Abstract
The endocannabinoid system plays an important role in numerous physiological processes including mood, appetite, and pain sensation. A critical compound in maintaining cannabinoid tone is the endocannabinoid anandamide (AEA). We have recently shown that AEA is metabolized by several different human cytochromes P450 (P450) to form a number of metabolites, one of which exhibits increased biological activity. CYP3A4, one of the major P450s involved in the metabolism of AEA, produces four major metabolites. One of these metabolites, 5,6-epoxyeicosatrienoic acid ethanolamide (5,6-EET-EA), exhibits a much higher affinity than AEA for the cannabinoid 2 receptor (CB-2), which leads to a marked decrease in intracellular cAMP levels in cells expressing CB-2. There are multiple human alleles of CYP3A4, and the CYP3A4.4 allele has been shown to exhibit a significant decrease in activity. Recombinant CYP3A4*4 was expressed in Escherichia coli and was demonstrated to produce 60% less 6-hydroxytestosterone than the wild-type (WT) 3A4 in a reconstituted system. The metabolism of AEA by the WT and the CYP3A4.4 variant was investigated. The mutant produced 60% less of the four EET-EA metabolites than the WT. The mutant also produced a new peak on liquid chromatography-mass spectrometry not seen with the WT, which corresponded to 19-hydroxyeicosatetraenoic acid-ethanolamide. In addition, the mutant produces four novel peaks at m/z 380, which correspond to the addition of two oxygen atoms, possibly to form a peroxide bond. These data indicate that individuals expressing the CYP3A4.4 allele may exhibit significant variations in the metabolism of AEA as well as any other compounds resembling AEA.
Introduction
The endocannabinoid system is a multifaceted signaling system that is located in many tissues in the body. Numerous results have illuminated how this complex system assists in controlling a wide variety of psychological and physiological processes including mood, appetite, inflammation, and pain sensation (Richardson et al., 1998; Di Marzo et al., 2004).
Two (possibly three) receptors (CB1 and CB2) have been discovered that bind either Δ9-tetrahydrocannabinol or endogenous cannabinoids. CB1 has been found in numerous areas in the body such as the central nervous system, the cardiovascular system, and the peripheral nervous system (Felder et al., 2006). CB2 receptors are located primarily in cells in the immune system (Ducobu, 2005; Ducobu and Sternon, 2005; Vickers and Kennett, 2005; Felder et al., 2006).
The cannabinoid receptors are targets for a family of lipid transmitters. The first of these signaling molecules to be discovered was arachidonoylethanolamide [anandamide (AEA)], which is a derivative of arachidonic acid (Rodríguez de Fonseca et al., 2005). It has been found in numerous locations including the brain, plasma, and peripheral tissues. A novel pathway for AEA metabolism by human cytochromes P450 (P450s) has been discovered (Snider et al., 2007). The primary P450 enzyme responsible for the oxidation of AEA in the liver is CYP3A4. This enzyme produces the four-monoepoxide molecules 5,6-, 8,9-, 11,12-, and 14,15-epoxyeicosatrienoic acid ethanolamides (EET-EAs). CYP2D6 and also CYP4F2 metabolize AEA to produce the same four metabolites along with a fifth metabolite, 20-hydroxyeicosatetraenoic acid (HETE) (Snider et al., 2008). It has been hypothesized that the epoxygenated and hydroxylated compounds may be more potent ligands for cellular receptors than the parent AEA, and, in fact, 5,6-EET-EA has been demonstrated to have a higher affinity for the CB2 receptor than its parent AEA (Snider et al., 2009).
As indicated previously, P450 CYP3A4 is responsible for the majority of P450-catalyzed oxygenations of AEA in the liver. This enzyme has been shown to possess many single nucleotide polymorphisms in its coding region. One of these polymorphisms is CYP3A4*4, which has a frequency of 3.3% in the Chinese population (Wang et al., 2005). A protein product of this allele has been shown to possess decreased catalytic activity. This allele also been reported to be correlated with an increased risk of subarachnoid hemorrhage in Japan with a P value for this correlation of 0.0006 (Yamada et al., 2008). This study provides insights into the ability of CYP3A4.4 to catalyze the metabolism of AEA and elucidates its altered pattern of AEA metabolites.
Materials and Methods
Reagents.
Anandamide, the various anandamide metabolite standards, and epoxide hydrolase were purchased from Cayman Chemical (Ann Arbor, MI). Catalase, NADPH, l-α-dilauroyl-phosphatidylcholine, l-α-dioleyl-sn-glycero-3-phosphatidylcholine, and l-α-phosphatidylserine were purchased from Sigma-Aldrich (St. Louis, MO). All other chemicals were of the highest quality and available from commercial sources.
Overexpression and Purification of CYP3A4, NADPH-Dependent Cytochrome P450 Reductase, and Cytochrome b5.
The plasmid for the overexpression of CYP3A4 was a generous gift from Dr. James Halpert (University of California at San Diego, San Diego, CA) (Domanski et al., 1998). CYP3A4 was expressed as a truncated form, in which the hydrophobic membrane-spanning domain was removed (Δ3–21) and several positively charged residues were introduced into the N terminus to increase the expression yield. CYP3A4, cytochrome b5, and cytochrome P450 reductase were expressed and purified as described previously (Hanna et al., 1998).
Site-Directed Mutagenesis.
The mutation of CYP3A4 was accomplished using the QuikChange site-directed mutagenesis kit according to the manufacturer's protocol (Stratagene, La Jolla, CA). The forward mutagenic primer was 5′-TTATGAAAAGTGCCGTCTCTATAGCTGAGGATG-3′, and the reverse primer was 5′-CATCCTCAGCTATAGAGACGGCACTTTTCATAA-3′. The site-specific mutation was confirmed by DNA sequencing at the University of Michigan Sequencing Core facility.
Testosterone Metabolism.
CYP3A4 protein was reconstituted with reductase and cytochrome b5 (1:2:1 ratio), a 10-mg mixture of l-α-dilauroyl-phosphocholine, l-α-dioleyl-sn-glycero-3-phosphocholine, and l-α-phosphatidylserine (1:1:1), for 1 h on ice. For the wild-type and mutant proteins, 50 pmol of protein was used. Incubations were performed in reaction mixtures (1.0 ml) containing 50 mM HEPES buffer, pH 7.4, and 50 U of catalase. Reactions were initiated by the addition of NADPH to give a final concentration of 1.2 mM. Control reactions were done in the absence of NADPH. Incubations were conducted for 10, 20, and 40 min in triplicate. Samples were terminated and extracted with ethyl acetate and dried under nitrogen. They were then resuspended in 150 μl of 65% methanol, and 90-μl fractions were injected into a Shimadzu high-performance liquid chromatograph (Shimadzu, Columbia, MD). The metabolites were resolved isocratically on a C18 reverse-phase column (Varian, Inc., Palo Alto, CA) equilibrated with 62% methanol and eluted using a flow rate of 1 ml/min. Metabolites were detected at 254 nm and were quantitated using a standard curve constructed with known amounts of 6-β-OH-testosterone.
Anandamide Metabolism Assays.
CYP3A4 was reconstituted with reductase and phospholipid as stated previously. Studies were performed using incubation mixtures (0.5 ml) containing 50 mM HEPES buffer, pH 7.4, and 50 U of catalase. Experimental reactions were initiated by the addition of NADPH to give a final concentration of 1.2 mM. Control reactions were done in the absence of NADPH. Samples were incubated at 37°C. The reactions were terminated by the addition of 3 ml of nitrogen-purged ethyl acetate. Samples were vortexed for 1 min and then centrifuged for 5 min at 1200 rpm to separate the organic layer. This layer was removed, dried under a constant stream of nitrogen, and resuspended in 160 μl of methanol, and 20-μl fractions were subjected to electrospray ionization (ESI)-liquid chromatography/mass spectrometry (ESI-LC/MS) analysis as described below.
For determinations of the Km and Vmax values, the incubation conditions were optimized for time and protein concentration and performed within the linear range for metabolite formation. For these studies, 30 pmol of CYP3A4 protein in the reconstituted system was incubated with AEA at the concentrations indicated for 20 min. After extraction with nitrogen-purged ethyl acetate and drying, the extracts were resuspended in 160 μl of methanol, and 20-μl aliquots were injected for analysis by ESI-LC/MS. The AEA peak was used as the internal standard because under reaction conditions the amount metabolized was always less than 5% of the total amount (Supplemental Fig. 1). Standard curves for 5,6-, 8,9-, 11,12-, and 14,15-EET-EA were generated by injecting known amounts of the standards (Cayman Chemical) into the ESI-LC/MS system. For reactions containing recombinant epoxide hydrolase (EH), CYP3A4.4 (50 pmol) was incubated in the above reaction mixture with recombinant EH (25 pmol) for 2 h at 37°C.
ESI-LC/MS Analysis of AEA Metabolism.
Samples (20 μl of each) were injected onto a Hypersil ODS column (5 μm, 4.6 × 100 mm; Thermo Fisher Scientific, Waltham, MA) that had been equilibrated with 75% solvent B (0.1% acetic acid in methanol) and 25% solvent A (0.1% acetic acid in water). The metabolites were resolved using the following gradient: 0 to 5 min, 75% B; 5 to 20 min, 75 to 100% B; 20 to 25 min, 100% B; 25 to 26 min, 100 to 75% B; and 26 to 30 min, 75% B. The flow rate was 0.3 ml/min. The column effluent was directed into the LCQ mass analyzer (Thermo Fisher Scientific). The ESI conditions were as follows: sheath gas, 90 arbitrary units; auxiliary gas, 30 arbitrary units; capillary temperature, 200°C; and spray voltage, 4.5 KV. Data were acquired in positive ion mode using the Xcalibur software package (Thermo Fisher Scientific) with one full scan from 300 to 500 m/z followed by one data-dependent scan of the most intense ion.
Data Analysis.
Nonlinear regression analyses of the data were performed using GraphPad Prism (version 5 for Mac; Graph Pad Software, Inc., La Jolla, CA).
Docking of Anandamide into CYP3A4.
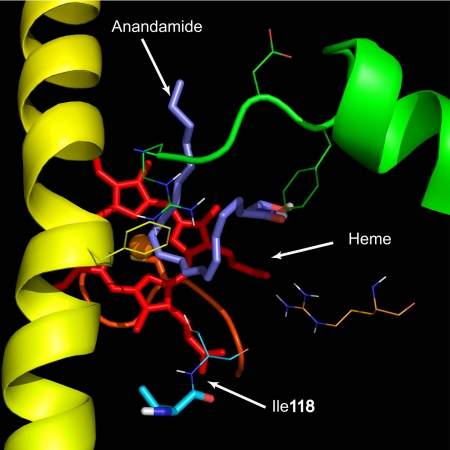
AEA was docked into the active site of CYP3A4 as a flexible ligand using AutoDock software 4.0 (Morris et al., 1996). The coordinates of CYP3A4 were obtained from the Protein Data Bank (code 1TQN). The water molecules were removed from the coordinates before docking. The coordinates for the AEA were constructed using ChemOffice Suite 2008 (CambridgeSoft, Cambridge, MA), and the geometry of AEA was optimized with the semiempirical quantum mechanical method AM1. Because of the large number of rotatable bonds in AEA, the docked conformations were not well clustered. Therefore, prediction of the metabolites of AEA is poor. The selected pose of AEA shown in Fig. 3 represents one possible conformation of the bound AEA that leads to the formation of 11,12-EET-EA.
Fig. 3.
Molecular modeling results showing AEA (purple) bound into the active site of CYP3A4. The coordinates for CYP3A4 were obtained from the Protein Data Bank, and AEA was docked into CYP3A4 using Autodock. Residue 118 is located in close proximity of the heme (red). Mutation of this residue possibly changes the conformation of the active site, thus affecting CYP3A4 activity.
Metabolism of the AEA Metabolites with m/z 380 by Prostaglandin D Synthase.
CYP3A4.4 protein (240 pmol) was reconstituted with reductase and cytochrome-b5, as stated previously, in a final volume of 152 μl. To eight 0.5-ml reaction mixtures 20 μl each of the reconstitution mix was added to samples containing 50 mM HEPES, pH 7.4, 100 μM AEA, and 50 U of catalase. NADPH was added to each of the reactions to give a final concentration of 1.2 mM, and the samples were incubated at 37°C for 20 min. Reactions were terminated by the addition of 2 ml of nitrogen-purged ethyl acetate, and the samples were extracted and dried as described previously. All eight samples were resuspended in 15 μl of 50% methanol each and pooled. The pooled samples were then added to 100 mM Tris buffer, pH 8.0, containing 2 mM MgCl2 and 2 mM GSH in a final volume of 0.5 ml. In the control samples, 10 μl of 100 mM Tris was added, and in experimental samples, 10 μl of 0.5 μg/μl of prostaglandin D synthase in 100 mM Tris was added. Samples were incubated for 30 min at room temperature, and the reactions were terminated with ethyl acetate. Samples were then analyzed by LC/MS as indicated previously.
Spectral Properties.
To analyze the spectral binding of AEA to CYP3A4 and the CYP3A4.4 mutant, the spectra were measured using methods published previously (Estabrook and Werringloer, 1978; Shebley and Hollenberg, 2007). CYP3A4 and the 3A4.4 polymorphic form were diluted in 50 mM KPi, pH 7.4, to a final concentration of 1 μM in a final volume of 2 ml, and equal volumes of each were placed in the reference and sample cuvettes. A difference spectrum between the two cuvettes was recorded from 350 to 700 nm on a UV-2501PC spectrophotometer (Shimadzu, Kyoto, Japan). Increasing concentrations of AEA (1–100 μM) in methanol were added to the sample cuvette, whereas the reference cuvette received equal volumes of methanol. The difference spectra were determined after each addition, and the spectral Ks values were calculated as described previously.
Measurements of Rates of Electron Transfer by Stopped-Flow Spectrophotometry.
The rates of reduction of the wild-type and the mutant P450 were determined at 25°C using a Hi-Tech SF61DX2 stopped-flow spectrophotometer (Hi-Tech Scientific, Salisbury, Wiltshire, UK) as described previously (Zhang et al., 2008). The CYP3A4 (3 μM) was reconstituted with 3 μM cytochrome P450 reductase (CPR) and 10 μg/ml lipid in 50 mM HEPES buffer, pH 7.4, in a final volume of 2 ml on ice for 60 min. After reconstitution for 60 min, the protein samples were bubbled with CO gas for 6 min and then loaded into a syringe of the stopped-flow spectrophotometer. CO-saturated HEPES buffer, pH 7.4, containing 0.1 mM NADPH was loaded into a second syringe. The kinetic traces at 450 nm were recorded after rapid mixing of the contents of both syringes. Four shots with CYP3A4 were performed and scanned for 120 s each. Four shots were performed using CYP3A4.4, and they were scanned for 1200 s each.
Results
Mutation of Residue 118 from Leucine to Valine in Human CYP3A4 Causes a Decrease in Catalytic Activity.
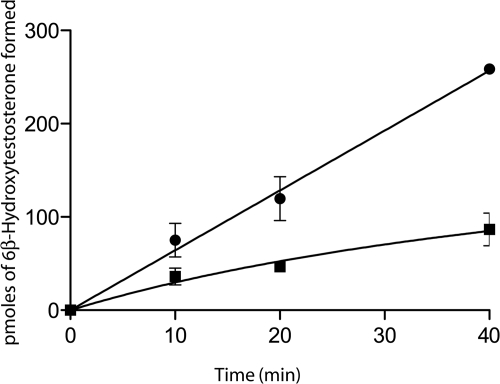
To investigate the activity of the mutant compared with the wild type, we analyzed its ability to metabolize testosterone to form 6-β-OH-testosterone. Using 50-pmol samples of wild-type and mutant P450s, we determined that the mutant possessed approximately 33% of the activity of the wild type (Fig. 1).
Fig. 1.
Formation of 6-β-OH-testosterone by CYP3A4 and the I228V mutant. Reaction mixtures contained 50 pmol of the wild-type CYP3A4 (●) or the I118V variant (■) P450 proteins in the reconstituted system and 100 μM testosterone. Reactions were incubated and then analyzed after 0, 20, and 40 min as described under Materials and Methods. The amount of product formed was determined based on a standard curve generated using 6-β-OH-testosterone.
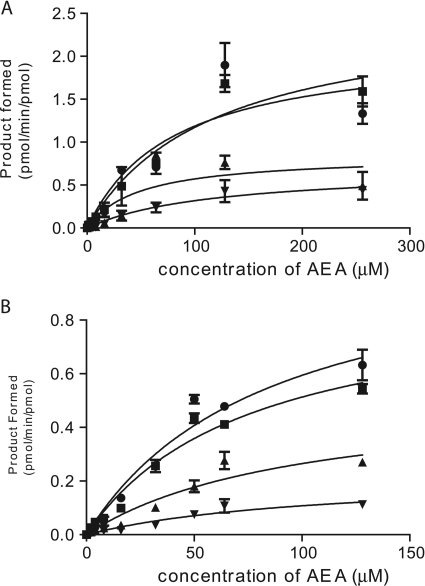
As shown previously, CYP3A4 metabolizes AEA to form four monoepoxide molecules (Snider et al., 2007, 2008). These metabolites are 5,6-, 8,9-, 11,12-, and 14,15-EET-EA, and they are formed in a time- and protein concentration-dependent manner. Using expressed human CYP3A4*1 and CYP3A4*4 purified from Escherichia coli, we performed kinetic analyses on the formation of these metabolites from AEA at concentrations ranging from 1 to 125 μM. Using optimized conditions for the incubation time and protein concentration, we determined that formations of 5,6-, 8,9-, 11,12-, and 14,15-EET-EA all exhibited Michaelis-Menten kinetics (Fig. 2). The values calculated for Km and Vmax from these data are presented in Table 1. These data demonstrate that the CYP3A4*4 polymorphism causes approximately 11/2 to 3-fold decreases in the Vmax values for the formation of the four primary metabolites of AEA. Except for the 8,9-EET-EA metabolite, for which the Km increased by a factor of 2, the Km values for all of the other metabolites were relatively unchanged.
Fig. 2.
Kinetics for the formation of the metabolites of AEA by human recombinant CYP3A4.1 and CYP3A4.4. Reaction mixtures contained 30 pmol of CYP3A4.1 (A) or CYP3A4.4.4 (B) and the reconstituted system as described under Materials and Methods. The AEA concentrations were 1 to 256 μM (A) and 1 to 128 μM (B), and the reaction mixtures were incubated for 20 min at 37°C. The metabolites of AEA that were quantified were 14,15-EET-EA (●), 11,2-EET-EA (■), 8,9-EET-EA (▴), and 5,6-EET-EA (▾). The amounts of products formed were determined based on standard curves generated for each metabolite, and the rate data (the average of six experiments) were fitted to a one-enzyme Michaelis-Menten model using Prism software.
TABLE 1.
Kinetic values for the metabolism of anandamide by CYP3A4.1 and CYP3A4.4
Km and Vmax values were determined from the data in Fig. 2 after 20-min incubations at 37°C of 0.5-ml reaction mixtures containing reconstituted CYP3A4.1 or CYP3A4 as described under Materials and Methods. Reaction mixtures contained 50 mM HEPES buffer, pH 7.4, AEA (1–256), and NADPH (1.2 mM), and the data represent the average of six experiments. Rates of formations were determined as described under Materials and Methods.
| CYP3A4 Allele | Product Formed | V max | K m | Vmax/Km |
|---|---|---|---|---|
| s −1 | μM | s−1/μM | ||
| *1 | 5,6-EET-EA | 0.69 | 118 | 0.006 |
| *4 | 5,6-EET-EA | 0.23 | 108 | 0.002 |
| *1 | 8,9-EET-EA | 0.85 | 48 | 0.018 |
| *4 | 8,9-EET-EA | 0.53 | 98 | 0.005 |
| *1 | 11,12-EET-EA | 2.56 | 118 | 0.022 |
| *4 | 11,12-EET-EA | 0.90 | 76 | 0.012 |
| *1 | 14,15-EET-EA | 2.13 | 80 | 0.027 |
| *4 | 14,15-EET-EA | 1.10 | 85 | 0.013 |
CYP3A4.4 Variant Exhibits an Increase in the Formation of Dioxygenated Products and an Additional Monooxygenated Metabolite of AEA.
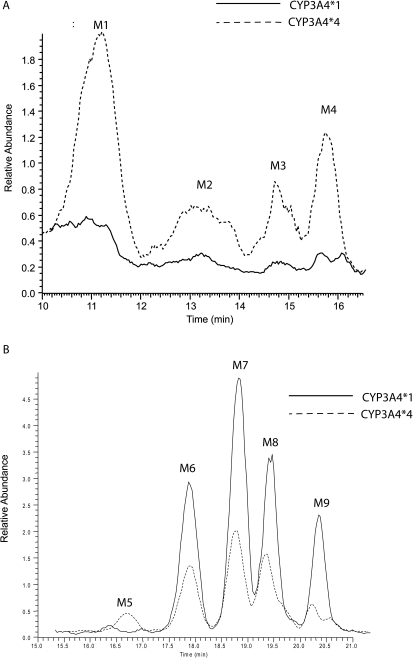
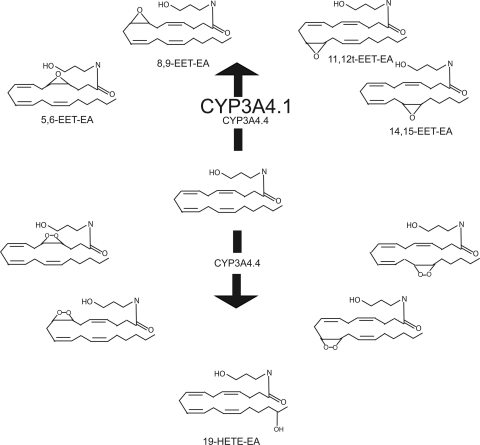
As reported previously, wild-type CYP3A4 primarily forms four epoxide metabolites of AEA (Snider et al., 2007). Because the 118 residue is near the active site and might be predicted to alter the binding of AEA (Fig. 3), we investigated the possibility that the amounts of the products or the ratios of metabolites formed during AEA metabolism might differ from those with the CYP3A4 wild type. The ratio of metabolites for the original epoxides was unchanged; however, AEA metabolism by the mutant protein resulted in the formation of one additional monooxygenated product with an m/z at 364, indicated as M5 in Fig. 4B, and four previously unreported dioxygenated products with m/z values at 380, indicated as M1 to M4 in Fig. 4A. The monooxygenated product is most likely 19-HETE-EA on the basis of its retention time on the C18 column (Nithipatikom et al., 2001). Wild-type CYP3A4 does not produce this metabolite; however, it has previously been observed as a metabolite of AEA that is formed by human liver microsomes (Snider et al., 2007).
Fig. 4.
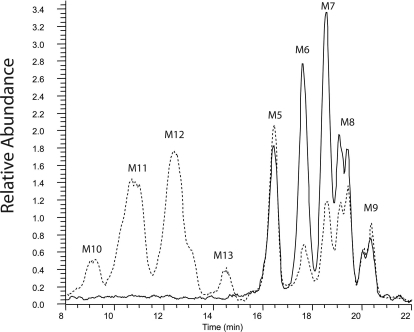
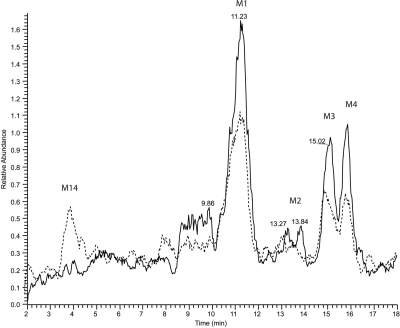
Metabolism of AEA by CYP3A4.1 and CYP3A.4. Reaction mixtures containing 30 pmol of CYP3A4.1 or CYP3A4.4 in the reconstituted system described under Materials and Methods and containing 64 μM AEA were incubated for 20 min at 37°C. Peaks were normalized to the AEA peak (data not shown). Reactions done in the absence of NADPH contained only the AEA peak (data not shown). A, polymorphic CYP3A4.4 produces significantly more of the M1 to M4 dioxygenated metabolites with m/z values at 380. B, polymorphic CYP3A4.4 also produces a novel product (M5) compared with the wild type and significantly less of the M6 to M9 products with m/z values at 364.
The four dioxygenated products (M1–M4) were formed in quantities comparable to those for the monooxygenated products (M6–M9) by the CYP3A4.4 mutant protein; however, the dioxygenated products were produced in relatively small quantities by the wild type compared with the epoxygenated products. The peaks for the dioxygenated products produced by the mutant protein were more than 10 times greater than those formed by the wild type. We initially hypothesized that these dioxygenated products were formed by a subsequent oxygenation of the monooxygenated products. To investigate whether these products might have been produced by a second oxygenation of the monooxygenated products (M6–M9), we incubated the monooxygenated products with CYP3A4.4 to determine whether any of the dioxygenated products observed would be formed. After 40-min incubations, we examined the samples by LC/MS and observed that the incubations with these substrates did not produce any of the dioxygenated products (M1–M4) (data not shown). These results led us to hypothesize that the dioxygenated products may be produced directly from AEA by the CYP3A4.4 mutant protein.
Novel Products M1 to M5 Formed from AEA by CYP3A4.4 Are Not Epoxides.
The possibility that the novel monooxygenated product and that the dioxygenated products might be epoxides was then investigated. CYP3A4.4 was incubated with AEA in the presence of added EH (Cayman Chemical) as described previously (Snider et al., 2007). As illustrated in Fig. 5, the M5 peak with m/z 364 did not decrease when AEA was incubated in the presence of EH; however, we did observe the expected decreases in peaks M6 to M9 as demonstrated previously (Snider et al., 2007). In addition, we observed no decrease in the intensity of the M1 to M4 peaks (results not shown), which indicates that these products are not epoxides.
Fig. 5.
Investigation of the novel M5 peak as an epoxide. Reactions were performed using the reconstituted system as described under Materials and Methods. Reaction mixtures contained CYP3A4.4 (50 pmol) with (– – –) and without (——) epoxide hydrolase (25 pmol) and were incubated for 2 h at 37°C. Peaks were normalized to the AEA peak (data not shown). The addition of epoxide hydroxylase did not cause a decrease in the signal for M5 but did result in the expected significant decreases in the peaks for M6, M7, and M8, which have previously been shown to be epoxides, with the concomitant formation of the four new diol products (M10–M13) with m/z values at 382 (Snider et al., 2007).
Novel M1 to M4 Products Formed from AEA by CYP3A4.4 Are Further Metabolized by Prostaglandin D Synthase.
Because M1 to M4 were unaffected by EH, we hypothesized that these novel peaks might be the result of formation of a peroxide bond on AEA. To test this hypothesis these metabolites were incubated with prostaglandin D synthase (PGDS), which catalyzes the cleavage of the peroxide bond present in prostaglandin H2 (Watanabe et al., 1980). For these studies, the CYP3A4.4 M1 to M4 metabolites of AEA were formed by incubation of the AEA with CYP3A4.4, isolated, and incubated with PGDS for 30 min and then examined by LC/MS. The M1 to M4 product peaks decreased after incubation with PGDS and a new peak (M14) with m/z 381 was observed (Fig. 6). These results suggest that the peroxide bond was broken to form an alcohol as well as a ketone bond, as has been observed for the metabolism of the peroxide bond in prostaglandin H2 by PGDS. These results indicate that these novel peaks (M1–M4) may possess peroxide bonds.
Fig. 6.
Incubation of the novel metabolites of CYP3A4.4 having peaks at m/z 380 with prostaglandin D synthase. Reactions were performed as described under Materials and Methods. Reaction mixtures were incubated with (– – –) or without (——) 5 μg of PGDS for 30 min at room temperature. Peaks were normalized to the AEA peak (data not shown). The addition of PGDS causes a decrease in peaks M1 to M4 and the formation of a new peak (M14) (m/z 381).
Mutation of Ile118 to Val Changes the Environment of the Heme.
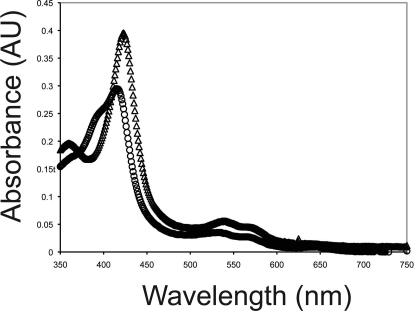
Analysis of the crystal structure of CYP3A4 suggested that the Ile188 to Val mutation might have an effect on the heme because the side chain of Ile118 is only 3.8 Å away from the pyrrole D ring of the heme (Fig. 3). Therefore, we analyzed the UV-visible spectra of both the wild-type and the I118V mutant (Fig. 7). The spectra of both were determined in the presence and absence of AEA. The spectrum of wild-type CYP3A4 exhibits a mix of both high- and low-spin heme iron with the absorption maximum being at 424 nm. Spectral titration of the wild-type enzyme with AEA gave a spectral Ks of 1.8 μM for AEA (data not shown). The mutant exhibited a predominantly low-spin spectrum with a maximum at 427 nm, and there was no change in the UV spectrum after the addition of AEA. Therefore, we were unable to determine a Ks for AEA binding to the mutant.
Fig. 7.
UV-visible spectra of CYP3A4.1 and CYP3A4.4 at 1 μM. Spectra were measured from 350 to 700 nm on a UV-visible spectrophotometer as described under Materials and Methods. CYP3A4.1 (○) exhibited a mix between high- and low-spin forms and CYP3A4.4 was (▵) more low-spin.
Mutation of Residue 118 from Ile to Val in CYP3A4 Causes a Decrease in the Rate of Reduction.
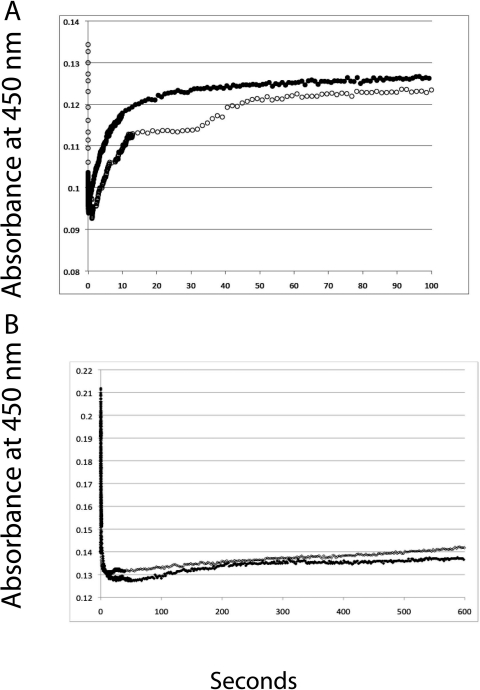
To investigate the basis for the lowered catalytic activity for the monooxygenation of testosterone and AEA by the mutant, we measured the rate of electron transfer from CPR to the ferric CYP3A4. These experiments were done in the presence and absence of 100 μM testosterone. The results are shown in Fig. 8. The wild-type CYP3A4A without testosterone (Fig. 8A, ○) demonstrates a biphasic kinetic trace. Fitting of the kinetic trace with two exponentials gives the apparent rate constants (kobs) of 0.15 for the fast phase and 0.0026 s−1 for the slow phase. In contrast, the mutant protein (Fig. 8B) shows monophasic kinetics with a kobs of 0.0002 s−1, which is approximately 750-fold slower than the fast phase of the wild type. This result demonstrates a very significant decrease in the rate of the first electron transfer from CPR to the mutant. In addition, the wild type demonstrates an increase in rate of transfer in the presence of testosterone (Fig. 8A, ●), whereas the mutant protein is unaffected by the presence of testosterone (Fig. 8B).
Fig. 8.
Rates for the reduction of CYP3A4 wild type and mutant as determined by stopped-flow spectrophotometry. The reconstituted CYP3A4.1 (A) and CYP3A4.4 (B) and CPR solution (3 mM each) in the presence (●) or absence (○) of 100 μM testosterone were rapidly mixed with 0.1 mM NADPH solution in the stopped-flow spectrometer as described under Materials and Methods. The reduction of the P450s was monitored at 450 nm.
Discussion
The results presented here demonstrate that the I118V mutation has a profound effect on the metabolism of AEA by CYP3A4. Not only does the mutation lower its activity for the metabolism of AEA, but it also causes changes in the ratios of the monooxygenated metabolites formed, leading to the formation of a new monooxygenated metabolite and four new dioxygenated metabolites. Besides changes in AEA metabolism, we also observed that the mutation results in a lowered rate of metabolism for testosterone, as well as lower rates of metabolism for other CYP3A4 substrates such as verapamil, tamoxifen, and ritonavir (data not shown).
UV spectral analysis of CYP3A4.1 and CYP3A4.4 indicates that this single nucleotide in CYP3A4.4 also leads to a significant change in the interactions between the apoprotein and the heme. The spectral results showing that the variant has an increased low-spin character suggest that the bond at the axial position between the heme and the apoprotein may have been strengthened significantly. The results from the stopped-flow studies also indicate that the rate of the flow of electrons from the reductase to the P450 for the reduction by the ferric iron by the first electron has been hindered significantly. This hindrance is most likely due to the modulation of the redox potential of the heme by the I118V mutation because of its proximity to the heme. This mutation could also result in greater access of water to the active site. These data support the possibility that changes in the environment of the heme due to the mutation may be responsible in part for the significant decrease in catalytic activity.
Understanding this and other P450 polymorphisms may prove to be very helpful in understanding disease states in numerous tissues and their responses to drugs as well as in the development of novel therapeutic agents. Previous studies have demonstrated that AEA is metabolized to give the five primary metabolites identified here in several different tissues, most importantly in the liver and the brain. In the liver and brain, AEA is metabolized to form multiple products. It has been further demonstrated that CYP3A4 is involved in the formation of these products in both tissues. Studies have shown that the addition of antibodies to CYP3A4 to brain microsomes results in a significant decrease in the formation of AEA metabolites (Bornheim et al., 1995; Snider et al., 2007). The novel monooxygenated product produced by the I118V mutant corresponds to the 19-HETE-EA product observed previously after AEA metabolism by murine BV-2 microglial cells (Snider et al., 2009).
The other four novel products produced by the I118V mutant may potentially be very important biological metabolites. Our results demonstrate that these products result from the addition of two oxygens. Because these products are not further metabolized by epoxide hydrolase, they are not products formed by the epoxidation of two different double bonds. This observation led us to hypothesize that these molecules might be due to the formation of a peroxide bond similar to that in prostaglandin H2, which is formed from arachidonic acid by the enzyme cyclooxygenase. Prostaglandin H2 is a precursor for many different molecules that are important for numerous functions such as inflammation, renal function, blood clotting, and the maintenance of the gastrointestinal tract (Simmons et al., 2004). Thus, we have hypothesized that these four products (M1–M4) are the result of CYP3A4 forming peroxide bonds at each of the four double bonds of AEA. The partial disappearance of all four of these metabolites after incubation with prostaglandin D synthase supports the suggestion that these metabolites are peroxides (Fig. 9).
Fig. 9.
Metabolic products of AEA formed by CYP3A4.1 and CYP3A4.4. As shown here, both CYP3A4.1 and CYP3A4.4 produce the four epoxide molecules 5,6-, 8,9-, 11,12-, and 14,15-EET-EA. In addition, CYP3A4.4 also catalyzes the formation of one monooxygenated product (believed to be 19-HETE-EA) as well as four dioxygenated products that are thought to be the peroxides shown here.
AEA as well as some of its metabolites have been shown to bind to multiple types of receptors. Progress has been made to elucidate the compounds that bind to these receptors and the physiological results of binding. The two most studied cannabinoid receptors, CB1 and CB2, have different binding affinities for AEA and its metabolites. CB1 is found in the brain and in other tissues (Palmer et al., 2000; Kozak and Marnett, 2002; Snider et al., 2009) and has been demonstrated to be involved in the cannabinoid signaling pathway that protects against stroke in mice (Parmentier-Batteur et al., 2002; Marsicano et al., 2003; Panikashvili et al., 2005). The second receptor, CB2, is involved in inflammation, and agonists targeting CB2 are currently being developed as therapies to reduce inflammation and pain (Whiteside et al., 2005). It has recently been shown that the metabolism of AEA by CYP3A4 can form at least one product, 5,6-EET-EA, which has a significantly increased binding affinity for CB2. This binding has also been shown to be very selective for CB2 over CB1 (Snider et al., 2009). Two other receptors that seem to play roles in AEA signaling are the vanilloid receptors and a non-CB1/non-CB-2 cannabinoid receptor. These receptors are believed to play roles in mediating AEA-dependent vasorelaxant effects (De Petrocellis et al., 2001; Herradón et al., 2007).
CYP3A4 produces a variety of different mono- and dioxygenated metabolites from AEA. These results demonstrate the formation of nine different oxygenated metabolites from AEA. Studies aimed to elucidate the structures of the five new metabolites of AEA reported here, their presence in various tissues, and their physiological roles are important for a comprehensive understanding of the endocannabinoid signaling system. Future studies will be needed to investigate the possible biological activity of the new CYP3A4 metabolites and the consequences of the CYP3A4.4 mutation on the regulation of the cannabinoid signaling system.
Supplementary Material
Acknowledgments.
We thank Drs. Mike Tarasev and David Ballou for help with stopped-flow experiments.
This work was supported in part by the National Institutes of Health National Institute on Drug Abuse [Grant DA007268-18] (Biology of Drug Abuse Postdoctoral Training Fellowship; to M.P.-H.); and the National Institutes of Health National Cancer Institute [Grant CA016954] (to P.F.H.). N.T.S. was supported by the Michigan Institute for Clinical and Health Research Postdoctoral Translational Scholars Program [Award UL1-RR024986].
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/dmd.110.033712.
The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
- CB
- cannabinoid receptor
- AEA
- anandamide
- P450
- cytochrome P450
- EET
- epoxyeicosatrienoic acid
- EA
- ethanolamide
- HETE
- hydroxyeicosatetraenoic acid
- OH
- hydroxy
- ESI
- electrospray ionization
- LC/MS
- liquid chromatography/mass spectrometry
- EH
- epoxide hydrolase
- CPR
- cytochrome P450 reductase.
References
- Bornheim LM, Kim KY, Chen B, Correia MA. (1995) Microsomal cytochrome P450-mediated liver and brain anandamide metabolism. Biochem Pharmacol 50:677–686 [DOI] [PubMed] [Google Scholar]
- De Petrocellis L, Bisogno T, Maccarrone M, Davis JB, Finazzi-Agro A, Di Marzo V. (2001) The activity of anandamide at vanilloid VR1 receptors requires facilitated transport across the cell membrane and is limited by intracellular metabolism. J Biol Chem 276:12856–12863 [DOI] [PubMed] [Google Scholar]
- Di Marzo V, Bifulco M, De Petrocellis L. (2004) The endocannabinoid system and its therapeutic exploitation. Nat Rev Drug Discov 3:771–784 [DOI] [PubMed] [Google Scholar]
- Domanski TL, Liu J, Harlow GR, Halpert JR. (1998) Analysis of four residues within substrate recognition site 4 of human cytochrome P450 3A4: role in steroid hydroxylase activity and α-naphthoflavone stimulation. Arch Biochem Biophys 350:223–232 [DOI] [PubMed] [Google Scholar]
- Ducobu J. (2005) The endocannabinoid system and the regulation of the metabolism. J Pharm Belg 60:84–88 [PubMed] [Google Scholar]
- Ducobu J, Sternon J. (2005) Rimonabant (Acomplia), specific inhibitor of the endocannabinoid system. Rev Med Brux 26:165–168 [PubMed] [Google Scholar]
- Estabrook RW, Werringloer J. (1978) The measurement of difference spectra: application to the cytochromes of microsomes. Methods Enzymol 52:212–220 [DOI] [PubMed] [Google Scholar]
- Felder CC, Dickason-Chesterfield AK, Moore SA. (2006) Cannabinoids biology: the search for new therapeutic targets. Mol Interv 6:149–161 [DOI] [PubMed] [Google Scholar]
- Hanna IH, Teiber JF, Kokones KL, Hollenberg PF. (1998) Role of the alanine at position 363 of cytochrome P450 2B2 in influencing the NADPH- and hydroperoxide-supported activities. Arch Biochem Biophys 350:324–332 [DOI] [PubMed] [Google Scholar]
- Herradón E, Martín MI, López-Miranda V. (2007) Characterization of the vasorelaxant mechanisms of the endocannabinoid anandamide in rat aorta. Br J Pharmacol 152:699–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak KR, Marnett LJ. (2002) Oxidative metabolism of endocannabinoids. Prostaglandins Leukot Essent Fatty Acids 66:211–220 [DOI] [PubMed] [Google Scholar]
- Marsicano G, Goodenough S, Monory K, Hermann H, Eder M, Cannich A, Azad SC, Cascio MG, Gutiérrez SO, van der Stelt M, et al. (2003) CB1 cannabinoid receptors and on-demand defense against excitotoxicity. Science 302:84–88 [DOI] [PubMed] [Google Scholar]
- Morris GM, Goodsell DS, Huey R, Olson AJ. (1996) Distributed automated docking of flexible ligands to proteins: parallel applications of AutoDock 2.4. J Comput Aided Mol Des 10:293–304 [DOI] [PubMed] [Google Scholar]
- Nithipatikom K, Grall AJ, Holmes BB, Harder DR, Falck JR, Campbell WB. (2001) Liquid chromatographic-electrospray ionization-mass spectrometric analysis of cytochrome P450 metabolites of arachidonic acid. Anal Biochem 298:327–336 [DOI] [PubMed] [Google Scholar]
- Palmer SL, Khanolkar AD, Makriyannis A. (2000) Natural and synthetic endocannabinoids and their structure-activity relationships. Curr Pharm Des 6:1381–1397 [DOI] [PubMed] [Google Scholar]
- Panikashvili D, Mechoulam R, Beni SM, Alexandrovich A, Shohami E. (2005) CB1 cannabinoid receptors are involved in neuroprotection via NF-κB inhibition. J Cereb Blood Flow Metab 25:477–484 [DOI] [PubMed] [Google Scholar]
- Parmentier-Batteur S, Jin K, Mao XO, Xie L, Greenberg DA. (2002) Increased severity of stroke in CB1 cannabinoid receptor knock-out mice. J Neurosci 22:9771–9775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson JD, Kilo S, Hargreaves KM. (1998) Cannabinoids reduce hyperalgesia and inflammation via interaction with peripheral CB1 receptors. Pain 75:111–119 [DOI] [PubMed] [Google Scholar]
- Rodríguez de Fonseca F, Del Arco I, Bermudez-Silva FJ, Bilbao A, Cippitelli A, Navarro M. (2005) The endocannabinoid system: physiology and pharmacology. Alcohol Alcohol 40:2–14 [DOI] [PubMed] [Google Scholar]
- Shebley M, Hollenberg PF. (2007) Mutation of a single residue (K262R) in P450 2B6 leads to loss of mechanism-based inactivation by phencyclidine. Drug Metab Dispos 35:1365–1371 [DOI] [PubMed] [Google Scholar]
- Simmons DL, Botting RM, Hla T. (2004) Cyclooxygenase isozymes: the biology of prostaglandin synthesis and inhibition. Pharmacol Rev 56:387–437 [DOI] [PubMed] [Google Scholar]
- Snider NT, Kornilov AM, Kent UM, Hollenberg PF. (2007) Anandamide metabolism by human liver and kidney microsomal cytochrome P450 enzymes to form hydroxyeicosatetraenoic and epoxyeicosatrienoic acid ethanolamides. J Pharmacol Exp Ther 321:590–597 [DOI] [PubMed] [Google Scholar]
- Snider NT, Nast JA, Tesmer LA, Hollenberg PF. (2009) A cytochrome P450-derived epoxygenated metabolite of anandamide is a potent cannabinoid receptor 2-selective agonist. Mol Pharmacol 75:965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider NT, Sikora MJ, Sridar C, Feuerstein TJ, Rae JM, Hollenberg PF. (2008) The endocannabinoid anandamide is a substrate for the human polymorphic cytochrome P450 2D6. J Pharmacol Exp Ther 327:538–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers SP, Kennett GA. (2005) Cannabinoids and the regulation of ingestive behaviour. Curr Drug Targets 6:215–223 [DOI] [PubMed] [Google Scholar]
- Wang A, Yu BN, Luo CH, Tan ZR, Zhou G, Wang LS, Zhang W, Li Z, Liu J, Zhou HH. (2005) Ile118Val genetic polymorphism of CYP3A4 and its effects on lipid-lowering efficacy of simvastatin in Chinese hyperlipidemic patients. Eur J Clin Pharmacol 60:843–848 [DOI] [PubMed] [Google Scholar]
- Watanabe K, Shimizu T, Iguchi S, Wakatsuka H, Hayashi M, Hayaishi O. (1980) An NADP-linked prostaglandin D dehydrogenase in swine brain. J Biol Chem 255:1779–1782 [PubMed] [Google Scholar]
- Whiteside GT, Gottshall SL, Boulet JM, Chaffer SM, Harrison JE, Pearson MS, Turchin PI, Mark L, Garrison AE, Valenzano KJ. (2005) A role for cannabinoid receptors, but not endogenous opioids, in the antinociceptive activity of the CB2-selective agonist, GW405833. Eur J Pharmacol 528:65–72 [DOI] [PubMed] [Google Scholar]
- Yamada Y, Metoki N, Yoshida H, Satoh K, Kato K, Hibino T, Yokoi K, Watanabe S, Ichihara S, Aoyagi Y, et al. (2008) Genetic factors for ischemic and hemorrhagic stroke in Japanese individuals. Stroke 39:2211–2218 [DOI] [PubMed] [Google Scholar]
- Zhang H, Hamdane D, Im SC, Waskell L. (2008) Cytochrome b5 inhibits electron transfer from NADPH-cytochrome P450 reductase to ferric cytochrome P450 2B4. J Biol Chem 283:5217–5225 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.