Abstract
DNA methylation patterns are established during embryonic development and faithfully copied through somatic cell divisions. Based on our understanding of DNA methylation and other interrelated epigenetic modifications, a comprehensive view of the epigenetic landscape and cancer epigenome is evolving. The cancer methylome is highly disrupted, making DNA methylation an excellent target for anti-cancer therapies. During the last few decades, an increasing number of drugs targeting DNA methylation have been developed in an effort to increase efficacy, stability and to decrease toxicity. The earliest and the most successful epigenetic drug to date, 5-Azacytidine, is currently recommended as the first-line treatment for high risk myelodysplastic syndromes (MDS) patients. Encouraging results from clinical trials have prompted further efforts to elucidate epigenetic alterations in cancer and subsequently develop new epigenetic therapies. This review delineates the latest cancer epigenetic models, recent discovery of hypomethylation agents and their application in the clinic.
Keywords: DNA methylation, DNA methyltransferase inhibitor, Epigenetic therapy, 5-Azacytidine
The epigenetic landscape
Primary heritable genetic information is dictated by the DNA sequence which is packed into chromatin. The basic repeating unit of chromatin is a nuclesome consisting of 146–147 bp of DNA wrapped around a histone octamer. Both DNA and histone proteins can undergo various post-synthetic modifications. These modifications, along with histone variants and nucleosome remodelers, have the ability to strongly influence chromatin structure and transcriptional regulation without changing the underlying DNA sequence, a process called epigenetic regulation. Epigenetic regulation can be roughly separated into three interrelated layers: nucleosome positioning, histone modifications and DNA methylation[1].
DNA methylation is probably the most extensively studied epigenetic mark and plays an important role in genomic imprinting, in X-chromosome inactivation and in the silencing of retrotransposon, repetitive elements and tissue specific genes. In mammalian cells, a methyl group is covalently added to cytosine in the context of CpG (cytosine-phosphate-guanine) dinucleotides in somatic cells and also CpHpG nucleotide sequences in embryonic stem cells (ES)[2]. In somatic cells, most CpG dinucleotides are methylated except those located in CpG islands[3], which are defined as regions of DNA greater than 500 base pairs (bp) with a GC content no less than 55% and a ratio of observed and expected CpG that is greater than than 0.65[4]. Nearly 60% of mammalian gene promoters are located in CpG islands[5]. DNA methylation of these CpG-rich promoters silences gene expression by changing the accessibility of DNA to transcription factors (TFs) or by recruiting additional silencing-associated proteins[6]. Nevertheless, there is also growing evidence suggesting that even in CpG-poor regions, such as the Oct4 promoter, DNA methylation still plays a role in gene regulation[7–8].
DNA methylation patterns are established and maintained by DNA methyltransferases (DNMTs). During early embryogenesis, de novo DNA methylation is mediated by DNMT3A and DNMT3B associated with DNMT3L, which lacks a methyltransferase domain[9–11]. To maintain DNA methylation patterns in daughter cells, UHRF1 (ubiquitin-like, containing PHD and RING finger domains 1) recognizes hemimethylated DNA and directs DNMT1 to methylate the appropriate cytosine in newly synthesized DNA strands during successive replications[12–13]. Recent studies have proposed an updated model, suggesting that DNMT3A/B are also required for the maintenance of DNA methylation patterns in somatic cells, especially of repeat regions and imprinted genes (Figure 1a)[6, 14].
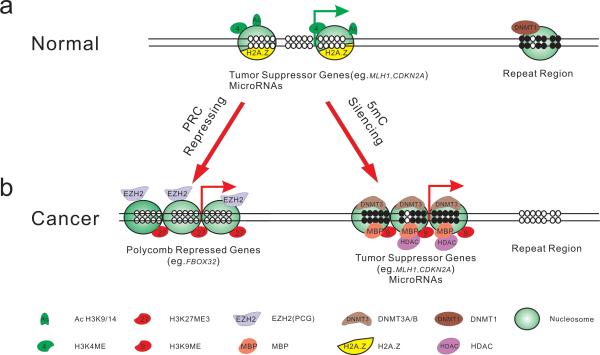
Figure 1. Epigenetic Regulation in Normal and Cancer Cells.
Shown are schematic promoters. Arrows represent transcription start site (TSS); filled circles represent methylated CpG dinucleotide and empty circles represent unmethylated CpG dinucleotide. In normal cells (a), genes such as MLH1, CDKN2A are generally unmethylated and packaged with active modified histone proteins (e.g. H3K4me3) as well as histone variants (e.g. H2A.Z). These epigenetic modifications constitute an “open” chromatin structure which, with nucleosome depleted region (NDR), favoring transcription. In other genomic regions, such as in the repetitive elements, the CpG sites are methylated and thereby maintain a closed chromatin structure.
In cancer cells (b), epigenetic modifications are disrupted. Besides cancer specific hypomethylation (e.g. in repetitive sequence), there are two interrelated epigenetic mechanisms to repress gene expression. Some genes (e.g.FBXO32) could be recognized by polycomb proteins, such as EZH2, which catalyses H3K27 methylation, and are consequently repressed. By contrast, CpG sites within gene promoters could undergo de novo methylation by DNMT3A/B, which are compartmentalized to these regions to complete the methylation process. The methylated CpG sites attract methyl-binding proteins such as MBD, which is coupled with HDAC proteins to remove histone acetylation as well as histone methyltransferase to methylate H3K9. Associated with all repressive factors shown, nucleosomes cover the promoter region, generating a tightly closed chromatin status to shut down gene expression.
In contrast to DNA methylation, the patterns of histone modifications are more labile. The N-termini of histones can undergo various post-synthetic modifications, including a diverse combination of methylation, acetylation, ubiquitylation, phosphorylation and sumoylation. In recent years, the development of modification-specific antibodies in Chromatin immunoprecipitation (ChIP), coupled with microarray (ChIP-chip) or direct sequencing technology (ChIP-seq), has revolutionized our ability to assess the global distribution of histone modifications and their roles in gene regulation[15–17]. For instance, H3K4me3 has been found to occur near the transcription start sites (TSS) of genes or miRNA and is positively correlated with transcription level[18–21]. Similarly, H3K9/14 acetylation is highly associated with gene transcriptional activation[22]. By contrast, H3K9me and H3K27me are enriched in the vicinity of suppressed or silenced gene promoters (Figure 1a)[15]. The enzymes that catalyze these modifications and the crosstalk between these complexes have been well summarized in several important review papers[23–26].
Other crucial components of the epigenetic landscape are histone variants which partially determine nucleosome stability and ultimately affect gene expression. H2A.Z is commonly localized in promoter regions. The differences in the amino acid sequence of H2A.Z compared to H2A might affect the protein's interactions with the H3/H4 tetramer, therefore altering nucleosome stability[27–28]. In mammalian cells, the role of H2A.Z in gene regulation is not fully understood; however, there is increasing evidence suggesting that H2A.Z assists in the transcription initiation of a special group of genes, such as endoplasmic reticulum (ER) target genes[29–32].
The interplay between histone modifications, histone variants and DNA methylation constitutes a global regulation network. Extensive studies have already elucidated the biological and functional interactions among these three epigenetic layers. For example, DNA methylation anti-correlates with the active mark H3K4me3, H3ac or H2A.Z[33–35]. Meanwhile, highly methylated promoter regions are bound by methylcytosine binding proteins such as MeCP2, which subsequently recruits histone deacetylase (HDAC) to deacetylate histones, thus further reinforcing the suppressive nature of promoters (Figure 1)[36–37]. The interplay between DNA methylation and histone modifications can be further observed in ES cells. Some studies have proposed histone modifications and associated proteins, such as EZH2 or G9a, take part in establishing de novo DNA methylation patterns during early development[38–41].
Cancer epigenetics
It has been well documented that epigenetic alterations are involved in cancer initiation and progression in addition to abnormal genetic events. Early studies that measured the global 5-methylcytosine content of tumors showed that hypomethylation was a common feature of carcinogenesis, leading to abnormal chromosomal instability and transcriptional regulation[42–44]. However, the majority of cancer epigenetic studies subsequently concentrated on focal CpG island hypermethylation in cancer and revealed many tumor suppressor genes, cellular functional genes and miRNAs silenced by promoter DNA methylation[45–48]. Recent genome-wide studies have demonstrated distinct DNA methylation patterns in cancerous tissues compared to their normal counterparts[49–52].
The detailed mechanisms by which these discrete regions undergo hyper- or hypomethylation are still unclear. Early evidence suggested that elevated DNA methyltransferase levels might trigger hypermethylation of tumor suppressor gene promoters which would consequentially result in cancer cell proliferation[53]. In addition to this “selection” model, an alternative mechanism has been proposed that takes aadvantage of the current genome-wide epigenetic studies in stem cells. Investigators have suggested that the establishment of aberrant epigenetic profiles in cancer undergoes a process that is similar to epigenetic reprogramming during development.[54] During cancer initiation, the promoters of genes, which are repressed by histone H3K27me3 in normal differentiated cells, might become methylated and thereby set up for long term silencing. This so-called “epigenetic switch” could be regulated by the cooperation of polycomb proteins and DNMTs[55–57]. (Figure 1b)
In cooperation with DNA methylation, other epigenetic mechanisms also exhibit abnormal regulation in cancer. For example, histone deacetylases (HDACs) are often found to be overexpressed in various types of cancer, resulting in histone deacetylation around the TSS region and the formation of a more compact structure to silence genes[58–59]. In addition, H3K4me is also selectively demethylated by histone lysine demethylase, LSD1, which is upregulated in cancer, making it a potential drug target[60–61]. In some loci, Polycomb-group (PcG) proteins associated with H3K27me work independently of DNA methylation to aberrantly repress genes in cancer cells[62–65]. Nucleosome occupancy is also switched from an “open” to a “covered” status in gene regulation elements in neoplastic cells[66–67]. (Figure 1b)
In vitro study of DNMT inhibitors (DNMTi)
Epigenetic modifications play a crucial role in regulating normal cells; however, these processes are disrupted during tumorigenesis. The relatively reversible character of epigenetic alterations, in contrast to genetic changes, has inspired the development of therapeutic strategies targeting various epigenetic components. Among them, DNA methylation and its associated enzymes have been well studied. The understanding of their fundamental mechanism of action and correlation with other epigenetic modifications makes them attractive drug targets.
Nucleoside analogues
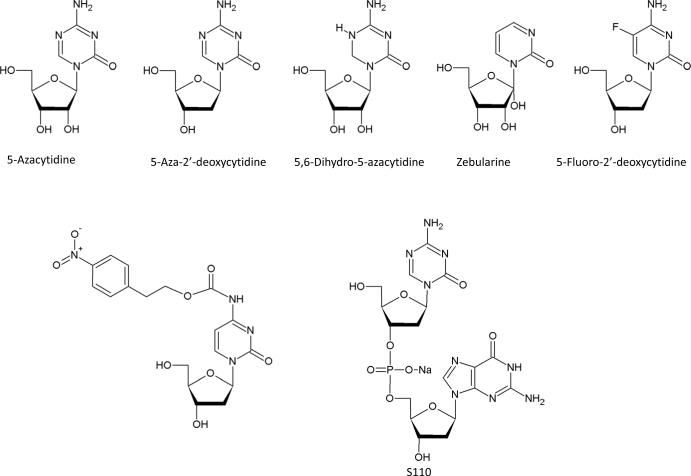
5-Azacytidine (5-Aza-CR) and 5-aza-2′-deoxycitidine (5-Aza-CdR) are the two most potent DNMT inhibitors, and have been approved by the FDA for the treatment of myeloid malignancies (Figure 2a). They were first synthesized as cytotoxic agents. In 1980s, these compounds were found to have hypomethylating activity after incorporation into the DNA of actively replicating tumor cells[68–70].
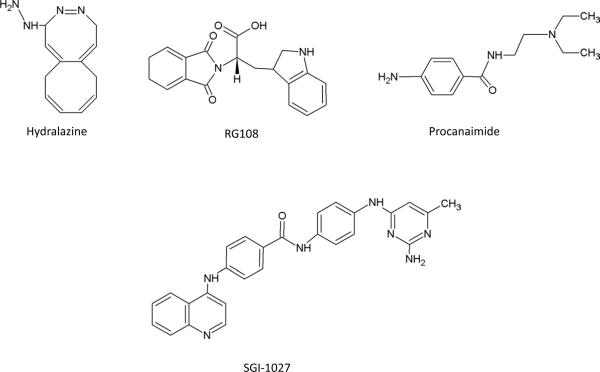
Figure 2. Chemical Structure of DNMT inhibitors.
A. nucleoside analogues
B. non-nucleoside analogues.
Upon transport into cells by the human concentrative nucleoside transporter 1 (hCNT1), 5-Aza-CR and 5-Aza-CdR are phosphorylated by different kinases, converting them to their active triphosphate forms, 5-Aza-CTP and 5-Aza-dCTP respectively (Figure 3a)[71–72]. 5-Aza-CR can be incorporated into both RNA and DNA following the reduction of 5-Aza-CDP by ribonucleotide reductase, whereas 5-Aza-CdR incorporates into DNA following its phosphorylation to 5-Aza-dCTP[73]. The incorporated 5-azanucleoside disrupts the interaction between DNA and DNMTs through the nitrogen in the 5 position of the modified pyrimidine and traps DNMTs for proteosomal degradation (Figure 3b)[74–75]. The depletion of DNMTs results in the passive loss of cytosine methylation in the daughter cells after replication. This is associated with reduced H3K9me3, increased H3ac and H3K4me3 modifications around gene promoter regions, as well as the formation of a nucleosome-deficient region[66, 76]. A recent genome-wide study of epigenetic landscape alterations after 5-Aza-CR treatment has further validated this concept[77]. The demethylation function of 5-Aza-CR and 5-Aza-CdR is most evident at low drug concentration because the drugs exhibit greater cytotoxicity, interfere with DNA synthesis and cause DNA damage at higher concentrations[78]. The S-phase is required for the selective and effective incorporation of these two drugs into the DNA of rapidly proliferating cells, thereby limiting unwanted hypomethylation in normal non-cycling cells.
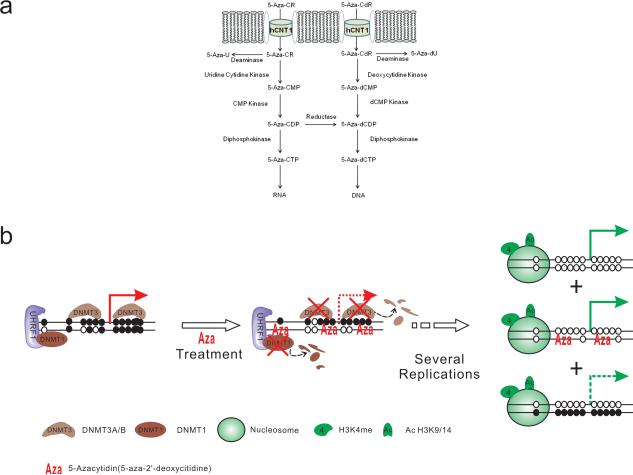
Figure 3. Metabolism pathway and action mechanism of 5-Azanucleosides.
A. Human Concentrated Nucleoside Transporters 1 (hCNT1) facilitates the entry of 5-Aza-CR and 5-Aza-CdR into cells where they are phosphorylated by uridine-cytidine kinase and deoxycytidine kinase respectively. 5-Aza-CMP and 5-Aza-dCMP are subsequently phosphorylated into their active triphosphate forms. 5 Aza-dCR can be incorporated solely into DNA, whereas 5-Aza-CR can be incorporated into RNA as well as DNA following the reduction of its 5-Aza-CDP form to the 5-Aza-dCDP form. Incorporation of 5-Azanucleosides into DNA induces hypomethylation of the daughter DNA strands whereas incorporation of 5-Aza-CR into RNA disrupts crucial cellular process such as protein translation and causes ribosomal disassembly. 5-Azanucleosides, however, are also very unstable and can be deaminated to inactive uridine.
B. Schematic working model of 5-Azanucleosides. During DNA replication, 5-Azanucleosides are incorporated into DNA and trap DNMTs, which are subsequently targeted for proteosomal degradation. DNA containing azanucleosides are hemimethylated after the first round of DNA replication and become fully demethylated after several rounds of replication. Using hypomethylation agents, the silenced epigenetic modifications could be switched to an active status (eg.H3K4me3 and AcH3)
Besides inhibiting DNMTs, 5-Aza-CR incorporates into RNA and interrupts normal cellular processes by inducing ribosomal disassembly and preventing the translation of oncogenic proteins[73, 79]. The ability of 5-Aza-CR to be incorporated into DNA and RNA increases its side effects in vitro and in vivo because it can function in resting as well as in dividing cells[80]. Both 5-Aza-CR and 5-Aza-CdR, however, are easily hydrolyzed in aqueous solution and subject to deamination by cytidine deaminase. The instabilities of these compounds inevitably present a challenge in their clinical applications.
To improve the stability and efficacy of 5-Azanucleosides, several other cytidine analogues have been developed (Figure 2a). For example, zebularine, a cytidine analogue that lacks an amino group in the 4 position of the pyrimidine ring, can inhibit both DNMTs and cytidine deaminase after oral administration[81–83]. Studies have shown that zebularine induces hypomethylation in breast cancer cell lines and reactivates silenced tumor suppressor genes[84–85]. The inefficient metabolic activation of this compound has, however, delayed its clinical use as a single agent. As an inhibitor of cytidine deaminase, its co-administration has increased the efficacy of 5-Aza-CdR[82]. The cytidine analogue 5-Fluoro-2′-deoxycytidine (5-F-CdR) had also been demonstrated to have hypomethylating activity in mouse cells as well as human breast and lung carcinoma cells[68, 86]. Clinical studies further showed that co-administration of 5-F-CdR with a cytidine deaminase inhibitor, tetrahydrouridine (THU), improved its stability[87]. The therapeutic potential of another stable analogue, dihydro-5-azacytidine (DHAC), was also assessed for the treatment of malignant mesothelioma. Results on the compound's clinical efficacy, however, have not been consistent[88].
Recently, the effort to improve the stability of DNMTi includes the development of prodrugs of the nucleoside analogues. A preclinical study shows that NPEOC-DAC, a prodrug of 5-Aza-CdR containing 2-(p-nitrophenyl)ethoxycarbonyl (NPEOC) group at the 4 position of the pyrimidine ring, can be incorporated into DNA and inhibit DNMTs following its activation by human carboxylesterase 1 in a liver cancer cell line. The NPEOC moiety protects 5-Aza-CdR from deamination, but the compound itself is less potent when administered at the same concentration as 5-Aza-CdR. Moreover, the activity of NPEOC-DAC depends on carboxylesterases which are not expressed in all tissues. Further studies will be required to explore the use of this compound in combination therapy[89]. Alternatively, S110, a dinucleotide containing the 5-azacytosine ring, has also been shown to improve efficacy of 5-Aza-CdR by protecting it from deamination. The compound is well tolerated and can reduce the level of DNA methylation in CDKN2A promoter region in xenografts[90].
Non-nucleoside analogues
Unlike cytidine analogues, non-nucleoside DNMTis (Figure 2b) do not require incorporation into DNA and thus might exhibit less cytotoxicity. Some of the compounds assessed for their potential to induce hypomethylation in solid tumors are hydralazine and procainamide, the widely used vasodilator and antiarrhythmic agents respectively[91]. Hydralazine has been reported to block the activity of DNMTs by the interaction of its nitrogens with the Lys-162 and Arg-240 residues of the enzyme, whereas procainamide acts similarly as a competitive inhibitor by preferentially binding to DNMT1[92–93]. These compounds, however, have limited DNA hypomethylation activity in living cells[94]. The small molecule RG108 also shows the potential to reactivate tumor suppressor genes in human colon cancer cells[95–96]. Recently, a lipophilic, quinoline-based compound, SGI-1027, was demonstrated to be a novel DNMTi in vitro. In RKO cells, SGI-1027 causes the degradation of DNMT1 and the demethylation of the CDKN2A gene promoter as well as reactivates silenced genes[97].
An alternative strategy to inhibit DNMT1 includes the use of short chain oligodeoxynucleotides and microRNAs. MG98 is a 20 bp antisense oligonucleotide that specifically binds to the 3' UTR of human DNMT1 mRNA to prevent its translation. Despite promising result in preclinical studies, the clinical use of MG98 has not yet been validated[98]. MicroRNA miR29a, which targets DNMT3A/B directly and DNMT1 indirectly in a similar way to MG98, has the ability to reduce global DNA methylation and reactivates CDKN2B[99].
Clinical data on DNMT inhibitors
Accumulated in vitro evidence has demonstrated that DNMT inhibitors have the ability to reverse abnormal DNA methylation, indirectly affect histone modifications and eventually restore normal gene expression profiles in various cancer cell lines. These successful results encouraged the use of DNMT inhibitors to develop novel clinical regimens. In Table 1, we have summarized several recent important clinical trials.
Table 1.
Selected Recent Clinical Trials with DNMT Inhibitors.
| Drug(s) | Indication(s) | Author (Year) | Total Patients | Results | Comments | Ref. |
|---|---|---|---|---|---|---|
| Azacitidine | MDS | Silverman (2002/2006) | 191 | 7% CR, 16% PR, 37% HI | Low dose treatment improved quality of life and survival rate; delayed progression time to AML( CALGB9221). | [102,104] |
| MDS | Fenaux (2009) | 358 | 17% CR, 12% PR, 42% SD | Improved OS compared to CCR in high risk MDS patients (AZA-001). | [105] | |
| MDS | Lyons (2009) | 151 | 44%, 45%, 56% HI | RBC transfusion independence achieved in 3 alternative dosing regimens. | [109] | |
| MDS | Martin (2009) | 25 | 27% ORR | Response rates are comparable to CALGB 9221 and AZA-001 trials but shorter OS in this 5 days treatment regimen. | [110] | |
| MDS/AML | Muller-Thomas (2009) | 32 | 15.6% CR, 25% HI, 34.4% SD | Survival advantage in responder group of high-risk MDS | [111] | |
| MDS | Musto (2010) | 74 | 10.8% CR, 9.5% PR, 20.3% HI | Survival benefits in responder group. | [108] | |
| Decitabine | MDS | Kantarjian (2006) | 170 | 9% CR, 13% HI, 17% ORR | RBC transfusion independence and delayed progression time toward AML (D-0007). | [112] |
| MDS | Kantarjian (2007) | 95 | 34% CR | Better response rate at low dose. | [114] | |
| MDS | Steensma (2009) | 99 | 17% CR, 18% HI, 32% ORR | Alternative dosing and significant difference in ORR compared to single-center trial by Kantarjian et al, (ADOPT). | [115] | |
| Refractory Solid Tumor | Stewart (2009) | 31 | Relative reduction of tumor size but no correlation between PBMC DNA methylation and tumor DNA methylation. | [117] | ||
| MG98 | MDS/AML | Klisovic (2009) | 23 | 26% SD | No clinical benefits observed in administered doses and no correlation between DNMT1 downregulation with SD. | [119] |
| Advanced Solid Malignancies | Plummer (2009) | 33 | Suppression of DNMT1 expression in PBMC in all doses with mild toxicity observed. | [121] | ||
| Azacitidine in Combination | ||||||
| Azacitidine + Valproic Acid + all-trans-Retinoic Acid | AML/MDS | Soriano (2007) | 53 | 22% CR, 42% ORR | The increase of histone acetylation level is not correlated with VPA dose level. | [122] |
| Azacitidine + Thalidomide | MDS/AML | Raza (2008) | 40 | 42% HI, 14% SD | High expression of cellular proliferation genes was observed in non-responders. | [135] |
| Azacitidine + Valproic Acid | Advanced cancer | Braiteh (2008) | 55 | 25%SD | Safe dosage of azacitidine up to 75mg/m2 in advanced malignancies. | [124] |
| Azacitidine + Sodium Phenylbutyrate | Refractory Solid Tumor | Lin (2009) | 27 | No clear benefits seen in the three dosing regimens tested. | [126] | |
| Azacitidine + Valproic Acid | MDS | Voso (2009) | 62 | 30.7% CR/PR, 15.4% HI, 38.5% SD | Increasing therapeutic level of VPA promoted efficacy of azacitidine | [123] |
| Azacitidine + Entinostat | MDS/AML | Fandy (2009) | 30 | 42%ORR | Lack of association between reversal of DNA methylation and clinical response. | [125] |
| Azacitidine + Allogeneic SCT | MDS/AML | Jabbour (2009) | 17 | High response rate in patients with recurrent disease after HSCT. | [136] | |
| Azacitidine + Lenalidomide | MDS | Sekeres (2010) | 18 | 44% CR, 17% HI, 67% ORR | Higher CR is associated with normal cytogenetics and lower methylation levels | [129] |
| Azacitidine + Cytarabine | MDS/AML | Borthakur (2010) | 34 | Limited clinical activity in patients with relapsed, refractory AML. | [137] | |
| Azacitidine + Etanercept | MDS/CMML | Scott (2010) | 32 | 72% ORR | Increased response rate and duration of response observed. | [138] |
| Decitabine in Combination | ||||||
| Decitabine + Valproic Acid | Advanced leukemia | Garcia-Manero (2006) | 54 | 19% CR | Lower pretreatment CDKN2B methylation level correlates with higher response rate. | [127] |
| Decitabine +Carboplatin | Solid Tumor | Appleton (2007) | 33 | A correlation between different doses of decitabine with PBC DNA demethylation. | [132] | |
| Decitabine + Valproic Acid | AML | Blum (2007) | 25 | 44% ORR | VPA increased treatment-related toxicity; Re-expression of ER associated with clinical response. | [128] |
| Decitabine + Gemtuzumab ozogamicin | AML | Chowdhury (2009) | 12 | 42% CR | Treatment facilitated subsequent HSCT. | [131] |
| Decitabine + Allogeneic SCT | MDS | De Padua Silva (2009) | 17 | No increased toxicity due to decitabine. | [139] | |
| Decitabine + Imatinib Mesylate | CML | Oki (2007) | 28 | 32% CR, 4% PR, 7% HI | Higher activity in patients without BCR-ABL mutations. | [130] |
| Decitabine + IL-2 | Metastatic Melanoma; Renal Carcinoma | Gollob (2006) | 21 | Reexpreesion of immunomodulatory genes. | [133] | |
Abbreviations: CR, complete remission; PR, partial response; HI, hematologic improvement; ORR, overall response rate; SD, stable disease. CCR, conventional care regimens; RBC, red blood cells; HSCT, hematopoietic stem cells transplantation; PBMC, peripheral blood mononuclear cell; PBC, peripheral-blood cells.
Single agent therapy
5-Azacytidine (Vidaza, Azacitidine)
In early clinical trials, 5-Aza-CR was administered at maximum tolerated doses (MTD) to patients with osteogenic sarcoma or other cancer related diseases, which showed unfavorable toxicity[100–101]. Increasing knowledge of the action mechanism of 5-Aza-CR indicated that lower dosages of 5-Aza-CR could act as DNMT inhibitors with minimal effect on DNA synthesis[102]. 5-Aza-CR was approved by the FDA for the treatment of MDS based on the positive results from the GALGB9221 clinical trial[103]. A reported 60% of 5-Aza-CR treated patients exhibited various levels of response, whereas only 5% of patients in the supportive group showed hematological improvement (HI). 5-Aza-CR also benefited patients by delaying progression time to acute myeloid leukemia (AML), improving the quality of life and prolonging overall survival in Refractory Anemia with Excess Blasts (RAEB) or Refractory Anemia with Excess of Blasts in Transformation (RAEB-T) subgroups[104–105].
To further validate the efficiencies of 5-Aza-CR in MDS patients, the European AZA-001 trial was conducted for intermediate-2- and high-risk MDS patients. In this study, patients treated with 5-Aza-CR showed a significant improvement in median overall survival (OS) than patients who received conventional care regimens (CCR): 24.5 months versus 15.0 months. It was the first time that 5-Aza-CR treatment was demonstrated to prolong OS in high-risk MDS patients[106]. Detailed analysis of AZA-001 indicated that low-dose cytarabine, one of conventional care treatments for patients with higher-risk MDS, yielded poor efficiency and more toxicity as compared to 5-Aza-CR[107]. Fenaux et al. also demonstrated that 5-Aza-CR benefited elder AML patients by prolonging the OS from 16.0 months to 24.5 months while reducing side effects[107]. Based on the outcome of AZA-001 trial, the National Comprehensive Cancer Network (NCCN) recommended 5-Aza-CR as the preferred therapy for patients with high-risk MDS. In addition to high risk MDS, 5-Aza-CR can also be used as a potentially effective treatment for patients with low-risk MDS[108–109].
However, the current FDA approved 7-day 5-Aza-CR treatment course requires weekend treatment and is inconvenient for both patients and care providers. To overcome this problem, Lyons et al.[110] designed three alternative regimens which avoided weekend treatment. Patients who received any one of the three regimens showed similar hematological improvement as the previously approved 7-day 5-Aza-CR regimen and also a higher transfusion independent rate. More patients who needed red blood cell (RBC) transfusion at baseline became independent of RBC transfusion. Another Phase II trial administered an alternative 5-day 5-Aza-CR intravenous schedule and reported a 27% partial response (PR) + complete remission (CR) rate, comparable to the 7-day subcutaneous regimen[111]. However, future studies are required to illuminate the survival benefit of these modified regimens. Although one group reported that a limited number of treatment cycles could achieve a 50% overall response rate according to the new International Working Group(IWG) criteria, most clinical trials indicate that prolonged exposure to 5-Aza-CR will benefit patients[112].
5-aza-2'-deoxycitidine (Decitabine)
Although 5-Aza-CdR was also approved by the FDA for MDS therapy, there is no clear evidence indicating 5-Aza-CdR improves patients overall survival. In the US-registered trial (D-0007), patients treated with 5-Aza-CdR had a 17% overall response rate (ORR), which is significantly higher than the best supportive care (BSC) group (0%). When comparing OS between the 5-Aza-CdR and control arms, no statistical improvement (14.0 vs 14.9 months) was observed, although clinical benefits such as RBC transfusion-independence or elongation of median time to AML progression were seen after 5-Aza-CdR treatment[113]. Wijermans et al. presented a similar negative result of the OS advantage of 5-Aza-CdR for elder MDS or CMML patients[114]. To increase the outpatient CR rate of 5-Aza-CdR, several clinical trials explored alternative schedules. Kantarjian et al[115] showed that 5-day intravenous schedule with the highest dose-intensity yielded the highest CR rate (39%). In a follow up study, ADOPT trial reported a 32% ORR rate, suggesting that this 5-day schedule is as effective as the approved inpatient regimen[116]. Currently, 5-Aza-CdR is being investigated in other cancer types to assess the best conditions for administration[117–119].
MG98
To date, several Phase I/II clinic trials have been conducted to find out the tumor types sensitive to MG98 and an appropriate working dosage. Although some of the patients already showed the decreases in DNMT1 levels, no consistent correlation of DNMT1 level and dosage has been observed[120–121]. New evidence indicated that a 7 day continuous dosing of MG98 was well tolerated for patients with advanced solid tumors but no objective clinical response was reported[122].
Combination therapy
HDAC inhibitors (HDACi) are a fast growing family of epigenetic drugs that increase acetylation of histone proteins and cytoplasmic proteins such as p53. As previously discussed, reversing abnormal epigenetic gene silencing is coordinated by increasing promoter histone acetylation levels and DNA demethylation. Based on these observations, numerous clinical trials have been conducted to study the synergistic effects of DNMT inhibitors and HDACis (Table 1).
To date, several groups have combined 5-Aza-CR with valproic acid (VPA), an HDACi, to treat MDS and AML. Soriano et al.[123] conducted a phase I/II clinical trial in which patients were treated with 5-Aza-CR and VPA daily for 7 days. The 42% ORR indicates that this combination strategy has clinical effectiveness. Another phase II study sequentially administered 5-Aza-CR following VPA and observed a 30.7% CR+PR. As plasma VPA concentration increased, (>50ug/ml vs <50ug/ml), a better median survival rate was observed (18.7 vs 10 months), indicating HDACi has the potential to increase 5-Aza-CR efficacy[124]. Combination therapy of 5-Aza-CR and VPA has also been investigated in breast cancer, colon cancer and other advanced cancers[125]. Other HDACis have also been combined with 5-Aza-CR. For example, 46% of patients with MDS and AML, who received 5-Aza-CR supplemented with entinostat, another potential HDACi, show promising outcomes. Among them 3 patients had complete remission, 4 patients had partial response and 7 patients had hematologic improvement[126]. In a phase I study, researchers co-administered 5-Aza-CR and sodium phenylbutyrate, a first generation HDACi, to patients with refractory solid tumors. The three dose schedule administered in this study showed mild toxicity albeit little benefits for the patients[127].
Similar to 5-Aza-CR, 5-Aza-CdR has also been tested for its therapeutic efficacy in combination with HDACi. 5-Aza-CdR combined with VPA achieved a 22% objective response in patients with leukemia, although in this case, the higher level of VPA did not correlate with clinical activity[128]. A later study confirmed this conclusion, demonstrating that 5-Aza-CdR itself had promising clinical activity for elderly patients with AML. Adding 25mg/kg/d of VPA neither enhanced 5-Aza-CdR's efficacy nor did it increase CDKN2B re-expression[129].
5-Aza-CR and 5-Aza-CdR have also been combined with other conventional therapies. For example, in a recent study combining 5-Aza-CR with lenalidomide, another FDA approved MDS chemotherapy drug, results showed an impressive 67% ORR[130]. 5-Aza-CdR has been combined with carboplatin, imatinib, or with gemtuzumab ozogamicin (the monoclonal antibody drug) for treatment of various cancer types[131–133]. The encouraging results from these trials would broaden the application of DNMT inhibitors to various tumor types, but further randomized clinical trials with control arms might be needed to demonstrate the feasibility of these approaches.
Validating epigenetic targets in clinical trials
The ability of both 5-Aza-CR and 5-Aza-CdR to reduce global DNA methylation in vivo has been validated in multiple clinical trials in which they have been used as single agents or combined with other anticancer therapies[118, 123, 125–126, 129, 134–140]. The reversal of DNA methylation around individual gene promoters, such as CDKN2B and MAGE1A has also been confirmed in various tumor types[126, 128, 133]. However, the majority of these studies did not show any clear association between the level of induced demethylation and the clinical response. Two recent papers reported that responders to 5-Aza-CR have more significant decreases in global methylation level and methylated promoters compared to non-responders as detected by the Infinium HumanMethylation array[111, 130].
Conclusion and future directions
This review summaries recent advances and future prospect for epigenetic therapy through DNA methylation. After decades of development, DNMT inhibitors are now been used in cancer therapies. Progress made in epigenetic research will no doubt further aid “bench to bedside” translation. A densely methylated region is strongly associated with other silencing mechanisms, including chromatin proteins, and consequently, the use of a single hypomethylating agent might not be enough to reverse epigenetic aberrations in cancer. This concept has already guided the clinical application of DNMT inhibitors and HDACi combination therapy. Further systematic exploration of the overall epigenetic changes after drug induced DNA demethylation might lead to the discovery of other drug targets with the goal of minimizing toxicity and maximizing patients' response. Such examples might be inhibitors to LSD1, which removes the active H3K4me mark, or inhibitors of EZH2, which plays an important role in gene suppression in cancer[141–142]. In addition to targeting DNMT1, new drugs could be designed to inhibit DNA methyltransferases 3A and 3B with the accumulated understanding of their functions in maintaining methylation patterns in somatic cells. The elucidation of differences between 5-Aza-CR and 5-Aza-CdR responses might help in understanding the different outcomes in clinical trials and prompt trials to compare them head-to-head[80].
Analyzing available clinical data might also provide better prognostic factors for patient survival or predict markers for patients' sensitivity to DNMT inhibitor treatment. It has been suggested that there is no positive correlation between high DNMT treatment response and the patients' baseline methylation[126]. In fact, patients who seem to respond to hypomethylation therapy best are those who carry markers such as lower methylation levels in the CDKN2B promoter or higher miR29[128, 143]. Ultimately, efforts to create a comprehensive picture of the epigenetic landscape should help in understanding of cancer pathology and the molecular bases of tumorigenesis in clinical settings, laying out the foundation for future drug development to improve the survival and quality of life of cancer patients.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant R01 “Mechanisms of de novo methylation in cancer” (CA082422-12) and a grant from “Stand Up To Cancer”.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 4.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci U S A. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Leung FC. An evaluation of new criteria for CpG islands in the human genome as gene markers. Bioinformatics. 2004;20:1170–1177. doi: 10.1093/bioinformatics/bth059. [DOI] [PubMed] [Google Scholar]
- 6.Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet. 2009;10:805–811. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hattori N, et al. Epigenetic control of mouse Oct-4 gene expression in embryonic stem cells and trophoblast stem cells. J Biol Chem. 2004;279:17063–17069. doi: 10.1074/jbc.M309002200. [DOI] [PubMed] [Google Scholar]
- 8.Irizarry RA, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okano M, et al. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 10.Bourc'his D, et al. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 11.Jia D, et al. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arita K, et al. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455:818–821. doi: 10.1038/nature07249. [DOI] [PubMed] [Google Scholar]
- 13.Bostick M, et al. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 14.Jeong S, et al. Selective anchoring of DNA methyltransferases 3A and 3B to nucleosomes containing methylated DNA. Mol Cell Biol. 2009;29:5366–5376. doi: 10.1128/MCB.00484-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 16.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schones DE, Zhao K. Genome-wide approaches to studying chromatin modifications. Nat Rev Genet. 2008;9:179–191. doi: 10.1038/nrg2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guenther MG, et al. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ozsolak F, et al. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 21.Liang G, et al. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc Natl Acad Sci U S A. 2004;101:7357–7362. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schuettengruber B, et al. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 24.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 25.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 26.Li B, et al. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 27.Zlatanova J, Thakar A. H2A.Z: view from the top. Structure. 2008;16:166–179. doi: 10.1016/j.str.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 28.Jin C, et al. H3.3/H2A.Z double variant-containing nucleosomes mark `nucleosome-free regions' of active promoters and other regulatory regions. Nat Genet. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gevry N, et al. Histone H2A.Z is essential for estrogen receptor signaling. Genes Dev. 2009;23:1522–1533. doi: 10.1101/gad.1787109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Creyghton MP, et al. H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell. 2008;135:649–661. doi: 10.1016/j.cell.2008.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sutcliffe EL, et al. Dynamic histone variant exchange accompanies gene induction in T cells. Mol Cell Biol. 2009;29:1972–1986. doi: 10.1128/MCB.01590-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marques M, et al. Reconciliating the positive and negative roles of histone H2A.Z in gene transcription. Epigenetics. 2010:5. doi: 10.4161/epi.5.4.11520. [DOI] [PubMed] [Google Scholar]
- 33.Zilberman D, et al. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008;456:125–129. doi: 10.1038/nature07324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okitsu CY, Hsieh CL. DNA methylation dictates histone H3K4 methylation. Mol Cell Biol. 2007;27:2746–2757. doi: 10.1128/MCB.02291-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zemach A, et al. Genome-Wide Evolutionary Analysis of Eukaryotic DNA Methylation. Science. 2010 doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 36.Nan X, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 37.Jones PL, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 38.Ooi SK, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vire E, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 40.Tachibana M, et al. G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. EMBO J. 2008;27:2681–2690. doi: 10.1038/emboj.2008.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong KB, et al. DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. EMBO J. 2008;27:2691–2701. doi: 10.1038/emboj.2008.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eden A, et al. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- 43.Pogribny IP, Beland FA. DNA hypomethylation in the origin and pathogenesis of human diseases. Cell Mol Life Sci. 2009;66:2249–2261. doi: 10.1007/s00018-009-0015-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu H, et al. Hypomethylation-linked activation of PAX2 mediates tamoxifen-stimulated endometrial carcinogenesis. Nature. 2005;438:981–987. doi: 10.1038/nature04225. [DOI] [PubMed] [Google Scholar]
- 45.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 46.Lujambio A, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A. 2008;105:13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toyota M, et al. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68:4123–4132. doi: 10.1158/0008-5472.CAN-08-0325. [DOI] [PubMed] [Google Scholar]
- 48.Saito Y, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 49.Ordway JM, et al. Identification of novel high-frequency DNA methylation changes in breast cancer. PLoS One. 2007;2:e1314. doi: 10.1371/journal.pone.0001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rauch TA, et al. High-resolution mapping of DNA hypermethylation and hypomethylation in lung cancer. Proc Natl Acad Sci U S A. 2008;105:252–257. doi: 10.1073/pnas.0710735105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Figueroa ME, et al. MDS and secondary AML display unique patterns and abundance of aberrant DNA methylation. Blood. 2009;114:3448–3458. doi: 10.1182/blood-2009-01-200519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noushmehr H, et al. Identification of a CpG Island Methylator Phenotype that Defines a Distinct Subgroup of Glioma. Cancer Cell. 2010 doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kautiainen TL, Jones PA. DNA methyltransferase levels in tumorigenic and nontumorigenic cells in culture. J Biol Chem. 1986;261:1594–1598. [PubMed] [Google Scholar]
- 54.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 55.Ohm JE, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schlesinger Y, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 57.Gal-Yam EN, et al. Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. Proc Natl Acad Sci U S A. 2008;105:12979–12984. doi: 10.1073/pnas.0806437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Halkidou K, et al. Upregulation and nuclear recruitment of HDAC1 in hormone refractory prostate cancer. Prostate. 2004;59:177–189. doi: 10.1002/pros.20022. [DOI] [PubMed] [Google Scholar]
- 59.Song J, et al. Increased expression of histone deacetylase 2 is found in human gastric cancer. APMIS. 2005;113:264–268. doi: 10.1111/j.1600-0463.2005.apm_04.x. [DOI] [PubMed] [Google Scholar]
- 60.Schulte JH, et al. Lysine-specific demethylase 1 is strongly expressed in poorly differentiated neuroblastoma: implications for therapy. Cancer Res. 2009;69:2065–2071. doi: 10.1158/0008-5472.CAN-08-1735. [DOI] [PubMed] [Google Scholar]
- 61.Lim S, et al. Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis. 2010;31:512–520. doi: 10.1093/carcin/bgp324. [DOI] [PubMed] [Google Scholar]
- 62.Kondo Y, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet. 2008;40:741–750. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- 63.Varambally S, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 64.Varambally S, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–1699. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Friedman JM, et al. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 2009;69:2623–2629. doi: 10.1158/0008-5472.CAN-08-3114. [DOI] [PubMed] [Google Scholar]
- 66.Lin JC, et al. Role of nucleosomal occupancy in the epigenetic silencing of the MLH1 CpG island. Cancer Cell. 2007;12:432–444. doi: 10.1016/j.ccr.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.He HH, et al. Nucleosome dynamics define transcriptional enhancers. Nat Genet. 2010;42:343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jones PA, Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20:85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- 69.Taylor SM, Jones PA. Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell. 1979;17:771–779. doi: 10.1016/0092-8674(79)90317-9. [DOI] [PubMed] [Google Scholar]
- 70.Vesely J, Alois C. 5-Azacytidine: mechanism of action and biological effects in mammalian cells. Pharmac. Ther. A. 1978;2:813–840. [Google Scholar]
- 71.Rius M, et al. Human concentrative nucleoside transporter 1-mediated uptake of 5-azacytidine enhances DNA demethylation. Mol Cancer Ther. 2009;8:225–231. doi: 10.1158/1535-7163.MCT-08-0743. [DOI] [PubMed] [Google Scholar]
- 72.Issa JP, Kantarjian HM. Targeting DNA methylation. Clin Cancer Res. 2009;15:3938–3946. doi: 10.1158/1078-0432.CCR-08-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stresemann C, Lyko F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. Int J Cancer. 2008;123:8–13. doi: 10.1002/ijc.23607. [DOI] [PubMed] [Google Scholar]
- 74.Ghoshal K, et al. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol Cell Biol. 2005;25:4727–4741. doi: 10.1128/MCB.25.11.4727-4741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 75.Kuo HK, et al. 5-Azacytidine induced methyltransferase-DNA adducts block DNA replication in vivo. Cancer Res. 2007;67:8248–8254. doi: 10.1158/0008-5472.CAN-07-1038. [DOI] [PubMed] [Google Scholar]
- 76.Nguyen CT, et al. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2'-deoxycytidine. Cancer Res. 2002;62:6456–6461. [PubMed] [Google Scholar]
- 77.Komashko VM, Farnham PJ. 5-azacytidine treatment reorganizes genomic histone modification patterns. Epigenetics. 2010:5. doi: 10.4161/epi.5.3.11409. [DOI] [PubMed] [Google Scholar]
- 78.Qin T, et al. Mechanisms of resistance to 5-aza-2'-deoxycytidine in human cancer cell lines. Blood. 2009;113:659–667. doi: 10.1182/blood-2008-02-140038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li LH, et al. Cytotoxicity and mode of action of 5-azacytidine on L1210 leukemia. Cancer Res. 1970;30:2760–2769. [PubMed] [Google Scholar]
- 80.Hollenbach PW, et al. A comparison of azacitidine and decitabine activities in acute myeloid leukemia cell lines. PLoS One. 2010;5:e9001. doi: 10.1371/journal.pone.0009001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yoo CB, et al. Long-term epigenetic therapy with oral zebularine has minimal side effects and prevents intestinal tumors in mice. Cancer Prev Res (Phila Pa) 2008;1:233–240. doi: 10.1158/1940-6207.CAPR-07-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lemaire M, et al. Inhibition of cytidine deaminase by zebularine enhances the antineoplastic action of 5-aza-2'-deoxycytidine. Cancer Chemother Pharmacol. 2009;63:411–416. doi: 10.1007/s00280-008-0750-6. [DOI] [PubMed] [Google Scholar]
- 83.Yoo CB, et al. Zebularine: a new drug for epigenetic therapy. Biochem Soc Trans. 2004;32:910–912. doi: 10.1042/BST0320910. [DOI] [PubMed] [Google Scholar]
- 84.Billam M, et al. Effects of a novel DNA methyltransferase inhibitor zebularine on human breast cancer cells. Breast Cancer Res Treat. 2010;120:581–592. doi: 10.1007/s10549-009-0420-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Flotho C, et al. The DNA methyltransferase inhibitors azacitidine, decitabine and zebularine exert differential effects on cancer gene expression in acute myeloid leukemia cells. Leukemia. 2009;23:1019–1028. doi: 10.1038/leu.2008.397. [DOI] [PubMed] [Google Scholar]
- 86.Beumer JH, et al. Pharmacokinetics, metabolism, and oral bioavailability of the DNA methyltransferase inhibitor 5-fluoro-2'-deoxycytidine in mice. Clin Cancer Res. 2006;12:7483–7491. doi: 10.1158/1078-0432.CCR-06-1250. [DOI] [PubMed] [Google Scholar]
- 87.Beumer JH, et al. Concentrations of the DNA methyltransferase inhibitor 5-fluoro-2'-deoxycytidine (FdCyd) and its cytotoxic metabolites in plasma of patients treated with FdCyd and tetrahydrouridine (THU) Cancer Chemother Pharmacol. 2008;62:363–368. doi: 10.1007/s00280-007-0603-8. [DOI] [PubMed] [Google Scholar]
- 88.Kratzke RA, et al. Response to the methylation inhibitor dihydro-5-azacytidine in mesothelioma is not associated with methylation of p16INK4a: results of cancer and leukemia group B 159904. J Thorac Oncol. 2008;3:417–421. doi: 10.1097/JTO.0b013e318168da0a. [DOI] [PubMed] [Google Scholar]
- 89.Byun HM, et al. 2'-Deoxy-N4-[2-(4-nitrophenyl)ethoxycarbonyl]-5-azacytidine: a novel inhibitor of DNA methyltransferase that requires activation by human carboxylesterase 1. Cancer Lett. 2008;266:238–248. doi: 10.1016/j.canlet.2008.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chuang JC, et al. S110, a 5-Aza-2'-Deoxycytidine-Containing Dinucleotide, Is an Effective DNA Methylation Inhibitor In vivo and Can Reduce Tumor Growth. Mol Cancer Ther. 2010 doi: 10.1158/1535-7163.MCT-09-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Segura-Pacheco B, et al. Global DNA hypermethylation-associated cancer chemotherapy resistance and its reversion with the demethylating agent hydralazine. J Transl Med. 2006;4:32. doi: 10.1186/1479-5876-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Singh N, et al. Molecular modeling and molecular dynamics studies of hydralazine with human DNA methyltransferase 1. ChemMedChem. 2009;4:792–799. doi: 10.1002/cmdc.200900017. [DOI] [PubMed] [Google Scholar]
- 93.Song Y, Zhang C. Hydralazine inhibits human cervical cancer cell growth in vitro in association with APC demethylation and re-expression. Cancer Chemother Pharmacol. 2009;63:605–613. doi: 10.1007/s00280-008-0773-z. [DOI] [PubMed] [Google Scholar]
- 94.Chuang JC, et al. Comparison of biological effects of non-nucleoside DNA methylation inhibitors versus 5-aza-2'-deoxycytidine. Mol Cancer Ther. 2005;4:1515–1520. doi: 10.1158/1535-7163.MCT-05-0172. [DOI] [PubMed] [Google Scholar]
- 95.Suzuki T, et al. Design, synthesis, inhibitory activity, and binding mode study of novel DNA methyltransferase 1 inhibitors. Bioorg Med Chem Lett. 2010;20:1124–1127. doi: 10.1016/j.bmcl.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 96.Stresemann C, et al. Functional diversity of DNA methyltransferase inhibitors in human cancer cell lines. Cancer Res. 2006;66:2794–2800. doi: 10.1158/0008-5472.CAN-05-2821. [DOI] [PubMed] [Google Scholar]
- 97.Datta J, et al. A new class of quinoline-based DNA hypomethylating agents reactivates tumor suppressor genes by blocking DNA methyltransferase 1 activity and inducing its degradation. Cancer Res. 2009;69:4277–4285. doi: 10.1158/0008-5472.CAN-08-3669. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 98.Amato RJ. Inhibition of DNA methylation by antisense oligonucleotide MG98 as cancer therapy. Clin Genitourin Cancer. 2007;5:422–426. doi: 10.3816/CGC.2007.n.029. [DOI] [PubMed] [Google Scholar]
- 99.Garzon R, et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Velez-Garcia E, et al. Twice weekly 5-azacytidine infusion in dissmeinated metastatic cancer: a phase II study. Cancer Treat Rep. 1977;61:1675–1677. [PubMed] [Google Scholar]
- 101.Srinivasan U, et al. Phase II study of 5-azacytidine in sarcomas of bone. Am J Clin Oncol. 1982;5:411–415. doi: 10.1097/00000421-198208000-00011. [DOI] [PubMed] [Google Scholar]
- 102.Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov. 2006;5:37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- 103.Silverman LR, et al. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: a study of the cancer and leukemia group B. J Clin Oncol. 2002;20:2429–2440. doi: 10.1200/JCO.2002.04.117. [DOI] [PubMed] [Google Scholar]
- 104.Kornblith AB, et al. Impact of azacytidine on the quality of life of patients with myelodysplastic syndrome treated in a randomized phase III trial: a Cancer and Leukemia Group B study. J Clin Oncol. 2002;20:2441–2452. doi: 10.1200/JCO.2002.04.044. [DOI] [PubMed] [Google Scholar]
- 105.Silverman LR, et al. Further analysis of trials with azacitidine in patients with myelodysplastic syndrome: studies 8421, 8921, and 9221 by the Cancer and Leukemia Group B. J Clin Oncol. 2006;24:3895–3903. doi: 10.1200/JCO.2005.05.4346. [DOI] [PubMed] [Google Scholar]
- 106.Fenaux P, et al. Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: a randomised, open-label, phase III study. Lancet Oncol. 2009;10:223–232. doi: 10.1016/S1470-2045(09)70003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fenaux P, et al. Prolonged survival with improved tolerability in higher-risk myelodysplastic syndromes: azacitidine compared with low dose ara-C. Br J Haematol. 2010 doi: 10.1111/j.1365-2141.2010.08082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Santini V. Azacitidine in lower-risk myelodysplastic syndromes. Leuk Res. 2009;33(Suppl 2):S22–26. doi: 10.1016/S0145-2126(09)70230-9. [DOI] [PubMed] [Google Scholar]
- 109.Musto P, et al. Azacitidine for the treatment of lower risk myelodysplastic syndromes: a retrospective study of 74 patients enrolled in an Italian named patient program. Cancer. 2010;116:1485–1494. doi: 10.1002/cncr.24894. [DOI] [PubMed] [Google Scholar]
- 110.Lyons RM, et al. Hematologic response to three alternative dosing schedules of azacitidine in patients with myelodysplastic syndromes. J Clin Oncol. 2009;27:1850–1856. doi: 10.1200/JCO.2008.17.1058. [DOI] [PubMed] [Google Scholar]
- 111.Martin MG, et al. A phase II study of 5-day intravenous azacitidine in patients with myelodysplastic syndromes. Am J Hematol. 2009;84:560–564. doi: 10.1002/ajh.21482. [DOI] [PubMed] [Google Scholar]
- 112.Muller-Thomas C, et al. A limited number of 5-azacitidine cycles can be effective treatment in MDS. Ann Hematol. 2009;88:213–219. doi: 10.1007/s00277-008-0583-8. [DOI] [PubMed] [Google Scholar]
- 113.Kantarjian H, et al. Decitabine improves patient outcomes in myelodysplastic syndromes: results of a phase III randomized study. Cancer. 2006;106:1794–1803. doi: 10.1002/cncr.21792. [DOI] [PubMed] [Google Scholar]
- 114.Pierre WijerMans SS, Baila Liliana, Platzbecker Uwe, Giagounidis Aristoteles, Selleslag Dominik, Labar Boris, Salih Helmut, Beeldens Filip, Muus Petra, de Witte Theo, Lübbert Michael., MD, PhD Low Dose Decitabine Versus Best Supportive Care in Elderly Patients with Intermediate or High Risk MDS Not Eligible for Intensive Chemotherapy: Final Results of the Randomized Phase III Study, (06011) of the EORTC Leukemia and German MDS Study Groups. blood. 2008 abstract 226. [Google Scholar]
- 115.Kantarjian H, et al. Results of a randomized study of 3 schedules of low-dose decitabine in higher-risk myelodysplastic syndrome and chronic myelomonocytic leukemia. Blood. 2007;109:52–57. doi: 10.1182/blood-2006-05-021162. [DOI] [PubMed] [Google Scholar]
- 116.Steensma DP, et al. Multicenter study of decitabine administered daily for 5 days every 4 weeks to adults with myelodysplastic syndromes: the alternative dosing for outpatient treatment (ADOPT) trial. J Clin Oncol. 2009;27:3842–3848. doi: 10.1200/JCO.2008.19.6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Steensma DP. Decitabine treatment of patients with higher-risk myelodysplastic syndromes. Leuk Res. 2009;33(Suppl 2):S12–17. doi: 10.1016/S0145-2126(09)70228-0. [DOI] [PubMed] [Google Scholar]
- 118.Stewart DJ, et al. Decitabine effect on tumor global DNA methylation and other parameters in a phase I trial in refractory solid tumors and lymphomas. Clin Cancer Res. 2009;15:3881–3888. doi: 10.1158/1078-0432.CCR-08-2196. [DOI] [PubMed] [Google Scholar]
- 119.Schrump DS, et al. Phase I study of decitabine-mediated gene expression in patients with cancers involving the lungs, esophagus, or pleura. Clin Cancer Res. 2006;12:5777–5785. doi: 10.1158/1078-0432.CCR-06-0669. [DOI] [PubMed] [Google Scholar]
- 120.Klisovic RB, et al. A phase I biological study of MG98, an oligodeoxynucleotide antisense to DNA methyltransferase 1, in patients with high-risk myelodysplasia and acute myeloid leukemia. Clin Cancer Res. 2008;14:2444–2449. doi: 10.1158/1078-0432.CCR-07-1320. [DOI] [PubMed] [Google Scholar]
- 121.Winquist E, et al. Phase II trial of DNA methyltransferase 1 inhibition with the antisense oligonucleotide MG98 in patients with metastatic renal carcinoma: a National Cancer Institute of Canada Clinical Trials Group investigational new drug study. Invest New Drugs. 2006;24:159–167. doi: 10.1007/s10637-006-5938-1. [DOI] [PubMed] [Google Scholar]
- 122.Plummer R, et al. Phase I study of MG98, an oligonucleotide antisense inhibitor of human DNA methyltransferase 1, given as a 7-day infusion in patients with advanced solid tumors. Clin Cancer Res. 2009;15:3177–3183. doi: 10.1158/1078-0432.CCR-08-2859. [DOI] [PubMed] [Google Scholar]
- 123.Soriano AO, et al. Safety and clinical activity of the combination of 5-azacytidine, valproic acid, and all-trans retinoic acid in acute myeloid leukemia and myelodysplastic syndrome. Blood. 2007;110:2302–2308. doi: 10.1182/blood-2007-03-078576. [DOI] [PubMed] [Google Scholar]
- 124.Voso MT, et al. Valproic acid at therapeutic plasma levels may increase 5-azacytidine efficacy in higher risk myelodysplastic syndromes. Clin Cancer Res. 2009;15:5002–5007. doi: 10.1158/1078-0432.CCR-09-0494. [DOI] [PubMed] [Google Scholar]
- 125.Braiteh F, et al. Phase I study of epigenetic modulation with 5-azacytidine and valproic acid in patients with advanced cancers. Clin Cancer Res. 2008;14:6296–6301. doi: 10.1158/1078-0432.CCR-08-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fandy TE, et al. Early epigenetic changes and DNA damage do not predict clinical response in an overlapping schedule of 5-azacytidine and entinostat in patients with myeloid malignancies. Blood. 2009;114:2764–2773. doi: 10.1182/blood-2009-02-203547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lin J, et al. A phase I dose-finding study of 5-azacytidine in combination with sodium phenylbutyrate in patients with refractory solid tumors. Clin Cancer Res. 2009;15:6241–6249. doi: 10.1158/1078-0432.CCR-09-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Garcia-Manero G, et al. Phase 1/2 study of the combination of 5-aza-2'-deoxycytidine with valproic acid in patients with leukemia. Blood. 2006;108:3271–3279. doi: 10.1182/blood-2006-03-009142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Blum W, et al. Phase I study of decitabine alone or in combination with valproic acid in acute myeloid leukemia. J Clin Oncol. 2007;25:3884–3891. doi: 10.1200/JCO.2006.09.4169. [DOI] [PubMed] [Google Scholar]
- 130.Sekeres MA, et al. Phase I combination trial of lenalidomide and azacitidine in patients with higher-risk myelodysplastic syndromes. J Clin Oncol. 2010;28:2253–2258. doi: 10.1200/JCO.2009.26.0745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Oki Y, et al. Phase II study of low-dose decitabine in combination with imatinib mesylate in patients with accelerated or myeloid blastic phase of chronic myelogenous leukemia. Cancer. 2007;109:899–906. doi: 10.1002/cncr.22470. [DOI] [PubMed] [Google Scholar]
- 132.Chowdhury S, et al. Decitabine combined with fractionated gemtuzumab ozogamicin therapy in patients with relapsed or refractory acute myeloid leukemia. Am J Hematol. 2009;84:599–600. doi: 10.1002/ajh.21478. [DOI] [PubMed] [Google Scholar]
- 133.Appleton K, et al. Phase I and pharmacodynamic trial of the DNA methyltransferase inhibitor decitabine and carboplatin in solid tumors. J Clin Oncol. 2007;25:4603–4609. doi: 10.1200/JCO.2007.10.8688. [DOI] [PubMed] [Google Scholar]
- 134.Gollob JA, et al. Phase I trial of sequential low-dose 5-aza-2'-deoxycytidine plus high-dose intravenous bolus interleukin-2 in patients with melanoma or renal cell carcinoma. Clin Cancer Res. 2006;12:4619–4627. doi: 10.1158/1078-0432.CCR-06-0883. [DOI] [PubMed] [Google Scholar]
- 135.Yang AS, et al. DNA methylation changes after 5-aza-2'-deoxycytidine therapy in patients with leukemia. Cancer Res. 2006;66:5495–5503. doi: 10.1158/0008-5472.CAN-05-2385. [DOI] [PubMed] [Google Scholar]
- 136.Raza A, et al. Combination of 5-azacytidine and thalidomide for the treatment of myelodysplastic syndromes and acute myeloid leukemia. Cancer. 2008;113:1596–1604. doi: 10.1002/cncr.23789. [DOI] [PubMed] [Google Scholar]
- 137.Jabbour E, et al. Low-dose azacitidine after allogeneic stem cell transplantation for acute leukemia. Cancer. 2009;115:1899–1905. doi: 10.1002/cncr.24198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Borthakur G, et al. Report of a phase 1/2 study of a combination of azacitidine and cytarabine in acute myelogenous leukemia and high-risk myelodysplastic syndromes. Leuk Lymphoma. 2010;51:73–78. doi: 10.3109/10428190903318329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Scott BL, et al. Prolonged responses in patients with MDS and CMML treated with azacitidine and etanercept. Br J Haematol. 2010;148:944–947. doi: 10.1111/j.1365-2141.2009.08061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.De Padua Silva L, et al. Feasibility of allo-SCT after hypomethylating therapy with decitabine for myelodysplastic syndrome. Bone Marrow Transplant. 2009;43:839–843. doi: 10.1038/bmt.2008.400. [DOI] [PubMed] [Google Scholar]
- 141.Huang Y, et al. Polyamine analogues targeting epigenetic gene regulation. Essays Biochem. 2009;46:95–110. doi: 10.1042/bse0460007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tan J, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Blum W, et al. Clinical response and miR-29b predictive significance in older AML patients treated with a 10-day schedule of decitabine. Proc Natl Acad Sci U S A. 2010;107:7473–7478. doi: 10.1073/pnas.1002650107. [DOI] [PMC free article] [PubMed] [Google Scholar]