Abstract
We investigated the relationship of functional neurocircuitries and dopamine receptor D1 (DRD1) polymorphisms in schizophrenics during a working memory task. Participants performed the Serial Item Recognition Paradigm memory task during functional magnetic resonance imaging acquisition. We performed a data-driven multivariate analysis (partial least squares) to characterize brain network (covariance) patterns. Genetic testing identified two main genotypes. Accuracy did not differ between the groups. Covariance patterns of different areas (including the dorsolateral prefrontal cortex and the inferior parietal lobule) were inversely related between the two genotypes. Two groups of schizophrenic patients with similar symptomatology and performance on a working memory task, but with distinct dopamine receptor genotypes, may use distinct neural systems to retrieve information.
Keywords: functional MRI, genes, partial least squares, schizophrenia
Introduction
A hallmark of schizophrenia is cognitive impairment, including working memory dysfunction. Functional magnetic resonance imaging (fMRI) studies of working memory have shown differential activations in specific areas including the dorsolateral prefrontal cortex, the anterior cingulate, inferior parietal lobule, basal ganglia and superior frontal gyrus [1–5]. It has been suggested that genetic differences may explain the phenomenon [6]. The dopamine receptor D1 (DRD1) is one of the candidate genes for schizophrenia and was also found to be essential for working memory [6,7].
In our earlier study, we found that the DRD1 polymorphic site located one kilobase upstream from the initiation codon (DdeI) was predictive of clinical response to clozapine in schizophrenia; [8] thus, we investigated whether this polymorphic site was also associated with specific functional neurocircuitries in the same population.
In a cohort of people with schizophrenia, we used partial least squares (PLS) as a data-driven tool to see whether patients with different dopaminergic genotypes recruited the same circuitry to perform a working memory task. PLS methods in neuroimaging identify areas of the brain with the strongest signal coherence [9,10]. Rather than hypothesizing a specific relationship between fMRI signal changes and performance in the experimental conditions, a PLS analysis determines what relationship is there between the signal changes and the conditions and performance covariates, and where in the brain that relationship is in evidence.
Materials and methods
Participants
Informed consent was obtained from all participants. University of California, Irvine institutional review board approved the study protocol. Twenty-one medically stable chronic schizophrenic patients were recruited for the study. Nineteen of these participants were included in the final analysis (two were homozygous for GG allele comprising too small a sample to analyze), and they were all right handed. Six women and 13 men participated. The patients were on average 43 years of age (SD, ± 10.5 years) and had been ill for 13.9 years (± 9.1 years). All except for two patients had been ill for at least 5 years. The Positive and Negative Syndrome Scale (PANSS) and the Premorbid Verbal IQ Estimates [11] are detailed in Table 1. All participants were on stable doses of atypical antipsychotic drugs, except for two on conventional antipsychotic agents. Six participants were also on mood stabilizers, four on antidepressants, and two on antiparkinson agents, all with doses consistent with the Food and Drug Administration approved package inserts. The PANSS scores and medications are typical for patients with chronic schizophrenia.
Table 1.
Positive and negative syndrome scale (PANSS) scores and pre-morbid verbal IQ estimates (PVIE) specifics
| PANSS total | PANSS positive | PANSS negative | PANSS general | PVIE | |
|---|---|---|---|---|---|
| Mean | 75.3 | 16.2 | 20.1 | 39.0 | 108.8 |
| Range | 56–104 | 10–28 | 12–26 | 31–50 | 87.11–119.04 |
Genetic methods
DNA was extracted from blood samples using the high-salt method [12] and polymerase chain reaction (PCR) was used to amplify the upstream DRD1 polymorphic site. The DRD1 polymorphism that was used is recognized by the restriction enzyme DdeI and is located about 1 kb upstream of the initiation codon. PCR amplification was done according to Cichon et al. [13] and DdeI restriction digest was performed according to the enzyme manufacturer’s instructions (Fermentas Inc., Hamilton, Ontario, Canada). The DdeI restriction fragments were visualized using 2% agarose gel electrophoresis. No studies have yet been carried out to determine whether this site alters promoter function. Participants were divided into groups on the basis of DRD1 DdeI polymorphism, which could be AA, AG, or GG.
MRI data collection
All imaging data were collected on a 1.5T Philips scanner (Marconi/Picker Eclipse model, Philips Health Care, And-over, Massachusetts, USA). The fMRI scans consisted of a T2*-weighted gradient echo planar imaging sequence (24 cm Field of View (FOV), 28 slices, 5-mm thick with no gap, axially oriented; TR=3 s, TE=40 ms, 90° flip angle, 80 frames per scan) tuned to blood oxygenation level dependent (BOLD) signal. During the fMRI scans, participants performed a serial item recognition paradigm, a working memory task based on Manoach et al. [3]. The serial item recognition paradigm has been repeatedly reported to activate the dorsolateral prefrontal cortex in healthy participants and people with schizophrenia [2,14]. Three runs (240 s each) of the working memory task were collected within the same scanning session.
The task included three conditions in a blocked design: a baseline condition, a low memory load condition and a high memory load condition. In the baseline condition blocks, participants were presented a series of arrows and asked to indicate the direction in which the arrow pointed (left or right). In both memory load conditions blocks, participants were presented with a set of numbers (presented simultaneously for 5 s), then presented with a series of 10 probe trials each consisting of a single number presented for 2 s. Participants indicated whether the probe was in the memory set of numbers, or not. In the low memory load condition, there were only two numbers in the memory set; in the high memory load condition, there were five. The memory sets were different in every block and every run. Each run consisted of nine blocks, beginning and ending with a baseline condition block.
The preprocessing steps included motion detection and correction, coregistration and normalization to a Montreal Neurological Institute brain template (Montreal Neurological Institute, Montreal, Quebec, Canada), and smoothing with an 8-mm FWHM 3D Gaussian filter (Wellcome Trust Centre for Neuroimaging, London, UK). The preprocessing steps were performed with the SPM99 software (http://www.fil.ion.ucl.ac.uk/spm/software/spm99/), using default settings where applicable [15,16]. The motion-corrected, normalized and smoothed images were the input to the PLS analysis.
Partial least squares: image analysis
The PLS analyses used PLS version 5.0701151 (http://www.rotman-baycrest.on.ca/). To quantitatively determine differences between circuitry of the two genotypes, a combined analysis was performed analyzing the two groups together. Separate analyses by genotype group were also performed to confirm results.
A blocked analysis of the fMRI data was used. Averages of the last six frames (18 s) of the baseline blocks were used as a baseline. Average images from the last six frames of each block during the low and high memory load conditions were included so that the hemodynamic response function could best reflect the participant’s efforts at recall, rather than at the stimuli presentation. The preceding baseline block was subtracted from each memory condition block to provide a measure of BOLD signal change.
The main goal of PLS is to identify areas of the brain presenting the same activations at the same time (covariance) [9,17]. The strongest covariance within each block is expected to describe the brain pattern related to the specific task. Singular value decomposition is performed on correlations between accuracy values averaged within each block and BOLD values at each voxel. This operation generates simultaneously a ‘singular image’ which is the brain image with the covarying voxels correlated with accuracy, a ‘correlation profile’ which is a plot expressing the relationship between accuracy and activation change in each memory condition for each group, and a ‘singular value’ which is a scalar number accounting for the covariance. The numerical weights within the singular image are termed saliencies; saliencies can be positive or negative. Singular image, singular value and correlational profile describe an overall pattern termed a latent variable (LV). Each singular value decomposition produces a number of LVs. The first LV accounts for the largest proportion of covariance, and thus is the primary pattern in the dataset; the second LV accounts for the next largest proportion, etc.
The significance of the singular value (i.e. whether the LV accounts for an amount of covariance that is unlikely to have arisen by chance – in essence, the strength of the LV signal relative to random noise) is determined by permutation sampling. This involves randomly reassigning the participants across groups [17]. The reliability of the saliences in the singular image (i.e. which voxels’ saliences in the singular image are significantly different from zero) is determined by the bootstrap method [17]. The bootstrap procedure involves sampling the dataset with replacement to derive estimates of standard errors of the LV saliencies for each voxel.
Results
Genetic data and demographic variables
Nine and 10 participants carried the AA and AG genotypes, respectively, in the DRD1. Two participants carried the GG genotype and were excluded from analysis owing to the inability to perform significance tests on such a small sample size. The distribution of males and females within the different groups was not significant (Fisher’s exact test, P > 0.05). The mean age, symptom severity as measured by the PANSS subscores, and duration of illness did not significantly differ across genotypes (P > 0.05).
Behavioral analysis by genotype
The mean proportion ± SD of correct answers was 0.90 ± 0.18 and 0.93 ± 0.07 for the baseline (arrows) condition, 0.80 ± 0.23 and 0.90 ± 0.09 for the low memory condition and 0.81±0.21 and 0.81±0.11 for the high memory load in the two genotypes, respectively. The main effect of condition was significant [F(2,34)=5.98, P < 0.006], confirming the overall decrease in accuracy with increasing load. The effect of genotype group on performance was not significant [F(1,17) < 1]; accuracy in both groups was equivalent and there was no interaction by genotype with memory load. This was also true for the response time measures: response times slowed with increasing load [F(2,34)=97, P <0.00001], whereas the effect of genotype group was not significant [F(1,17)=1.5, P <0.25].
Partial least squares results
This analysis was performed to contrast the brain behavior between the two groups. Of the four LVs identified by the analysis, only the first one was significant (P < 0.02 by permutation testing) and stable, and it accounted for 37% of the cross-block covariance between behavior and fMRI data.
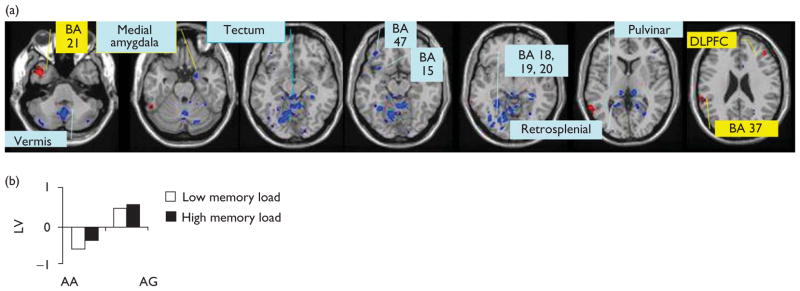
The significant LV indicated the two groups had very different patterns of BOLD signal relationship with accuracy. The network (singular image thresholded for significant voxels) and the correlation profile are plotted in Fig. 1a and b, respectively. The LV shows a different profile by genotype grouping. The voxels shown in yellow/red are positively weighted on the underlying profile, whereas the voxels shown in blue are negatively weighted on the underlying profile for the LV.
Fig. 1.
The voxels are thresholded at 3.0 SD. (a) Yellow areas tend to be more engaged as accuracy increases in group AG, but not in AA in both memory conditions; blue areas represent the inverse pattern. (b) The correlational profile describes the correlation between voxels and accuracy values.
In Fig. 1a, yellow areas (positive weights) represent areas where increasing and covarying activation levels correlated with increased accuracy values within the AG group compared with the AA group; these included the temporal pole, the dorsolateral prefrontal cortex (Brodmann area 46), the inferior parietal lobule (Brodmann area 40) and the Brodmann areas 6, 21, 22 and 37. Blue areas (negative weights), in contrast, represent brain patterns that correlated with increased accuracy within the AA group relative to the AG group; these included the tectum, retrosplenial, vermis, medial amygdala, posterior inferior temporal lobe, hippo-campal area, anterior insula (Brodmann area 15), Brodmann area 7, 8, 18 and 20.
The correlations specific to the dorsolateral prefrontal cortex, temporal pole, tectum and vermis are inverted for the two genotypes. The same areas show the opposite relationships with accuracy and memory between the two genotypic groups: what is positively correlated for the AA group is negatively correlated for the AG group, and vice versa. Each group analyzed individually also showed the pattern it showed in the combined group analysis.
Discussion
The patients with schizophrenia presented a similar symptomatology and working memory performance, but fell into two genotypic groups. The data-driven analysis showed that the strongest patterns of BOLD coherence identified primarily differences between the two genotypes. It appears that patients used distinct neural systems to retrieve information based on their DRD1 genotype. The AG genotype may engage a more conscious visual and verbal attentional neocortical system compared with the AA genotype to achieve an increasing level of accuracy. In contrast, the AA genotype seems to adopt a more tectopulvinar pathway, involving an unconscious visual pathway part of the posterior attentional system compared with the other.
Our study corroborated previous findings showing differential activations during working memory tasks in the dorsolateral prefrontal cortex, the anterior cingulate area, the inferior parietal lobule and the basal ganglia [1–5]. It is possible that the pathology cannot be simplified to a reduced or increased activation during working memory, but to specific underlying neuronal pathways determined by specific genetic mechanisms [6].
In our study, the higher the accuracy, the more the dorsolateral prefrontal cortex seemed to be engaged in the AG and not in the AA genotype, although their performance was equivalent. This finding may contribute to aspects of the inverted U model for dopaminergic regulation perhaps based on genotype. According to the model, the dorsolateral prefrontal cortex is increasingly engaged as the working memory task becomes more difficult until it reaches a maximum capacity [18]. At the highest memory efforts, dorsolateral prefrontal cortex activation declines. Although both healthy participants and patients follow this model, the curve for people with schizophrenia peaks and falls off with lower memory load, reflecting lower working memory capacity [1].
These analyses were performed on patients with schizophrenia only. As such, the results relate to working memory issues within the context of the disease. They should not be interpreted as indicating causal relationships with the clinical diagnosis per se. Finding a genetic influence on the circuitry underlying memory performance within the patient sample, however, indicates the feasibility of using this integrated imaging-genetics approach to tease apart potentially different subtypes of the diagnosis.
A data-driven analysis was necessary to answer the question of whether or not there was a significant difference in the neurocircuitry of patients with a specific genotype. This was a rich dataset, with many possible outcomes: one group may have accounted for the most coherent pattern, whereas the other group was more variable and had no significant coherent pattern, or shown a coherent pattern in an entirely different group of brain regions. Or, the two groups may have shown the same pattern, either increasing or decreasing with memory load condition and performance. A data-driven approach extracts the most important patterns, and when followed by permutation testing and bootstrapping, indicates the reliability of those findings.
Permutation testing has been found to be particularly suitable for neuroimaging datasets with small sample sizes where parametric assumptions may not be met [19]. Our findings reflect brain-behavior patterns in a small group; however, our sample size is similar to the majority of neuroimaging studies that use multivariate analysis combined with permutation testing [10,17,20,21]. Studies with a larger sample can investigate imaging-genetics patterns including a larger number of polymorphisms and genes to be able fully characterize common features of schizophrenia.
This preliminary study indicates that the DRD1 DdeI marker, which was previously identified as a potential pharmacogenetic target, may also be a useful imaging-genetics biomarker. The effect of the GG genotype is currently undetermined: It may be more like the AG group, or it may show a different pattern altogether.
Conclusion
The DRD1 (DdeI) genotype is associated with different brain patterns in schizophrenic patients. The AG genotype uses a specific network that includes the dorsolateral prefrontal cortex positively correlated with accuracy. The AA genotype, in contrast, has a negative correlation in the dorsolateral prefrontal cortex with accuracy, and has positive correlations in a different network. Therefore, even though the two groups achieve the same performance, the circuitry is possibly associated with DRD1 genotypes. This study may provide imaging-genetics insights to develop targeted therapies reflecting an individual genetic and physiological variations.
Acknowledgments
This research was supported by U24 RR021992 to the FBIRN and by P20 RR020837-01 to the Transdisciplinary Imaging Genetics Center, from the National Center for Research Resources/National Institutes of Health. The authors acknowledge and thank Randy McIntosh, PhD, University of Toronto, for his advice on PLS analysis, Hal Stern, PhD, University of California, Irvine, for his input on these discussions. They also thank Patrick MacLeod, M.D., Nigel Livingston, PhD, University of Victoria for their invaluable assistance. Sources of support: U24 RR021992 and P20 RR020837-01 from the National Center for Research Resources/National Institutes of Health.
References
- 1.Callicott JH, Mattay VS, Verchinski BA, Marenco S, Egan MF, Weinberger DR. Complexity of prefrontal cortical dysfunction in schizophrenia: more than up or down. Am J Psychiatry. 2003;160:2209–2215. doi: 10.1176/appi.ajp.160.12.2209. [DOI] [PubMed] [Google Scholar]
- 2.Johnson MR, Morris NA, Astur RS, Calhoun VD, Mathalon DH, Kiehl KA, Pearlson GD. A functional magnetic resonance imaging study of working memory abnormalities in schizophrenia. Biol Psychiatry. 2006;60:11–21. doi: 10.1016/j.biopsych.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 3.Manoach DS, Press DZ, Thangaraj V, Searl MM, Goff DC, Halpern E, et al. Schizophrenic subjects activate dorsolateral prefrontal cortex during a working memory task, as measured by fMRI. Biol Psychiatry. 1999;45:1128–1137. doi: 10.1016/s0006-3223(98)00318-7. [DOI] [PubMed] [Google Scholar]
- 4.Karlsgodt KH, Glahn DC, van Erp TG, Therman S, Huttunen M, Manninen M, et al. The relationship between performance and fMRI signal during working memory in patients with schizophrenia, unaffected co-twins, and control subjects. Schizophr Res. 2007;89:191–197. doi: 10.1016/j.schres.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 5.Schneider F, Habel U, Reske M, Kellermann T, Stocker T, Shah NJ, et al. Neural correlates of working memory dysfunction in first-episode schizophrenia patients: an fMRI multi-center study. Schizophr Res. 2007;89:198–210. doi: 10.1016/j.schres.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 6.Williams GV, Castner SA. Under the curve: critical issues for elucidating D1 receptor function in working memory. Neuroscience. 2006;139:263–276. doi: 10.1016/j.neuroscience.2005.09.028. [DOI] [PubMed] [Google Scholar]
- 7.Hirvonen J, van Erp TG, Huttunen J, Aalto S, Nagren K, Huttunen M, et al. Brain dopamine D1 receptors in twins discordant for schizophrenia. Am J Psychiatry. 2006;163:1747–1753. doi: 10.1176/ajp.2006.163.10.1747. [DOI] [PubMed] [Google Scholar]
- 8.Potkin SG, Basile VS, Jin Y, Masellis M, Badri F, Keator D, et al. D1 receptor alleles predict PET metabolic correlates of clinical response to clozapine. Mol Psychiatry. 2003;8:109–113. doi: 10.1038/sj.mp.4001191. [DOI] [PubMed] [Google Scholar]
- 9.McIntosh AR, Bookstein FL, Haxby JV, Grady CL. Spatial pattern analysis of functional brain images using partial least squares. NeuroImage. 1996;3 (3 Pt 1):143–157. doi: 10.1006/nimg.1996.0016. [DOI] [PubMed] [Google Scholar]
- 10.McIntosh AR, Lobaugh NJ. Partial least squares analysis of neuroimaging data: applications and advances. NeuroImage. 2004;23 (Suppl 1):S250–S263. doi: 10.1016/j.neuroimage.2004.07.020. [DOI] [PubMed] [Google Scholar]
- 11.Grober E, Sliwinski M. Development and validation of a model for estimating premorbid verbal intelligence in the elderly. J Clin Exp Neuropsychol. 1991;13:933–949. doi: 10.1080/01688639108405109. [DOI] [PubMed] [Google Scholar]
- 12.Lahiri DK, Nurnberger JI., Jr A rapid non-enzymatic method for the preparation of HMW DNA from blood for RFLP studies. Nucleic Acids Res. 1991;19:5444. doi: 10.1093/nar/19.19.5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cichon S, Nothen MM, Erdmann J, Propping P. Detection of four polymorphic sites in the human dopamine D1 receptor gene (DRD1) Hum Mol Genet. 1994;3:209. doi: 10.1093/hmg/3.1.209. [DOI] [PubMed] [Google Scholar]
- 14.Manoach DS. Prefrontal cortex dysfunction during working memory performance in schizophrenia: reconciling discrepant findings. Schizophr Res. 2003;60:285–298. doi: 10.1016/s0920-9964(02)00294-3. [DOI] [PubMed] [Google Scholar]
- 15.Friston KJ, Ashburner J, Frith CD, Poline J-B, Heather JD, Frackowiak RSJ. Spatial registration and normalization of images. Human Brain Mapping. 1995;2:165–189. [Google Scholar]
- 16.Poline JB, Worsley KJ, Holmes AP, Frackowiak RS, Friston KJ. Estimating smoothness in statistical parametric maps: variability of P values. J Comput Assist Tomogr. 1995;19:788–796. doi: 10.1097/00004728-199509000-00017. [DOI] [PubMed] [Google Scholar]
- 17.McIntosh AR, Chau WK, Protzner AB. Spatiotemporal analysis of event-related fMRI data using partial least squares. NeuroImage. 2004;23:764–775. doi: 10.1016/j.neuroimage.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 18.Callicott JH, Mattay VS, Bertolino A, Finn K, Coppola R, Frank JA, et al. Physiological characteristics of capacity constraints in working memory as revealed by functional MRI. Cereb Cortex. 1999;9:20–26. doi: 10.1093/cercor/9.1.20. [DOI] [PubMed] [Google Scholar]
- 19.Nichols TE, Holmes AP. Nonparametric permutation tests for functional neuroimaging: a primer with examples. Hum Brain Mapp. 2002;15:1–25. doi: 10.1002/hbm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caplan JB, McIntosh AR, De Rosa E. Two distinct functional networks for successful resolution of proactive interference. Cereb Cortex. 2007;17:1650–1663. doi: 10.1093/cercor/bhl076. [DOI] [PubMed] [Google Scholar]
- 21.Della-Maggiore V, Sekuler AB, Grady CL, Bennett PJ, Sekuler R, McIntosh AR. Corticolimbic interactions associated with performance on a short-term memory task are modified by age. J Neurosci. 2000;20:8410–8416. doi: 10.1523/JNEUROSCI.20-22-08410.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]