The structure and mechanism of diisopropyl fluorophosphatase (DFPase) have been studied using a variety of methods, including isotopic labelling, X-ray crystallography and neutron crystallography. The neutron structure of DFPase, mechanistic studies and subsequent rational design efforts are described.
Keywords: neutron crystallography, DFPase, enzymes, rational design, phosphotriesterases, mechanism
Abstract
Diisopropyl fluorophosphatase (DFPase) is a calcium-dependent phosphotriesterase that acts on a variety of highly toxic organophosphorus compounds that act as inhibitors of acetylcholinesterase. The mechanism of DFPase has been probed using a variety of methods, including isotopic labelling, which demonstrated the presence of a phosphoenzyme intermediate in the reaction mechanism. In order to further elucidate the mechanism of DFPase and to ascertain the protonation states of the residues and solvent molecules in the active site, the neutron structure of DFPase was solved at 2.2 Å resolution. The proposed nucleophile Asp229 is deprotonated, while the active-site solvent molecule W33 was identified as water and not hydroxide. These data support a mechanism involving direct nucleophilic attack by Asp229 on the substrate and rule out a mechanism involving metal-assisted water activation. These data also allowed for the re-engineering of DFPase through rational design to bind and productively orient the more toxic S P stereoisomers of the nerve agents sarin and cyclosarin, creating a modified enzyme with enhanced overall activity and significantly increased detoxification properties.
1. Introduction
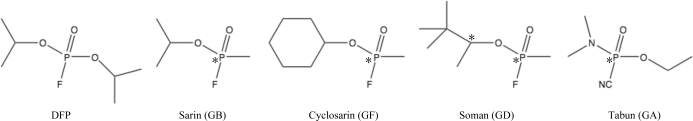
Diisopropyl fluorophosphatase (DFPase; 314 amino acids; 35 kDa; EC 3.1.8.2) from the squid Loligo vulgaris is a potent Ca2+-dependent phosphotriesterase that is capable of hydrolyzing a range of organophosphorus compounds that act as irreversible acetylcholinesterase (AChE) inhibitors. Its substrates include diisopropyl fluorophosphate (DFP) and a range of highly toxic G-type organophosphorus (OP) nerve agents such as tabun (GA), sarin (GB), soman (GD) and cyclosarin (GF) (Blum et al., 2006 ▶; Scharff et al., 2001 ▶; Fig. 1 ▶). The hydrolytic reaction catalyzed by DFPase leads to the formation of a phosphate or phosphonate and a fluoride ion, resulting in detoxification of the organophosphorus agent. DFPase is a promising candidate for enzymatic decontamination, as the enzyme can be expressed in bulk amounts, shows a high tolerance for organic solvents and is thermally stable.
Figure 1.
Structures of the DFPase substrates DFP, GB, GF, GD and GA. Stereocentres are indicated by asterisks.
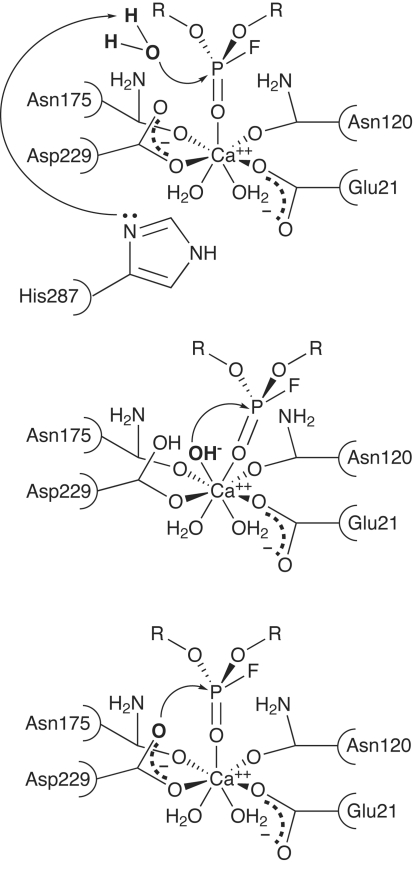
A number of mechanisms have been proposed for DFPase. The first published structure of DFPase showed the presence of His287 in the vicinity of the active site, which was suggested to act as a general base in the reaction (Fig. 2 ▶ a). However, mutational studies on His287 showed little change in overall activity even on replacing His287 with aliphatic residues such as leucine. A series of functional and structural studies to elucidate the mechanism of DFPase are described here, including mutants of active-site residues, isotopic labelling and the 2.2 Å neutron structure of DFPase. A reaction mechanism involving direct nucleophilic attack by Asp229 and a phosphoenzyme intermediate is proposed. On the basis of these studies, a series of mutants engineered to augment the activity and detoxification properties of DFPase are described.
Figure 2.
Potential reaction mechanisms for DFPase. (a) Mechanism with His287 acting as a general base to activate water for nucleophilic attack on the phosphorus. (b) Mechanism involving metal-activated water. (c) Mechanism involving direct nucleophilic attack by Asp229 and a subsequent phosphoenzyme intermediate.
2. Materials and methods
DFPase was expressed in Escherichia coli and purified as described previously (Hartleib et al., 2001 ▶). The protein was crystallized at room temperature (RT) by vapour diffusion of a 2 mM protein solution in 10 mM Tris pH 7.5, 2 mM CaCl2 against 11% PEG 4000, 0.1 M MES pH 6.5. For this study, a single crystal measuring 2.4 × 0.5 × 0.36 mm was mounted in a glass capillary and the labile H atoms were exchanged for deuterium by vapour diffusion against deuterated mother liquor for one week prior to screening. Crystals of DFPase grew in space group P212121, with unit-cell parameters a = 43.4, b = 83.3, c = 87.5 Å, and diffracted neutrons to 2.2 Å resolution. A total of 37 images were collected over a period of about one month at the PCS at Los Alamos National Laboratory, with approximately 24 h exposure per image (Blum et al., 2007 ▶). The data were indexed and integrated using a version of d*TREK modified for neutron diffraction data and scaled using SCALA. X-ray and neutron diffraction data were used together for joint refinement in nCNS (Adams et al., 2009 ▶). X-ray structure factors for the joint refinement were obtained from the RT 1.86 Å data set of DFPase (PDB entry 2gvw; Blum et al., 2006 ▶). The structure was refined using positional, individual temperature-factor, simulated-annealing and occupancy refinement protocols in nCNS. The data-collection and refinement statistics are summarized in Table 1 ▶. The final refined structure has been deposited as PDB entry 3byc (Blum et al., 2009 ▶).
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the last shell.
| Neutron (3byc) | X-ray (2gvw) | |
|---|---|---|
| Source | PCS, Los Alamos | |
| Settings | 37 | |
| Space group | P212121 | |
| Unit-cell parameters (Å) | a = 43.4, b = 83.3, c = 87.5 | |
| Resolution (Å) | 10.0–2.2 (2.32–2.20) | |
| Reflections (measured/unique) | 64711/10999 | |
| Redundancy | 5.9 | |
| Completeness (%) | 81.8 (72.9) | |
| Rmerge | 0.199 (0.397) | |
| Wavelength range (Å) | 0.8–4.5 | |
| 〈I/σ(I)〉 | 1.9 (1.8) | |
| Mean(I)/s.d. | 4.0 (2.2) | |
| Refinement (joint refinement) | ||
| Resolution (Å) | 20–2.2 | 20–1.84 |
| No. of reflections | 13829 | 25773 |
| Rcryst/Rfree | 0.264/0.316 | 0.231/0.251 |
| No. of atoms | ||
| Total | 5567 | |
| Protein | 4827 | |
| Ions | 2 | |
| Water | 738 (246 D2O) | |
| B factors (Å2) | ||
| Overall | 25.9 | |
| Protein | 24.2 | |
| Ions | 9.9 | |
| Water | 37.3 | |
| R.m.s.d. | ||
| Bond lengths (Å) | 0.0047 | |
| Bond angles (°) | 1.025 | |
| Ramachandran plot | ||
| Most favoured (%) | 81.8 | |
| Allowed (%) | 17.4 | |
| Generous (%) | 0.8 | |
| Disallowed (%) | 0.0 |
3. Results and discussion
3.1. Structure and active-site environment
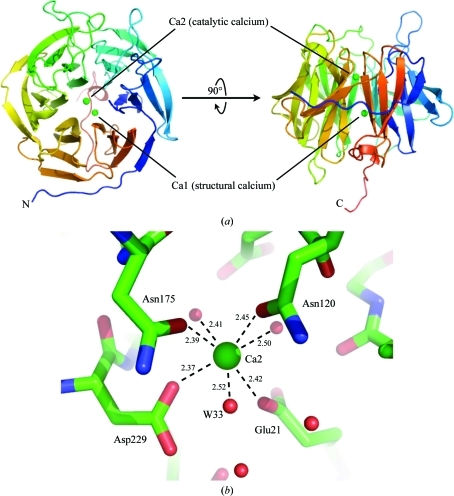
Numerous DFPase structures have been reported, including X-ray structures of the holoenzyme reported at 1.86 Å resolution at RT (PDB code 2gvw) and 2.1 Å resolution at 100 K (PDB code 1e1a), an RT perdeuterated X-ray structure at 2.1 Å resolution (PDB code 3kgg) and an atomic resolution structure at 0.85 Å resolution (PDB code 1pjx) (Blum et al., 2006 ▶, 2010 ▶; Scharff et al., 2001 ▶; Koepke et al., 2003 ▶). In addition, several structures of active-site mutants and a structure in complex with the designed substrate analogue DcPPA have been solved (Blum et al., 2006 ▶; Blum & Chen, 2010 ▶). The protein adopts a six-bladed β-propeller fold surrounding a central water tunnel and coordinates two calcium ions (Fig. 3 ▶ a). The high-affinity calcium Ca1 located in the central water tunnel is required for structural integrity and shows a rare Ca–N coordination by the side chain of His274 (Scharff et al., 2001 ▶; Koepke et al., 2003 ▶). The lower affinity calcium ion Ca2 is located at the base of the active-site pocket and is necessary for catalytic activity. Ca2 is coordinated by Glu21, Asn120, Asn175 and Asp229. The calcium ion is further coordinated below the active site by two water molecules that link to the central water tunnel of the enzyme and by one solvent molecule (W33) in the active-site pocket (Fig. 3 ▶ b).
Figure 3.
Fold of DFPase and catalytic calcium environment. (a) Structure of DFPase. The two calcium ions, Ca1 and Ca2, are shown as green spheres. (b) Coordination environment around the catalytic calcium. Distances are shown in Å.
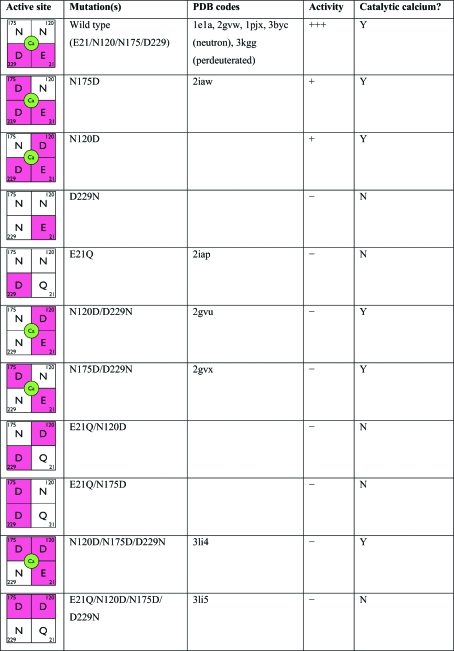
The catalytic calcium-coordinating residues have been extensively mutated (Katsemi et al., 2005 ▶; Blum & Chen, 2010 ▶). In general, alterations to these calcium-coordinating residues have a major effect on binding and activity (Fig. 4 ▶). Of the ten mutants created, only N175D and N120D showed residual (<5%) enzymatic activity against DFP, while the remaining eight mutants were inactive. The inactivity of these mutants can be attributed to the loss of the catalytic calcium when there is only one negative charge remaining in the active site (D229N, E21Q). The crystal structure of E21Q (PDB code 2iap; Scharff et al., 2001 ▶) shows that despite the loss of the catalytic calcium ion, the overall fold of the protein is unaffected. Glu21 appears to play a crucial role in calcium coordination, as all mutations of Glu21, regardless of the number of negative charges in the vicinity, result in the loss of calcium binding.
Figure 4.
Active-site mutations of DFPase. The calcium-binding environment in the active site is indicated in the first column, with negatively charged residues marked in red and calcium shown in green.
However, five of the mutants still contained bound calcium, so their lack of activity cannot be explained by loss of the catalytic calcium. The electrostatic environment around the catalytic calcium is finely tuned and is crucial for enzymatic activity. Mutations that rearrange the distribution of charges in the active site while preserving the charge balance, such as the double mutants N120D/D229N (PDB code 2gvu; Blum et al., 2006 ▶) and N175D/D229N (PDB code 2gvx; Blum et al., 2006 ▶), bind calcium and have an active site that is virtually identical to that of the wild type, but show little or no substrate binding and are inactive. Although they contain two negative charges, the inactive mutants E21Q/N120D and E21Q/N175D appear to lack the catalytic calcium ion as a consequence of the substitution at Glu21. Calcium binding is dependent on the presence of at least two negatively charged residues and the presence of residue Glu21. However, the presence of bound calcium is necessary but not sufficient for enzymatic activity, which requires a Glu21/Asp229 pair on one face of the active site.
3.2. Mechanistic studies
To distinguish whether the reaction proceeds through a metal-activated water (Fig. 2 ▶ b) or through direct nucleophilic attack by the protein (Fig. 2 ▶ c), DFP was reacted with DFPase in 18O-labelled water under single- and multiple-turnover conditions. If the reaction proceeds via metal-activated water, the product would be expected to incorporate 18O from the labelled solvent under single- and multiple-turnover conditions. Conversely, if the reaction proceeds via a phosphoenzyme intermediate, in other words a transient covalent adduct to the protein, the product would incorporate unlabelled 16O from the protein under single-turnover conditions. Under multiple-turnover conditions, the majority of the product would be expected to contain 18O. These two mechanisms can be distinguished by mass spectrometry of the products.
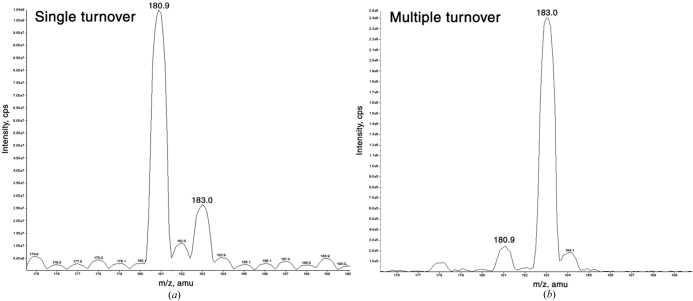
The primary product generated under single-turnover conditions had a molecular mass of 180.9 Da, corresponding to the incorporation of 16O from the protein into the diisopropyl phosphate product and not 18O from the solvent. As expected, the product generated after multiple turnovers had a mass of 183.0 Da, which is consistent with the eventual incorporation of 18O into the product (Fig. 5 ▶). The enzyme would also be expected to incorporate 18O after multiple turnovers. In order to demonstrate this, a complementary experiment was performed in which the enzyme was allowed to react with an excess of substrate in 18O-labelled water. The enzyme was digested with endoproteinase Glu-C and the resulting fragments were profiled by mass spectroscopy. One peptide was identified with an altered mass of 1018.1 Da compared with the control sample peptide of 1013.6 Da and had sequence GGADGMDFDE, corresponding to the fragment containing Asp229. In this fragment, Asp229 is the only residue exposed to the active site and thus is the most likely candidate for the nucleophile. The altered peptide mass is consistent with incorporation of 18O into the two carboxyl O atoms of the Asp229 side chain. Although these experiments demonstrated that a phosphoenzyme intermediate was formed, a number of investigators have suggested that metal-assisted water activation was a potential mechanism, as the catalytic calcium coordinated solvent in the active site.
Figure 5.
Mass-spectrometric analysis of single- and multiple-turnover products of the reaction of DFPase with DFP in H2 18O. (a) Single-turnover conditions showing a predominant product of 180.9 Da. (b) Multiple-turnover conditions showing a product of 183.0 Da. Reprinted with permission from Blum et al. (2006 ▶). Copyright 2006 American Chemical Society.
3.3. Neutron structure
To further elucidate the mechanism, the structure of DFPase was studied by neutron crystallography. DFPase was an excellent candidate for neutron crystallography, as crystals typically grew to sizes exceeding 1 mm in their longest dimension. Although DFPase crystals diffracted X-rays to ultrahigh resolution (0.85 Å) at a synchrotron, it was not possible to discern the protonation states of critical residues in the active site. Neutron crystallography is one of the only techniques that can experimentally determine the positions of H atoms in a three-dimensional structure even at moderate resolution and provides complementary information to the locations of heavier atoms obtained from X-ray crystallographic analyses (Niimura & Bau, 2008 ▶). The scattering power of deuterium is comparable to those of heavier atoms such as N, C, O and S in proteins, so typically the labile H atoms are exchanged for deuteriums prior to the start of data collection. The unique information available from neutron crystallography has provided important insights into the mechanisms of enzymes (Adachi et al., 2009 ▶; Bennett et al., 2006 ▶; Blakeley et al., 2008 ▶; Blum et al., 2009 ▶; Coates et al., 2001 ▶, 2008 ▶; Katz et al., 2006 ▶; Kossiakoff & Spencer, 1981 ▶; Kovalevsky et al., 2008 ▶; Tamada et al., 2009 ▶; Yagi et al., 2009 ▶).
Neutron diffraction data for DFPase crystals were collected to 2.2 Å resolution at the PCS in Los Alamos and the structure was refined using joint X-ray and neutron refinement to yield the final structure (Table 1 ▶). The nuclear density maps in the active site revealed a number of features that were pertinent to the reaction mechanism (Blum et al., 2009 ▶). The 2F o − F c maps show a deprotonated Asp229, which is consistent with its role as a potential nucleophile. The active-site solvent molecule W33 coordinating the catalytic calcium has the characteristic elongated nuclear density of water (Fig. 6 ▶ a). This was confirmed by means of a nuclear OMIT density map. By omitting the D atoms of W33 from the structure-factor calculation and performing a simulated-annealing run on the remaining atoms, two difference peaks appeared at equal intensity with the geometry of a water molecule. Therefore, the solvent molecule coordinated to the catalytic calcium is water and not hydroxide, so a mechanism involving metal-assisted water activation is unlikely. The orientation of the water is unusual, with a Ca—O—H angle of 53°, and the H makes a close (2.08 Å) contact to the Ca (Fig. 6 ▶ b). This is far smaller than the angle of 75° seen in small-molecule calcium hydrates, which have also been studied by neutron crystallography (Einspahr & Bugg, 1980 ▶). The orientation of W33, although strained, appears to be a consequence of the environment of the active site, including a hydrogen bond to the nearby water W140, minimizing even more unfavourable interactions.
Figure 6.
(a) 2F o − F c nuclear density map contoured at 1.5σ showing elongated density for solvent molecule W33, consistent with water, and deprotonated Asp229. (b) Detail of the Ca2–W33 interaction. The Ca—O—H(D) angle is 53°; distances are indicated in Å.
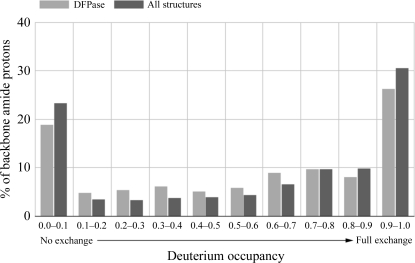
The backbone hydrogen/deuterium exchange characteristics of DFPase show that overall the internal regions of the protein were more resistant to exchange than the areas closer to the surface. One characteristic of the backbone exchange pattern is that β-strand main-chain amide regions involved in hydrogen bonding to adjacent β-strands do not exchange, while in many cases the adjacent main-chain amide facing the solvent is fully exchanged. A more quantitative analysis of backbone amide H atoms can be obtained by analyzing the backbone amide deuterium occupancies in DFPase and in neutron structures in the PDB where the relative hydrogen/deuterium occupancies are reported (Fig. 7 ▶). In general, around half of the backbone H atoms are fully exchanged, while around one-third are not exchanged during the time course of the experiment. Solvent molecules are found to be D2O, including those in the central water tunnel, and each of the solvent molecules could be unequivocally oriented. The central water tunnel is a highly restricted environment; for exchange to occur, the waters must pass through the coordination sphere of the structural calcium Ca1.
Figure 7.
Backbone amide H/D-exchange characteristics. A histogram of deuterium occupancies versus percentage of residues is shown for DFPase and for structures in the PDB for which the occupancies of backbone amide H atoms were refined.
3.4. Re-engineering DFPase
The DFPase neutron structure and the isotopic labelling studies support a reaction mechanism involving nucleophilic attack of Asp229 on the P atom of the substrate, resulting in the formation of a phosphoenzyme intermediate that is subsequently hydrolyzed (Fig. 2 ▶ c). This mechanistic information serves as a starting point for rational design efforts to increase the overall activity and detoxification properties of the enzyme. Furthermore, the available structures of DFPase show a rigid active-site environment and a cocrystal structure with the substrate analogue DcPPA shows little conformational change in the active site upon binding, making DFPase an example of a lock-and-key enzyme (Blum et al., 2006 ▶).
The G-type nerve agents each contain a chiral centre (or two chiral centres in soman; Fig. 1 ▶), with one stereoisomer (S P) being more toxic than the other (R P). Wild-type DFPase preferentially binds and hydrolyzes the less toxic stereoisomers of the G-type nerve agents. For these re-engineering efforts, a model of the phosphoenzyme intermediate was generated for the R P and S P stereoisomers of cyclosarin. The active site was modified to accommodate the more toxic S P stereoisomer. A more open binding pocket on one face of the active site was created by introducing shorter side chains at residues 37, 144 and 146 and the opposite face was restricted by introducing a larger side chain at residue 195. The two resulting mutants were E37A/Y144A/R146A/T195M and E37D/Y144A/R146A/T195M (Melzer et al., 2009 ▶). The activities of these two mutants were tested and their X-ray structures were solved. Both showed an overall enhanced activity against racemic mixtures of cyclosarin (GF) and sarin (GB). Notably, the mutations have incorporated stereoselectivity into DFPase, with the more toxic S P stereoisomer being the preferred substrate. In particular, the mutant E37A/Y144A/R146A/T195M showed a nearly 200-fold enhancement of stereoselectivity for the S P stereoisomer of cyclosarin (GF) relative to wild-type DFPase. The mutants have greater specific activities against DFP, GB and GF and as the S P stereoisomer is quickly hydrolyzed the detoxification properties are significantly enhanced (Table 2 ▶). These modified versions of DFPase serve as a starting point for efforts to further improve and expand its repertoire of substrates.
Table 2. Stereoselectivity of engineered DFPase mutants. 1 U = 1 µmol min−1 .
| Wild type | E37A/Y144A/R146A/T195M | E37D/Y144A/R146A/T195M | |
|---|---|---|---|
| DFP | |||
| Specific activity (U) | 247 ± 10 | 390 ± 21 | 429 ± 21 |
| kcat/Km (M−1 s−1) | 5.6 × 104 | 1.4 × 105 | 9.6 × 104 |
| Sarin (GB) | |||
| Specific activity (U, racemic mixture) | 110 ± 6 | 862 ± 32 | 418 ± 3 |
| (S) kcat/Km (M−1 s−1) | 4.2 × 104 | 2.3 × 105 | 7.1 × 104 |
| (R) kcat/Km (M−1 s−1) | 4.7 × 104 | 4.5 × 104 | 2.4 × 104 |
| Ratio S/R | 0.89 | 5.11 | 2.96 |
| Relative to wild type | 5.74 | 3.33 | |
| Cyclosarin (GF) | |||
| Specific activity (U, racemic mixture) | 198 ± 15 | 258 ± 13 | 143 ± 8 |
| (S) kcat/Km (M−1 s−1) | 1.7 × 104 | 4.9 × 105 | 2.4 × 104 |
| (R) kcat/Km (M−1 s−1) | 7.2 × 105 | 1.3 × 105 | 2.7 × 104 |
| Ratio S/R | 0.02 | 3.77 | 0.89 |
| Relative to wild type | 189 | 44.5 | |
4. Conclusions
The neutron structure of DFPase reveals that the putative nucleophile Asp229 is deprotonated and that the active-site solvent molecule W33 is water. This is consistent with a mechanism that proceeds through a phosphoenzyme intermediate, as opposed to a metal-activated water, although there is calcium bound in the active site. Furthermore, the unusual orientation of W33 relative to the calcium was revealed by the use of neutrons and could not be seen in the ultrahigh-resolution X-ray structure of DFPase. The proposed mechanism, as derived from the neutron structure and isotopic labelling studies, has successfully led to a designed DFPase with improved activity and stereoselectivity against the more toxic isomers of sarin and cyclosarin (Melzer et al., 2009 ▶).
4.1. Outlook for neutron crystallography
Neutron crystallography is a rapidly growing technique in structural biology, although it remains technically demanding. Until recently, elucidating neutron structures required large amounts of sample in order to grow large crystals (typically >1 mm3) and long data-collection periods. Recent developments in instrumentation and data-collection techniques at both reactor and spallation neutron sources (Blakeley et al., 2008 ▶; Langan et al., 2008 ▶) have significantly reduced sample-size requirements and data-acquisition times. Concurrently, recent improvements in structure-refinement software and strategies, such as the use of X-ray and neutron crystallographic data in maximum-likelihood refinement in nCNS and PHENIX, have helped to address the limitations of low data-to-parameter ratios and low data resolutions that are often associated with neutron crystallographic data (Adams et al., 2009 ▶). The addition of H atoms and their occupancies to the model significantly increases the number of parameters being refined, as H atoms typically make up nearly half of all atoms in a protein.
The neutron structure of DFPase represents a major technical advance in neutron crystallography; the data were collected in a relatively short period of time from one of the smallest crystals used for data collection (0.43 mm3 in volume; Blum et al., 2007 ▶). Since then, data sets have been collected from perdeuterated crystals as small as ∼0.1 mm3 in volume (Blakeley et al., 2008 ▶). Complete data sets have been collected in 72 h or less and protocols for the perdeuteration of heterologously expressed proteins as a means of increasing the signal-to-noise ratio have been developed (Gamble et al., 1994 ▶; Shu et al., 2000 ▶; Liu et al., 2007 ▶; Blum et al., 2010 ▶). A new generation of neutron sources and detectors have recently been commissioned, including the BIX-4 diffractometer in Japan and the TOPAZ detector at the SNS in Oak Ridge. The LADI-III detector at the ILL in Grenoble has been refurbished and the MaNDi diffractometer is being constructed at the SNS.
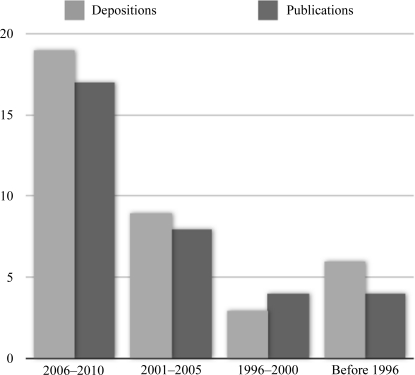
While data-collection times have been significantly reduced, the increased interest in neutron crystallography has meant that the few sources in the world are now oversubscribed. This is illustrated by a graph of the number of macromolecular neutron structure depositions in the PDB and their associated publications, which numbered 37 entries and 33 publications as of April 2010 (Fig. 8 ▶). In addition, there are a number of early structures that have not been deposited in the PDB. Although at present the number of PDB entries for neutron structures is quite small compared with the 56 000 X-ray structures currently in the database, and still far smaller than the 600 membrane-protein structures, the technique is growing quickly. These developments will allow neutron diffraction to address progressively more complex issues and mechanistic problems, such as membrane proteins and larger macromolecular complexes.
Figure 8.
Neutron structures and associated publications in the PDB by year. Source: PDB, April 2010.
Acknowledgments
This project was funded by the Hessian Ministry of Science and Culture and the German Ministry of Defence under Grants E/E590/6Z004/4F170 and E/UR3G/6G115/6A801. The PCS is funded by the Office of Science and the Office of Biological and Environmental Research of the US Department of Energy. MM and PL were partly supported by an NIH–NIGMS-funded consortium (1R01GM071939-01) between Los Alamos National Laboratory and Lawrence Berkeley National Laboratory to develop computational tools for neutron protein crystallography.
References
- Adachi, M. et al. (2009). Proc. Natl Acad. Sci. USA, 106, 4641–4646.
- Adams, P. D., Mustyakimov, M., Afonine, P. V. & Langan, P. (2009). Acta Cryst. D65, 567–573. [DOI] [PMC free article] [PubMed]
- Bennett, B., Langan, P., Coates, L., Mustyakimov, M., Schoenborn, B., Howell, E. E. & Dealwis, C. (2006). Proc. Natl Acad. Sci. USA, 103, 18493–18498. [DOI] [PMC free article] [PubMed]
- Blakeley, M. P., Ruiz, F., Cachau, R., Hazemann, I., Meilleur, F., Mitschler, A., Ginell, S., Afonine, P., Ventura, O. N., Cousido-Siah, A., Haertlein, M., Joachimiak, A., Myles, D. & Podjarny, A. (2008). Proc. Natl Acad. Sci. USA, 105, 1844–1848. [DOI] [PMC free article] [PubMed]
- Blum, M.-M. & Chen, J. C.-H. (2010). Chem. Biol. Interact.187, 373–379. [DOI] [PubMed]
- Blum, M.-M., Koglin, A., Rüterjans, H., Schoenborn, B., Langan, P. & Chen, J. C.-H. (2007). Acta Cryst. F63, 42–45. [DOI] [PMC free article] [PubMed]
- Blum, M.-M., Löhr, F., Richardt, A., Rüterjans, H. & Chen, J. C.-H. (2006). J. Am. Chem. Soc.128, 12750–12757. [DOI] [PubMed]
- Blum, M.-M., Mustyakimov, M., Rüterjans, H., Kehe, K., Schoenborn, B. P., Langan, P. & Chen, J. C.-H. (2009). Proc. Natl Acad. Sci. USA, 106, 713–718. [DOI] [PMC free article] [PubMed]
- Blum, M.-M., Tomanicek, S. J., John, H., Hanson, B. L., Rüterjans, H., Schoenborn, B. P., Langan, P. & Chen, J. C.-H. (2010). Acta Cryst. F66, 379–385. [DOI] [PMC free article] [PubMed]
- Coates, L., Erskine, P. T., Wood, S. P., Myles, D. A. A. & Cooper, J. B. (2001). Biochemistry, 40, 13149–13157. [DOI] [PubMed]
- Coates, L., Tuan, H., Tomanicek, S., Kovalevsky, A., Mustyakimov, M., Erskine, P. & Cooper, J. (2008). J. Am. Chem. Soc.130, 7235–7237. [DOI] [PMC free article] [PubMed]
- Einspahr, H. & Bugg, C. E. (1980). Acta Cryst. B36, 264–271.
- Gamble, T. R., Clauser, K. R. & Kossiakoff, A. A. (1994). Biophys. Chem.53, 15–25. [DOI] [PubMed]
- Hartleib, J., Geschwindner, S., Scharff, E. I. & Rüterjans, H. (2001). Biochem. J.353, 579–589. [DOI] [PMC free article] [PubMed]
- Katsemi, V., Lücke, C., Koepke, J., Löhr, F., Maurer, S., Fritzsch, G. & Rüterjans, H. (2005). Biochemistry, 44, 9022–9033. [DOI] [PubMed]
- Katz, A. K., Xinmin, L., Carrell, H. L., Hanson, B. L., Langan, P., Coates, L., Schoenborn, B. P., Glusker, J. P. & Bunick, G. J. (2006). Proc. Natl Acad. Sci. USA, 103, 8342–8347. [DOI] [PMC free article] [PubMed]
- Koepke, J., Scharff, E. I., Lücke, C., Rüterjans, H. & Fritzsch, G. (2003). Acta Cryst. D59, 1744–1754. [DOI] [PubMed]
- Kossiakoff, A. A. & Spencer, S. A. (1981). Biochemistry, 20, 6462–6474. [DOI] [PubMed]
- Kovalevsky, A. Y., Katz, A. K., Carrell, H. L., Hanson, L., Mustyakimov, M., Fisher, S. Z., Coates, L., Schoenborn, B. P., Bunick, G. J., Glusker, J. P. & Langan, P. (2008). Biochemistry, 47, 7595–7597. [DOI] [PMC free article] [PubMed]
- Langan, P., Fisher, Z., Kovalevsky, A., Mustyakimov, M., Sutcliffe Valone, A., Unkefer, C., Waltman, M. J., Coates, L., Adams, P. D., Afonine, P. V., Bennett, B., Dealwis, C. & Schoenborn, B. P. (2008). J. Synchrotron Rad.15, 215–218. [DOI] [PMC free article] [PubMed]
- Liu, X., Hanson, B. L., Langan, P. & Viola, R. E. (2007). Acta Cryst. D63, 1000–1008. [DOI] [PubMed]
- Melzer, M., Chen, J. C.-H., Heidenreich, A., Gäb, J., Koller, M., Kehe, K. & Blum, M.-M. (2009). J. Am. Chem. Soc.131, 17226–17232. [DOI] [PubMed]
- Niimura, N. & Bau, R. (2008). Acta Cryst. A64, 12–22. [DOI] [PubMed]
- Scharff, E. I., Koepke, J., Fritzsch, G., Lücke, C. & Rüterjans, H. (2001). Structure, 9, 493–502. [DOI] [PubMed]
- Shu, F., Ramakrishnan, V. & Schoenborn, B. P. (2000). Proc. Natl Acad. Sci. USA, 97, 3872–3877. [DOI] [PMC free article] [PubMed]
- Tamada, T., Kinoshita, T., Kurihara, K., Adachi, M., Ohhara, T., Imai, K., Kuroki, R. & Tada, T. (2009). J. Am. Chem. Soc.131, 11033–11040. [DOI] [PubMed]
- Yagi, D., Yamada, T., Kurihara, K., Ohnishi, Y., Yamashita, M., Tamada, T., Tanaka, I., Kuroki, R. & Niimura, N. (2009). Acta Cryst. D65, 892–899. [DOI] [PubMed]