Abstract
Aurora-B is a component of the Chromosomal Passenger Complex (CPC) required for correct spindle-kinetochore attachments during chromosome segregation and for cytokinesis. The chromatin factors that recruit the CPC to centromeres are unknown, however. Here we show that phosphorylation of Histone-H3 Thr-3 (H3T3ph) by Haspin is necessary for CPC accumulation at centromeres, and that the CPC subunit Survivin binds directly to H3T3ph. A non-binding Survivin-D70A/D71A mutant does not support centromeric CPC concentration and both Haspin depletion and Survivin-D70A/D71A mutation diminish centromere localization of MCAK and mitotic checkpoint signaling in taxol. Survivin-D70A/D71A mutation and microinjection of H3T3ph-specific antibody both compromise centromeric Aurora-B functions but do not prevent cytokinesis. Therefore, H3T3ph generated by Haspin positions the CPC at centromeres to regulate selected targets of Aurora-B during mitosis.
The CPC (containing Aurora-B, INCENP, Survivin and Borealin) is found on chromosome arms in prophase, concentrates at inner centromeres during prometaphase, and transfers to the central spindle at anaphase (1). Aurora-B phosphorylates several substrates at these locations, including Histone-H3 Ser-10 (H3S10ph) on chromosome arms, Mitotic Centromere-Associated Kinesin (MCAK) at inner centromeres, Centromere Protein-A Ser-7 (CENP-AS7ph) at outer centromeres and the KNL1/Mis12 complex/Ndc80 network at kinetochores (1–6). Current models suggest that centromeric Aurora-B responds to lack of tension across sister kinetochores that are incorrectly attached to the spindle. Bipolar kinetochore attachment forces may pull kinetochore substrates away from inner centromeric Aurora-B, leading to substrate dephosphorylation, selective stabilization of correct microtubule attachments, and eventually to satisfaction of the spindle checkpoint (Fig. S1) (7). Despite its central importance, it is not understood how Aurora-B accumulates at centromeres (8–10).
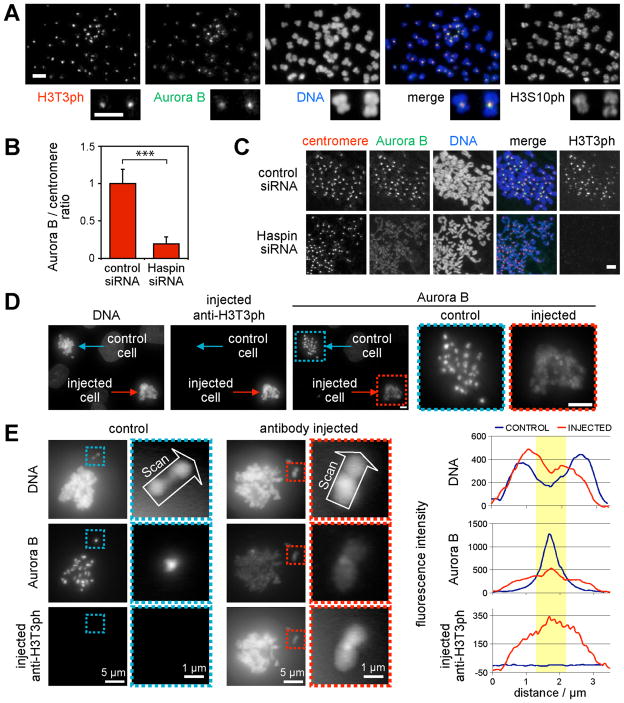
Immunofluorescence microscopy of mitotic cells shows that H3T3ph and Aurora-B localize similarly at inner centromeres (Fig. 1A, Fig. S2). We therefore tested whether Haspin, which is responsible for generating H3T3ph in mitosis (11, 12), is required for CPC localization. Upon Haspin RNAi in HeLa cells, a marked reduction in Aurora-B at centromeres (>5-fold) and an increase on chromosome arms were observed (Fig. 1B, C). In contrast, Haspin knockdown had little effect on the amounts of CPC subunits in mitosis, and did not disrupt CPC formation (Fig. S3). Similar results were obtained in U2OS cells, where Haspin RNAi delocalized all CPC components from centromeres (Fig. 2A, Fig. S4). Microinjection of H3T3ph-specific antibody into mitotic LLC-PK cells caused a similar delocalization of Aurora-B (Fig. 1D, E), indicating that Haspin acts through H3T3ph to position the CPC during mitosis and not through effects at other cell cycle stages.
Fig. 1. Haspin RNAi or microinjection of anti-H3T3ph delocalizes Aurora-B from mitotic centromeres.
(A) Immunofluorescence microscopy of nocodazole-arrested HeLa chromosome spreads reveals colocalization of H3T3ph and Aurora-B.
(B, C) Cells were transfected with siRNA, and prepared as in (A). The Aurora-B/centromere autoantigen intensity ratio was determined by immunofluorescence at approximately 40 centromeres in 9 cells per condition (B). Means + SD are shown (N=3). *** p<0.001 by Student t test. Example images are shown in (C).
(D, E) Nocodazole and MG132-treated LLC-PK cells were microinjected with anti-H3T3ph. After ~2 h, cells were subject to immunofluorescence staining for Aurora-B and with anti-rabbit antibodies to reveal microinjected antibody. Line scans of chromosomes are shown in (E). Yellow highlights the centromere. Scale bars = 5 μm unless noted.
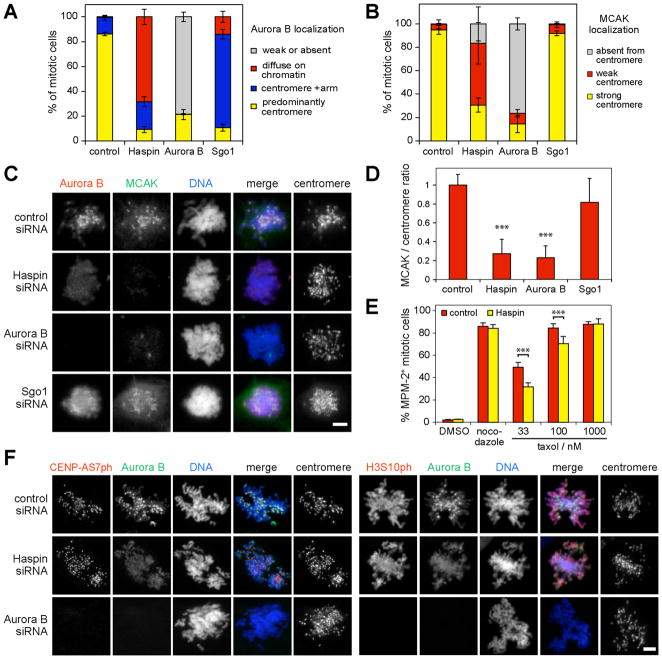
Fig. 2. Haspin RNAi delocalizes centromeric MCAK and compromises the spindle checkpoint, but has minor effects on Aurora-B activity toward CENP-AS7 and H3S10.
(A, B) U2OS cells were transfected with siRNA, treated with nocodazole for 1–2 h, and subject to immunofluorescence staining. Approximately 100 mitotic cells in each condition were classified according to the localization of Aurora B (A) or MCAK (B). Means +/− SD are shown (N=3).
(C) Example images of cells treated as above.
(D) Following treatment as above, the MCAK/centromere autoantigen intensity ratio was determined at approximately 30 centromeres in 10–11 mitotic cells per condition. Means + SD are shown (N=3).
(E) HeLa cells were transfected with siRNA, treated with thymidine for 24 h, and released into medium containing the compounds indicated. After 18 h, mitotic indices were determined by flow cytometry with MPM-2 antibody. Means + SD are shown (N=4). In (D and E), *** p<0.001 by Bonferroni’s multiple comparison test compared to corresponding control.
(F) Immunofluorescence microscopy of CENP-AS7ph and H3S10ph in mitotic siRNA-transfected U2OS cells. Scale bars = 5 μm.
To determine whether the reduced CPC accumulation at centromeres following Haspin depletion was functionally significant, we first focused on MCAK, a microtubule depolymerizing kinesin whose inner centromeric localization is dependent on Aurora-B (3, 4, 13). Both Aurora-B and Haspin RNAi reduced the centromeric enrichment of MCAK in ≥70% of U2OS cells (Fig. 2B–D) and HeLa cells (Fig. S5). Artificial retargeting of Aurora-B to centromeres with CENP-B-INCENP (7) largely restored MCAK localization in Haspin-depleted cells (Fig. S6). Disruption of CPC activity also compromises the spindle checkpoint, particularly in low doses of taxol (14–18). Consistent with this, HeLa cells transfected with Haspin siRNA were less efficiently arrested in mitosis by taxol than controls (Fig. 2E). Thus, Haspin appears to influence MCAK localization and checkpoint signaling at centromeres by controlling Aurora-B localization.
Two experiments suggest that these effects are not a consequence of defects in chromosome cohesion caused by Haspin RNAi (12). First, Haspin overexpression, which increases cohesion between chromosome arms (12), also influenced Aurora-B localization (Fig. S7). Second, loss of cohesion induced by depletion of Sgo1 (12, 19, 20) rendered Aurora-B localization somewhat more diffuse, but it remained enriched at centromeres in >80% of nocodazole-treated cells and centromeric MCAK was little changed (Fig. 2A–D). Therefore, as reported elsewhere (12, 19–22), loss of centromeric CPC and MCAK is not an inevitable consequence of cohesion loss.
In contrast, Haspin RNAi had minor effects on total CENP-AS7ph and H3S10ph (Fig. S8A), though both were reduced by Aurora-B RNAi, as expected (2, 5, 14). Haspin RNAi also did not detectably alter total T232 autophosphorylation (Fig. S8B) or in vitro kinase activity of immunoprecipitated Aurora-B (Fig. S8C). H3S10ph, CENPAS7ph and Aurora-B T232ph immunofluorescence remained present in individual U2OS and HeLa cells in which Aurora-B and MCAK were delocalized (Fig. 2F, Fig. S8D–F). Thus, Aurora-B kinase activation and some of its chromosomal functions appear less sensitive to delocalization of the CPC from inner centromeres than localization of MCAK.
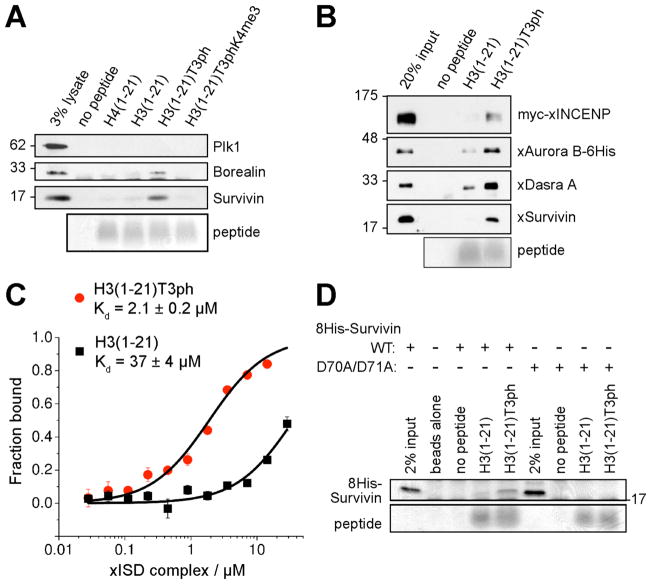
We tested if H3T3ph plays a direct role in CPC binding at inner centromeres. Biotinylated peptides encompassing residues 1 to 21 of Histone-H3 with various side chain modifications were used in “pull-down” experiments from mitotic HeLa cell lysates. Survivin bound strongly to H3(1-21) peptides containing H3T3ph but poorly to unmodified H3(1-21), H4(1-21) or peptides containing H3S10ph or H3T11ph (Fig 3A, Fig. S9A). Borealin, which directly binds Survivin (10, 23), also bound preferentially to H3T3ph peptides (Fig. 3A). Enrichment of Aurora-B on H3T3ph peptides was detectable but weaker than that of Survivin or Borealin (Fig. S9A), perhaps because not all CPC components are associated in cell lysates (23). Tri-methylation of H3K4 adjacent to T3ph (H3T3phK4me3) strongly diminished association of CPC components with H3T3ph peptides (Fig. 3A, Fig. S9A). Two different recombinant forms of the CPC (the full complex, and “xISD” containing INCENP(1-58), Survivin and Dasra-A, a Xenopus homolog of Borealin) also bound preferentially to H3T3ph peptides, confirming that H3T3ph enhances direct binding of the CPC to H3 almost 20-fold (Fig. 3B, C).
Fig. 3. The CPC binds H3T3ph through Survivin.
(A) Lysates of nocodazole-arrested HeLa cells were incubated with bead-immobilized histone peptides. Protein binding was visualized by immunoblotting, and peptide loading by Coomassie Blue staining.
(B) Binding of recombinant Xenopus CPC to H3 peptides analyzed as above.
(C) Binding of Xenopus ISD complex to H3 peptides measured by fluorescence anisotropy. Means +/− SD are shown (N=3).
(D) Binding of recombinant human 8His-Survivin to H3 peptides visualized by Coomassie Blue staining.
Purified recombinant human Survivin also preferentially bound H3T3ph peptides (Fig. 3D, Fig. S9B), consistent with previous reports that Survivin is a key factor targeting the CPC to centromeres (8, 17, 18, 24). Similar to previous observations (18, 25), in HeLa cells depleted of endogenous Survivin, the mutation D70A/D71A within the BIR domain prevented Survivin from concentrating at centromeres in mitosis, causing it to become diffusely distributed on chromatin (Fig. S10A–C). Survivin-D70A/D71A still localized to the central spindle, however (Fig. S10D). Recombinant Survivin-D70A/D71A showed no binding to H3 peptides in vitro (Fig. 3D), suggesting that the failure of Survivin-D70A/D71A to concentrate at centromeres is due to a failure to recognize H3T3ph.
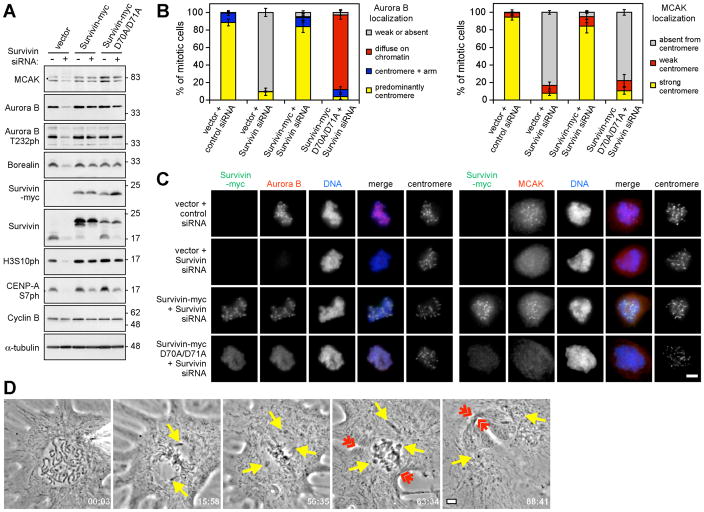
We then tested whether Survivin-D70A/D71A mimicked the effects of Haspin depletion on CPC functions. Indeed, Survivin-D70A/D71A supported CPC formation (Fig. S11A) and substantial Aurora-B autophosphorylation, H3S10 phosphorylation, and CENP-AS7 phosphorylation, but did not restore normal localization of Aurora-B or MCAK to inner centromeres (Fig. 4A–C, Fig S11B, C), or support spindle checkpoint activity in low doses of taxol (Fig. S12A). Survivin-D70A/D71A rescued cytokinesis failure caused by Survivin depletion (Fig. S12B), but it did not allow normal correction of improper spindle-kinetochore attachments, and an increase in chromosome alignment defects was evident compared to cells expressing wild-type Survivin (Fig. S12C, D). Similarly, in chicken DT40 cells expressing Survivin-D72A/D73A (equivalent to D70A/D71A in human Survivin) and in human cells containing Survivin mutated at other sites in the BIR domain, the CPC failed to localize to centromeres and defects in spindle checkpoint activity in taxol were reported, yet mitotic progression and cytokinesis could still occur (17, 18). Finally, microinjection of H3T3ph-specific antibody into LLC-PK or Xenopus S3 cells also caused mitotic defects consistent with loss of Aurora-B activity at centromeres, while leaving intact its function in cytokinesis (Fig. 4D, Movies S1 to S4), further supporting a role for H3T3ph in centromeric CPC function.
Fig. 4. CPC function at centromeres requires Survivin binding to H3T3ph.
(A) Lysate immunoblotting of nocodazole-arrested HeLa cells stably expressing Survivin-myc or D70A/D71A mutant and transfected with siRNA as indicated.
(B) The localization of Aurora-B and MCAK was classified by immunofluorescence microscopy in approximately 100 HeLa cells per condition. Means +/− SD are shown (N=3).
(C) Example images of cells treated as in (B).
(D) A Xenopus S3 cell microinjected with anti-H3T3ph exits mitosis in the presence of unaligned chromosomes (yellow arrows) with clear formation of a cleavage furrow (red arrows). Time is in min:s. Three of three injected S3 cells behaved similarly. See Movie S2. Scale bars = 5 μm.
Our results are consistent with a model in which H3T3ph generated by Haspin enhances the binding of the CPC to centromeric chromatin, and suggest a molecular basis for the importance of the Survivin BIR domain (8, 17, 18, 25). BIR domains in other molecules use the equivalent of residue D71 to bind the N-terminal Ala of the N-terminal “IAP-binding motif” (IBM) of proteins such as Smac/DIABLO (26). Survivin binds Smac/DIABLO weakly, but the relevance of this interaction is unclear (27, 28). Instead, the IBM-binding region of the Survivin BIR domain may recognize the N-terminal Ala-Arg-Thr-Lys sequence of Histone-H3 when T3 is phosphorylated, raising the possibility that other BIR domains act as phospho-specific protein recognition modules.
H3T3ph does not appear to be required for chromatin association or activity of the CPC per se. Rather, H3T3ph contributes to accurate positioning of the CPC at inner centromeres. In this respect, the CPC may resemble heterochromatin protein HP1 which binds Histone-H3 methylated at K9, but can also interact with chromatin by K9 methylation-independent mechanisms (29). In addition, H3T3ph prevents H3 from competitively inhibiting Aurora-B autophosphorylation, potentially facilitating Aurora-B activation at inner centromeres (30). Therefore, Haspin might act through H3T3ph both to position and modulate activation of Aurora-B at centromeres.
Loss of inner centromeric CPC accumulation has similar differential effects on centromeric Aurora-B functions whether caused by Haspin depletion or Survivin D70A/D71A mutation. Our results imply that proximity to inner centromeric Aurora-B is not the sole determinant of substrate phosphorylation at centromeres, likely because substrates have different susceptibilities to phosphorylation and dephosphorylation by Aurora-B and phosphatases (Fig. S1). It may be valuable to further test the idea that Aurora-B target sites are “programmed” to respond differently to microtubule-dependent displacement from the inner centromere, allowing sophisticated regulation of kinetochore/centromere functions in response to tension.
Supplementary Material
Acknowledgments
We thank W.C. Earnshaw, H. Funabiki, M.A. Lampson, S.M.A. Lens, J.P. Noel, S.S. Taylor, L. Wordeman and Y. Zheng for cDNAs and antibodies, C. Crafter of AstraZeneca for ZM447439, and H. Funabiki and A. Kelly for sharing unpublished results. J.D. and J.M.G.H. have patents pending on Haspin inhibitors. This work was funded by grants from the American Cancer Society (RSG-05-13401-GMC to J.M.G.H) and the National Institutes of Health (R01-GM074210 to J.M.G.H., R01-GM050412 to G.J.G. and R01-GM063045 to P.T.S.).
References and Notes
- 1.Ruchaud S, Carmena M, Earnshaw WC. Nat Rev Mol Cell Biol. 2007;8:798. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- 2.Hsu JY, et al. Cell. 2000;102:279. doi: 10.1016/s0092-8674(00)00034-9. [DOI] [PubMed] [Google Scholar]
- 3.Andrews PD, et al. Dev Cell. 2004;6:253. doi: 10.1016/s1534-5807(04)00025-5. [DOI] [PubMed] [Google Scholar]
- 4.Lan W, et al. Curr Biol. 2004;14:273. doi: 10.1016/j.cub.2004.01.055. [DOI] [PubMed] [Google Scholar]
- 5.Zeitlin SG, Shelby RD, Sullivan KF. J Cell Biol. 2001;155:1147. doi: 10.1083/jcb.200108125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Welburn JP, et al. Mol Cell. 2010;38:383. doi: 10.1016/j.molcel.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu D, Vader G, Vromans MJ, Lampson MA, Lens SM. Science. 2009;323:1350. doi: 10.1126/science.1167000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vader G, Kauw JJ, Medema RH, Lens SM. EMBO Rep. 2006;7:85. doi: 10.1038/sj.embor.7400562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klein UR, Nigg EA, Gruneberg U. Mol Biol Cell. 2006;17:2547. doi: 10.1091/mbc.E05-12-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeyaprakash AA, et al. Cell. 2007;131:271. doi: 10.1016/j.cell.2007.07.045. [DOI] [PubMed] [Google Scholar]
- 11.Dai J, Sultan S, Taylor SS, Higgins JMG. Genes Dev. 2005;19:472. doi: 10.1101/gad.1267105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dai J, Sullivan BA, Higgins JMG. Dev Cell. 2006;11:741. doi: 10.1016/j.devcel.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 13.Ohi R, Sapra T, Howard J, Mitchison TJ. Mol Biol Cell. 2004;15:2895. doi: 10.1091/mbc.E04-02-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ditchfield C, et al. J Cell Biol. 2003;161:267. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carvalho A, Carmena M, Sambade C, Earnshaw WC, Wheatley SP. J Cell Sci. 2003;116:2987. doi: 10.1242/jcs.00612. [DOI] [PubMed] [Google Scholar]
- 16.Lens SMA, et al. EMBO J. 2003;22:2934. doi: 10.1093/emboj/cdg307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lens SMA, et al. Mol Biol Cell. 2006;17:1897. doi: 10.1091/mbc.E05-08-0727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yue Z, et al. J Cell Biol. 2008;183:279. doi: 10.1083/jcb.200806118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salic A, Waters JC, Mitchison TJ. Cell. 2004;118:567. doi: 10.1016/j.cell.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 20.McGuinness BE, Hirota T, Kudo NR, Peters JM, Nasmyth K. PLoS Biol. 2005;3:e86. doi: 10.1371/journal.pbio.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawashima SA, et al. Genes Dev. 2007;21:420. doi: 10.1101/gad.1497307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang H, et al. J Cell Biol. 2007;177:413. doi: 10.1083/jcb.200701122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gassmann R, et al. J Cell Biol. 2004;166:179. doi: 10.1083/jcb.200404001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vong QP, Cao K, Li HY, Iglesias PA, Zheng Y. Science. 2005;310:1499. doi: 10.1126/science.1120160. [DOI] [PubMed] [Google Scholar]
- 25.Cao L, et al. Biochem Biophys Res Commun. 2006;346:400. doi: 10.1016/j.bbrc.2006.05.131. [DOI] [PubMed] [Google Scholar]
- 26.Srinivasula SM, Ashwell JD. Mol Cell. 2008;30:123. doi: 10.1016/j.molcel.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song Z, Yao X, Wu M. J Biol Chem. 2003;278:23130. doi: 10.1074/jbc.M300957200. [DOI] [PubMed] [Google Scholar]
- 28.Sun C, Nettesheim D, Liu Z, Olejniczak ET. Biochemistry. 2005;44:11. doi: 10.1021/bi0485171. [DOI] [PubMed] [Google Scholar]
- 29.Vermaak D, Malik HS. Annu Rev Genet. 2009;43:467. doi: 10.1146/annurev-genet-102108-134802. [DOI] [PubMed] [Google Scholar]
- 30.Rosasco-Nitcher SE, Lan W, Khorasanizadeh S, Stukenberg PT. Science. 2008;319:469. doi: 10.1126/science.1148980. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.