] [Google Scholar]
- 66.Laude AJ, Prior IA. Plasma membrane microdomains: organization, function and trafficking (Review) Molecular Membrane Biology. 2004;21:193–205. doi: 10.1080/09687680410001700517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **67.Kusumi A, Nakada C, Ritchie K, Murase K, Suzuki K, Murakoshi H, Kasai RS, Kondo J, Fujiwara T. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: High-speed single-molecule tracking of membrane molecules. Annu Rev Biophys Biomol Struct. 2005;34:351–378. doi: 10.1146/annurev.biophys.34.040204.144637. Review of single particle tracking experiments focusing on the importance of high speed sampling. [DOI] [PubMed] [Google Scholar]
- 68.Bortz AB, Kalos MH, Lebowitz JL. A new algorithm for Monte Carlo simulations of Ising spin systems. Journal of Computational Physics. 1975;17:10–18. [Google Scholar]
- *69.Ziff RM, Gulari E, Barshad Y. Kinetic phase transitions in an irreversible surface-reaction model. Physical Review Letters. 1986;56:2553–2556. doi: 10.1103/PhysRevLett.56.2553. A classic paper analyzing 2-D reaction diffusion systems. This paper forms the basis for the widely used ZGB model. [DOI] [PubMed] [Google Scholar]
- 70.Lam R, Basak T, Vlachos DG, Katsoulakis MA. Validation of mesoscopic theories and their application to computing effective diffusivities. Journal of Chemical Physics. 2001;115:11278–11288. [Google Scholar]
- 71.Suzuki K, Ritchie K, Kajikawa E, Fujiwara T, Kusumi A. Rapid hop diffusion of a g-protein-coupled receptor in the plasma membrane as revealed by single-molecule techniques. Biophys J. 2005;88:3659–3680. doi: 10.1529/biophysj.104.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ritchie K, Shan XY, Kondo J, Iwasawa K, Fujiwara T, Kusumi A. Detection of non-Brownian diffusion in the cell membrane in single molecule tracking. Biophys J. 2005;88:2266–2277. doi: 10.1529/biophysj.104.054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *73.Murase K, Fujiwara T, Umemura Y, Suzuki K, Iino R, Yamashita H, Saito M, Murakoshi H, Ritchie K, Kusumi A. Ultrafine membrane compartments for molecular diffusion as revealed by single molecule techniques. Biophys J. 2004;86:4075–4093. doi: 10.1529/biophysj.103.035717. Single particle tracking with ultrahigh speed cameras reveal compartmentalization of the plasma membrane. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Murakoshi H, Iino R, Kobayashi T, Fujiwara T, Ohshima C, Yoshimura A, Kusumi A. Single-molecule imaging analysis of Ras activation in living cells. Proc Natl Acad Sci U S A. 2004;101:7317–7322. doi: 10.1073/pnas.0401354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wilson BS, Pfeiffer JR, Oliver JM. Observing FcεRI signaling from the inside of the mast cell membrane. J Cell Biol. 2000;149:1131–1142. doi: 10.1083/jcb.149.5.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]